Abstract
A challenge facing metabolomics in the analysis of large human cohorts is the cross-laboratory comparability of quantitative metabolomics measurements. In this study, 14 laboratories analyzed various blood specimens using a common experimental protocol provided with Biocrates AbsoluteIDQ p400HR kit, to quantify up to 408 metabolites. The specimens included human plasma and serum from male and female donors, mouse and rat plasma as well as NIST SRM 1950 reference plasma. The metabolite classes covered range from polar (e.g. amino acids and biogenic amines), to nonpolar (e.g. diacyl- and triacyl-glycerols), and span 11 common metabolite classes. The manuscript describes a strict system suitability testing (SST) criteria used to evaluate each laboratory’s readiness to perform the assay, and provides the SST Skyline documents for public dissemination. The study found approximately 250 metabolites were routinely quantified in the sample types tested, using Orbitrap instruments. Inter-laboratory variance for the NIST SRM-1950 has a median of 10% for amino acids, 24% for biogenic amines, 38% for acylcarnitines, 25% for glycerolipids, 23% for glycerophospholipids, 16% for cholesteryl esters, 15% for sphingolipids, and 9% for hexoses. Comparing to consensus values for NIST SRM-1950, nearly 80% of comparable analytes demonstrated bias of <50% from the reference value. The findings of this study result in recommendations of best practices for system suitability, quality control, and calibration. We demonstrate that with appropriate controls, high-resolution metabolomics can provide accurate results with good precision across laboratories, and the p400HR therefore is a reliable approach for generating consistent and comparable metabolomics data.
Graphical Abstract
A multitude of publications exist on the broad-spectrum metabolomic analysis of biofluids based on liquid chromatography-mass spectrometry (LC-MS), but standardization remains a challenge for translational and epidemiological metabolomics, which is important for cross-study and cross-cohort comparison.1, 2 Demonstration of inter-laboratory comparability of quantitative metabolomics measurements would seem to be an analytical prerequisite to allow reproducible measurements on a population-wide scale, since no single laboratory can possibly address all the analyses which will be needed for such measurements to have a long-term impact on our knowledge of human health and disease. Two recent community whitepapers have highlighted the challenges and exciting opportunities possible for improvement in human health, should the metabolomics community be able to embrace harmonization in biobanking and analysis methods for global precision medicine initiatives.3, 4 Important recent efforts in the metabolomics5, 6 and proteomics7 space have demonstrated significant progress in cross-laboratory standardization of analytical methods, with inter-laboratory precision generally well below that of biological variance when using well-controlled and predefined assays. Reference materials also play an important role in the ability to build standardized methods and reporting standards across laboratories.8 A recent report including the analysis of the NIST SRM-1950 plasma sample by a variety of lipidomics methods from more than 30 laboratories established consensus concentrations for more than 300 lipids, yet more than 1000 additional lipids were reported inconsistently, highlighting yet more work to be done in the area of analytical harmonization.9
The AbsoluteIDQ p400HR assay quantifies over 400 metabolites from eleven analyte groups: amino acids, biogenic amines, acylcarnitines, monosaccharides (hexose), diglycerides, triglycerides, lysophosphatidylcholines, phosphatidylcholines, sphingomyelins, ceramides, and cholesteryl esters.10 The kit includes calibration standards, internal standards, and quality control (QC) samples. Selective analyte detection is accomplished by the Q Exactive Orbitrap™ family of high-resolution, accurate-mass mass spectrometers (Thermo Fisher Scientific). There are four separate mass spectrometric analyses of each sample. For the analysis of acylcarnitines, monosaccharides (hexoses), diglycerides, triglycerides, lysophosphatidylcholines, phosphatidylcholines, sphingomyelins, ceramides, and cholesteryl esters samples are quantified using Flow Injection Analysis methods (FIA-MS) at different m/z ranges. Sample analysis of amino acids and biogenic amines is performed by two UHPLC (ultra-high pressure liquid chromatography) methods (one with full scan MS and one with parallel reaction monitoring or PRM) using a reversed phase analytical column. A significant difference between this kit and previous targeted metabolomics kits is the use of accurate mass, high-resolution mass spectrometers. The ring trial described herein was coordinated by an independent group of academic and corporate metabolomics research laboratories, with data collected by 14 labs (including Biocrates and Thermo Fisher Scientific, see a map of geographic distribution in Figure S-1). This particular ring trial focused on the analysis of plasma from humans and rodents, since this is the most common matrix utilized in translational medicine studies. A few serum samples from select individuals were also analyzed. It may be possible to analyze a wide variety of other matrices using the p400HR kit, but those were not addressed herein.
In establishing an international ring trial, we sought to empirically evaluate the intra- and inter-laboratory precision and accuracy of the AbsoluteIDQ p400HR metabolite quantification kit, conceptually similar to prior ring trials for the AbsoluteIDQ p180 and Bile Acids kits from Biocrates.5, 6 A training kit, one 96-well plate p400HR kit, and the ring trial samples (described in Materials and Methods) were distributed to the participating laboratories throughout North America and Europe. The reported ring trial of Siskos et al. for the p180 kit demonstrated <20% interlaboratory variance for more than 80% of the metabolites measured across six laboratories, but this kit has limited lipid coverage and utilized triple quadrupole MS systems. The current effort seeks to more than double the metabolite coverage, and performs a first-of-its-kind metabolomics ring trial utilizing high-resolution mass spectrometers. Although training and technical assistance was available from Biocrates for the kits and Thermo Fisher Scientific for instrumentation, each laboratory was independently responsible for sample preparation, instrument setup, data collection, and data analysis. Data was submitted in a blinded fashion to an automated online repository at Duke University, where each laboratory was assigned a random numerical identifier. Data aggregation was performed at Duke University and redistributed to all laboratories. Each laboratory followed the kit Standard Operating Procedure (SOP) included in the guidance for the ring trial. The laboratories were blinded to sample identifications, and all labs agreed to remain blinded to laboratory ID (other than their own) throughout data acquisition and publication of the results. While the primary goal of this study was to evaluate the p400HR kit as a method of performing reproducible and accurate quantification with high resolution liquid chromatography-mass spectrometry, we believe there are findings which will be helpful in establishing recommendations for best practices for broad success and analytical harmonization in targeted metabolomics.
Materials and Methods
All AbsoluteIDQ p400HR kits include a very detailed Standard Operating Procedure (SOP) protocol with detailed documentation for sample preparation, instrument setup, system suitability testing, and data analysis. Additionally, ring trial participants received a Ring Trial Guidance document (Supplementary Information), which contained additional information on specific topics such as troubleshooting, data upload, and data analysis.
Safety Precautions
Study participants were instructed to handle human biofluid samples at BioSafety Level 2 (universal precautions). All participants handled volatile organic solvents in accordance with OSHA recommendations and disposed of chemical waste appropriately. Otherwise standard laboratory safety precautions were followed.
Biological Specimens and Chemical Reagents
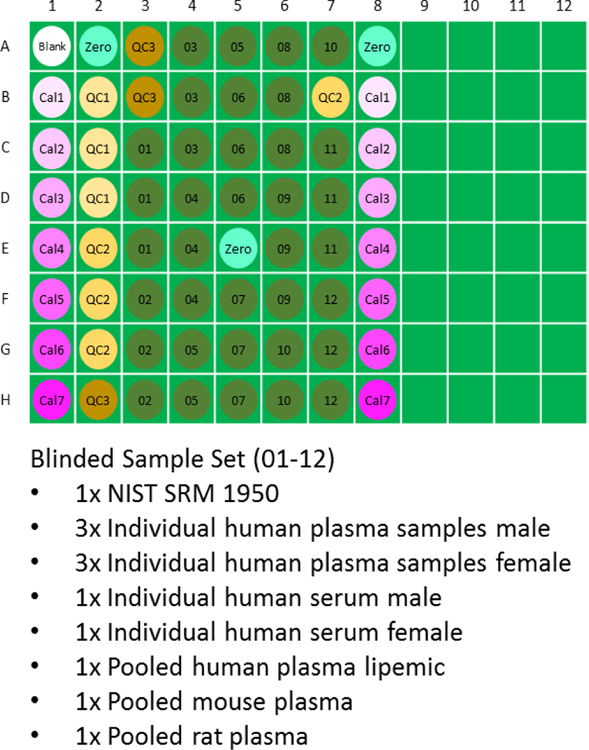
LC-MS grade acetonitrile, methanol, water, and formic acid were obtained independently by each laboratory. A set of 12 plasma/serum samples, 60 µL per aliquot (see Figure 1), was organized by Biocrates. The human plasma/serum samples numbered from 1 to 9 (lipemic and individual male and female) were collected and aliquoted by in.vent Diagnostika GmbH (Berlin, Germany) as instructed by Biocrates. Each individual gave his/her informed consent for the blood collection and these documents are available upon request. The NIST SRM 1950 sample was ordered from the National Institute of Standards and Technology (NIST, Gaithersburg, Maryland, USA) and aliquoted at Biocrates. The pooled rat plasma and pooled mouse plasma samples were acquired from Sera Laboratories International Ltd. (West Sussex, United Kingdom) and aliquoted at Biocrates. All samples were blinded during the ring trial measurements, i.e. the sample description was not revealed to the participants. The sample sets and kit reagents (calibrators, internal standards (ISTD), Quality Control samples, and testmix) were distributed to the participants on dry ice shortly before the scheduled measurement. For U.S. participants, a single shipment of test materials was made to the Duke University and materials were subsequently distributed. Materials tested and the plate layout utilized in the study are shown in Figure 1.
Figure 1.
96-well plate layout utilized for the p400HR ring trial. All samples were delivered to each lab frozen in cryovials on dry ice, and prepared by each laboratory according to a standard operating procedure (SOP). In the Figure, “Cal” represents calibrators (low =1 to high =7), “QC” represents spiked quality control samples (low =1, mid =2, high=3), and samples labeled 01–12 were blinded to each lab. The blinded sample set is listed beneath the plate layout.
Instrument Calibration
As detailed in the kit SOP, a specialized 2-step calibration procedure was utilized for this study. Positive ion calibration was first performed with the Pierce LTQ ESI Positive Ion Calibration Solution (“Thermo Cal Mix”, Pierce Cat #88322). A custom calibration routine was then carried out by mixing the Biocrates Flow-Injection test mix 1:10 v/v with the Thermo Cal Mix, in order to improve calibration accuracy in the low mass range. Calibration was performed immediately prior to acquisition of the system suitability data. Please note that instrument calibration and System Suitability Testing is recommended prior sample preparation, since the samples are only stable for a fixed period of time (approximately 48–72 hours) after prepared.
System Suitability Testing (SST)
Evaluation of the instrument performance prior to sample analysis was assessed by a common system suitability test across all laboratories. Evaluated metrics included mass accuracy (ppm), peak intensity (response), retention time, and chromatographic peak shape. Separate test mixtures were provided with the kit for LC-MS and FIA-MS SST evaluation. Participants were asked to set the LC-MS system up for the p400HR assay and analyze the test mixes three times (by LC-MS and then by FIA-MS) prior to preparing samples for analysis. For evaluating this data, the organizers provided each laboratory with two Skyline v4.1 documents (www.skyline.ms), one for each method and test mix. These Skyline files contained the test mix (system suitability) data from two laboratories against which the labs could choose to compare the results of the SST sample in order to measure relative instrument suitability. Participants uploaded these Skyline files as part of the data submission for the Ring Trial, and the Import->Document function in Skyline was utilized to combine the files from all laboratories. There was not an a priori cutoff criteria established for SST testing as part of the study because insufficient data was available prior to the ring trial to establish such criteria. The UHPLC-MS and FIA-MS aggregate SST files from all ring trial participants have been made available on the PanoramaPublic repository under the project https://panoramaweb.org/p400HR_SST_RingTrial.url.
Sample Preparation
Within each laboratory, samples (described above) were prepared using the AbsoluteIDQ® p400HR kit (Biocrates Innsbruck, Austria) in strict accordance with their detailed protocol. Briefly, 10 µL of the supplied internal standard solution was first added to each well of the 96-well extraction plate with exception of the blank well position A1, followed by 10 µL of each blank, calibration standard, Biocrates QC, or sample. Laboratories utilized the appropriate wells in a predefined layout (Figure 1) as directed in the ring trial instructions. The plate was then dried under a gentle stream of nitrogen for 30 minutes. The samples were derivatized with 50 µL of phenyl isothiocyanate solution at room temperature, loosely covered for 20 minutes, then dried under N2 for 1 hr. Metabolites were then extracted with 5 mM ammonium acetate in methanol (300 µL per well, 30 minutes shaking at 450 rpm), and centrifuged for collection through a filter plate. Samples were diluted with either water for the UHPLC analysis (1:1) or running solvent (a proprietary mixture provided by Biocrates) for flow injection analysis (5:1).
Sample Analysis
Separation of amino acids and biogenic amines was performed using UHPLC with a C18 column (Biocrates, Part 9120052121032) and guard column (Biocrates, Part 9120052121049). The laboratories utilized a variety of UHPLC systems, but all utilized the same analytical columntype, LC solvents, and gradient composition. Analytes were separated using a gradient from 0.2% formic acid in water, to 0.2% formic acid in acetonitrile. Total UHPLC analysis time was approximately 6 minutes per sample. Acylcarnitines, monosaccharides (hexose), diglycerides, triglycerides, lysophosphatidylcholines, phosphatidylcholines, sphingomyelins, ceramides, and cholesteryl esters were analyzed by flow injection analysis (FIA) with total analysis time of approximately 3.8 minutes per sample. Biocrates provided the FIA mobile phase buffer (Part 9120052121018), which was diluted into LC-MS grade methanol for use with the kit, per manufacturer instructions. Using electrospray ionization in positive ion mode, samples for both UHPLC and flow injection analysis were introduced directly into Q Exactive™ Orbitrap MS systems operating in the full scan or parallel reaction monitoring (PRM) mode. Four different Q Exactive instrument platforms were represented in the ring trial, including at least two data sets each from Q Exactive, Q Exactive Focus, Q Exactive Plus, and Q Exactive HF mass spectrometers. The kit is not currently deployed on the Orbitrap Tribrid™ systems. Acquisition methods and tune parameters for all instruments were provided by Biocrates as part of the p400HR kit. Because of the wide variety of LC systems used, each lab was responsible for programming their own LC methods, closely following the kit SOP. System suitability testing (described above) was used to ensure each laboratory properly programmed the method.
Data Analysis
The LC-MS data were imported into the QuanBrowser module of the Thermo Xcalibur software (Thermo Fischer Scientific) for peak integration and quantification, then imported into MetIDQ ™software package (Biocrates AG). Quantification was performed using the ratio of analyte to stable-isotope internal standard to calculate response, and response was calibrated against a seven-point calibration curve. Linear regression with 1/x2 weighting was used for curve fits. During this process, laboratories were instructed to remove calibration points for which the calculated amount vs. the theoretical amount was >20 % for the lowest calibration point and >15 % for the higher level calibration standards, to maintain consistency with FDA guidance for bioanalytical methods. The accurate mass FIA-MS data were analyzed directly using Biocrates MetIDQ™ software. Data was extracted with 5 ppm mass accuracy for analytes and internal standards; quantification for lipids and other FIA-MS analytes was calculated using stable-isotope dilution to class-based internal standards (followed by single-point normalization described below). Each laboratory exported data as a *.metidq project file, and uploaded to the Express data repository at Duke University (https://discovery.genome.duke.edu/express/). Individual *.metidq projects were imported into a single MetIDQ repository and then exported as an aggregate. The LC-MS based quantification (amino acids and biogenic amines) was exported without additional normalization of any type, as the µM concentration values delivered by each laboratory. As detailed in the ring trial publication of the AbsoluteIDQ p180 kit, we adopted the approach of exporting the FIA-MS quantitative values after normalization to the QC2 (medium level quality control) sample on each p400HR plate.6 Subsequent data analysis was performed in Excel (Microsoft) and JMP Pro v14 (SAS Institute, Cary, NC). Percent coefficient of variation (%CV) was calculated as standard deviation divided by average for each analyte within (intra-) and among (inter-) laboratories.
Results and Discussion
System Suitability
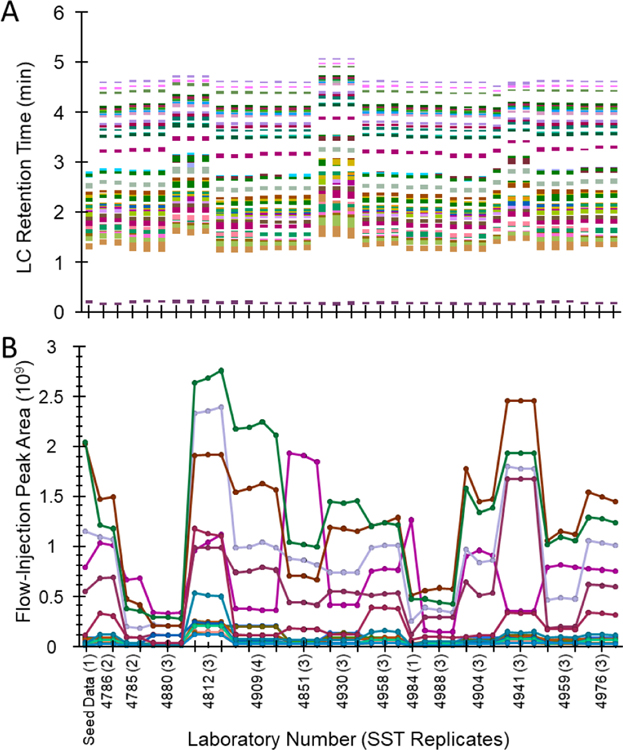
System suitability testing (SST) is essential for evaluating instrument performance prior to sample analysis, and optimally will be done prior to sample preparation. In this study, 41 analytes in the LC-MS test mix and 17 analytes in the FIA-MS test mix were used to evaluate instrument performance, including signal abundance, mass accuracy, retention time, and peak shapes. Data collected for the LC-MS and FIA-MS test mixes within each laboratory was imported into Skyline v4.1 for interpretation as an aggregate. The UHPLC-MS and FIA-MS aggregate SST files have been made available on the PanoramaPublic repository https://panoramaweb.org/p400HR_SST_RingTrial.url. Skyline enables easy visualization and comparison of retention time, peak shape, intensity, and mass accuracy across all laboratories. Raw values for each of these parameters were exported and analyzed in JMP Pro using Principal Component Analysis (PCA), restricted maximum likelihood (REML) method. Mass accuracy during the SST analysis was observed to be less than 2 ppm for LC-MS and less than 4 ppm for FIA-MS analysis for all laboratories. Peak shape and retention time in the LC-MS were highly reproducible (Figure 2A). Two laboratories were found to be preliminary outliers based on PCA of the retention time data (Figure S-2). One laboratory (4904) showed extensive chromatographic peak tailing for taurine, but the reason could not be readily determined. Since quantification is based on abundance ratio to internal standard, these laboratories were not excluded from downstream data analysis even though performance may have ultimately been slightly improved by troubleshooting the source of retention time shift and peak tailing. In the FIA-MS SST analyses, all instrument platforms displayed appropriate flow-injection peak shapes and low background before and after peak elution. Figure 2B shows the peak area for each of the 17 FIA-MS SST compounds; PCA analysis of this data (Figure S-3) shows one laboratory (4812) as an outlier, due to overall higher peak intensity compared to the other laboratories. Higher overall instrument response would not be expected to result in poorer kit performance, so this laboratory was not excluded from downstream analysis. Based on combined analysis of the system suitability data, no participating laboratories were excluded.
Figure 2.
System Suitability Test (SST) visualizations from Skyline. (A) Observed retention time for UHPLC-MS SST injections across all participating laboratories, each color representing a different analyte out of the total 41 used. (B) Observed intensities for each of 17 FIA-MS SST analytes. The blinded laboratory ID is the four digit code and the number of SST replicates returned per laboratory is shown in parenthesis.
In Figure 2B most analytes increase or decrease together, indicating generally ‘higher’ or ‘lower’ total instrument signal, however some trendlines do suggest certain machines have analyte- or class-specific bias (either higher or lower response). Considering all instruments are from the Q-Exactive line, it might be expected that such drastic changes in intensity may lead to wide inter-laboratory variance in concentration values. However, as will be shown in the following sections, all instrumentation in the study gave fundamentally good results with the kit, presumably due to the benefits of external calibration and use of stable isotope internal standards. Therefore, we believe the SST Skyline documents available on Panorama will serve as a reasonable set of boundary SST conditions for laboratories interested in using the p400HR kit.
Data Aggregation Across Laboratories
Data was exported from MetIDQ for UHPLC-MS and FIA-MS analyses, as concentration values (µM) for all sample matrices. Data from all 408 possible analytes in all four injection modes (two LC-MS and two FIA-MS) were combined and the data were organized such that each sample matrix is grouped together. The laboratories are identified by their unique four-digit code (termed “Project Number”). Values reported as “<LOD” (below the lower limit of detection) in the individual laboratory exports were treated as missing values and blank spaces were left in the data table. The compiled Ring Trial quantitative data for all 36 samples, 14 laboratories, and 408 analytes is reported in Table S-1 (supplementary information). This data matrix represents a total of over 131,000 quantitative metabolite measurements performed in the context of this ring trial, excluding calibrators and quality control samples.
Compounds Reliably Detected
The p400HR platform can detect and quantify up to 408 compounds across eleven different compound classes. We first sought to utilize the aggregate data from all laboratories and all matrices to determine the number of analytes that a user might reasonably expect to detect using the kit. We chose a robust threshold for analytes ‘reliably detected’ by allowing only 20% missing values across all laboratories, within a single sample type; a rationale can easily be made for other missing value thresholds, therefore the missingness for each analyte and each matrix is detailed in Table S-1. A missing value indicates either that no peak was observed or the quantified value was calculated to be less than the defined lower limit of detection (at times, due to high blank background in FIA-MS/MS).
While more sophisticated methods could certainly be used, this method is simple to understand and to implement and is consistent with previous data processing practices in large-scale metabolomics studies.11 Since three individual male and female plasma samples were prepared in triplicate by each laboratory, there were 126 possible measurements for each analyte for those matrices, while for the other sample types there was a single sample, each prepared three times, for a total of 42 measurements (Table 1, Column 1). To simplify data analysis and attempt to observe broad trends, the data was summarized by observations made within each of the eight major metabolite classes targeted by the kit. The top row of Table 1 lists these classes, as well as the total number of metabolites targeted by the kit in each class, listed in parentheses.
Table 1.
Analyte detection by class and sample type. The table lists the number of possible analytes detected in each analyte class by the p400HR kit (top row). Each column then represents the number of analytes reliably detected across all laboratories (<20% missing values) in each sample matrix tested. Amino Acids and Biogenic Amines were measured by LC-HRMS, other classes by FIA-MS/MS. Note samples from three individuals for male plasma and female plasma were tested, thus the higher number of potential observations for those matrices.
| Sample Type (total measurements) | Amino Acids (21) | Biogenic Amines (21) | Acylcarnitines (55) | Glycerolipids (60) | Glycerophospholipids (196) | Cholesteryl Esters (14) | Sphingolipids (40) | Total Hexose (1) | Total Analytes (408) |
|---|---|---|---|---|---|---|---|---|---|
| Plasma, male (126) | 21 | 10 | 17 | 33 | 81 | 12 | 30 | 1 | 205 |
| Plasma, female (126) | 21 | 10 | 13 | 32 | 76 | 10 | 28 | 1 | 191 |
| Plasma, lipemic (42) | 21 | 12 | 13 | 50 | 84 | 10 | 31 | 1 | 222 |
| Pooled Plasma, NIST SRM-1950 (42) | 21 | 8 | 16 | 47 | 79 | 11 | 27 | 1 | 210 |
| Serum, male (42) | 21 | 11 | 17 | 36 | 84 | 13 | 31 | 1 | 214 |
| Serum, female (42) | 21 | 12 | 13 | 38 | 91 | 12 | 30 | 1 | 218 |
| Plasma, rattus norvegicus (42) | 21 | 12 | 7 | 42 | 70 | 9 | 16 | 1 | 178 |
| Plasma, mus musculus (42) | 20 | 13 | 9 | 43 | 80 | 11 | 20 | 1 | 198 |
Table 1 gives a conservative estimation of the number of analytes which a user of the p400HR might expect to detect, based on only 20% missing data across all 14 laboratories for each sample matrix type, as compiled from the complete data set reported in Table S-1. For instance, all laboratories essentially observed 100% complete data for amino acids, reporting 21 analytes for all sample matrices with very little missing data. The lone exception is aspartic acid (Asp) in mouse plasma, which was measured at an average of 3.0 µM with 26% missing data (the analyte was quantified in 31 out of 42 possible measurements), likely due to the poor relative stability of this analyte. For biogenic amines, a median of 11 metabolites were reported without missing data, out of 21 possible analytes. Consistent measurement of amino acids and amines in plasma samples is similar to the results obtained in the interlaboratory study of the p180 kit.5 Histamine and serotonin seem more likely to be observed in the animal model (rat, mouse) matrices than in human samples using the kit. While the p400HR kit targets 15 extra acylcarnitines compared to the p180 kit, we did not find that more acylcarnitines (AC) were typically measured, with a median of 13 AC reliably detected at 20% missingness (21 analytes at 40% missing). Interestingly, the laboratories found that roughly half as many AC were reliably detected in rat and mouse samples than in the human samples. Total hexoses were reproducibly detected in all matrices and laboratories, with data completeness of 100%. Forty different sphingolipid species (sphingomyelins (SM) and ceramides (Cer)) are targeted by the kit, and Table 1 shows that for all human samples tested, between 27 and 31 sphingolipid species were reliably detected depending on sample type. Slightly fewer SM species appear to be routinely detected in rat and mouse plasma. Similar to AC, the expanded set of glycerophospholipids targeted by the p400HR kit, including phosphatidylcholine (PC) and lysophosphatidylcholine (LPC), did not seem to result in a higher number of reliably measured compounds in plasma or serum, with an average of 80 L/PC species routinely measured above LOD (40% of the total targeted glycerophospholipids).
The p400HR kit targets a wide variety of lipid species, with diacyl- and triacylglycerols (DAG, TAG), ceramide (Cer), and cholesteryl esters (ChoE) as new additions to Biocrates targeted metabolomics kits. Additionally, because of the high mass-resolving power of the Q Exactive, the p400HR measurements are able to independently resolve nominally-isobaric phosphatidylcholine (PC) lipids (such as PC(34:4), 754.53 m/z vs. PC-O(35:4), 754.567 m/z), which are reported together in the p180 kit.11 Out of 60 potential glycerolipids (DAG+TAG), the study found that most labs were routinely able to measure the majority of them in all tested matrices (Table 1). ChoE measurements are equally reliable with an average of 12 (out of 14) ChoE species measured reliably in human blood samples. Overall, out of 408 possible metabolites measured, the aggregate data from all labs shows that in the human samples between 178 and 222 metabolites were measured with <20% missing data across all laboratories (Table 1). It is important to state that within any one laboratory, the number of analytes was significantly higher, with an ‘average’ sample measurement consisting of 261±22 (mean ± stdev) metabolites above the LOD (Table S-1). Much of the difference in ‘analytes detected’ when aggregating across laboratories seems to stem from differences in lower limits of detection for individual metabolites in each lab, which is addressed below.
Inter- and Intra-laboratory Variance
Inter- and intra-laboratory variance were evaluated utilizing for each of the sample matrices. Metrics including inter-lab CV (% relative standard deviation) as well as the average, median, min and max intra-lab CV and total missing values were calculated for all matrices and analytes (Table S-2). In order to calculate metrics for typical performance, it is important to remove significantly outlying measurements. Prior to these calculations, the quantitative data was analyzed across all sample types using Principal Components Analysis (PCA) to detect sample or laboratory outliers, with each analyte class analyzed separately in order to detect potential problems in a single analyte subtype or class. The data from Table S-1 was used as input in to PCA in JMP v14. The data revealed the majority of variance by sample type, and an example is shown in Figure S-4A, where mouse and rat plasma clearly differentiate in LysoPC content from the human samples. Figure S-4B shows the same situation for acylcarnitines. Based on this analysis, all laboratories were included in subsequent variance analysis.
Because of the widespread utilization of the NIST SRM-1950 pooled plasma reference material in the metabolomics community, we selected to perform a detailed examination of inter- and intra-laboratory variance using the three preparations of this sample that were performed in each of 14 labs (for a total of 42 possible measurements). Concentration values from Table S-1 for analytes with a minimum of 31 measurements reported (out of 42 possible, i.e. >80% of the time) were used for statistical analysis of all analyte classes; the subset report for NIST SRM-1950 in detail, with values from each laboratory as well as the inter-and intra-laboratory metrics, is reported in Table S-3. Using this robust set of 208 analytes, Table 2 shows the compiled results for each analyte class as the median CV (min CV and max CV is also reported in order to describe the range of analyte variance within each class). As expected, intra-laboratory imprecision was low for all analyte classes, with the best performance observed for amino acids (5.7% median %CV), while all analyte classes were below 15% CV. Inter-laboratory imprecision varied between analyte classes, with the median %CV for amino acids, cholesteryl esters, sphingolipids, and total hexoses below 20% CV between labs; biogenic amines, glycerolipids and glycerophospholipids below 25% between labs; and acylcarnitines showing median of 38% CV between labs. In summary, the p400HR platform demonstrated good inter-laboratory precision (<25% CV), with the exception of a few lower-abundance acylcarnitines and lipid species.
Table 2.
Inter- and Intra-laboratory performance for the NIST SRM-1950 by analyte class, for 208 analytes which were observed 80% of the time or more across 14 laboratories. n* refers to the number of analytes used from each class in the reproducibility calculation.
| Analyte Class (n*) | Platform | Median Inter-Lab %CV (min, max) | Median Intra-Lab %CV (min, max) |
|---|---|---|---|
| Amino acids (21) | UHPLC | 10.2 (7.2, 30.1) | 5.7 (3.8, 9.7) |
| Biogenic Amines (8) | UHPLC | 24.0 (8.2, 66.4) | 7.8 (4.5, 16.0) |
| Acylcarnitines (16) | FIA | 38.2 (8.9, 113) | 11.2 (5.1, 15.7) |
| Glycerolipids (47) | FIA | 24.8 (18.6, 306) | 13.0 (9.3, 20.0) |
| Glycerophospholipids (79) | FIA | 22.6 (11.3, 181) | 8.5 (5.6, 41.0) |
| Cholesteryl Esters (11) | FIA | 15.9 (12.0, 108) | 7.9 (7.7, 9.9) |
| Sphingolipids (27) | FIA | 15.1 (12, 58.7) | 9.2 (7.5, 20.9) |
| Hexoses (1) | FIA | 9.4 | 5.4 |
Notably, some acylcarnitine measurements were quite reproducible between labs (e.g. AC(0:0) = 8.9% CV, AC(2:0) = 10.9%) while many others performed comparably poorly (e.g. AC(5:0) = 63% CV, AC(16:0) = 84% CV) or had high levels of missing data and were not reported (e.g. AC(5:1), 95% missing data and AC(5:1-DC), 88% missing data). Investigations revealed that in the SST mixture analyses, acylcarnitines generally showed higher mass errors than the other compounds, many times approaching 4 ppm. Reanalysis of a subset of raw data in Skyline demonstrated that when analyzing plasma samples, high ion flux (due to AGC target value setting of 3e6) in the acylcarnitine mass windows was observed and presumably due to space-charging effects the measured mass error for acylcarnitine analytes and internal standards was often greater than the targeted extraction window of 5 ppm, which one might expect under conditions of large ion flux in a narrow mass window in an Orbitrap™ mass analyzer.12 Therefore future versions of the kit might benefit from utilizing lower AGC target values and/or tandem MS for measurement of acylcarnitines. It is also important to consider that many of the acylcarnitines targeted, such as hydroxyl and dicarboxylic acid forms, are considered ‘exotic’ or trace level acylcarnitines and may be expected to have higher variability and to be found in fewer biological samples.
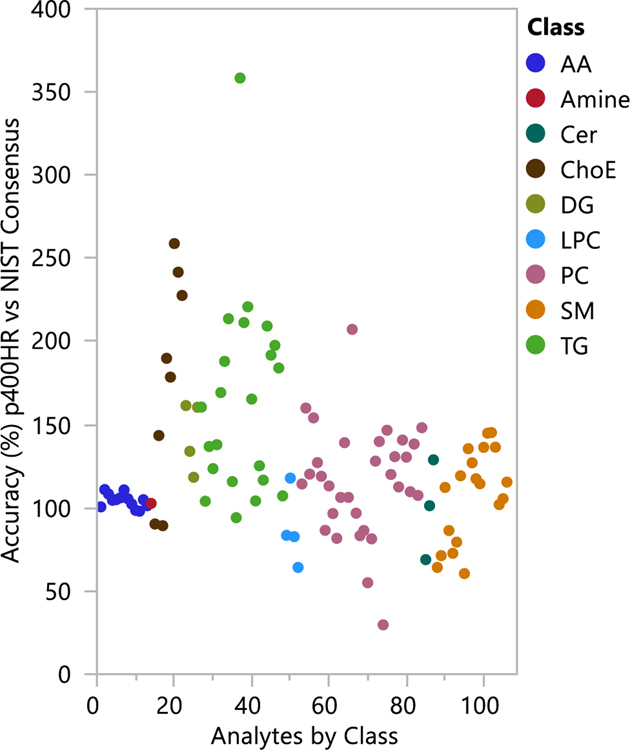
Accuracy of the p400HR Kit
We compared the quantitative values obtained for each analyte measured in this ring trial to the reference values or consensus values from the NIST SRM-1950 material, to determine the accuracy of the p400HR kit.9 We only considered analytes measured in at least five laboratories in the Bowden lipidomics harmonization paper with RSD<40%, and those analytes measured in at least 80% of the ring trial laboratories also with RSD<40%. There were 254 lipids in Bowden et al with these criteria, plus the amino acids and creatinine from the NIST SRM-1950 certificate of analysis. There were 163 lipids in our p400HR results with <20% missing values for NIST SRM-1950. The accuracy of the p400HR kit was calculated versus the reference (amino acids) or consensus (lipids) values for the NIST SRM-1950, and results are reported in Table S-4 and plotted in Figure 3. Analytes are colored by class in Figure 3, divided into amino acids (AA), amines, and the various lipid classes. Amino acids showed better accuracy than lipids on the whole; nonetheless, 79% of all analytes measured by the kit with good reproducibility between labs have accuracy between 50–150% (i.e. <50% bias) when compared to the established consensus value for the NIST SRM-1950. Root mean square (RMS) bias by analyte class demonstrated that some classes do perform better than others; RMS bias for amino acids was 5.9%, lysoPC 23%, PC 36%, ceramides 25%, SM 28%, and DG 47%. Larger or more complex lipids showed higher bias on average, with TG and ChoE having the highest deviation from the consensus (RMS bias 88% and 98%, respectively). These measurements demonstrate superior performance of the LC-HRMS portion of the platform compared to FIA-HRMS for the purposes of accuracy.
Figure 3.
Accuracy of the p400HR ring trial data compared to consensus data for the NIST SRM-1950 reference plasma, expressed as a percentage of the consensus value, and colored by analyte class. 106 compounds, including 92 lipids, were compared to the NIST consensus. 79% of compared analytes demonstrated accuracy of 50–150% (<50% bias) relative to the reference value.
Limits of Detection for FIA-MS Analysis
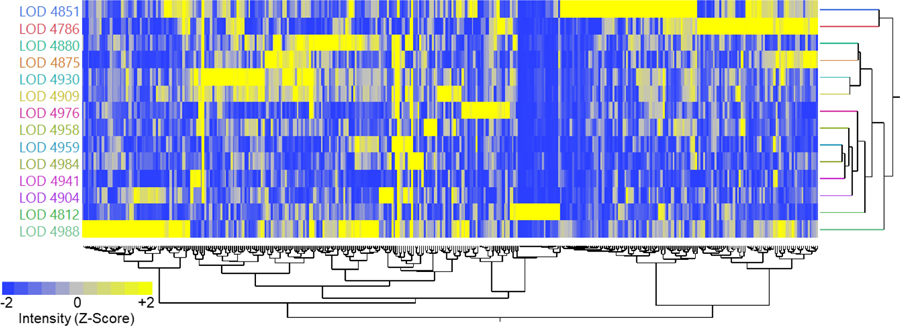
As discussed above, it seemed that differences in detectability of lower-abundance metabolites (Table 1) seemed to potentially be caused by differences in lower limit of detection between labs. Based on the Biocrates SOP, the lower limit of detection is defined within the flow injection analysis (FIA) as three times the level of signal in the blank, which is extracted and analyzed immediately prior to running the samples. Therefore, an important parameter for having good sensitivity for acylcarnitines and lipids with the p400HR kit is to have a clean background. To compare the laboratories without weighting one analyte more than the rest, the lower limit of detection (LOD) for each analyte was z-score normalized such that the mean=0 and standard deviation=1. This metric allows the easy visualization of whether each laboratory was within or outside the normal distribution for LOD values, with the assertion that a higher-than average LOD due to higher background noise in the FIA signal may lead to a higher proportion of ‘missing’ values for low abundance analytes. A principal components analysis (PCA, Figure S-5) and a 2D hierarchical clustering analysis (Figure 4) were performed in order to observe trends in LOD between labs. Bright yellow color in Figure 4 denotes laboratories that are 2 standard deviations or more above the mean LOD for that analyte; therefore, bright yellow analytes are those with higher background. Eleven out of the 14 laboratories clustered tightly in the PCA (Figure S-5), yet 2D clustering (Figure 4) shows that while two of the outlier laboratories (4851 and 4786) share a modest number of lipids with high background, most of the lipid signals show no discernible trend.
Figure 4.
2D Hierarchical clustering, after Z-Score transformation, of the LOD values observed during FIA-MS experiments from each lab. Bright yellow indicates 2 or more standard deviations above the mean for that analyte, interpreted as a measurement with higher than average background for the accurate mass region corresponding to a particular lipid signal. The important finding is that stochastic background from undetermined sources, which vary between labs, may play a key role for inter-laboratory reproducibility for low abundance lipid measurements.
Based on not observing any consistent patterns in Figure 4, the critical finding of this analysis is that stochastic laboratory-based contamination for FIA-MS analysis, with unclear sources, may be a real barrier to reproducible analyses between laboratories in flow-injection based lipidomics workflows for analytes which are at or near the lower limit of detection. Meanwhile, LC-MS analysis appeared to be more reproducible. One may speculate that differences in glassware, plasticware, solvents, and atmospheric contaminants may all play a role in these inter-laboratory LOD differences. This study did not address, and it remains unclear, how much longitudinal variability (even within a single lab) background contamination will cause for p400HR kit and for similar workflows. It is also unclear, and has not been addressed in any systematic way, what role background contamination may play in limiting lipid analysis reproducibility in LC-MS based workflows.
Inter-Laboratory Variance is Typically Less Than Biological Variance
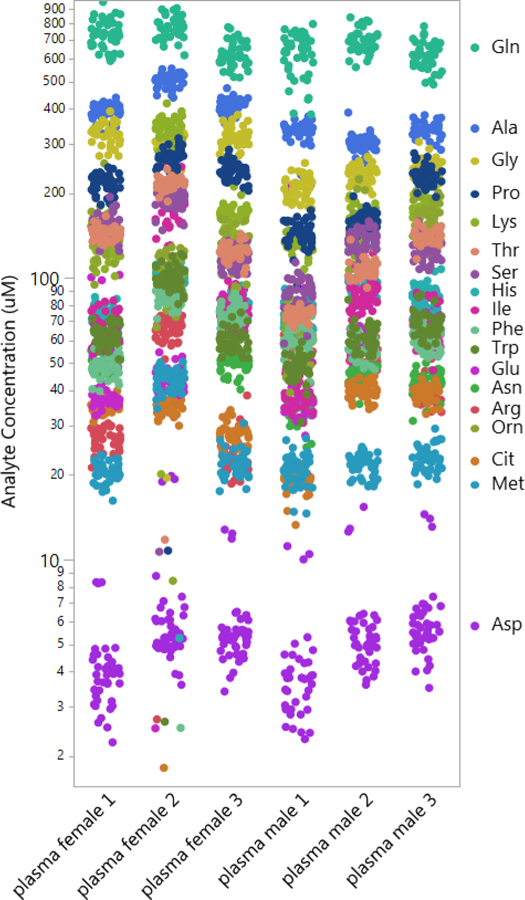
It is advantageous for an assay to be capable of a broad detection of endogenous metabolites with a potential large dynamic range across different matrices. Micromolar metabolite concentrations from all matrices tested, in all participating laboratories, is returned in Table S-1. Performance metrics across all sites is compiled in Table S-2. As an example, Figure 5 shows the raw concentration values obtained for amino acids from all the laboratories, spanning concentration values from approximately 4 µM (aspartic acid) to 600 µM (glutamine). This visual analysis demonstrates that the variance between laboratories is smaller than the variance observed between these randomly selected human subjects. Similarly, an unbiased 2D hierarchical cluster analysis (Figure S-6) including all samples measured in the study shows that clustering occurs primarily by sample type, not by laboratory. These analyses support the hypothesis that broad-spectrum targeted metabolomics represents a viable methodology for detecting metabolite differences that may occur between individuals due to factors such as disease, diet, or medication.
Figure 5.
Quantitative amino acid measurements performed across 14 laboratories using the p400HR kit for six representative human samples, three male and three female. Each color represents an individual analyte. The technical reproducibility between laboratories enables facile visualization of differences in amino acid concentration between individual human subjects.
Sources of Variance and Outliers
In order to investigate sources of potential variability and gain understanding of areas where targeted metabolomics analyses can be improved, a variety of meta-analyses were performed focusing on analytes which showed single-analyte or single-lab outliers. In the case of tyrosine, 13 out of 14 laboratories reported values for NIST SRM-1950 of 61±5 µM, while lab 4904 returned a value of 0.7±0.1 µM, nearly 100-fold different than other groups (Figure S-7 Panel A). Values for other analytes from this laboratory were in line with measurements from other groups, but the SST showed a large retention time shift for this analyte in lab 4904, highlighting the need for analyte-specific system suitability testing, in line with FDA guidance (https://www.fda.gov/downloads/drugs/guidances/ucm134409.pdf). Creatinine measurements in NIST SRM-1950 are also informative (Figure S-7 Panel B). In this case, a single measurement from a single laboratory (4988) was observed as an outlier, with values nearly double all other measurements and more than 12 standard deviations outside the mean (60±3 µM) from the other 41 measurements across 14 labs. Intriguingly, further investigation revealed this was not because of poor sample preparation or a missed injection, as other measurements in the same injection were within specification. Most likely, this error occurred because of incorrect automated integration of analyte peak or under-integration of the internal standard, highlighting the need for further development in software to both improve automated data processing and identify outliers for manual intervention. Such developments will be critical for accurate measurements in precision medicine metabolomics initiatives where large data streams make manual data curation impractical.
Conclusion
The results of this ring trial demonstrate that high resolution mass spectrometers, specifically in this case Q Exactive platforms, are able to provide reproducible and accurate targeted metabolomics data, given proper usage of system suitability testing, adherence to protocol documentation, and usage of calibration curves and stable isotope internal standards. The Biocrates p400HR kit is an example of such an approach, which provides broad metabolome coverage, good reproducibility between laboratories, and generally accurate results. An important step taken in this ring trial not highlighted in previous studies of this type is the importance of cross-laboratory quantitative comparisons of the system suitability data in order to make sure that the LC-MS system is properly configured and calibrated for making the measurements. We have provided the Skyline files containing the LC-MS and FIA-MS system suitability data from the study, so that future users of the p400HR kit might directly compare their SST data to that of the ring trial participants prior to starting sample preparation. Moreover, the ring trial implemented the use of a training/validation kit which had to be analyzed prior to the ring trial sample kit in order to familiarize each laboratory with sample preparation, data collection, and data analysis procedures. We believe this type of training potentially improved participants’ performance in the ring trial, and demonstrates that it would be reasonable to consider metabolomics proficiency testing in future metabolomics studies similar to that used by the CDC in newborn screening, particularly those studies and platforms geared towards translational medicine.13
Utilizing the p400HR kit, the variance in intra-laboratory repeat measurements and intra-laboratory measurements were typically far below what was observed between-sample biological variance for the three male and three female plasma samples. While n=3 is far too few to estimate population variance, this dataset nonetheless suggests that with a kit such as the p400HR and appropriate between-plate control samples (such as the NIST SRM-1950), the analysis of population-based metabolomics studies should be comparable between analysts and laboratories. More broadly, we observed better performance in both precision and accuracy for those analytes measured using external calibration and LC-HRMS than those measured with stable-isotope dilution (single point) quantification by FIA-HRMS (Figure S-8), suggesting that multi-point calibration curves and chromatographic separation should be used to obtain the most accurate data, when analytically and financially feasible. Clearly this is a community-wide challenge in lipidomics with few purified reference materials available, however our data supports the idea that the NIST SRM-1950 or similar reference materials should serve as valuable single-point external calibrators in lipidomics studies. The NIST SRM-1950 is commercially available and could theoretically make a significant impact in harmonization efforts if each metabolomics study extracted, analyzed, and published this data in parallel.
Supplementary Material
ACKNOWLEDGMENT
The authors gratefully acknowledge Helen Karuso for her support of this study. This work was performed in part using a Q-Exactive Plus tandem mass spectrometer acquired via the S10 grant 1 S10OD012266–01A1 (MA Moseley). FMF acknowledges support from 1R01CA218664–01, a CF Foundation RDP grant, NIH 1U2CES030167–01, the CMaT NSF Research Center (EEC-1648035), and the NIH MoTrPAC Consortium (1U24DK112341–01).
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website. The supplemental document p400HR_AChem_SupplementalInformation.pdf includes Ring Trial Guidance Document distributed to each laboratory for the purposes of performing the ring trial, Supplemental Table Descriptions, Supplemental Figures, and Supplemental Figure descriptions. Supplemental Tables.xlsx includes supplemental Tables S-1 through S-4.
Certain commercial equipment or instruments are identified in the paper to specify adequately the experimental procedures. Such identification does not imply recommendations or endorsement by NIST; nor does it imply that the equipment or instruments are the best available for the purpose.
REFERENCES
- 1.Kosmides AK; Kamisoglu K; Calvano SE; Corbett SA; Androulakis IP, Metabolomic fingerprinting: challenges and opportunities. Crit Rev Biomed Eng 2013, 41 (3), 205–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tzoulaki I; Ebbels TM; Valdes A; Elliott P; Ioannidis JP, Design and analysis of metabolomics studies in epidemiologic research: a primer on -omic technologies. Am J Epidemiol 2014, 180 (2), 129–39. [DOI] [PubMed] [Google Scholar]
- 3.Beger RD; Dunn W; Schmidt MA; Gross SS; Kirwan JA; Cascante M; Brennan L; Wishart DS; Oresic M; Hankemeier T; Broadhurst DI; Lane AN; Suhre K; Kastenmuller G; Sumner SJ; Thiele I; Fiehn O; Kaddurah-Daouk R; for “Precision, M.; Pharmacometabolomics Task Group”-Metabolomics Society, I., Metabolomics enables precision medicine: “A White Paper, Community Perspective”. Metabolomics 2016, 12 (10), 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kirwan JA; Brennan L; Broadhurst D; Fiehn O; Cascante M; Dunn WB; Schmidt MA; Velagapudi V, Preanalytical Processing and Biobanking Procedures of Biological Samples for Metabolomics Research: A White Paper, Community Perspective (for “Precision Medicine and Pharmacometabolomics Task Group”-The Metabolomics Society Initiative). Clin Chem 2018, 64 (8), 1158–1182. [DOI] [PubMed] [Google Scholar]
- 5.Pham HT; Arnhard K; Asad YJ; Deng L; Felder TK; St John Williams L; Kaever V; Leadley M; Mitro N; Muccio S; Prehn C; Rauh M; Rolle-Kampczyk U; Thompson JW; Uhl O; Ulaszewska M; Vogeser M; Wishart DS; Koal T, Inter-Laboratory Robustness of Next-Generation Bile Acid Study in Mice and Humans: International Ring Trial Involving 12 Laboratories. J. Appl. Lab Med 2016, 01 (02), 129–142. [DOI] [PubMed] [Google Scholar]
- 6.Siskos AP; Jain P; Romisch-Margl W; Bennett M; Achaintre D; Asad Y; Marney L; Richardson L; Koulman A; Griffin JL; Raynaud F; Scalbert A; Adamski J; Prehn C; Keun HC, Interlaboratory Reproducibility of a Targeted Metabolomics Platform for Analysis of Human Serum and Plasma. Anal Chem 2017, 89 (1), 656–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collins BC; Hunter CL; Liu Y; Schilling B; Rosenberger G; Bader SL; Chan DW; Gibson BW; Gingras AC; Held JM; Hirayama-Kurogi M; Hou G; Krisp C; Larsen B; Lin L; Liu S; Molloy MP; Moritz RL; Ohtsuki S; Schlapbach R; Selevsek N; Thomas SN; Tzeng SC; Zhang H; Aebersold R, Multi-laboratory assessment of reproducibility, qualitative and quantitative performance of SWATH-mass spectrometry. Nat Commun 2017, 8 (1), 291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Phinney KW; Ballihaut G; Bedner M; Benford BS; Camara JE; Christopher SJ; Davis WC; Dodder NG; Eppe G; Lang BE; Long SE; Lowenthal MS; McGaw EA; Murphy KE; Nelson BC; Prendergast JL; Reiner JL; Rimmer CA; Sander LC; Schantz MM; Sharpless KE; Sniegoski LT; Tai SS; Thomas JB; Vetter TW; Welch MJ; Wise SA; Wood LJ; Guthrie WF; Hagwood CR; Leigh SD; Yen JH; Zhang NF; Chaudhary-Webb M; Chen H; Fazili Z; LaVoie DJ; McCoy LF; Momin SS; Paladugula N; Pendergrast EC; Pfeiffer CM; Powers CD; Rabinowitz D; Rybak ME; Schleicher RL; Toombs BM; Xu M; Zhang M; Castle AL, Development of a Standard Reference Material for metabolomics research. Anal Chem 2013, 85 (24), 11732–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bowden JA; Heckert A; Ulmer CZ; Jones CM; Koelmel JP; Abdullah L; Ahonen L; Alnouti Y; Armando AM; Asara JM; Bamba T; Barr JR; Bergquist J; Borchers CH; Brandsma J; Breitkopf SB; Cajka T; Cazenave-Gassiot A; Checa A; Cinel MA; Colas RA; Cremers S; Dennis EA; Evans JE; Fauland A; Fiehn O; Gardner MS; Garrett TJ; Gotlinger KH; Han J; Huang Y; Neo AH; Hyotylainen T; Izumi Y; Jiang H; Jiang H; Jiang J; Kachman M; Kiyonami R; Klavins K; Klose C; Kofeler HC; Kolmert J; Koal T; Koster G; Kuklenyik Z; Kurland IJ; Leadley M; Lin K; Maddipati KR; McDougall D; Meikle PJ; Mellett NA; Monnin C; Moseley MA; Nandakumar R; Oresic M; Patterson R; Peake D; Pierce JS; Post M; Postle AD; Pugh R; Qiu Y; Quehenberger O; Ramrup P; Rees J; Rembiesa B; Reynaud D; Roth MR; Sales S; Schuhmann K; Schwartzman ML; Serhan CN; Shevchenko A; Somerville SE; St John-Williams L; Surma MA; Takeda H; Thakare R; Thompson JW; Torta F; Triebl A; Trotzmuller M; Ubhayasekera SJK; Vuckovic D; Weir JM; Welti R; Wenk MR; Wheelock CE; Yao L; Yuan M; Zhao XH; Zhou S, Harmonizing lipidomics: NIST interlaboratory comparison exercise for lipidomics using SRM 1950-Metabolites in Frozen Human Plasma. J Lipid Res 2017, 58 (12), 2275–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kanemoto N; Okamoto T; Tanabe K; Shimada T; Minoshima H; Hidoh Y; Aoyama M; Ban T; Kobayashi Y; Ando H; Inoue Y; Itotani M; Sato S, Antidiabetic and cardiovascular beneficial effects of a liver-localized mitochondrial uncoupler. Nat Commun 2019, 10 (1), 2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.St John-Williams L; Blach C; Toledo JB; Rotroff DM; Kim S; Klavins K; Baillie R; Han X; Mahmoudiandehkordi S; Jack J; Massaro TJ; Lucas JE; Louie G; Motsinger-Reif AA; Risacher SL; Alzheimer’s Disease Neuroimaging, I.; Alzheimer’s Disease Metabolomics, C.; Saykin AJ; Kastenmuller G; Arnold M; Koal T; Moseley MA; Mangravite LM; Peters MA; Tenenbaum JD; Thompson JW; Kaddurah-Daouk R, Targeted metabolomics and medication classification data from participants in the ADNI1 cohort. Sci Data 2017, 4, 170140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Makarov A; Denisov E; Kholomeev A; Balschun W; Lange O; Strupat K; Horning S, Performance evaluation of a hybrid linear ion trap/orbitrap mass spectrometer. Anal Chem 2006, 78 (7), 2113–20. [DOI] [PubMed] [Google Scholar]
- 13.De Jesus VR; Mei JV; Cordovado SK; Cuthbert CD, The Newborn Screening Quality Assurance Program at the Centers for Disease Control and Prevention: Thirty-five Year Experience Assuring Newborn Screening Laboratory Quality. Int J Neonatal Screen 2015, 1 (1), 13–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.