Abstract
The increasing accumulation of healthcare data provides researchers with ample opportunities to build machine learning approaches for clinical decision support and to improve the quality of health care. Several studies have developed conventional machine learning approaches that rely heavily on manual feature engineering and result in task-specific models for health care. In contrast, healthcare researchers have begun to use deep learning, which has emerged as a revolutionary machine learning technique that obviates manual feature engineering but still achieves impressive results in research fields such as image classification. However, few of them have addressed the lack of the interpretability of deep learning models although interpretability is essential for the successful adoption of machine learning approaches by healthcare communities. In addition, the unique characteristics of healthcare data such as high dimensionality and temporal dependencies pose challenges for building models on healthcare data. To address these challenges, we develop a gated recurrent unit-based recurrent neural network with hierarchical attention for mortality prediction, and then, using the diagnostic codes from the Medical Information Mart for Intensive Care, we evaluate the model. We find that the prediction accuracy of the model outperforms baseline models and demonstrate the interpretability of the model in visualizations.
Keywords: Health care, electronic health records, deep learning, recurrent neural networks, attention, interpretability, visualization
1. INTRODUCTION
Health care is undergoing a revolution. The accumulation of big data in health care such as electronic health records (EHR) and claims data has enabled researchers to use machine learning techniques to learn from these data and build clinical decision support tools. Such capabilities facilitate timely and accurate predictions of medical risks of patients with regard to readmissions or deaths, and thus improve the quality and efficiency of clinical interventions. Several studies have demonstrated that health information technologies have improved the quality of health care [10; 11; 17].
Although healthcare data analytics offer promising opportunities for improving health care, key challenges remain unsolved regarding how to appropriately model healthcare data. Such data, such as EHR, have unique characteristics, including high dimensionality and temporal dependencies. With regard to the former, for example, a standard coding system of diagnosis, the International Classification of Diseases (ICD-9-CM), contains approximately 20,000 codes; and with regard to the latter, for example, the existence of one disease might increase the risk of one patient having certain diseases in the future. In addition, representing healthcare data is not a trivial task since medical histories vary greatly in length and contain hierarchical structures. That is, a typical healthcare record of one patient is a variable-sized set of discrete elements such as diagnostic codes with variable numbers.
An ideal model for healthcare data should not only appropriately model the unique structure of healthcare data but also maintain a balance between accuracy and interpretation. Conventional predictive models of clinical decision support rely heavily on feature engineering, such as ranking and selecting features based on certain criteria [12; 15; 31], which results in classifier-dependent features and task-specific models. An alternative to models that rely on feature engineering is deep learning, which has attracted considerable attention because of its impressive accuracy in a wide range of tasks [1; 20; 33] and its ability to generalize high-level representation of raw data even without domain knowledge [22]. However, resistance to and criticism of deep learning approaches still persist, which primarily results from its lack of interpretability. That is, to infer the importance of individual features on model outputs, people find it difficult to follow the reasoning of deep learning models. For clinical decision support, lack of model interpretability will meet resistance to such models by medical communities. Fortunately, the interpretability problem can be mitigated by the attention mechanism [5], which mimics the visual attention of human cognition by learning a set of attention weights that represent the relative importance of individual features on certain time steps or locations to the final prediction.
Researchers have recently begun to apply deep learning in health care. Some pioneer studies have focused on identifying patterns from healthcare data with deep learning. Lasko et al. were the first to apply deep learning in health care, and they demonstrated the capability of deep learning to generalize patterns from serum uric acid measurements with autoencoders [21]. Miotto et al. utilized denoising autoencoders to produce a compact and general representation of electronic health records that demonstrated better prediction accuracy when used in downstream analysis than the representation produced by conventional dimensional reduction approaches [26]. Some recent studies have characterized the temporal dependencies of EHR data using sophisticated gated recurrent neural networks, but these models suffer from lack of interpretation. Pham et al. used a sophisticated model built on long short-term memory (LSTM) to model the interaction between diagnosis and medication [30]. Nguyen et al. employed convolutional neural networks to extract “clinical motifs” from medical records and predict clinical outcomes [27]. Choi et al. were the first to introduce attention mechanism in health care, and they proposed a model that uses two recurrent neural networks (RNN) that separately produce attention weights [7]. Our work continues to explore the possibilities and advantages of introducing the attention mechanism to facilitate interpretation of prediction results.
To generate interpretable predictions of clinical outcomes, our work develops a gated recurrent unit (GRU)-based RNN with hierarchical attention (GRNN-HA). We use the GRNN-HA to address the following challenges: (1) handling the high dimensionality of medical codes, (2) modeling the temporal dependencies of healthcare events, (3) characterizing the hierarchical structure of healthcare data, and (4) improving the interpretability of predictive models. To handle the high dimensionality of medical codes, our model learns a low-dimensional representation of medical codes with word2vec [25]. To model temporal dependencies, we use bidirectional GRUs to encode temporal information from healthcare data. To characterize the hierarchical structure of healthcare data and to improve the interpretability of models, our model organizes visit- and code-level information hierarchically and learns hierarchical attention weights on both levels dependently to represent the visit- and code-level importance, respectively. We validate our model on MIMIC-III (Medical Information Mart for Intensive Care) [16], which is a publicly available database that contains rich information about critical care from a large hospital, and compare mortality predictions of our model to those of several baseline models. To illustrate the interpretable results generated by our model, we also provide visualizations.
2. METHODS
2.1. Recurrent Neural Networks and GRU
A recurrent neural network (RNN) belongs to a class of neural networks suitable for modeling sequential data, which is enabled by the recurrent structure of RNNs in which the activation of the hidden state of the current time step is dependent on that of the previous time step. Specifically, given a sequence x= (x1, x2, …, xt), an RNN updates its current hidden state ht as
in which is a nonlinear activation function that can be either as simple as sigmoid or as sophisticated as LSTM [14]. The purpose of introducing sophisticated activation functions is to overcome the vanishing gradient problem, which causes difficulties of training an RNN to capture long-term dependencies [2; 13].
To overcome the vanishing gradient problem, we apply a gated recurrent unit (GRU)-based RNN that encodes input sequences of variable length as vectors of fixed length. The GRU, proposed by Cho et al. [6], has been proven to outperform LSTM in a set of tasks [9]. Although similar to LSTM, GRU provides a simpler alternative, using two types of gates, the reset gate rt and the update gate Zt, to control the information flow of hidden states without a separate memory gate. The reset gate determines part of the past information, which will remain in the current hidden state, and the update gate determines part of the new information, which will be added to the current hidden state. The detailed calculation of GRU is defined by the following equations:
in which σ(∙) is a sigmoid function, xt is the sequence input at time t, ht is the value of the hidden state at time t, and is the candidate value of the hidden state at time t. The computation of candidate hidden states is similar to that of a traditional RNN, except that the information of ht-1 is gated by rt. ht is calculated as a linear interpolation between ht-1 and .
2.2. Low-dimensional Representation of Medical Codes
Motivated by considerable successful applications of word2vec [25] in learning word representations for a wide range of natural language processing (NLP) tasks [6; 18; 32], we extend word2vec to learn a representation of diagnostic codes in ICD-9. ICD codes are often sequential in healthcare datasets. For example, in the MIMIC data database, ICD codes assigned to different hospital admissions are chronically ordered, and the ICD codes assigned to one hospital admission are ordered based on priority. Because of the sequentiality of ICD codes, we observe an analogy between them and natural languages. That is, we can naturally view the sequence of ICD codes of one admission as a sentence and an ICD code as a word and then apply the continuous bag-of-words (CBOW) architecture of word2vec. CBOW constructs representations of words by optimizing the prediction of a center word from its context words. With wo denoting a center word and wI denoting a context word of the center word, the objective of CBOW is to maximize
which is computed based on
in which Vw and Vw’ are two vector representations of word w, respectively, and C is the number of words in the vocabulary. The vector representations of words learned by word2vec tend to retain the semantic meanings of words so that words with similar meanings are close to each other in the vector space.
2.3. Hierarchical Attention
Our work applies a hierarchical attention model with a similar structure to that proposed by [35] to predict the outcome of a patient based on longitudinal diagnostic codes. The architecture of the model is illustrated in Figure 1. Suppose that a patient has L hospital visits, each of which is assigned T diagnostic codes. We denote each diagnostic code as wit, which represents the t-th ICD code in the i-th visit of this patient, with i ∈ [1, L] and t ∈ [1, T]. We represent wit with a low-dimensional vector through
in which V is an embedding matrix obtained through word2vec as described in 2.2. Then we encode each ICD code using a bidirectional GRU that summarizes the information of a sequence of ICD codes in one hospital visit from both directions through
Figure 1:
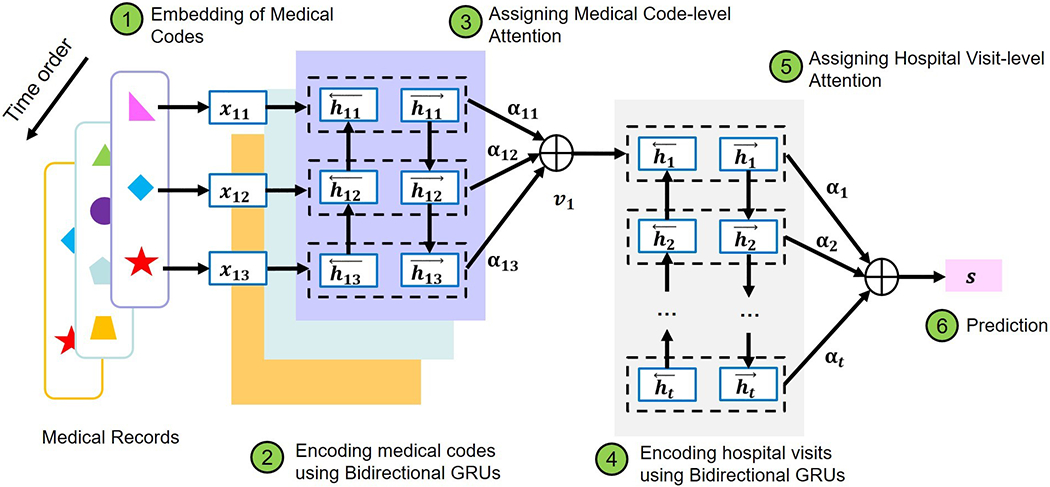
The architecture of the GRNN-HA model
Bidirectional GRUs increase the amount of input information by enabling future information to be accessible by the current state. The purpose of introducing bidirectional GRUs is to mimic the behavior of a physician examining a patient’s medical history both forwards and backwards.
Since each ICD code does not contribute equivalent information to one hospital visit, we introduce the code-level attention mechanism, which enables the model to pay more attention to informative ICD codes than other ICD codes. To implement the code-level attention mechanism, we first compute the hidden representation of hit, denoted as uit, to derive the attention of individual ICD codes αit as the similarity of uit and a code-level context vector uc, and then encode each hospital visit vi as a weighted sum of hit through
For each visit vector vi, we also encode it with a bidirectional GRU as follows:
Similarly, since individual visits do not contribute equivalent information to predictions, we introduce the visit-level attention mechanism to render the model capable of focusing more on those visits that contribute more to the prediction of patient outcomes than other visits. To implement the visit-level attention mechanism, we introduce a visit-level context vector uv and use it to calculate the visit-level attention αi as follows:
Finally, to represent features, we use s and build a binary classifier as
and we use the cross-entropy as our loss function, defined as
in which pi is the predicted probability of a sample belonging to class i, and is the true probability of the sample belonging to that class.
3. EXPERIMENTS
3.1. Description of Dataset
The dataset we use in this study comes from MIMIC-III (Medical Information Mart for Intensive Care), which is a freely accessible critical care database [16]. MIMIC-III contains data associated with 46,250 distinct patients admitted to the critical care units of a large tertiary hospital. The types of data include demographics, vital signs, diagnostic codes, and laboratory tests. Because of the wide availability of diagnostic codes in healthcare data and the ease of demonstrating the results produced by the model without excessive medical domain knowledge, we focus on diagnostics codes in this study. To ensure enough medical information about each patient, we include 7,537 patients who had at least two hospital admissions in MIMIC-III and then extract the diagnostic codes of each chosen patient. In addition, we maintain both the temporal order of diagnostic codes from multiple admissions and the priority-based order of diagnostic codes within one admission. We use these sequential diagnostic codes to predict mortality. Whether a patient died or not is recorded in MIMIC-III, and we find that 1,462 out of the selected 7,537 patients had died. To maintain the original proportion of mortality in both the training and test data, we use stratified sampling to set one-third of the dataset as a test set and the rest as a training set. We have repeated this stratified sampling three times to generate three sets of test and training data.
3.2. Model Implementation and Training
Here we describe the details of model implementation and training. We implement both the GRU-based RNN with hierarchical attention (GRNN-HA) and a GRU-based RNN without attention (GRNN) using keras 1.2.0 [8] with Theano 0.8.2 as the backend [3]. By training the CBOW version of word2vec on the training dataset, we obtain 200-dimentional representations of diagnostic codes. Then we use stratified sampling to set one-fifth of the training set for validation. We use the validation set to tune the hyperparameters of the models. The hyperparameters we have adjusted include the number of hidden units in GRUs at the visit and code levels. We vary the number of hidden units in GRUs over a range of 50 to 200 with a step size of 10. For model training, we minimize the loss function by using mini-batch stochastic gradient descent with a momentum of 0.9 and a batch size of 12. To evaluate the accuracy of mortality prediction given a relatively low mortality rate in our dataset, we use the Matthews correlation coefficient (MCC) in addition to commonly used measures, including the area under the receiver operating characteristic curve (AUROC) and the F1 score. MCC is an appropriate indicator of the quality of binary classification even when the sizes of both classes in a dataset significantly differ [24].
3.3. Baseline Models
Here we describe how we build our baseline models. To construct features for the baseline models, we derive n-grams from our data. An n-gram is a consecutive sequence of n items from sequential data such as text [4]. Because of their simplicity and ability to capture local contexts, N-gram-based features have been widely used in natural language processing [19; 23; 28]. To avoid model overfitting and computational infeasibility resulting from longer and more expressive n-grams, our study includes only the most frequent 5,000 unigrams and bigrams in the dataset to build a feature profile. After deriving the n-gram feature profile, we use either the number of occurrences of n-grams or the term frequency–inverse document frequency transformation (tf-idf) of n-grams as values for the feature profile. After constructing the features, we use logistic regression and support vector machines (SVM) as classifiers to build our baseline models. To select hyperparameters for the baseline models, we use a grid search over a parameter space through four-fold cross-validation on the training set. The hyperparameters of logistic regression include types of regularization and regularization parameters. We vary regularization over L1 and L2 and regularization parameter C over the logarithmic range of [10−3, 100]. Likewise, the hyperparameters of SVM include gamma (the inverse of the standard deviation of kernel function) and regularization penalty parameter C when we use the Radial Basis Function (RBF) as the kernel function. When we use linear SVM as the kernel function, the hyperparameters of SVM include only regularization penalty parameter C. We choose gamma over the logarithmic range of [10−2, 1] and regularization penalty parameter C over the logarithmic range of [10−2, 10]. We implement these baseline models with scikit-learn 0.18.1[29].
4. RESULTS
We show the results of mortality prediction on the MIMIC dataset by both the baseline and advanced methods in Table 1, in which “Logreg” is short for logistic regression. For all three metrics, the two advanced models, GRNN and GRNN-HA, demonstrate significantly better prediction results than those of the baseline models. Specifically, the average MCC of GRNN-HA is 11.71% higher than that of the highest average MCC of the baseline models. Even without the attention mechanism, the MCC of GRNN is still 7.32% higher than that of the highest average MCC of the baseline models. Using Steiger’s test [34], which is a statistical test for comparing two dependent correlations, we find the two comparisons of MCC are both statistically significant (p<0.05). The superior results generated by the two GRNN-based advanced models demonstrate the advantages of utilizing word2vec to handle the high dimensionality of medical codes and GRNN for appropriate modeling of the temporal dependencies of healthcare data. Comparing the prediction results of two GRNN-based models, we find that GRNN-HA has higher MCC and F1 score values than GRNN. The AUROC of GRNN-HA is slightly lower than that of GRNN. Since AUROC is not sensitive for binary classification on imbalanced datasets, we argue that the prediction results of GRNN-HA are better than those of GRNN. That is, in our case, the attention mechanism is able to improve prediction results. In addition to producing accurate predictions, GRNN-HA generates interpretable results that are suitable for visualization. Figure 2 provides an example visualization for the results of the prediction about one patient in one of the test sets. This patient has three recorded hospital admissions, shown as three horizontal bars colored in blue of different intensities. The intensity of the color is proportional to the corresponding visit-level attention weight. The multiple diagnostic codes associated with each hospital admission are represented by lines of various lengths, the vertical order of which, from top to bottom, corresponds to the priority-based order of these codes. These lines, representing multiple diagnostics associated with one admission, are contained within the horizontal bar of the corresponding hospital admission. The length of a horizontal line is proportional to the attention weight of the diagnostic code to which the line corresponds.
Table 1:
The results of mortality predictions
| Model | MCC | F1 score | AUROC |
|---|---|---|---|
| Logreg + count | 0.3664±0.01 35 | 0.4604±0.02 51 | 0.7884±0.02 28 |
| Logreg + tf-idf | 0.3509±0.02 03 | 0.4457±0.03 30 | 0.7979±0.00 94 |
| SVM + count | 0.3234±0.02 41 | 0.4157±0.06 79 | 0.7726±0.02 67 |
| SVM + tf-idf | 0.3549±0.02 18 | 0.4404±0.02 27 | 0.7899±0.01 18 |
| GRNN + word2vec | 0.4396±0.04 86 | 0.5074±0.06 13 | 0.8650±0.01 00 |
| GRNN-HA + word2vec | 0.4835±0.05 85 | 0.5766±0.06 23 | 0.8603±0.01 67 |
Figure 2:
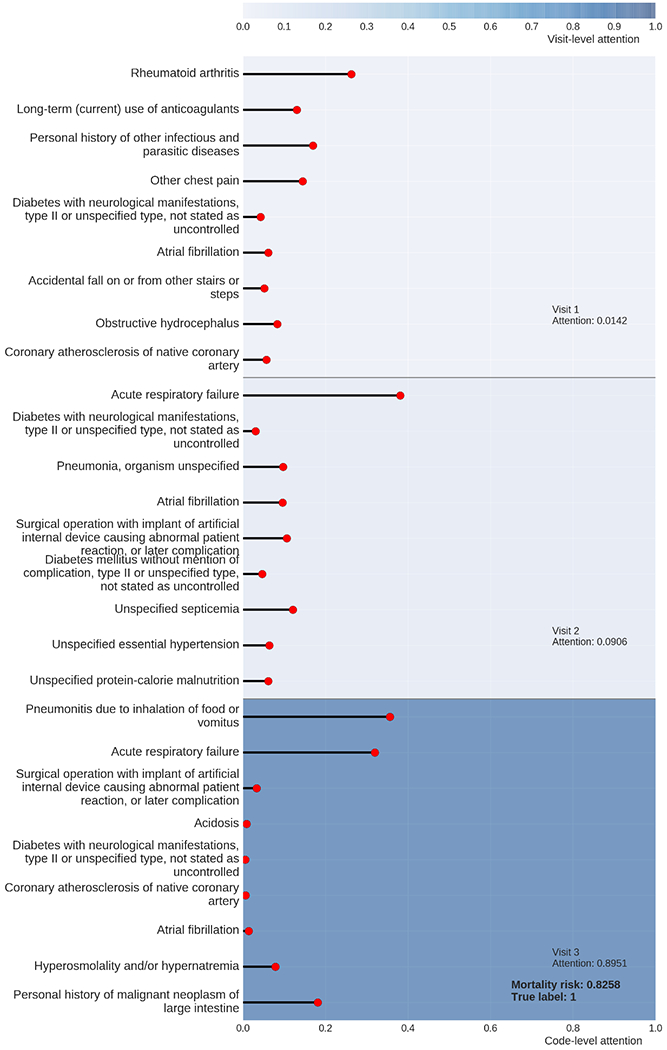
The visualization of the prediction result of a patient by GRNN-HA.
From Figure 2, we observe that among the three hospital admissions, Visit 3, the last visit of this patient, dominates visit-level attention. This finding is reasonable because recent clinical conditions are usually more informative of a clinical outcome than early conditions. We also observe that two diagnostic codes, pneumonia and acute respiratory failure, dominate the code-level attention of the last hospital admission. To validate whether the interpretation of prediction by GRNN-HA is reasonable, we read the clinical notes linked to this patient and find out that the patient suffered mainly from acute respiratory failure, which is probably the consequence of multifocal pneumonia with suspected sepsis. Therefore, by assigning high attention weights of pneumonia and acute respiratory failure, GRNN-HA determines that this patient has a high risk of mortality, which is consistent with the clinical notes written by the medical experts.
To further evaluate the interpretability of the GRNN-HA model, we illustrate the distribution of attention weights of several representative and frequent diagnostic codes from the prediction results of the test dataset in Figure 3. Since the diagnostic codes receiving high attention weights tend to have more impact on the final prediction of mortality, we hypothesize that generally, diagnostic codes with high attention weights should represent diseases that are more life-threatening. Among the six diagnostic codes whose distribution we select to visualize, congestive heart failure and acute respiratory failure, which are life-threatening conditions, have attention weights with distribution shifted more to the right of the x-axis than the remaining ones (Figure 3a and 3c). The remaining conditions include chronic conditions such as diabetes mellitus and tobacco use disorder. Despite their risk to health, these conditions tend not to be determinants of mortality (Figure 3d and 3e). Although pneumonia receives a high attention weight in the example shown in Figure 2, most attention weights of pneumonia of patients from test set are below 0.2. This observation implies that GRNN-HA infers the attention weight of one diagnostic code in the context of medical history and suggests the potential of this model for personalized clinical decision support.
Figure 3:
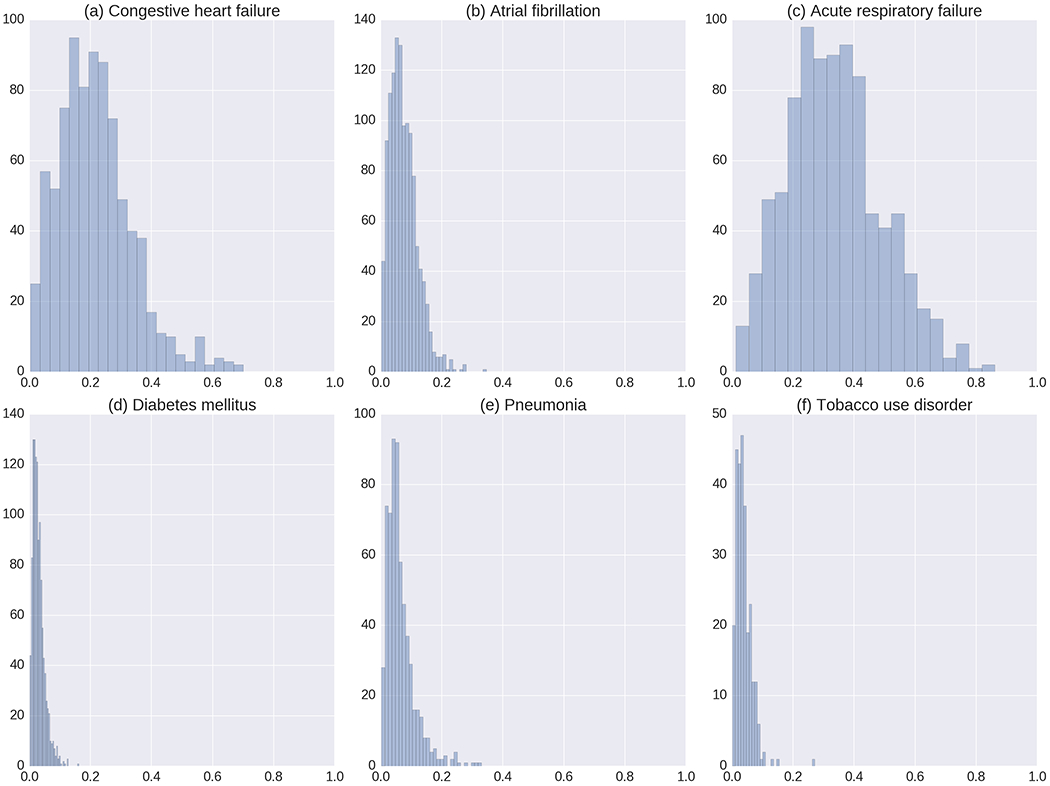
The distributions of the code-level attention weights of the following six diseases: (a) congestive heart failure, unspecified, (b) atrial fibrillation, (c) acute respiratory failure, (d) diabetes mellitus without mention of complication, type II or unspecified type, not stated as uncontrolled, (e) pneumonia, organism unspecified, and (d) tobacco use disorder.
5. DISCUSSION
To predict mortality on diagnostic codes from the MIMIC 3 database, our work uses a GRU-based RNN with hierarchical attention. This model frees researchers from manual feature engineering that usually results in classifier-dependent features and task-specific models and achieves both high prediction accuracy and interpretability that make it suitable for clinical decision support. The interpretability of the model relies on attention weights, determined from relative importance of diagnostic codes on prediction, assigned to individual diagnostic codes and hospital visits. We notice that the attention weights for one specific condition form a distribution rather than a fixed value. This observation suggests that the model embraces the idea of personalized clinical decision support by inferring relative importance of one diagnostic code based on the context of the medical history of a patient. We envision an application scenario of the GRNN-HA model in which the GRNN-HA model scans the medical history of a new patient admitted into a hospital, generates a risk assessment, and provides an annotated visualization for physicians that can help them prioritize the order in which they will see patients.
We will address several limitations in our work in the future. First, the dataset may not be an optimal choice for our model to fully utilize longitudinal information because the MIMIC3 database contains limited longitudinal medical information of patients. That is, most of the patients in the dataset contain only two or three admissions. In addition, the MIMIC3 dataset may not capture information about patients’ visits to other hospitals. Second, the number of samples is relatively small with respect to the need for large training sets of deep learning methods. Although the GRNN-HA significantly outperforms baseline models with respect to available data, we still expect to see improvement in its prediction accuracy and more reasonable attention weights if we have a larger sample size and longer medical histories of patients. Finally, we analyze only diagnostic codes for ease of interpretation of results. In future work, we aim to extend our analysis over other data types such as vital signs and laboratory tests.
CCS CONCEPTS.
• Applied computing → Life and medical sciences →Health informatics; • Computing methodologies → Artificial intelligence
ACKNOWLEDGMENTS
The authors are grateful to Li Tong and Janani Venugopalan for their valuable comments and suggestions.
REFERENCES
- [1].Bahdanau D, Cho K, and Bengio Y, 2014. Neural machine translation by jointly learning to align and translate. arXiv preprint arXiv:1409.0473. [Google Scholar]
- [2].Bengio Y, Simard P, and Frasconi P, 1994. Learning long-term dependencies with gradient descent is difficult. IEEE Transactions on neural networks 5, 2, 157–166. [DOI] [PubMed] [Google Scholar]
- [3].Bergstra J, Breuleux O, Bastien F, Lamblin P, Pascanu R, Desjardins G, Turian J, Warde-Farley D, and Bengio Y, 2010. Theano: A CPU and GPU math compiler in Python. In Proc. 9th Python in Science Conf, 1–7. [Google Scholar]
- [4].Brown PF, Desouza PV, Mercer RL, Pietra VJD, and Lai JC, 1992. Class-based n-gram models of natural language. Computational linguistics 18, 4, 467–479. [Google Scholar]
- [5].Cho K, Courville A, and Bengio Y, 2015. Describing multimedia content using attention-based encoder-decoder networks. IEEE Transactions on Multimedia 17, 11, 1875–1886. [Google Scholar]
- [6].Cho K, Van Merriënboer B, Gulcehre C, Bahdanau D, Bougares F, Schwenk H, and Bengio Y, 2014. Learning phrase representations using RNN encoder-decoder for statistical machine translation. arXiv preprint arXiv:1406.1078. [Google Scholar]
- [7].Choi E, Bahadori MT, Schuetz A, Stewart WF, and Sun J, 2016. RETAIN: Interpretable Predictive Model in Healthcare using Reverse Time Attention Mechanism. arXiv preprint arXiv:1608.05745. [Google Scholar]
- [8].Chollet F, 2015. Keras. [Google Scholar]
- [9].Chung J, Gulcehre C, Cho K, and Bengio Y, 2014. Empirical evaluation of gated recurrent neural networks on sequence modeling. arXiv preprint arXiv:1412.3555. [Google Scholar]
- [10].Free C, Phillips G, Watson L, Galli L, Felix L, Edwards P, Patel V, and Haines A, 2013. The effectiveness of mobile-health technologies to improve health care service delivery processes: a systematic review and meta-analysis. PLoS Med 10, 1, e1001363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Frisse ME and Holmes RL, 2007. Estimated financial savings associated with health information exchange and ambulatory care referral. Journal of biomedical informatics 40, 6, S27–S32. [DOI] [PubMed] [Google Scholar]
- [12].He D, Mathews SC, Kalloo AN, and Hutfless S, 2014. Mining high-dimensional administrative claims data to predict early hospital readmissions. Journal of the American Medical Informatics Association 21, 2, 272–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hochreiter S, 1998. The vanishing gradient problem during learning recurrent neural nets and problem solutions. International Journal of Uncertainty, Fuzziness and Knowledge-Based Systems 6, 02, 107–116. [Google Scholar]
- [14].Hochreiter S and Schmidhuber J, 1997. Long short-term memory. Neural computation 9, 8, 1735–1780. [DOI] [PubMed] [Google Scholar]
- [15].Jensen PB, Jensen LJ, and Brunak S, 2012. Mining electronic health records: towards better research applications and clinical care. Nature Reviews Genetics 13, 6, 395–405. [DOI] [PubMed] [Google Scholar]
- [16].Johnson AE, Pollard TJ, Shen L, Lehman L.-w.H., Feng M, Ghassemi M, Moody B, Szolovits P, Celi LA, and Mark RG, 2016. MIMIC-III, a freely accessible critical care database. Scientific data 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Jones SS, Rudin RS, Perry T, and Shekelle PG, 2014. Health information technology: an updated systematic review with a focus on meaningful use. Annals of internal medicine 160, 1, 48–54. [DOI] [PubMed] [Google Scholar]
- [18].Karpathy A and Fei-Fei L, 2015. Deep visual-semantic alignments for generating image descriptions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 3128–3137. [DOI] [PubMed] [Google Scholar]
- [19].Kešelj V, Peng F, Cercone N, and Thomas C, 2003. N-gram-based author profiles for authorship attribution. In Proceedings of the conference pacific association for computational linguistics, PACLING, 255–264. [Google Scholar]
- [20].Krizhevsky A, Sutskever I, and Hinton GE, 2012. Imagenet classification with deep convolutional neural networks. In Advances in neural information processing systems, 1097–1105. [Google Scholar]
- [21].Lasko TA, Denny JC, and Levy MA, 2013. Computational phenotype discovery using unsupervised feature learning over noisy, sparse, and irregular clinical data. PloS one 8, 6, e66341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].LeCun Y, Bengio Y, and Hinton G, 2015. Deep learning. Nature 521, 7553, 436–444. [DOI] [PubMed] [Google Scholar]
- [23].Marafino BJ, Davies JM, Bardach NS, Dean ML, Dudley RA, and Boscardin J, 2014. N-gram support vector machines for scalable procedure and diagnosis classification, with applications to clinical free text data from the intensive care unit. Journal of the American Medical Informatics Association 21, 5, 871–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Matthews BW, 1975. Comparison of the predicted and observed secondary structure of T4 phage lysozyme. Biochimica et Biophysica Acta (BBA)-Protein Structure 405, 2, 442–451. [DOI] [PubMed] [Google Scholar]
- [25].Mikolov T, Chen K, Corrado G, and Dean J, 2013. Efficient estimation of word representations in vector space. arXiv preprint arXiv:1301.3781. [Google Scholar]
- [26].Miotto R, Li L, Kidd BA, and Dudley JT, 2016. Deep Patient: An Unsupervised Representation to Predict the Future of Patients from the Electronic Health Records. Scientific Reports 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Nguyen P, Tran T, Wickramasinghe N, and Venkatesh S, 2016. Deepr: A Convolutional Net for Medical Records. arXiv preprint arXiv:1607.07519. [DOI] [PubMed] [Google Scholar]
- [28].Pak A and Paroubek P, 2010. Twitter as a Corpus for Sentiment Analysis and Opinion Mining. In LREc. [Google Scholar]
- [29].Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, and Dubourg V, 2011. Scikit-learn: Machine learning in Python. Journal of Machine Learning Research 12, October, 2825–2830. [Google Scholar]
- [30].Pham T, Tran T, Phung D, and Venkatesh S, 2016. DeepCare: A Deep Dynamic Memory Model for Predictive Medicine. In Pacific-Asia Conference on Knowledge Discovery and Data Mining Springer, 30–41. [Google Scholar]
- [31].Rios A and Kavuluru R, 2013. Supervised extraction of diagnosis codes from EMRs: role of feature selection, data selection, and probabilistic thresholding. In Healthcare Informatics (ICHI), 2013 IEEE International Conference on IEEE, 66–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Rocktäschel T, Grefenstette E, Hermann KM, Kočiský T, and Blunsom P, 2015. Reasoning about entailment with neural attention. arXiv preprint arXiv:1509.06664. [Google Scholar]
- [33].Sainath TN, Mohamed A. r., Kingsbury B, and Ramabhadran B, 2013. Deep convolutional neural networks for LVCSR. In Acoustics, speech and signal processing (ICASSP), 2013 IEEE international conference on IEEE, 8614–8618. [Google Scholar]
- [34].Steiger JH, 1980. Tests for comparing elements of a correlation matrix. Psychological bulletin 87, 2, 245–251. [Google Scholar]
- [35].Yang Z, Yang D, Dyer C, He X, Smola A, and Hovy E, 2016. Hierarchical attention networks for document classification. In Proceedings of the 2016 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies. [Google Scholar]


