Abstract
Patient similarity measurement is an important tool for cohort identification in clinical decision support applications. A reliable similarity metric can be used for deriving diagnostic or prognostic information about a target patient using other patients with similar trajectories of health-care events. However, the measure of similar care trajectories is challenged by the irregularity of measurements, inherent in health care. To address this challenge, we propose a novel temporal similarity measure for patients based on irregularly measured laboratory test data from the Multiparameter Intelligent Monitoring in Intensive Care database and the pediatric Intensive Care Unit (ICU) database of Children’s Healthcare of Atlanta. This similarity measure, which is modified from the Smith Waterman algorithm, identifies patients that share sequentially similar laboratory results separated by time intervals of similar length. We demonstrate the predictive power of our method; that is, patients with higher similarity in their previous histories will most likely have higher similarity in their later histories. In addition, compared with other non-temporal measures, our method is stronger at predicting mortality in ICU patients diagnosed with acute kidney injury and sepsis.
Categories and Subject Descriptors
H.3.3 [Information Storage and Retrieval]: Retrieval models and rankings – similarity measures; J.3 [Applied Computing]: Life and medical sciences – health and medical information systems
General Term
Algorithm
Keywords: Patient similarity, temporal similarity measure, laboratory tests, acute kidney injury, sepsis
1. INTRODUCTION
As more and more U.S. health-care facilities adopt electronic health record (EHR) systems, the medical histories of millions of patients are being stored in electronic form. The immense amount of EHR data serves as a unique resource for clinical decision support. For example, to guide the treatment of a target patient, clinicians can derive diagnostic or prognostic information from patients with similar health trajectories.
Mining EHR data is a challenging task, partly because EHR data consists of irregularly sampled observations [1]. One source of irregularity is differences in the health status of patients, by which patients are typically monitored. Clinicians monitor a particular set of measurements based on a patient’s symptoms, or they may follow a patient more intensively when the patient’s health is deteriorating. Therefore, the timestamps, the order, and the frequency of measurements in addition to the results of these measurements contain hidden, but meaningful information that facilitates clinical decisions [2]. However, one of the main obstacles in the extraction of meaningful information is the lack of quantitative measures for identifying similarities between each pair of patients according to clinical event trajectories. Some previous existing similarity measures account for only static characteristics such as demographics or average numeric measurement values within a certain period [3, 4] while others are supervised methods that require large training sets and that are not applicable to individual case-based reasoning [5, 6]. Because of the lack of similarity measures for temporal data in EHR, we propose a novel similarity measure. In this paper, we will first describe our similarity measure and then demonstrate the performance with two evaluation experiments.
2. METHODS
2.1. Patient Data Extraction
The patient data we used in this study are from two databases, MIMIC-II (Multiparameter Intelligent Monitoring in Intensive Care) database and the pediatric Intensive Care Unit (ICU) database of Children’s Healthcare of Atlanta (CHOA). MIMIC-II is a publicly available database that contains rich temporal data such as laboratory test results, electronic clinical documentation, and numeric bedside monitor trends and waveforms of 32,424 ICU stays from 22,870 hospital admissions [7]. The dataset that we have access to from the CHOA database contains clinical information including visit information, procedures, laboratory testing, and microbiology testing that are collected in 5,739 ICU stays from 4,975 patients aged from birth to 21 years old during the year of 2013. We focus on the top ten most frequent laboratory tests in both the MIMIC-II and CHOA database. The top ten most frequent laboratory items in MIMIC-II are hematocrit, potassium, sodium, creatinine, platelets, urea nitrogen, chloride, bicarbonate, anion gap, and leukocytes. And the top ten most frequent laboratory items in the CHOA dataset are point-of-care (POC) glucose, oxygen saturation, arterial POC pH, arterial POC pCO2, arterial POC pO2, sodium, POC ionized calcium, potassium, calcium and glucose. Table 1 and 2 list the number of records for the top ten laboratory tests of MIMIC-II and the CHOA dataset, respectively. Each laboratory result is annotated as either normal or abnormal, based on thresholds that are determined by clinicians. We extract the time, the identifier, and the results of these tests from the last ICU stay of each patient, which we use to predict whether patients died in hospitals. We extract patients with a specific disease based on International Classification of Diseases, Ninth revision (ICD-9). The ICD-9 of acute kidney injury is 584, and the ICD-9 for sepsis and severe sepsis are 995.91 and 995.92.
Table 1.
Top 10 most frequent Laboratory Items in MIMIC-II
| Laboratory Item | ID in MIMIC 2 | # of Records |
||
|---|---|---|---|---|
| Total | Normal | Abnormal | ||
| Hematocrit | 50,383 | 596,604 | 73,402 | 523,202 |
| Potassium | 50,149 | 561,178 | 494,082 | 67,096 |
| Sodium | 50,159 | 528,229 | 444,852 | 83,377 |
| Creatinine | 50,090 | 526,270 | 315,795 | 210,475 |
| Platelets | 50,428 | 526,190 | 338,222 | 187,968 |
| Urea nitrogen | 50,177 | 522,118 | 228,110 | 294,008 |
| Chloride | 50,083 | 517,904 | 378,534 | 139,370 |
| Bicarbonate | 50,172 | 516,225 | 370,295 | 145,930 |
| Anion gap | 50,068 | 507,265 | 474,916 | 32,349 |
| Leukocytes | 50,468 | 506,625 | 293,538 | 213,087 |
| Total | 502,177 | 5,308,608 | 3,411,746 | 1,896,862 |
Table 2.
Top 10 most frequent Laboratory Items in the CHOA database
| Laboratory Item | # of Records |
||
|---|---|---|---|
| Total | Normal | Abnormal | |
| POC glucose | 58,639 | 19,711 | 38,928 |
| Oxygen Saturation | 49,260 | 21,487 | 27,773 |
| Arterial POC pH | 49,256 | 21,620 | 27,636 |
| Arterial POC pCO2 | 49,246 | 26,477 | 22,769 |
| Arterial POC Po2 | 49,246 | 5,617 | 43,629 |
| Sodium | 43,603 | 33,872 | 9,731 |
| POC ionized calcium | 43,194 | 28,614 | 14,580 |
| Potassium | 42,985 | 28,139 | 14,846 |
| Calcium | 42,727 | 29,555 | 13,172 |
| Glucose | 42,629 | 26,712 | 15,917 |
| Total | 470,785 | 241,804 | 228,981 |
2.2. Temporal Similarity Measure for Laboratory Tests
Each laboratory test is typically associated with zero to four records per day, and time intervals between consecutive tests are inconsistent not only within each test but also across various tests. Such inconsistency poses difficulty for measuring the similarity between the trajectories of laboratory tests from two patients with traditional methods such as the Euclidean distance. To address this difficulty, we view the trajectory of a patient’s laboratory results as a sequence that consists of time-stamped laboratory results as well as the time intervals of various lengths separating consecutive laboratory results, and we develop our similarity measure based on the assumption that the laboratory tests of similar patients share similar orders and results, and the time intervals between corresponding laboratory results will also be similar.
To calculate our patient similarity measure, we modify the well-known Smith-Waterman algorithm, originally designed to find an optimal local alignment for a pair of DNA sequences using dynamic programming with a pre-defined scoring system [8]. The modified algorithm builds the scoring matrix H(i,j), where H(i,j) corresponds to the highest similarity score of suffix T1[1..i] and T2[1..j]. The trajectories of two patients’ laboratory results are represented as T1 = {a1, a2…am}, T2 = {b1, b2…bn}, where m, n∈N, and both ai and bj represent observations that are either a set of laboratory results entered into EHR systems at the same time—a set of laboratory tests performed within a short period—or an inter-val of time. The equation of the modified Smith-Waterman algorithm is
| (1) |
To penalize alignments of patient histories based on not only exact laboratory test results but also their temporal patterns—orders and time intervals, we construct s(ai,bj), the similarity function of ai and bj, under three circumstances, as
| (2) |
where P is a positive score, ρ is a coefficient for time differences, Δt is the differences between two time- corresponding intervals, and s(ai,bj) is represented by three conditions: (i) If both observations are sets of laboratory results, it equals a positive score P if the Jaccard similarity coefficient [9] of the two sets of laboratory results passes a certain threshold c; otherwise, it equals a negative score; (ii) if both observations are time intervals, it equals a positive score minus a number linearly correlated with the absolute difference between the two time intervals; or (iii) if one observation is a time interval and the other is a set of laboratory results, it equals negative infinity because we forbid a match between a time interval and a set of laboratory results. We represent the gap-scoring scheme by w(ai,- ) or w(-, bj). When we cannot match an observation in one patient’s history to a corresponding observation in another patient’s history, a gap occurs. We penalize a gap according to the duration of a missing or extra observation, shown as ρ*Δt in the formula, and we view laboratory results as transient events. The definition of laboratory results can vary based on one of the following decisions: to record both the normal and abnormal results, to record only abnormal results, or to record changes in results by omitting the same results except for the first. Based on our preliminary results on a small set of patient records, we set the value of c as 0.5, P as 3, w(ai,−) and w(−,bj) as −1. And the value of ρ is chosen as the value of P divided by 20 because we do not award the match of two time intervals with the absolute difference more than 20 hours. We obtain optimal alignment between two trajectories of laboratory tests by backtracking from the global maximum of the matrix H(i,j) and proceeding until an element with score 0 is encountered. To calculate the final score of the two trajectories of laboratory tests, we normalize the optimum score obtained by comparing the trajectory of the target patient to the same trajectory:
| (3) |
We need to normalize the score to prevent a patient being assigned a higher similarity score because of his/her longer clinical event trajectory.
2.3. Testing for the Predictive Power of Using Information from Similar Patients
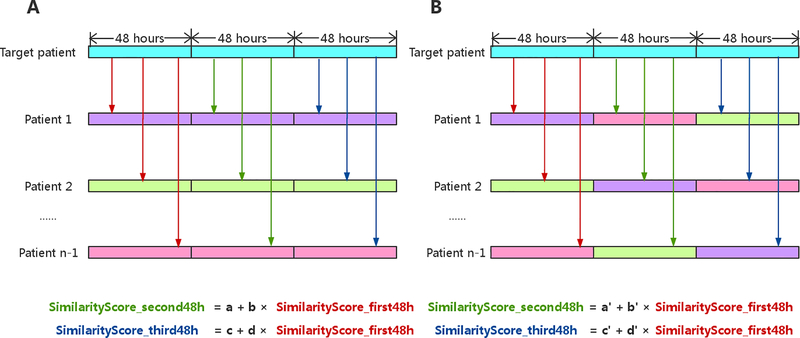
In an experiment that we designed to determine the predictive power of using information from similar patients retrieved with our temporal similarity measure, we tested the hypothesis that patients sharing similar laboratory-test trajectories tend to share similar laboratory-test trajectories in the future. In this experiment (figure 1), we randomly selected 300 adult patients and obtained laboratory results of their last ICU stays with their lengths of stay longer than 144 hours. To ensure sufficient laboratory test results and temporal patterns for each patient, we chose 48 hours and equally partitioned the last 144 hours into three 48-hour periods. We compared each of laboratory-test trajectory of the remaining patients to that of the target patient and calculated their similarity scores between the first 48 hours of the trajectory of the last ICU stay of the target patient and that of the similar patients. We repeated the same procedure for the second 48 hours and the third 48 hours. For each target patient, we built a simple linear model with calculated alignment scores of the last two windows (the second and third 48 hours) as dependent variables and alignment scores of the earlier time window (the first 48 hours) as independent variables. We computed p-values to check whether the slope was significantly nonzero to determine if our similarity measure demonstrated predictive power.
Figure 1.
The experiment of testing for the predictive power of using information from similar patients. (A) We select n patients and obtain the last 144 hours of laboratory results of their last ICU stays, and equally partitioned the 144 hours into three 48-hour periods. The trajectory of each patient is colored uniquely in this figure. We compared each of laboratory-test trajectory of the remaining patients to that of the target patient and calculated their similarity scores between the first 48 hours of the trajectory of the last ICU stay of the target patient and that of the similar patients, shown as red arrows. We repeat the same procedure for the second 48 hours and the third 48 hours, shown as green and blue arrows, respectively. For each target patient, we built a simple linear model with calculated alignment scores of the last two windows as dependent variables and alignment scores of the earlier time window as independent variables. (B) We randomly joining the later time window of one patient to the earlier time window of another patient to generate baseline for comparisons.
Demonstrating the Stronger Predictive Power of Our Temporal Similarity Measure is Better than That of Non-temporal Similarity Measure
To demonstrate the predictive power of our similarity measure, we compare it with two non-temporal measures. The first non-temporal measure represents each patient as a Boolean vector in which each element is either 0 (i.e., no abnormal results for a specific laboratory test during the time window) or 1 (i.e., the presence of abnormal laboratory results during the time window). We compute the Jaccard similarity coefficient with the following formula [9]:
| (4) |
The second non-temporal measure represents each patient as a Euclidean vector, and each element equals the number of abnormal test results during the time window. We compute the cosine similarity with the following formula [10]:
| (5) |
In the comparison of our similarity measure with the two non-temporal measures, we use the metrics of sensitivity, specificity, and F-measure (the harmonic mean of the sensitivity and the specificity), which determine the performance of the measures. Using K-nearest neighbor (KNN) algorithm with 4-fold cross validation, we test the capabilities of the three measures of predicting mortality based on the last 48-hour laboratory results. Because laboratory tests are informative of a diagnosis of acute kidney injury (AKI) [11], we focus on laboratory results from adult patients diagnosed with AKI during their last admission in the MIMIC-II database. AKI is a fatal syndrome characterized by the rapid loss of excretory functions of the kidneys, and patients with AKI require intensive treatment [12]. We also focus on pediatric patients diagnosed with sepsis in the CHOA dataset. Sepsis, known as a systemic inflammatory response to infection, is one of the leading causes of death in children [13, 14]. Certain common laboratory tests including pCO2, arterial pO2 are predictive for both diagnosis and prognosis of sepsis [13].
3. RESULTS
3.1. Visualization of an Alignment of Patients’ History
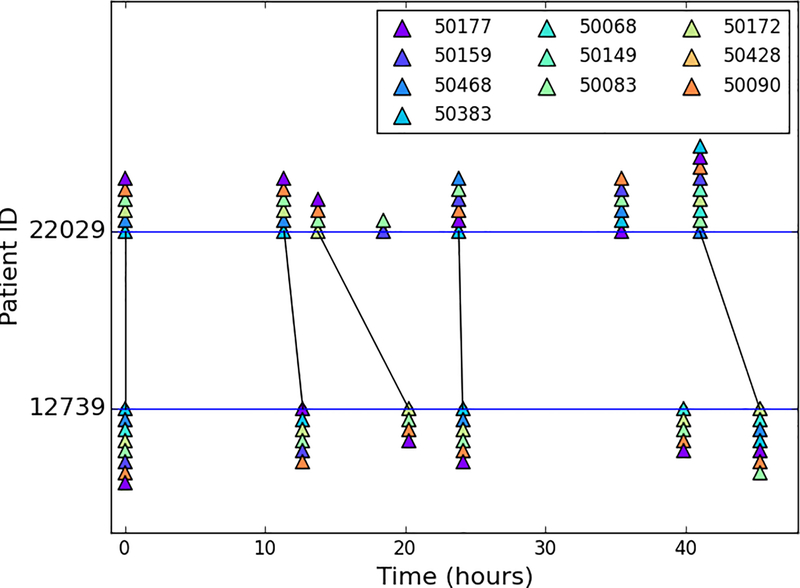
Figure 2 exemplifies the alignments of laboratory test trajectories within the last 48 hours of the last ICU stays of a target patient and a similar patient. Although the time intervals between the laboratory tests of the patients were irregular, our temporal similarity measure is able to align the two trajectories of laboratory results properly, which facilitates the comparison of the test results reported by clinicians while tolerating some missing or extra events.
Figure 2.
An example of visualizing the alignment between the laboratory test results of two patients with subject IDs 22029 and 12739. A corresponding set of laboratory results inferred by our novel similarity measure are connected by black lines. The patients’ last 48-hour laboratory results are shown as timelines with triangles of various colors representing abnormal results of a particular laboratory test. Each laboratory test is annotated as its corresponding identifier in MIMIC-II.
3.2. Predictive Power of Our Novel Similarity Measure
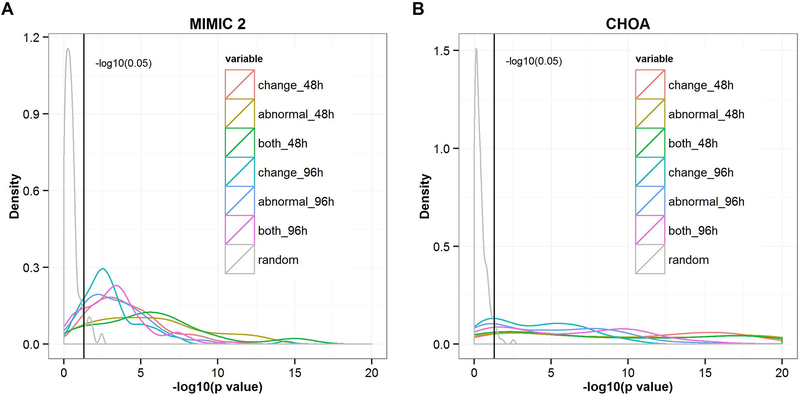
To examine the predictive power of our similarity measure, we hypothesize that the similarity score between the later time window of a target patient and that of another patient should be high if we observe a high similarity score between the earlier time window of the same target patient and that of the other patient. We constructed 300 simple linear regression models with the similarity scores of later time windows as dependent variables and the similarity scores of earlier time windows as independent variables given that each patient served as a target patient one time. Figure 3A and 3B illustrate the distribution of –log10(p-value) from 300 simple linear regression models using data from MIMIC-II and CHOA, respectively. All of the distributions of the -log10(p-value) generated by actual data significantly differ from the distribution of the –log10(p-value) generated by randomly joining the later time window of one patient to the earlier time window of another patient, which indicates that patients who share similar patterns in their past health data may have similar intrinsic characteristics; therefore, they are more likely to display similar patterns in the future.
Figure 3.
(A) The distribution of –log10 (p-value) generated from 300 simple linear regression models using data from MIMIC-II. (B) The distribution of –log10 (p-value) generated from 300 simple linear regression models using data from CHOA. “Change,” which refers to only changes in the results of certain laboratory tests (omitting the same results except for the first), is used to compute the similarity score; “abnormal” refers to only abnormal laboratory results; “both” refers to both normal and abnormal laboratory results; “48h” and “96h” refer to similarity scores generated from laboratory results within the second and third 48 hours, respectively, of the patients’ last 144-hour ICU stay and used as dependent variables. The black vertical lines indicate the value of –log10 (0.05).
Figure 3A and 3B also show that the density lines that represent the second 48 hours (red, yellow, and green lines) shift more to the right than those that represent the third 48 hours (cyan, blue, and purple lines), which indicates that as the gap between the earlier and later time windows decreases, the predictive power of future trajectories increases. In addition, we found that using only abnormal test results captures as much information as using both normal and abnormal test results, and more information than using changes in test results. Thus, we used only abnormal laboratory results to perform the following experiments. To demonstrate the reproducibility of our results, we repeated this procedure from the random sampling of 300 patients to generate p-value distributions three times.
3.3. Comparisons Between Our Temporal Similarity Measure and Other Non-temporal Similarity Measures
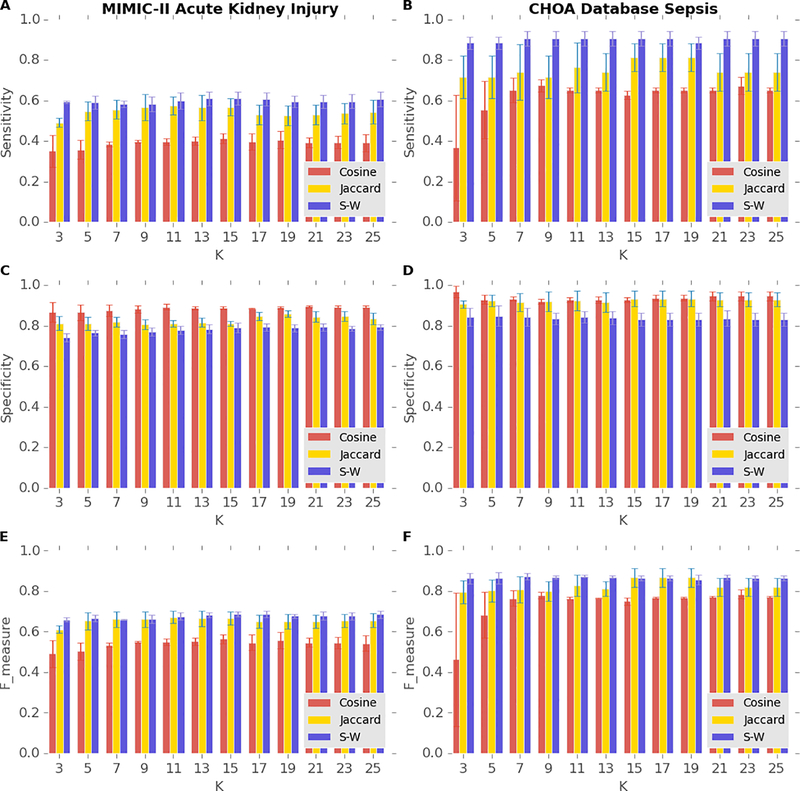
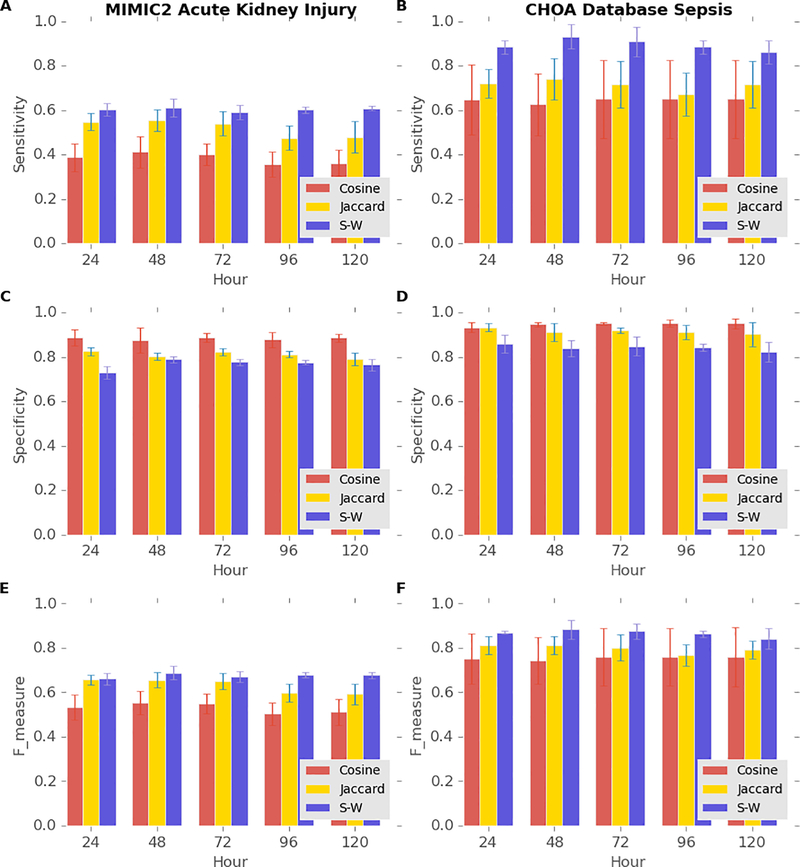
To demonstrate that our novel temporal similarity measure has better predictive power than that of non-temporal measures, we compare the sensitivity, the specificity, and the F-measure of our similarity measure with those of two non-temporal measures based on the laboratory results of AKI patients within the last 48 hours of their last ICU stays. The total number of patients who were diagnosed with AKI during their last admissions of their last ICU stays longer than 144 hours in MIMIC-II is 1,590, and the mortality rate of this population is 27%. Figure 4A and 4E illustrate that our similarity measure achieves both the highest sensitivity and the F-measure, and Figure 4C shows our similarity measure maintains a fairly high specificity with K varying from 3 to 25 among the three similarity measures. The total number of patients who were diagnosed with sepsis and severe sepsis in the CHOA dataset is 267, and the mortality rate of this population is 16%. Figure 4B and 4F shows that our similarity measure achieves the highest sensitivity and the highest F-measure in predicting mortality in septic patients in the CHOA dataset. The sensitivity of our similarity measure in predicting mortality in septic patients is noticeably higher than that of the measure in predicting mortality in AKI patients, probably resulting from the higher patient homogeneity of the CHOA dataset that contains only pediatric patients. The high sensitivity and F-measure of our similarity measures support the advantages of using temporal patterns instead of using only the presence of abnormal results for particular laboratory tests with the Jaccard similarity measure or only the number of specific abnormal laboratory results with the Cosine similarity measure. Our similarity measure did not achieve the highest specificity, partly because the mortality of both groups of patients were much lower than 50%; other similarity measures can still achieve high specificity by predicting that most of the patients will survive. Achieving high sensitivity is usually more desirable than achieving high specificity because methods with high sensitivity can help rule out negative outcomes. For example, when the trajectory of one patient is predicted to be similar to those of diseased patients, clinicians should monitor the health status of the patient more intensively and probably take preventive treatments. Therefore, we view our similarity measure as superior to the other non-temporal measures.
Figure 4.
Performance of predicting mortality in patients with AKI from MIMIC-II database (A, C, and E) and patients with sepsis from CHOA (B, D, and F) using the last 48-hour laboratory results with different similarity measures. Blue, purple, and light green bars indicate the performance of Jaccard similarity measures, cosine similarity, and our similar measure (S-W).
To investigate how the length of laboratory-test trajectory influence the predictive power of our similarity measure, we fix the K of KNN to 9 and vary the length of laboratory-test trajectory from 24 hours to 120 hours, with increment of 24 hours. Figure 5A, 5C, and 5E illustrate that the sensitivity, the specificity, and the F-measure of predicting mortality in AKI patients in MIMIC-II with the three similarity measures remain almost the same with various lengths of trajectory, while figure 5B shows that the sensitivity of predicting mortality in septic patients in the CHOA dataset with our similarity measure peaks when the length of trajectory used for prediction is 48 hours and then gradually decreases as the length of trajectory used for prediction elongates. Therefore, we do not necessarily need longer prediction time windows to improve the prediction performances, as opposed to our intuition.
Figure 5.
Performance of predicting mortality in patients with AKI from MIMIC-II database (A, C, and E) and patients with sepsis from CHOA (B, D, and F) using the various lengths of laboratory results with different similarity measures (K=9). Blue, purple, and light green bars indicate the performance of Jaccard similarity measures, cosine similarity, and our similar measure (S-W).
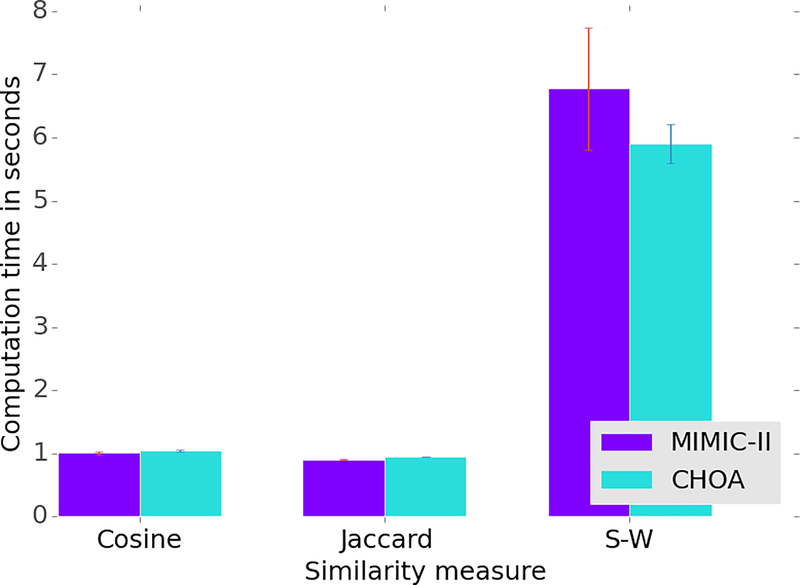
In addition to predictive power, we also compare the computation time of our similarity with that of non-temporal measures. To perform this task, we randomly selected 100 ICU stays of patients from both MIMIC-II and the CHOA dataset, respectively, and constructed a distance matrix of 48-hour laboratory-test trajectories of the 100 ICU stays with our similarity measure and two non-temporal measures, and the construction of the distance matrix requires computation of similarity for 4,950 pairs of ICU stays. And we repeated this procedure three times. Figure 6 shows that the time needed for our similarity measure is almost six times that needed for the two non-temporal measures, which is reasonable because the computation of our similarity measure is much more complex than the two non-temporal measures. In fact, we should recognize that the total computation time for constructing this distance matrix is just six seconds. We analyzed the runtime on a PC with Intel(R) Core i7–4702HQ Core Processor Unit at 2.20GHz and 16GB RAM running on Windows 10 using Python 3.4.
Figure 6.
Computation time of constructing a distance matrix of 100 patients using the last 48-hour laboratory results with different similarity measures.
4. DISCUSSION
This study introduces a temporal patient-similarity measure of health event trajectories that takes advantage of the irregular sampling procedures of ICU stays. Our study defines the irregular sampling procedures as those for which clinicians perform a particular set of laboratory tests in a particular order and at a particular frequency based on the patient’s health status. The results of two experiments with MIMIC-II data support our two major hypotheses: (1) Our novel similarity measure has predictive power, and (2) the similarity measure has better predictive power than other non-temporal measures.
As tremendous amounts of health data have been digitalized in EHR, similar patients can serve as reliable resources for clinical decision support. Traditional patient-similarity measures account for only non-temporal information by averaging measurements in specific, fixed time periods [3, 4]. Although these methods are intuitive, they prevent us from utilizing hidden temporal information. Our similarity measure provides an innovative way to compare two sequences of time-stamped clinical events with intuitive visualization. In the future, clinical experts can use this method to facilitate complex patient history interpretations.
Our study has a few limitations that need to be addressed in the future. First, one assumption, that patients are monitored more intensively when their health is deteriorating, should be experimentally validated. Second, our approach uses only the abnormality of a laboratory test instead of a numeric value because of a lack of notations for a normal range of value for specific tests. If the database of the future contains this information, we may be able to abstract a trend of the numeric values of laboratory tests. In the future, we will also to apply our method to other data types such as medications, procedures, and clinical notes. Third, since our method was tested in an ICU setting where patients are usually sicker and monitored more intensively than normal patients, it may be applicable to only short-term analysis. By targeting for just a short-term analysis, we penalize a match between two time intervals according to their absolute difference instead of more complex techniques such as warped time difference, which might hinder the application of this method to measurements spanning over multiple time scales: days for ICU care and years for outpatients. Finally, our method, which finds only similar patients, is not able to select informative features such as crucial events predictive of outcomes of our interest. In the future, we will extend our work by mining discriminative events prior to similarity analysis.
5. ACKNOWLEDGMENTS
The authors thank Dr. Chih-wen Cheng for helpful discussion through this study. The authors also thank Po-Yen Wu and Tong Li for assisting their valuable comments and suggestions. The authors thank Dr. Nikhil Chanani and Dr. Kevin Maher for providing the CHOA dataset used in this study. This research has been supported by grants from NIH (R01CA163256 and R01CA163256), CHOA Center for Pediatric Intervention (CPI), Georgia Cancer Coalition Award to Prof. MD Wang, Hewlett Packard, and Microsoft Research.
6. REFERENCES
- [1].Pivovarov R, Albers DJ, Sepulveda JL, and Elhadad N, “Identifying and mitigating biases in EHR laboratory tests,” Journal of biomedical informatics, vol. 51, pp. 24–34, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Hripcsak G, Albers DJ, and Perotte A, “Parameterizing time in electronic health record studies,” Journal of the American Medical Informatics Association, vol. 22, p. 797–804, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Lee J, Maslove DM, and Dubin JA, “Personalized Mortality Prediction Driven by Electronic Medical Data and a Patient Similarity Metric,” Plos One, vol. 10, p. e0127428, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Gottlieb A, Stein GY, Ruppin E, Altman RB, and Sharan R, “A method for inferring medical diagnoses from patient similarities,” BMC medicine, vol. 11, p. 194, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Sun J, Wang F, Hu J, and Edabollahi S, “Supervised patient similarity measure of heterogeneous patient records,” ACM SIGKDD Explorations Newsletter, vol. 14, pp. 16–24, 2012. [Google Scholar]
- [6].Ng K, Sun J, Hu J, and Wang F, “Personalized predictive modeling and risk factor identification using patient similarity,” AMIA Summits on Translational Science Proceedings, vol. 2015, p. 132, 2015. [PMC free article] [PubMed] [Google Scholar]
- [7].Saeed M, Villarroel M, Reisner AT, Clifford G, Lehman L-W, Moody G, et al. , “Multiparameter Intelligent Monitoring in Intensive Care II (MIMIC-II): a public-access intensive care unit database,” Critical care medicine, vol. 39, p. 952, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Smith TF and Waterman MS, “Identification of common molecular subsequences,” Journal of molecular biology, vol. 147, pp. 195–197, 1981. [DOI] [PubMed] [Google Scholar]
- [9].Jaccard P, Nouvelles recherches sur la distribution florale, 1908. [Google Scholar]
- [10].Baeza-Yates R and Ribeiro-Neto B, Modern information retrieval vol. 463: ACM press; New York, 1999. [Google Scholar]
- [11].Rahman M, Shad F, and Smith MC, “Acute kidney injury: a guide to diagnosis and management,” American family physician, vol. 86, pp. 631–639, 2012. [PubMed] [Google Scholar]
- [12].Bellomo R, Kellum JA, and Ronco C, “Acute kidney injury,” The Lancet, vol. 380, pp. 756–766, 2012. [DOI] [PubMed] [Google Scholar]
- [13].Goldstein B, Giroir B, and Randolph A, “International pediatric sepsis consensus conference: Definitions for sepsis and organ dysfunction in pediatrics*,” Pediatric critical care medicine, vol. 6, pp. 2–8, 2005. [DOI] [PubMed] [Google Scholar]
- [14].Watson RS, Carcillo JA, Linde-Zwirble WT, Clermont G, Lidicker J, and Angus DC, “The epidemiology of severe sepsis in children in the United States,” American journal of respiratory and critical care medicine, vol. 167, pp. 695–701, 2003. [DOI] [PubMed] [Google Scholar]