Abstract
Tuberculosis (TB) incidence in Nigeria is high, with a significant burden of TB/Human Immunodeficiency Virus (HIV). Genotyping and drug susceptibility of Mycobacterium tuberculosis Complex (MTBC) are important in order to improve the control of the disease. This study sought to determine drug susceptibility and genetic diversity of MTBC in the country. The sputum samples of 202 patients [133 (65.8%) males/69 (34.2%) females] were collected in the North Central zone of Nigeria and cultured using Lowenstein–Jensen medium. Immunochromatography for the primary identification and Drug Susceptibility Testing (DST) by proportion method, as well as IS6110 typing, regions of difference 1, 4, 9, 12, 702, and 711, and spoligotyping were carried out on the isolates. Following the DST on 202 isolates, 51 (25.2%) showed resistance to at least one drug. Multidrug resistance was observed in 29/202 (14.4%) cases. HIV positivity [37/202 (18.3%) patients] was associated with rifampicin 9/37 (24.3%) resistance (p = 0.012) as well as gender (p = 0.009). Of the 202 isolates, 150 (74.3%) were identified as the Cameroon sublineage, followed by the UgandaI, Haarlem, and West Africa 1 with 18 (8.9%), 10 (5%), and 6 (3%), respectively. The LAM10_CAM was the most prevalent genetic family [128/202 (63.4%)], with the shared international type 61 [111 (55%) isolates] the largest cluster. Gender (p = 0.038) and age (p = 0.015) had significant associations with the LAM10_CAM family but neither with HIV (p = 0.479) nor drug resistance. Rifampicin resistance in TB/HIV coinfected patient is a major concern in the study area. The Mycobacterium africanum lineage showed a marked decrease, and the need to educate females most at risk of TB/HIV coinfection is advocated.
Keywords: Tuberculosis, HIV, rifampicin resistance, LAM10_CAM, Nigeria
1. INTRODUCTION
The existence of Tuberculosis (TB) over the past millennia is not uncommon and outranks Human Immunodeficiency Virus (HIV) infection/acquired immunodeficiency syndrome among the 10 prominent deadliest infectious diseases worldwide. Globally, an estimated 10 million individuals suffered from TB (90% adults) in 2017 [1]. The increasing trend toward globalization, transnational migration, inadequate treatment of active pulmonary TB, injudicious prescriptions among physicians accompanied by uninformed drug selection as well as default among patients exposing Mycobacterium tuberculosis (MTB) to sublethal doses for shorter durations are potential targets for outbreaks of drug resistant (DR) TB [2,3].
Nigeria ranks not only among the 30 countries with a significant burden of TB, TB/HIV, and DR-TB but equally among the 14 countries accounting for more than 64% of the estimated number of incident TB cases worldwide in 2017 [1]. Nigeria also holds a record of several reports on the occurrence of DR-TB among patients across different settings. In Nigeria, there is an estimated 4.3% and 25% of patients with Multiple Drug Resistance (MDR) among new cases and previously treated cases, respectively [4]. The rising drug resistance imparts a significant threat on the control of the disease by the National Tuberculosis and Leprosy Control Programme. Therefore, it is of paramount importance to predict a reliable estimate on the magnitude of DR-TB in a bid to inform policy intervention programs for better management of the disease and for the monitoring of antimicrobial resistance.
Newer technologies, especially genomic techniques, have uncovered novel avenues in curbing TB [5]. Genotyping of MTB Complex (MTBC) identifies and distinguishes distinct (sub) lineages that are instrumental to track and control TB [6], taking into consideration the existence of stable host–pathogen interaction [7] and phylogeographic varsity of strains [8]. There is a strong association of the genetic lineages with pathogenicity and resistance [9–11], and the genetic background of the MTBC can provide an insight in the understanding of its prevalence and transmission [12].
Recently transmitted and reactivation of TB disease can be differentiated using genotyping of MTBC where isolates that share the same genotype are considered clustered and are assumed to be epidemiologically linked, while isolates with unique genotype not shared by others within the population are considered to have resulted from reactivation of latent infection, presumably acquired outside the population [13]. Genotyping has been used in epidemiologic studies to track specific isolates of MTBC and to understand the transmission dynamics of TB. Generation of phylogenetically informative data through genotyping methods has been developed to investigate multiple MTBC clinical samples from different sources [8]. The lineages of MTBC can be characterized using Large Sequence Polymorphisms (LSPs) [14]. Regions of Difference (RD) representing the loss of genetic material in Mycobacterium bovis Bacillus Calmette-Guérin (BCG) compared to MTB H37Rv has been revealed using comparative genomics, and the presence or absence of these regions could be useful in MTBC differentiation. Polymerase Chain Reaction (PCR) analysis of LSPs has shown some RD loci to be restricted to one MTBC strain or subspecies, while others are differentially distributed among the other members [15].
A number of methods such as classification, similarity search, and expert rule-based methods have emerged to accurately map genotyped isolates using Mycobacterial Interspersed Repetitive Units (MIRU) and/or spoligotypes to the major lineages [8,16,17]. Spoligotyping is a rapid and cost-effective PCR-based reverse hybridization technique that has been used to differentiate and identify specific genotypes of MTBC. Spoligotypes have evolved through the successive loss of spacer DNA sequences that separate short, tandemly repeated DNA sequences consisting of 36 bp in the direct repeat locus of MTB. Thus, spoligotyping identifies polymorphism in the presence or absence of 43 specific DNA spacer units in the direct repeat region of the MTBC strains [18]. Databases for strain lineage identification that involve spoligotype signature matching have been developed [19].
IS6110, an insertion sequence found exclusively within the MTBC, has been recognized as a multicopy target with increased sensitivity for the molecular detection of the members of the complex [20]. The IS6110 sequence in the genome of MTB has shown the stability required for use in molecular epidemiology. The element’s presence at different locations in the genome has been extensively used for epidemiological studies and has provided a suitable method for genotyping MTBC strains [21–23].
Recently, several studies have been carried out in various part of the world including Africa describing the genetic diversity of MTBC [24–26]. Therefore, this study sought to determine the drug susceptibility as well as the genetic diversity of MTBC causing TB in the North Central zone of Nigeria.
2. MATERIALS AND METHODS
2.1. Study Area, Specimen Collection, and Culture
About 202 pure isolates were obtained from patients across the North Central zone of Nigeria (Middle Belt), which includes the following states: Benue, Kogi, Kwara, Nasarawa, Niger, Plateau, and Federal Capital Territory. The isolates were obtained following sputum collection and decontamination using N-acetyl l-cysteine–sodium hydroxide (NALC/NaOH) method from suspected TB patients with the classical symptom of prolonged cough. The culture was carried out at the Zankli Research Center, Bingham University, Nasarawa State, Nigeria, using Lowenstein–Jensen (L–J) medium incubated at 37°C for 6–8 weeks, and the positive slants were reconfirmed by acid fast bacilli microscopy following Ziehl–Neelsen staining technique. Contaminated cultures were discarded.
2.2. Identification and Drug Susceptibility Testing
Primary identification of MTBC was carried out using the SD BIOLINE TB Ag MPT64 RAPID kit (Standard Diagnostics, Inc., Yongin, Korea) based on the manufacturer’s guide. In brief, a loopful of colonies were picked from the solid medium and suspended in 200 μL of the extraction buffer. Then 100 μL of the suspension was added to the sample well and left to flow chromatographically for 15 min. The appearance of a red band on the test window alongside with the control band was indicative of a positive result. Drug Susceptibility Testing (DST) was done by means of the proportion method on L–J medium against Isoniazid (INH; 0.2 μg/ml), Rifampicin (RIF; 40 μg/ml), Streptomycin (STR; 4 μg/ml), and Ethambutol (ETH; 2 μg/ml). The slopes were incubated at 37°C and monitored for growth at 4 and 6 weeks.
2.3. Genotyping MTBC Isolates
The preserved isolates from Nigeria in glycerol were shipped to the Bacteriology Laboratory of the Noguchi Memorial Institute for Medical Research (NMIMR)—Ghana, where approval from the Scientific Technical Committee and Institutional Review Board of the NMIMR was obtained for the genotyping of the isolates. The obtained DNA from heat-killed mycobacterial cell suspensions (95°C for 50 min) was subjected to molecular analyses. The primers used are shown in Supplementary Table A.
Supplementary Table A.
Primers used in this study
| Primers designation | 5′ → 3′ |
|---|---|
| IS6110 | |
| TB284 | GGACAACGCCGAATTGCG |
| TB850 | TAGGCGTCGGTGACAAAGGCCAC |
| AAGCGGTTGCCGCCGACCGACC | |
| RD 1 | CTGGCTATATTCCTGGGCCCGG |
| GAGGCGATCTGGCGGTTTGGGG | |
| ATGTGCGAGCTGAGCGATG | |
| RD 4 | TGTACTATGCTGACCCATGCG |
| AAAGGAGCACCATCGTCCAC | |
| CAAGTTGCCGTTTCGAGCC | |
| RD 9 | CAATGTTTGTTGCGCTGC |
| GCTACCCTCGACCAAGTGTT | |
| GGGAGCCCAGCATTTACCTC | |
| RD 12 | GTGTTGCGGGAATTACTCGG |
| AGCAGGAGCGGTTGGATATTC | |
| CAGCAGCAGGGTGTCATTGC | |
| RD 702 | GCAGCAGCACGATTCCTTGC |
| GATCGTCGCCGACCAGTGT | |
| GGTTGGCCACTACCAGAGAC | |
| RD 711 | GAACTCGCCGACTAGGTCG |
| CGACGAAGTGCGTGATTTCG | |
IS, insertion sequence; RD, region of difference.
2.3.1. IS6110 amplifications
Polymerase chain reaction detection of the insertion sequence IS6110 was carried out to confirm the MTBC. The PCR mix (50 μL) contained the following: 16.2 μL of H2O, 5 μL PCR buffer (10×), 10 μL Q solution, 2.5 μL MgCl2 (25 mM), 1 μL dNTP (10 mM), 2.5 μL primer F (10 pmol/μL), 2.5 μL primer R (10 pmol/μL), 0.3 μL Hot Start Taq DNA polymerase (5 U/μL), and 5 μL Coral dye (10×) and 5 μL of extracted mycobacterial DNA. A denaturation step was carried out at 96°C for 5 min, followed by 35 cycles of 95°C, 62°C, and 72°C for 1 min. The amplification was completed with one final extension cycle at 72°C for 10 min. Electrophoresis on 2% agarose gel was carried out on the PCR products and visualized under Ultraviolet (UV) light following ethidium bromide staining.
2.3.2. Deletion analyses and spoligotyping
The LSPs typing assay identifying RD 1, 4, 9, 12, 702, and 711 [14,27–29] was carried out on the mycobacterial DNA. The detection of the lineage-defining LSPs was carried out by PCR using the oligonucleotide primers (Supplementary Table A). In addition to a “forward” primer specific for the upstream region of each LSP, each reaction included two “reverse” primers (one internal to the deleted region and the other located immediately downstream of the LSP) [6].
Reactions were carried out in 25 μL volumes and included 15.4 μL of H2O, 2.5 μL PCR buffer (10×), 10 μL Q solution, 2.5 μL MgCl2 (25 mM), 0.5 μL dNTP (10 mM), 1 μL primer F (10 pmol/μL), 1 μL primer INT (10 pmol/μL), 1 μL primer DEL (10 pmol/μL), 0.1 μL Hot Start Taq DNA polymerase (5 U/μL), and 2.5 μL DNA. A denaturation step was carried out at 94°C for 2 min, followed by 35 cycles of 94°C for 10 s, 58°C for 10 s, and 72°C for 30 s. One final extension cycle at 72°C for 5 min completed the reaction. Electrophoresis on 2% agarose gel was carried out and visualized under UV light following ethidium bromide staining.
The isolates were sorted out based on the distinct phylogenetic lineages within the MTBC as previously defined [6]. Spoligotyping (Isogen Bioscience BV Maarssen, The Netherlands) was carried out on a membrane using the 43-spacer following manufacturer’s protocol [18]. The positive controls used were MTB H37Rv and Mycobacterium bovis BCG DNAs in parallel and distilled water as the negative control.
2.4. Data Analysis
The entire data were inputted in an Excel spreadsheet. The SpolDB4 database/MIRU-VNTRplus was used to analyze the spoligotype patterns in a binary format [30]. SPSS version 20.0 (IBM, Chicago, IL, USA) was used to analyze the association between the variables as age, gender, serology, drug resistance, and TB families. The p-value <0.05 were considered statistically significant.
3. RESULTS
3.1. Demography and HIV Association
Of the 202 isolates included in this study, 22 (10.9%) were isolated in 2009, 15 (7.4%) in 2010, two (1%) in 2012, 11 (5.4%) in 2013, 19 (9.4%) in 2016, and the majority 133 (65.8%) in 2017. One hundred and thirty-three (65.8%) of the 202 subjects were male and 69 (34.2%) were female. The age varies from 15 to 75 years with the majority [85/202 (42.1%)] between 25 and 34 years. Thirty-seven (18.3%) of the 202 subjects were HIV positive, 134 (66.3%) negative, and 31 (15.3%) did not know their status. Gender was significantly associated with the HIV status (p = 0.009), with 19 (51.4%) of the 37 HIV-infected patients being female (Table 1).
Table 1.
HIV association with gender, rifampicin resistance, and LAM10_CAM clades in the studied subjects
| HIV | Total | p-value | |||
|---|---|---|---|---|---|
| Positive (%) | Negative (%) | Unknown (%) | |||
| Gender | 0.009 | ||||
| Male | 18 (48.6) | 89 (66.4) | 26 (83.9) | 133 | |
| Female | 19 (51.4) | 45 (33.6) | 5 (16.1) | 69 | |
| Rifampicin | 0.012 | ||||
| Resistance | 9 (24.3) | 38 (23.4) | 1 (3.2) | 48 | |
| Sensitive | 28 (75.7) | 96 (71.6) | 30 (96.8) | 154 | |
| LAM10_CAM | 0.479 | ||||
| Yes | 21 (56.8) | 85 (63.4) | 22 (71) | 128 | |
| No | 16 (43.2) | 49 (36.6) | 9 (29) | 74 | |
| Total | 37 | 134 | 31 | 202 | |
3.2. Drug Resistance and HIV Association
The DST result on the 202 isolates showed that 103 (51%) were pan susceptible; 51 (25.2%) were resistant to at least one drug; and 61 (30.2%), 48 (23.8%), 48 (23.8%), and 14 (6.9%) isolates were resistant to STR, INH, RIF, and ETH, respectively. MDR (RIF and INH) was observed in 29/202 (14.4%) cases and resistance to the four drugs in 2/202 (1%) isolates. HIV positivity [37/202 (18.3%) patients] was associated with RIF 9/37 (24.3%) resistance (p = 0.012) (Table 1).
3.3. Distribution of Genetic Families
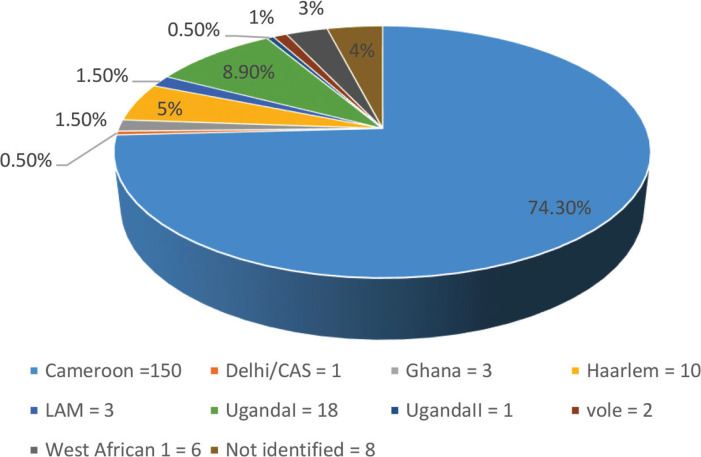
Of the 202 isolates, 186 (92.1%) were classified as MTB, six (3%) as Mycobacterium africanum, two (1%) as Mycobacterium microti, and eight (4%) not identified. The lineages were distributed as shown in Figure 1. Of the 202 isolates, 150 (74.3%) were identified as the Cameroon sublineage, followed by the UgandaI, Haarlem, and West African 1 with 18 (8.9%), 10 (5%), and 6 (3%), respectively.
Figure 1.
Distribution of the sublineages from the studied subjects.
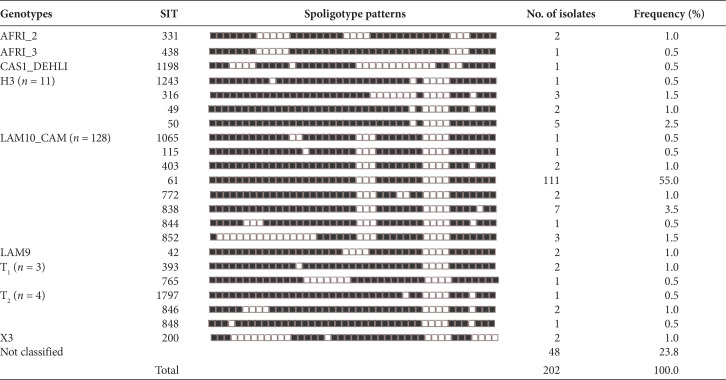
The spoligotype patterns recorded 58 different profiles, with 154 (76.2%) of the 202 isolates distributed 22 identified Shared International Types (SIT) according to SpolDB4 and the remaining 36 as orphans. Of the 202 isolates, 128 (63.4%) were LAM10_CAM and the most prevalent genetic family with the SIT 61 the largest cluster with 111 (55%) isolates. H3 was the second family most encountered with 11/202 (5.4%). Other genotypes included four (2%) T2, three (1.5%) T1, and two (1%) each of AFRI_2, LAM9, X3, and one (0.5%) each of AFRI_3 and CAS1_DEHLI. Forty-eight (23.8%) isolates with unknown genotype were equally recorded in this study (Table 2).
Table 2.
Distribution of genotypes, share types, and spoligotype patterns of 202 Mycobacterium tuberculosis complex isolates in North Central zone of Nigeria
3.4. Association of LAM10_CAM Family with Drug Resistance, Gender, Age, and HIV
The comparison between the largest family (LAM10_CAM) showed no significant association with individual drug resistance to STR (p = 0.287), INH (p = 0.841), RIF (0.841), and ETH (p = 0.281). However, gender with male 91/128 (71.1%) and age 25–34 years [46/128 (35.9%)] were significantly (p = 0.038 and 0.015, respectively) associated with the LAM10_CAM family (Table 3), but not with the HIV (p = 0.479) as shown in Table 1.
Table 3.
LAM10_CAM distribution of drug resistance, gender, and age of the studied subjects
| LAM10_CAM (%) | Other clades (%) | Total | p-value | |
|---|---|---|---|---|
| STR | 0.287 | |||
| S | 86 (67.2) | 55 (74.3) | 141 | |
| R | 42 (32.8) | 19 (25.7) | 61 | |
| INH | 0.841 | |||
| S | 97 (75.8) | 57 (77) | 154 | |
| R | 31 (24.2) | 17 (23) | 48 | |
| RIF | 0.841 | |||
| S | 97 (75.8) | 57 (77) | 154 | |
| R | 31 (24.2) | 17 (23) | 48 | |
| ETH | 0.281 | |||
| S | 121 (94.5) | 67 (90.5) | 188 | |
| R | 7 (5.5) | 7 (9.5) | 14 | |
| Gender | 0.038 | |||
| Male | 91 (71.1) | 42 (56.8) | 133 | |
| Female | 37 (28.9) | 32 (43.2) | 69 | |
| Age | 0.015 | |||
| 15–24 | 14 (10.9) | 14 (18.9) | 28 | |
| 25–34 | 46 (35.9) | 39 (52.7) | 85 | |
| 35–44 | 40 (31.3) | 13 (17.6) | 53 | |
| 45–54 | 17 (13.3) | 6 (8.1) | 23 | |
| ≥55 | 11 (8.6) | 2 (2.7) | 13 | |
| Total | 128 | 74 | 202 | |
STR, streptomycin; INH, isoniazid; RIF, rifampicin; ETH, ethambutol; S, sensitive; R, resistant.
4. DISCUSSION
This study carried out in the North Central zone of Nigeria has shown a high rate of MDR-TB (14.4%) compared to previous studies carried out in the country [31,32]. MDR-TB is a growing concern in Nigeria [31] and forms an important public health problem worldwide [33]. MDR-TB usually occurs as a consequence of injudicious adherence to TB-preventive measures or transmission of the MDR-TB strains. The burden of drug-resistant TB in Nigeria has been shown to be increasing and consequently can undermine control efforts, especially in resource-limited settings [34].
HIV coinfection was more prevalent in this study in females than males, contrary to a study carried out in Calabar—southeastern part of the country [35]. A significant association (p = 0.012) among HIV-positive patients with rifampicin resistance (24.3%) was recorded in this study. A similar association was observed by Adetunji et al. [36] in Oyo state, Nigeria, who showed a significant increase of rifampicin resistance (12%) among HIV patients with TB. Dinic et al. [37] also recorded a significant increase in MDR-TB including RIF (5.52%) in Lagos and Jos (two cities located in the Southwest and North Central zones of the country respectively). This is clearly above the World Health Organization predictions (0–4.3%) among HIV subjects. The authors concluded that there is a higher transmission of drug-resistant TB in HIV-infected patients in Nigeria than predicted. It has been shown that the weakening of the immune system due to HIV infection increases the burden of TB with several outcomes such as the reactivation of latent TB and equally the emergence of resistant TB strains, such as the MDR-TB [38]. The delay in the initiation of treatment or effective therapy, generally observed in developing countries, is a major contributor to MDR-TB increase, although TB resistance to drugs is multifactorial and clear insight can only be brought by specific investigations [39,40].
In this study, 82.2% were aged <45 years. TB and MDR-TB have been shown to affect the most productive age-group worldwide [41,42]. This has been shown previously in Nigeria [43]. Although INH resistance was associated with male in our study (data not shown), the risk for developing susceptible TB has been shown to be linked to male sex [44], contrary to a study who found MDR cases more likely to affect the male and younger age [45]. A study observed no difference in patients’ gender and MDR-TB [46], although conflicting results about vulnerability of female gender to MDR-TB as well as male more at risk have been reported [47–49].
The most prevalent spoligotype was SIT 61 (55%), which belongs to the LAM10_CAM family with 63.4% of isolates recorded in this study. A similar predominance of this family has previously been described among MTB isolates within the country [32,50–52], as well as from the neighboring country Cameroon [53,54] sharing a long boundary with Nigeria, and other several West African countries [55,56]. BCG vaccination, shaping this population selection, has been hypothesized [54].
This study recorded a further drastic decrease of Mycobacterium africanum, especially SIT 331, representing only 1% as opposed to 12.35% in a study carried out merely a decade ago in the southeastern part of the country [32]. Although Ani et al. [52] equally reported around the same period the unexpected low isolation of M. africanum in Jos within the middle belt of the country (only one isolate), LAM10_CAM has been successfully replacing the africanum sublineage not only in Nigeria. This trend observed earlier across Africa shifting toward its disappearance requires a careful reexamination of the prevalence of M. africanum in different parts of the continent [51].
This study has shown an association between the age and the LAM10_CAM family (p = 0.015). This association has equally been demonstrated in a similar study carried out in a neighboring country [53], as well as within the country [32], where the authors observed that the age-group range 25–34 years was significantly associated with shared-type pattern SIT 61. LAM10_CAM family was not associated with HIV in this study as shown earlier in Anambra State—Nigeria, where no spoligotype pattern was associated with the infection [51]. Equally, no relationship was observed between the LAM10_CAM family and drug resistance, similar to previous studies [51,53]. Since the hypothesis of the expansion of this family is as a result of vaccination selection as noted earlier, it might not yet have been subjected to drug-selective pressure. It is obvious that drug pressure is among the most important selection pressures currently taking place in MTBC, and molecular markers of drug resistance have been appropriately identified in the detection of positive selection [57–59]. The signature of positive selection as a result of the frequent independent occurrence of the same mutation known as convergent evolution [60] might not have yet occurred significantly in the LAM10_CAM family. However, considering drug misuse, defaulting, poor surveillance as well as all other factors likely to increase drug resistance such as drug administration based solely on smear microscopy still currently practiced widely in various African settings with the resultant consequences [61] will ineluctably result in the emergence of the Cameroon’s family drug-resistant strains.
5. CONCLUSION
Tuberculosis persists as important public health concern in Nigeria in which rifampicin resistance is of particular concern among HIV-infected patients. The abundance of the Cameroon lineage indicates an adaption, spread, and recent transmission with the risk of future evolution of drug resistance in this clade. A decreasing trend earlier noticed of the M. africanum lineages was observed and will require a wide evaluation. We advocate that education should be targeted toward females who happen to be the group most at risk of TB/HIV coinfection based on our findings. Improved early detection of HIV patients might help to curb the increase in MDR as well as its transmission in the studied communities. Moreover, clustered TB patients in this study probably result from recently acquired infection, and surveillance of this group is important in the control of the disease transmission.
CONFLICTS OF INTEREST
The authors declare they have no conflicts of interest.
AUTHOR CONTRIBUTIONS
B.D.T.P., D.Y.M. and L.L. conceived and designed the experiment. B.D.T.P., R.O. and L.M. performed the laboratory experiment. B.D.T.P., P.W.G. and N.Y.Y. analyzed the data. B.D.T.P., D.Y.M., P.W.G. and A.E.A. drafted the manuscript and wrote the paper. L.L. and A.E.A. substantially revised the manuscript. All the authors read and approved the final version.
FUNDING
This work was supported by the Bill and Melinda Gates Foundation to B.D.T.P under the Postdoctoral and Postgraduate Training in Infectious Diseases Research awarded to the Noguchi Memorial Institute for Medical Research (Global Health Grant number OPP52155). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
SUPPLEMENTARY TABLE
REFERENCES
- [1].World Health Organization (WHO) Global tuberculosis report 2018. Geneva, Switzerland: World Health Organization; 2018. [Google Scholar]
- [2].Sharma SK, Mohan A. Multidrug-resistant tuberculosis: a menace that threatens to destabilize tuberculosis control. Chest. 2006;130:261–72. doi: 10.1016/S0012-3692(15)50981-1. [DOI] [PubMed] [Google Scholar]
- [3].Pablos-Méndez A, Knirsch CA, Barr RG, Lerner BH, Frieden TR. Nonadherence in tuberculosis treatment: predictors and consequences in New York City. Am J Med. 1997;102:164–70. doi: 10.1016/S0002-9343(96)00402-0. [DOI] [PubMed] [Google Scholar]
- [4].World Health Organization (WHO) Global tuberculosis control: WHO report 2016. Geneva, Switzerland: World Health Organization; 2016. [Google Scholar]
- [5].Galagan JE. Genomic insights into tuberculosis. Nat Rev Genet. 2014;15:307–20. doi: 10.1038/nrg3664. [DOI] [PubMed] [Google Scholar]
- [6].Gagneux S, DeRiemer K, Van T, Kato-Maeda M, de Jong BC, Narayanan S, et al. Variable host–pathogen compatibility in Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2006;103:2869–73. doi: 10.1073/pnas.0511240103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Hirsh AE, Tsolaki AG, DeRiemer K, Feldman MW, Small PM. Stable association between strains of Mycobacterium tuberculosis and their human host populations. Proc Natl Acad Sci USA. 2004;101:4871–76. doi: 10.1073/pnas.0305627101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ferdinand S, Valétudie G, Sola C, Rastogi N. Data mining of Mycobacterium tuberculosis complex genotyping results using mycobacterial interspersed repetitive units validates the clonal structure of spoligotyping-defined families. Res Microbiol. 2004;155:647–54. doi: 10.1016/j.resmic.2004.04.013. [DOI] [PubMed] [Google Scholar]
- [9].Mokrousov I, Jiao WW, Sun GZ, Liu JW, Valcheva V, Li M, et al. Evolution of drug resistance in different sublineages of Mycobacterium tuberculosis Beijing genotype. Antimicrob Agents Chemother. 2006;50:2820–3. doi: 10.1128/AAC.00324-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kato-Maeda M, Shanley CA, Ackart D, Jarlsberg LG, Shang S, Obregon-Henao A, et al. Beijing sublineages of Mycobacterium tuberculosis differ in pathogenicity in the guinea pig. Clin Vaccine Immunol. 2012;19:1227–37. doi: 10.1128/CVI.00250-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Thwaites G, Caws M, Hong Chau TT, D’Sa A, Ngoc Lan NT, Thu Huyen MN, et al. Relationship between Mycobacterium tuberculosis genotype and the clinical phenotype of pulmonary and meningeal tuberculosis. J Clin Microbiol. 2008;46:1363–8. doi: 10.1128/JCM.02180-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Firdessa R, Berg S, Hailu E, Schelling E, Gumi B, Erenso G, et al. Mycobacterial lineages causing pulmonary and extrapulmonary tuberculosis, Ethiopia. Emerg Infect Dis. 2013;19:460–3. doi: 10.3201/eid1903.120256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kato-Maeda M, Metcalfe JZ, Flores L. Genotyping of Mycobacterium tuberculosis: application in epidemiologic studies. Future Microbiol. 2011;6:203–16. doi: 10.2217/fmb.10.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Gagneux S, Small PM. Global phylogeography of Mycobacterium tuberculosis and implications for tuberculosis product development. Lancet Infect Dis. 2007;7:328–37. doi: 10.1016/S1473-3099(07)70108-1. [DOI] [PubMed] [Google Scholar]
- [15].Huard RC, de Oliveira Lazzarini LC, Butler WR, van Soolingen D, Ho JL. PCR-based method to differentiate the subspecies of the Mycobacterium tuberculosis complex on the basis of genomic deletions. J Clin Microbiol. 2003;41:1637–50. doi: 10.1128/JCM.41.4.1637-1650.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Supply P, Allix C, Lesjean S, Cardoso-Oelemann M, Rüsch-Gerdes S, Willery E, et al. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J Clin Microbiol. 2006;44:4498–510. doi: 10.1128/JCM.01392-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Aminian M, Shabbeer A, Bennett KP. A conformal Bayesian network for classification of Mycobacterium tuberculosis complex lineages. BMC Bioinformatics. 2010;11:S4. doi: 10.1186/1471-2105-11-S3-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, Kuijper S, et al. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997;35:907–14. doi: 10.1128/jcm.35.4.907-914.1997. https://www.ncbi.nlm.nih.gov/pubmed/9157152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Brudey K, Driscoll JR, Rigouts L, Prodinger WM, Gori A, Al-Hajoj SA, et al. Mycobacterium tuberculosis complex genetic diversity: mining the fourth international spoligotyping database (SpolDB4) for classification, population genetics and epidemiology. BMC Microbiol. 2006;6:23. doi: 10.1186/1471-2180-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wang HY, Lu JJ, Chang CY, Chou WP, Hsieh JC, Lin CR, et al. Development of a high sensitivity TaqMan-based PCR assay for the specific detection of Mycobacterium tuberculosis complex in both pulmonary and extrapulmonary specimens. Sci Rep. 2019;9:113. doi: 10.1038/s41598-018-33804-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].McEvoy CRE, Falmer AA, Gey van Pittius NC, Victor TC, van Helden PD, Warren RM. The role of IS6110 in the evolution of Mycobacterium tuberculosis. Tuberculosis (Edinb) 2007;87:393–404. doi: 10.1016/j.tube.2007.05.010. [DOI] [PubMed] [Google Scholar]
- [22].Thierry D, Cave MD, Eisenach KD, Crawford JT, Bates JH, Gicquel B, et al. IS6110, an IS-like element of Mycobacterium tuberculosis complex. Nucleic Acids Res. 1990;18:188. doi: 10.1093/nar/18.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].van Embden JD, Cave MD, Crawford JT, Dale JW, Eisenach KD, Gicquel B, et al. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol. 1993;31:406–9. doi: 10.1128/jcm.31.2.406-409.1993. https://www.ncbi.nlm.nih.gov/pubmed/8381814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Uddin MKM, Ahmed M, Islam MR, Rahman A, Khatun R, Hossain MA, et al. Molecular characterization and drug susceptibility profile of Mycobacterium tuberculosis isolates from Northeast Bangladesh. Infect Genet Evol. 2018;65:136–43. doi: 10.1016/j.meegid.2018.07.027. [DOI] [PubMed] [Google Scholar]
- [25].Adesokan HK, Streicher EM, van Helden PD, Warren RM, Cadmus SIB. Genetic diversity of Mycobacterium tuberculosis complex strains isolated from livestock workers and cattle in Nigeria. PLoS One. 2019;14:e0211637. doi: 10.1371/journal.pone.0211637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Luo D, Chen Q, Xiong G, Peng Y, Liu T, Chen X, et al. Prevalence and molecular characterization of multidrug-resistant M. tuberculosis in Jiangxi province, China. Sci Rep. 2019;9:7315. doi: 10.1038/s41598-019-43547-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Brosch R, Gordon SV, Marmiesse M, Brodin P, Buchrieser C, Eiglmeier K, et al. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc Natl Acad Sci USA. 2002;99:3684–9. doi: 10.1073/pnas.052548299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].de Jong BC, Antonio M, Gagneux S. Mycobacterium africanum—review of an important cause of human tuberculosis in West Africa. PLoS Negl Trop Dis. 2010;4:e744. doi: 10.1371/journal.pntd.0000744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Warren RM, Gey van Pittius NC, Barnard M, Hesseling A, Engelke E, de Kock M, et al. Differentiation of Mycobacterium tuberculosis complex by PCR amplification of genomic regions of difference. Int J Tuberc Lung Dis. 2006;10:818–22. https://www.ncbi.nlm.nih.gov/pubmed/16850559. [PubMed] [Google Scholar]
- [30].Weniger T, Krawczyk J, Supply P, Niemann S, Harmsen D. MIRU-VNTRplus: a web tool for polyphasic genotyping of Mycobacterium tuberculosis complex bacteria. Nucleic Acids Res. 2010;38:W326–W31. doi: 10.1093/nar/gkq351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Lawson L, Yassin MA, Abdurrahman ST, Parry CM, Dacombe R, Sogaolu OM, et al. Resistance to first-line tuberculosis drugs in three cities of Nigeria. Trop Med Int Health. 2011;16:974–80. doi: 10.1111/j.1365-3156.2011.02792.x. [DOI] [PubMed] [Google Scholar]
- [32].Thumamo BP, Asuquo AE, Abia-Bassey LN, Lawson L, Hill V, Zozio T, et al. Molecular epidemiology and genetic diversity of Mycobacterium tuberculosis complex in the Cross River State, Nigeria. Infect Genet Evol. 2012;12:671–7. doi: 10.1016/j.meegid.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Wright A, Zignol M, Van Deun A, Falzon D, Gerdes SR, Feldman K, et al. Epidemiology of antituberculosis drug resistance 2002–07: an updated analysis of the Global Project on Anti-Tuberculosis Drug Resistance Surveillance. Lancet. 2009;373:1861–73. doi: 10.1016/S0140-6736(09)60331-7. [DOI] [PubMed] [Google Scholar]
- [34].Onyedum CC, Alobu I, Ukwaja KN. Prevalence of drug-resistant tuberculosis in Nigeria: a systematic review and meta-analysis. PLoS One. 2017;12:e0180996. doi: 10.1371/journal.pone.0180996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Kooffreh ME, Offor JB, Ekerette EE, Udom UI. Prevalence of tuberculosis in Calabar, Nigeria: a case study of patients attending the outpatients Department of Dr. Lawrence Henshaw Memorial Hospital, Calabar. Saudi J Health Sci. 2016;5:130–3. doi: 10.4103/2278-0521.195817. [DOI] [Google Scholar]
- [36].Adetunji SO, Donbraye E, Ekong MJ, Adetunji BI. Rifampicin-resistant tuberculosis among known HIV-infected patients in Oyo state, Nigeria. J Immunoass Immunoch. 2019;40:289–99. doi: 10.1080/15321819.2019.1583579. [DOI] [PubMed] [Google Scholar]
- [37].Dinic L, Akande P, Idigbe EO, Ani A, Onwujekwe D, Agbaji O, et al. Genetic determinants of drug-resistant tuberculosis among HIV infected patients in Nigeria. J Clin Microbiol. 2012;50:2905–9. doi: 10.1128/JCM.00982-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Rijal KR, Ghimire P, Rijal B, Bam DS. The pattern of anti-tuberculosis drug resistance in pulmonary tuberculosis patients. J Inst Med. 2005;27:26–8. doi: 10.3126/joim.v27i3.412. [DOI] [Google Scholar]
- [39].Nasiri MJ, Haeili M, Ghazi M, Goudarzi H, Pormohammad A, Imani Fooladi AA, et al. New insights in to the intrinsic and acquired drug resistance mechanisms in mycobacteria. Front Microbiol. 2017;8:681. doi: 10.3389/fmicb.2017.00681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].da Silveira Paro Pedro H, Nardi SMT, Pereira MIF, Oliveira RS, Suffys PN, Gomes HM, et al. Clinical and epidemiological profiles of individuals with drug-resistant tuberculosis. Mem Inst Oswaldo Cruz. 2015;110:235–41. doi: 10.1590/0074-02760140316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].World Health Organization (WHO) Stop TB partnership: 2010/2011 Tuberculosis Global Facts. 2010 Available from: http://www.who.int/tb/publications/2010/factsheet_tb_2010.pdf.
- [42].Demile B, Zenebu A, Shewaye H, Xia S, Guadie A. Risk factors associated with multidrug-resistant tuberculosis (MDR-TB) in a tertiary armed force referral and teaching hospital, Ethiopia. BMC Infect Dis. 2018;18:249. doi: 10.1186/s12879-018-3167-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Pokam BT, Asuquo AE, Abia-Bassey LN, Idasa MB, Umoh NO, Eko FO, et al. Multidrug resistance and demography of newly diagnosed tuberculosis patients in Cross River State, Nigeria. Int J Mycobacteriol. 2013;2:89–93. doi: 10.1016/j.ijmyco.2013.03.002. [DOI] [PubMed] [Google Scholar]
- [44].Artiom J, Evelina L. Social determinants of drug-susceptible and drug resistant tuberculosis. Eur Respir J. 2016;48:PA2749. doi: 10.1183/13993003.congress-2016.PA2749. [DOI] [Google Scholar]
- [45].Faustini A, Hall AJ, Perucci CA. Risk factors for multidrug resistant tuberculosis in Europe: a systematic review. Thorax. 2006;61:158–63. doi: 10.1136/thx.2005.045963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Ullah I, Javaid A, Tahir Z, Ullah O, Shah AA, Hasan F, et al. Pattern of drug resistance and risk factors associated with development of drug resistant Mycobacterium tuberculosis in Pakistan. PLoS One. 2016;11:e0147529. doi: 10.1371/journal.pone.0147529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Ejaz M, Siddiqui AR, Rafiq Y, Malik F, Channa A, Mangi R, et al. Prevalence of multi-drug resistant tuberculosis in Karachi, Pakistan: identification of at risk groups. Trans R Soc Trop Med Hyg. 2010;104:511–17. doi: 10.1016/j.trstmh.2010.03.005. [DOI] [PubMed] [Google Scholar]
- [48].Lomtadze N, Aspindzelashvili R, Janjgava M, Mirtskhulava V, Wright A, Blumberg HM, et al. Prevalence and risk factors for multidrug-resistant tuberculosis in the Republic of Georgia: a population-based study. Int J Tuberc Lung Dis. 2009;13:68–73. https://www.ncbi.nlm.nih.gov/pubmed/19105881. [PMC free article] [PubMed] [Google Scholar]
- [49].Mor Z, Goldblatt D, Kaidar-Shwartz H, Cedar N, Rorman E, Chemtob D. Drug-resistant tuberculosis in Israel: risk factors and treatment outcomes. Int J Tuberc Lung Dis. 2014;18:1195–201. doi: 10.5588/ijtld.14.0192. [DOI] [PubMed] [Google Scholar]
- [50].Lawson L, Zhang J, Gomgnimbou MK, Abdurrahman ST, Le Moullec S, Mohamed F, et al. A molecular epidemiological and genetic diversity study of tuberculosis in Ibadan, Nnewi and Abuja, Nigeria. PLoS One. 2012;7:e38409. doi: 10.1371/journal.pone.0038409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Uzoewulu GN, Lawson L, Nnanna IS, Rastogi N, Goyal M. Genetic diversity of Mycobacterium tuberculosis complex strains isolated from patients with pulmonary tuberculosis in Anambra State, Nigeria. Int J Mycobacteriol. 2016;5:74–9. doi: 10.1016/j.ijmyco.2015.06.008. [DOI] [PubMed] [Google Scholar]
- [52].Ani A, Bruvik T, Okoh Y, Agaba P, Agbaji O, Idoko J, et al. Genetic diversity of Mycobacterium tuberculosis complex in Jos, Nigeria. BMC Infect Dis. 2010;10:189. doi: 10.1186/1471-2334-10-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Kamgue Sidze L, Mouafo Tekwu E, Kuaban C, Assam Assam J, Tedom J, Niemann S, et al. Estimates of genetic variability of Mycobacterium tuberculosis complex and its association with drug resistance in Cameroon. Adv Infect Dis. 2013;3:55–9. doi: 10.4236/aid.2013.31007. [DOI] [Google Scholar]
- [54].Niobe-Eyangoh SN, Kuaban C, Sorlin P, Cunin P, Thonnon J, Sola C, et al. Genetic biodiversity of Mycobacterium tuberculosis complex strains from patients with pulmonary tuberculosis in Cameroon. J Clin Microbiol. 2003;41:2547–53. doi: 10.1128/JCM.41.6.2547-2553.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Godreuil S, Torrea G, Terru D, Chevenet F, Diagbouga S, Supply P, et al. First molecular epidemiology study of Mycobacterium tuberculosis in Burkina Faso. J Clin Microbiol. 2007;45:921–7. doi: 10.1128/JCM.01918-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Affolabi D, Anyo G, Faïhun F, Sanoussi N, Shamputa IC, Rigouts L, et al. First molecular epidemiological study of tuberculosis in Benin. Int J Tuberc Lung Dis. 2009;13:317–22. https://www.ncbi.nlm.nih.gov/pubmed/19275790. [PubMed] [Google Scholar]
- [57].Farhat MR, Shapiro BJ, Kieser KJ, Sultana R, Jacobson KR, Victor TC, et al. Genomic analysis identifies targets of convergent positive selection in drug-resistant Mycobacterium tuberculosis. Nat Genet. 2013;45:1183–9. doi: 10.1038/ng.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Zhang H, Li D, Zhao L, Fleming J, Lin N, Wang T, et al. Genome sequencing of 161 Mycobacterium tuberculosis isolates from China identifies genes and intergenic regions associated with drug resistance. Nat Genet. 2013;45:1255–60. doi: 10.1038/ng.2735. [DOI] [PubMed] [Google Scholar]
- [59].Osório NS, Rodrigues F, Gagneux S, Pedrosa J, Pinto-Carbó M, Castro AG, et al. Evidence for diversifying selection in a set of Mycobacterium tuberculosis genes in response to antibiotic- and nonantibiotic-related pressure. Mol Biol Evol. 2013;30:1326–36. doi: 10.1093/molbev/mst038. [DOI] [PubMed] [Google Scholar]
- [60].Coscolla M, Gagneux S. Consequences of genomic diversity in Mycobacterium tuberculosis. Semin Immunol. 2014;26:431–44. doi: 10.1016/j.smim.2014.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Pokam BT, Asuquo AE. Acid-fast bacilli other than mycobacteria in tuberculosis patients receiving directly observed therapy short course in Cross River State, Nigeria. Tuberc Res Treat. 2012;2012:1–4. doi: 10.1155/2012/301056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.