Abstract
Tuberculosis (TB) remains a main hurdle for national programs due to increase in drug resistance to antitubercular drugs. World Health Organization (WHO)-endorsed Line Probe Assay, Genotype MTBDRsl Ver 2.0, gives opportunity for rapid diagnosis and molecular characterization of different mutations in drug targets of fluoroquinolone (FQ) and second-line injectable drugs (SLID). We, retrospectively, analyzed the data of Genotype MTBDRsl Ver 2.0 from January 2018 to June 2018. A total of 863 isolates of Mycobacterium tuberculosis, 687 rifampicin resistant and 176 isoniazid resistant only, were screened for drug resistance in FQ and SLID. All the isolates were tested for Genotype MTBDRsl Ver 2.0 according to the manufacturer’s instructions. The FQ and SLID resistance were detected in 295 (34.2%) and 70 (8.1%) isolates, respectively. Among newly diagnosed and follow-up rifampicin-resistant TB (RR TB) patients, the FQ resistance was 25.8% and 44.5%, respectively. The most common mutation (42.7%) in FQ-resistant isolates was MUT3C in gyrA gene. Both SLID and FQ resistance were detected in 59 (6.8%) RR TB isolates. The mono SLID resistance was detected in 12 (1.7%) isolates of RR TB. Genotype MTBDRsl Ver 2.0 assay is a rapid and important tool for the diagnosis and molecular characterization of second-line drug resistance under programmatic conditions.
Keywords: Drug resistance, diagnosis, MTBDRsl Ver 2.0, Mycobacterium tuberculosis
1. INTRODUCTION
Drug-resistant tuberculosis (TB) remains a main hurdle for national tuberculosis control programs. As per the estimates of World Health Organization (WHO), in 2016, the multidrug-resistant (MDR) TB/rifampicin-resistant (RR) TB was 19% in previously treated patients and 4.1% in newly diagnosed TB patients [1]. The diagnosis of drug resistance for first-line antitubercular drugs is easy due to availability of WHO-endorsed molecular tests like Xpert MTB/RIF and line probe assays (LPAs). The second-line drug resistance detection is a challenge due to very limited WHO-endorsed tests [2]. The most reliable test for second-line drug resistance is phenotypic drug-susceptibility testing (DST) in liquid culture, which is laborious and time consuming.
The nucleic acid amplification methods have very high sensitivity and specificity and also recommended for detection of drug resistance to first-line drugs, mainly rifampicin and isoniazid. Recently, Genotype MTBDRsl Ver 2.0, LPA for fluoroquinolone (FQ) and second-line injectable drugs (SLID) drug resistance detection based on reverse hybridization technology, was endorsed by WHO for rapid detection of second-line drug resistance [3]. The previous version of this assay, that is, Genotype MTBDRsl Ver 1.0, includes the detection of FQ, SLID, and ethambutol. According to the meta-analysis by Theron et al. [4], the sensitivity and specificity of this assay for FQ and aminoglycoside resistance detection were 83.1% and 76.9%, respectively. The assay version 1.0 was redesigned and new targets were added for FQ (gyrB) and kanamycin (eis), and the ethambutol targets were removed [5]. The modified assay detects mutations in quinolone resistance determining region of gyrA and gyrB for FQ and rrs and eis for aminoglycosides. The assay includes 27 probes based on reverse hybridization technology for detection of FQ and aminoglycoside drug resistance. Gardee et al. [6] evaluated the assay and found it to have a sensitivity and specificity of 100% and 98.9%, respectively, for FQ drug resistance detection. In our earlier evaluation, we observed a sensitivity and specificity of 97.2% and 99.1%, respectively, for FQ resistance detection by Genotype MTBDRsl Ver 2.0 assay, whereas the sensitivity and specificity for detection of SLID resistance were 89.2–92.5% and 98.5–99.5%, respectively. The detection of extensively drug-resistant TB (XDR TB) was increased compared to the previous version of the assay [7].
Genotype MTBDRsl Ver 2.0 assay is included as a frontline test for second-line resistance detection in MDR TB/RR TB or isoniazid-resistant TB in India. In this study, we retrospectively analyzed the resistance pattern and molecular characterization of FQ and aminoglycoside in MDR TB/RR TB or isoniazid-resistant TB.
2. MATERIALS AND METHODS
2.1. Study Settings
This retrospective analysis was carried out for two centers of TB diagnosis under Revised National Tuberculosis Control Program (RNTCP), India—the Department of Medical Microbiology, Postgraduate Institute of Medical Education and Research (PGIMER), Chandigarh, and the Intermediate Reference Laboratory, Patiala, Punjab. Both the laboratories are certified by Central TB Division, India, for the testing of molecular and phenotypic drug sensitivity of Mycobacterium tuberculosis.
2.2. Clinical Isolates
A total of 863 consecutive drug-resistant isolates (either MDR TB/RR TB or isoniazid-resistant) received between August 2017 and March 2018 were analyzed. The information related to rifampicin and isoniazid resistance was extracted from RNTCP request form. Under programmatic (RNTCP) conditions in India, all the samples from rifampicin and isoniazid or both resistant patients (tested by the first-line LPA—Genotype MTBDRplus Ver 2.0, Hain Lifescience, or Xpert MTB/RIF) were subjected to second-line LPA (Genotype MTBDRsl Ver 2.0, Hain Lifescience) to check the additional resistance to FQs and aminoglycosides. These isolates were from sputum samples received for diagnosis of second-line drug resistance. The samples were processed by NALC NaOH method, followed by inoculation of 500 µl of processed sample in mycobacteria growth indicator tubes (MGIT) tube containing polymyxin B, amphotericin B, nalidixic acid, trimethoprim, azlocillin (PANTA) and growth supplement [8]. The positive culture tube from BACTEC MGIT 960 instrument was identified as M. tuberculosis by using SD Bioline MPT 64 Ag kit (Standard Diagnostics, Inc., Republic of Korea). The cultures were processed for Genotype MTBDRsl Ver 2.0 assay. The drug resistance characterization was done by second-line LPA (Genotype MTBDRsl Ver 2.0) only, and no phenotypic DST was performed.
2.3. Genotype MTBDRsl Ver 2.0 Assay
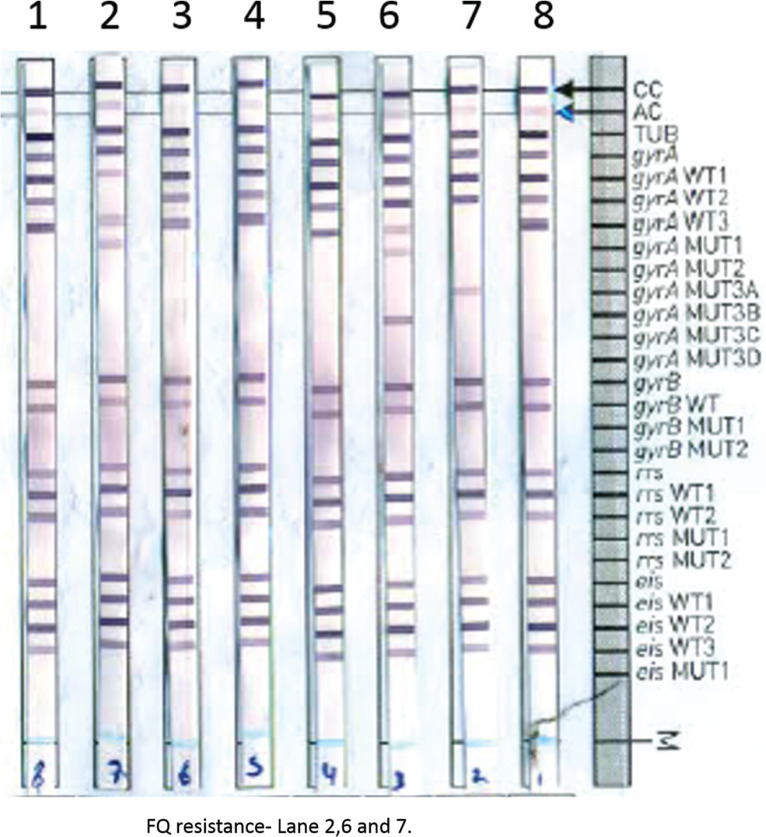
The Genotype MTBDRsl Ver 2.0 assay was performed as per the manufacturer’s instructions [5]. In brief, 1 ml of positive liquid culture was centrifuged and the pellet was taken for DNA extraction by GenoLyse kit Hain Lifescience (Nehren, Germany). Before amplification, the kit components AM-A and AM-B were mixed and then the extracted DNA was added and amplified. The hybridization was performed using TwinCubator/GT-Blot and the results were analyzed. The strip contains 27 probes to check internal controls, identification of M. tuberculosis complex, and drug targets gyrA, gyrB for FQ and rrs, eis for SLID. The missing of wild probe and presence of mutant probe are considered as resistant (Figure 1).
Figure 1.
Fluoroquinolones and aminoglycosides drug resistance detection by Genotype MTBDRsl Ver 2.0.
3. RESULTS
Among 863 drug-resistant isolates, 687 were RR and 176 were isoniazid-resistant but rifampicin-sensitive isolates. The age was ranging from 12 to 94 years and there were 319 (36.9%) females and 544 (63.03%) males. The isolates were obtained by culturing the sputum samples from newly diagnosed and follow-up during the therapy for both RR and/or isoniazid-resistant patients.
3.1. Second-Line Drug Resistance
Among the 863 isolates, 687 were RR including 375 (54.6%) newly diagnosed and 312 (45.4) follow-up isolates. The FQ resistance among RR isolates was detected in 265 (38.6%) isolates, of which FQ resistance among newly diagnosed and follow-up isolates were 97 (25.8%) and 139 (44.5%), respectively. Among 176 isoniazid-resistant isolates, 136 were newly diagnosed and 136 were follow-up isolates. The FQ resistance in isoniazid-resistant but rifampicin-sensitive isolates were 14 (10.3%) and 15 (11%) in newly diagnosed and follow-up isolates, respectively. The SLID resistance was found in 12 (1.7%) in RR isolates, 6 in RR newly diagnosed isolates, and 6 in RR follow-up samples. The SLID resistance was also detected in five isoniazid-resistant isolates, and four newly diagnosed and three follow-up samples were found resistant to SLID. Both FQ and SLID resistance were detected in 59 isolates, 6 in newly diagnosed and 53 in follow-up RR isolates (Table 1).
Table 1.
Second line drug resistance profile among rifampicin and isoniazid resistant isolates
| Resistance type of isolates | Total | FQ R | FQ S | SLID R | SLID S | FQ and SLID both |
|---|---|---|---|---|---|---|
| Rifampicin resistant newly diagnosed | 375 | 97 | 272 | 6 | 363 | 6 |
| Rifampicin resistant follow-up | 312 | 139 | 114 | 6 | 139 | 53 |
| Isoniazid mono-resistant newly diagnosed | 136 | 14 | 122 | 2 | 134 | 0 |
| Isoniazid mono-resistant follow-up | 40 | 15 | 25 | 3 | 37 |
FQ, fluoroquinolones; SLID, second line injectable drug; R, resistant; S, sensitive.
3.2. Molecular Characterization of Drug Resistance
Among 295 FQ-resistant isolates, 18 different types of banding pattern were observed. The most common banding pattern was MUT3C in 139 cases, including 126 RR samples and 13 isoniazid-resistant samples. The second most common banding pattern was gyrA MUT1 in 58 isolates, followed by gyrA MUT3A in 30 isolates. The following types of single-defined mutations were detected at gyrA codon 94: gyrA MUT3C (D94G, 139/324, 42.9%); gyrA MUT3A (D94A, 30/324, 9.2%); gyrA MUT3D (D94H, 3/324, 0.9%); and gyrA MUT3B (D94N/D94Y, 22/324, 6.8%) (Table 2). Other gyrA-defined mutations detected at codons 90 and 91 were gyrA MUT1 (A90V, 58/324, 17.9%) and gyrA MUT2 (S91P, 11/324, 3.4%) (Table 2). Four types of double mutations gyrA MUT3A and MUT3C, gyrA MUT3B and MUT3D, gyrA MUT3B and MUT3C, and gyrA MUT1 and MUT3A were found in 8/324 (2.4%) isolates. A total of 26 isolates showed missing of wt probe only, which includes 20, 4, and 2 in WT 3-, WT 2-, and WT 1-2-, respectively. Ten isolates (10/324, 3.1%) missing gyrB WT1- showed a low prevalence of gyrB mutations in this region. For SLID resistance, 58 defined mutations in rrs and eis and 18 undefined mutations in rrs and eis were detected. The most frequently observed mutation (32/73, 43.8%) for KM resistance was rrsMUT1 (A1401G); 10 of these isolates indicated the presence of both rrs wt and MUT1. The mutation rrsMUT2 (G1484T) was observed in 4/73 isolates (Table 2).
Table 2.
Genotype MTBDRsl Ver 2.0 hybridization pattern observed in resistant isolates
| Hybridization band(s) observed | Rifampicin resistant (N) | Isoniazid mono-resistant (N) | Total | ||
|---|---|---|---|---|---|
| Newly diagnosed | Follow-up | Newly diagnosed | Follow-up | ||
| gyrA gene | |||||
| gyrA MUT1 | 25 | 27 | 2 | 4 | 58 |
| WT/gyrA MUT1 | 1 | 3 | 0 | 0 | 4 |
| WT1-, WT2- | 1 | 0 | 1 | 2 | |
| gyrA MUT2 | 4 | 7 | 0 | 0 | 11 |
| WT2- | 1 | 2 | 1 | 0 | 4 |
| WT/gyrA MUT2 | 1 | 4 | 0 | 0 | 5 |
| gyrA MUT3A | 13 | 15 | 2 | 0 | 30 |
| gyrA MUT3B | 9 | 12 | 0 | 1 | 22 |
| gyrA MUT3A and MUT3C | 0 | 2 | 0 | 0 | 2 |
| gyrA MUT3B and MUT3D | 0 | 2 | 0 | 0 | 2 |
| gyrA MUT3C | 39 | 87 | 7 | 6 | 139 |
| WT/gyrA MUT3A | 0 | 1 | 0 | 0 | 1 |
| WT/gyrA MUT3B | 0 | 1 | 0 | 1 | 2 |
| WT/gyrA MUT3C | 6 | 6 | 1 | 2 | 15 |
| gyrA MUT3D | 0 | 3 | 0 | 0 | 3 |
| gyrA MUT3B and MUT3C | 1 | 1 | 0 | 0 | 2 |
| gyrA MUT1 and MUT3A | 1 | 1 | 2 | ||
| WT3- | 2 | 17 | 1 | 0 | 20 |
| rrs gene | |||||
| rrs MUT1 | 3 | 27 | 1 | 1 | 32 |
| WT/rrs MUT1 | 2 | 9 | 0 | 1 | 12 |
| WT/rrs MUT2 | 2 | 2 | 0 | 0 | 4 |
| eis gene | |||||
| eis MUT1 | 1 | 7 | 0 | 0 | 8 |
| eis WT2 absent | 4 | 10 | 1 | 0 | 15 |
| WT/eis MUT1 | 0 | 1 | 0 | 1 | 2 |
4. DISCUSSION
The rapid and accurate diagnosis of second-line drug resistance among RR and isoniazid mono-resistant cases is important for timely initiation of correct treatment regimen. WHO-endorsed test Genotype MTBDRsl gives rapid and accurate diagnosis for second-line drug resistance detection and has also been started in RNTCP in India. In this study, we demonstrate the use of Genotype MTBDRsl assay for second-line drug resistance detection under programmatic condition in North India. The overall FQ resistance was 38.6% among the RR isolates, which is similar to other studies from India that range from 24 to 59.6%, while the meta-analysis by Ho et al. [13] has shown FQ resistance in MDR patients ranging from 1 to 22%. Among newly diagnosed isolates, the FQ resistance was 25.8% in our study, which was slightly higher than the 22% reported in the recent drug resistance survey carried out in India [14]. Similarly, the FQ resistance in follow-up culture isolates was 44.6% compared to the reported 20.9% in the survey. The overall SLID resistance was 1.7%, which is less than that from other regions of India [15]. Both the FQ and SLID resistance were detected in 8.6% isolates, which is quite higher than reported prevalence of XDR TB worldwide [16]. In our study, we also included the isoniazid mono-resistant isolates and found that the FQ resistance in newly diagnosed and follow-up samples was almost ranging from 10 to 11%. The meta-analysis done by Ho et al. [11] had shown the prevalence of FQ resistance to be 0–4.4% among non-MDR TB patients [11]. The high FQ resistance was also noted in newly diagnosed MDR/RR TB cases, which might be due to the high transmission of the drug-resistant strains. The high rate of FQ resistance in this study is alarming for the national program and indicates injudicious use of FQ. The SLID resistance was detected in 1.7% of RR isolates, which was also quite less than the previously reported 7.7–17% [16,17].
The most common mutation detected by Genotype MTBDRsl in FQ-resistant isolates was a change at codon 94. Among codon 94 mutations, the most prominent mutation was gyrA MUT3C in 139 cases, which was comparable to the studies from South Africa, China, and India [6,7,18]. We found the rare gyrA MUT3D mutation in three cases that is absent in most of the studies, including the recent study from China [13]. We also found the gyrB WT1 missing in 3.1% cases, indicating the presence of gyrB mutations in this region. The heteroresistance reported from South Africa was also observed in 27 of our cases [6]. Among the rrs mutations, the most common was rrsMUT1 found in 32 cases, while rrsMUT2 was present in 4 cases. These observations are comparable to the previous studies from India and South Africa [6,7]. We also found the mutations in eis region and the most common was eisMUT2 absent in 15 cases. The limitation of this analysis lies in the absence of phenotypic DST and sequencing data for confirmation of different drug-resistant related mutations.
In conclusion, the WHO-endorsed rapid method for detection of second-line drug resistance, Genotype MTBDRsl assay, is also able to detect mutations with a short turnaround time, thus enabling early and precise combination therapy decisions.
CONFLICTS OF INTEREST
The authors declare they have no conflicts of interest.
AUTHORS’ CONTRIBUTION
SS, RY and PA contributed in conceptualization and writing original draft. RY, JC and NK contributed in data curation. RY, NK and ANA contributed in formal analysis. SS, RK, PA and NK contributed in project administration. NK and RY contributed in methodology. All authors contributed in review draft & editing.
FUNDING
This was a retrospective analysis of the routine testing under RNTCP programme and the consumables were provided by Central TB Division, India.
REFERENCES
- [1].World Health Organization . Global tuberculosis report. Geneva: WHO; 2017. Available from: http://apps.who.int/iris/bitstream/handle/10665/259366/9789241565516-eng.pdf?sequence=1. [Google Scholar]
- [2].World Health Organization . Companion handbook to the WHO guidelines for the programmatic management of drug-resistant tuberculosis. Geneva: WHO; 2014. Available from: http://apps.who.int/iris/bitstream/handle/10665/130918/9789241548809_eng.pdf?sequence=1. [PubMed] [Google Scholar]
- [3].World Health Organization . The use of molecular line probe assays for the detection of resistance to second-line anti-tuberculosis drugs. WHO Policy guidance. Geneva: WHO; 2016. Available from: http://www.who.int/tb/WHOPolicyStatementSLLPA.pdf. [Google Scholar]
- [4].Theron G, Peter J, Richardson M, Barnard M, Donegan S, Warren R, et al. The diagnostic accuracy of the GenoType® MTBDRsl assay for the detection of resistance to second-line anti-tuberculosis drugs. Cochrane Database Syst Rev. 2014:CD010705. doi: 10.1002/14651858.CD010705.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Hain Lifescience . GenoType MTBDRsl Ver 2.0 instructions for use. Document IFU-317A-01. Nehren, Germany: Hain Life Science; 2015. [Google Scholar]
- [6].Gardee Y, Dreyer AW, Koornhof HJ, Omar SV, da Silva P, Bhyat Z, et al. Evaluation of the GenoType MTBDRsl Version 2.0 assay for second-line drug resistance detection of Mycobacterium tuberculosis isolates in South Africa. J Clin Microbiol. 2017;55:791–800. doi: 10.1128/JCM.01865-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Yadav R, Saini A, Kaur P, Behera D, Sethi S. Diagnostic accuracy of GenoType® MTBDRsl VER 2.0 in detecting second-line drug resistance to M. tuberculosis. Int J Tuberc Lung Dis. 2018;22:419–24. doi: 10.5588/ijtld.17.0663. [DOI] [PubMed] [Google Scholar]
- [8].Levin W, Brandon GR, McMillen S. The culture method of laboratory diagnosis of tuberculosis. Am J Public Health Nations Health. 1950;40:1305–10. doi: 10.2105/ajph.40.10.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Parmar MM, Sachdeva KS, Dewan PK, Rade K, Nair SA, Pant R, et al. Unacceptable treatment outcomes and associated factors among India’s initial cohorts of multidrug-resistant tuberculosis (MDR-TB) patients under the revised national TB control programme (2007–2011): evidence leading to policy enhancement. PLoS One. 2018;13:e0193903. doi: 10.1371/journal.pone.0193903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Paramasivan CN, Rehman F, Wares F, Sundar Mohan N, Sundar S, Devi S, et al. First- and second-line drug resistance patterns among previously treated tuberculosis patients in India. Int J Tuberc Lung Dis. 2010;14:243–6. https://www.ncbi.nlm.nih.gov/pubmed/20074419. [PubMed] [Google Scholar]
- [11].Ho J, Jelfs P, Sintchenko V. Fluoroquinolone resistance in non-multidrug-resistant tuberculosis—a surveillance study in New South Wales, Australia, and a review of global resistance rates. Int J Infect Dis. 2014;26:149–53. doi: 10.1016/j.ijid.2014.03.1388. [DOI] [PubMed] [Google Scholar]
- [12].Singhal P, Dixit P, Singh P, Jaiswal I, Singh M, Jain A. A study on pre-XDR & XDR tuberculosis & their prevalent genotypes in clinical isolates of Mycobacterium tuberculosis in north India. Indian J Med Res. 2016;143:341–7. doi: 10.4103/0971-5916.182625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Gao Y, Zhang Z, Deng J, Mansjö M, Ning Z, Li Y, et al. Multi-center evaluation of GenoType MTBDRsl line probe assay for rapid detection of pre-XDR and XDR Mycobacterium tuberculosis in China. J Infect. 2018;77:328–34. doi: 10.1016/j.jinf.2018.06.014. [DOI] [PubMed] [Google Scholar]
- [14].Jain A, Dixit P, Prasad R. Pre-XDR & XDR in MDR and ofloxacin and kanamycin resistance in non-MDR Mycobacterium tuberculosis isolates. Tuberculosis (Edinb) 2012;92:404–6. doi: 10.1016/j.tube.2012.05.010. [DOI] [PubMed] [Google Scholar]
- [15].Ramachandran R, Nalini S, Chandrasekar V, Dave PV, Sanghvi AS, Wares F, et al. Surveillance of drug-resistant tuberculosis in the state of Gujarat, India. Int J Tuberc Lung Dis. 2009;13:1154–60. https://www.ncbi.nlm.nih.gov/pubmed/19723407. [PubMed] [Google Scholar]
- [16].Report of the First National Anti-tuberculosis Drug Resistance Survey. India: Central TB division; 2018. Available from: https://tbcindia.gov.in/showfile.php?lid=3315. [Google Scholar]
- [17].Lange C, Chesov D, Heyckendorf J, Leung CC, Udwadia Z, Dheda K. Drug-resistant tuberculosis: an update on disease burden, diagnosis and treatment. Respirology. 2018;23:656–73. doi: 10.1111/resp.13304. [DOI] [PubMed] [Google Scholar]
- [18].Sharma AK, Gupta N, Kala DK, Patni T, Dixit R, Verma S, Chandran A. A study on pattern of resistance to second line anti tubercular drugs among multi drug resistant tuberculosis patients. Indian J Tuberc. 2018;65:233–6. doi: 10.1016/J.Ijtb.2018.02.005. [DOI] [PubMed] [Google Scholar]