Abstract
Background:
Acute myeloid leukemia (AML) is a common hematological malignancy. Gemtuzumab ozogamicin (GO), a humanized anti-CD33 antibody conjugated with the potent anti-tumor antibiotic calicheamicin, represents a promising targeted therapy for AML. Annexin A5 (ANXA5) is a proposed marker for the clinical prognosis of AML to guide treatment choice.
Methods:
In total, 253 patients with pediatric AML were enrolled and divided into two treatment groups: conventional chemotherapy alone and conventional chemotherapy in combination with GO. Univariate, multivariate, and Kaplan–Meier survival analyses were conducted to assess risk factors and clinical outcomes, and to estimate hazard ratios (HRs) and their 95% confidence interval. The level of statistical significance was set at p < 0.05.
Results:
In the GO treatment group, high ANXA5 expression was considered a favorable prognostic factor for overall survival (OS) and event-free survival (EFS). Multivariate analysis showed that high ANXA5 expression was an independent favorable factor for OS (HR = 0.629, p = 0.084) and EFS (HR = 0.544, p = 0.024) distinct from the curative effect of GO treatment. When all patients were again divided into two groups, this time based on the median expression of ANXA5, patients undergoing chemotherapy combined with GO had significantly better OS (p = 0.0012) and EFS (p = 0.0003) in the ANXA5 high-expression group. Gene set enrichment analysis identified a relevant series of pathways associated with glutathione metabolism, leukocyte transendothelial migration, and hematopoietic cell lineage.
Conclusion:
The expression level of ANXA5 can help optimize the treatment regimen for individual patients, and patients with overexpression of ANXA5 may circumvent poor outcomes from chemotherapy combined with GO.
Keywords: acute myeloid leukemia, ANXA5, chemotherapy, gemtuzumab ozogamicin, prognosis
Introduction
Acute myeloid leukemia (AML) is a common hematologic malignancy characterized by excessive proliferation of hematopoietic stem and progenitor cells, accounting for approximately 20% of childhood leukemia cases.1 The diagnostic classification of AML includes a combination of morphology, immunology, cytogenetics, and molecular biology.2 Currently, the standard treatment protocol for AML is combination chemotherapy, and patients may also undergo allogeneic stem cell transplantation, especially in high-risk groups for induction failure or recurrence.3–5 Among patients over the age of 60 years, a complete remission (CR) rate of 40% to 60% can be achieved, though only 5% to 15% can be cured.1,6 For the large number of AML cases in children and adolescents, overall survival (OS) and CR rates are 67% and 93%, respectively.7 Although the diagnostic technology and treatment protocol of AML have gradually improved, the cure rate for pediatric AML lags behind pediatric acute lymphoblastic leukemia, mostly due to relapse.8,9 Therefore, there is an urgent need for optimizing therapeutic strategies to design personalized therapy for individual patients.
Gemtuzumab ozogamicin (GO), a humanized anti-CD33 antibody conjugated with the potent anti-tumor drug calicheamicin, represents a promising targeted therapy in AML.10 In May 2000, GO was granted accelerated approval by the US Food and Drug Administration (FDA) for AML patients in first relapse who were not candidates for conventional chemotherapy.11 However, GO has not demonstrated sufficient effect in different patient populations as a single agent; the overall response rate has been only 25–35%.12–14 In 2010, Pfizer voluntarily withdrew the drug after a phase III trial that failed to demonstrate superior efficacy of GO.15 Despite these setbacks, results from subsequent clinical trials administering GO at 3–6 mg/m2 per dose demonstrated clinical benefit compared with standard chemotherapy in children and adults with AML, and the US FDA announced re-approval of GO in September 2017.13,16
Annexin A5 (ANXA5) is a member of the annexin family, which binds phospholipids in a calcium-dependent manner, playing major roles in regulating cellular growth, differentiation, inflammation, and signaling.17,18 Under stress, ANXA5 is produced and released into the extracellular medium or bloodstream, functioning as a physiological anticoagulant, anti-inflammatory, and anti-apoptotic agent by protecting stressed or dying cells from contact with inflammatory cells.17,19 A previous study showed that abnormal expression patterns of ANXA5 are associated with proliferation, invasion, drug resistance, and tumor treatment.20–23 Furthermore, annexin family members have distinct prognostic roles in adult and pediatric AML. High expression levels of ANXA2, ANXA6, and ANXA7 have been associated with worse prognoses of patients with AML, whereas ANXA5 has been correlated with more favorable clinical outcomes.24,25 Nevertheless, the relevance of annexin family members as predictive molecular markers to guide treatment choice remains largely unexplored.
In this study, we compared GO-containing treatment regimens with non-GO regimens in pediatric patients with AML. From the Therapeutically Applicable Research to Generate Effective Treatment (TARGET) database, a gene expression pattern associated with ANXA5 expression was derived to investigate pediatric AML. ANXA5 was identified as a gene that may circumvent poor outcomes in pediatric patients with AML treated with GO. Finally, our study revealed that expression of ANXA5 can help optimize the treatment regimen for individual patients.
Materials and methods
Patients
A total of 253 patients aged 0–24 years and diagnosed with pediatric/adolescent AML were included in this study. The clinical data and treatment of the patients were analyzed retrospectively for receiving conventional chemotherapy (no-GO group, n = 95) or the same chemotherapy regimen combined with a dose of 3 mg/m2 GO (GO group, n = 158). The datasets used in this investigation were acquired from the publicly available TARGET database (https://ocg.cancer.gov/), and clinical outcome data were included in Children’s Oncology Group trials AAML0531 and AAML03P1.26,27 Data were downloaded and analyzed until 20 June 2019 in this study. Exclusion criteria were as follows: (1) samples without clinical data, (2) samples without complete gene expression data and survival period, and (3) patients with acute promyelocytic leukemia, Down syndrome, or secondary/treatment-related leukemia. As the data were obtained from a publicly available database, further approval from the local ethics committee was not required.
Gene expression profiling
We used Perl language (https://www.perl.org/; version 5.30.0) to extract the gene matrix file, including ANXA5 expression data and match gene expression array with clinical long-term follow-up data. Then, the Ensembl IDs were transformed into gene names based on the Ensembl database (http://www.ensembl.org). Patients with FLT3-ITD mutations were classified as either positive or negative.28,29 The Search Tool for the Retrieval of Interacting Genes (STRING 11.0; https://string-db.org/) was used to predict proteins interacting with a query protein.30 In the present study, the following parameters were selected: hiding disconnected nodes in the network; medium confidence score > 0.400; no more than 20 interactions. The ”pheatmap” package in R 3.5.2 software (https://www.r-project.org/) was used to construct a heat map of coexpression patterns of ANXA5. The Cancer Cell Line Encyclopedia (CCLE; https://portals.broadinstitute.org/ccle/about) dataset was used to analyze ANXA5 expression in cancer cell lines.31
Gene set enrichment analysis
Gene set enrichment analysis (GSEA) is a knowledge-based method that determines whether a particular set of functionally related genes shows statistically significant, concordant differences between two biological states.32 In this study, we used GSEA version 4.0.1 software (http://software.broadinstitute.org/gsea/). The 253 AML samples in this investigation were divided into a low- or high-expression group using ANXA5 median expression level as a cut-off point. To identify potential mechanisms underlying the effects of gene expression, the expression level of ANXA5 was used as a phenotype label, and gene set permutations were performed 1000 times for each analysis. Finally, the pathways enriched in each phenotype were sorted by normalized enrichment score (NES) and nominal p-value.
Statistical analysis
Statistical analysis was performed using GraphPad Prism software (version 8.02; GraphPad Software, Inc., La Jolla, CA, USA). Clinical and molecular characteristics of patients were described by median and/or range. Comparisons between continuous variables were analyzed using the Mann–Whitney U test, and the chi-square test or Fisher’s exact test were used to compare differences in proportions of variables among groups. OS was defined as the time period from diagnosis to death or the date of last follow up. EFS was defined as the time from diagnosis to relapse, induction failure, death in remission, or the date of last follow up. OS and EFS were estimated by Kaplan–Meier analysis and log-rank test. Univariate and multivariate Cox proportional hazard models were constructed to analyze the impact of clinical prognostic factors in pediatric AML, and to estimate the hazard ratios (HRs) and their 95% confidence interval (CI). The level of statistical significance was set at p < 0.05.
Results
Patient characteristics
To establish correlations between ANXA5 expression and various clinical characteristics in pediatric AML, we assigned patients who underwent chemotherapy combined with ±GO to one of two groups, according to median ANXA5 expression levels, respectively. Details on the clinical and molecular characteristics of patients in both groups are summarized in Table 1. The median age was 10.4 (range 0.1–23.5) years. In the no-GO treatment group, participants who exhibited downregulated ANXA5 had a higher percentage of WT1 mutation compared with upregulated ANXA5 expression (p = 0.016). In addition, high ANXA5 expression often had more MLL (p = 0.008) or CBFβ-MYH11 (p = 0.002) mutations and fewer RUNX1-RUNX1T1 (p = 0.004) fusions. Moreover, patients with lower ANXA5 expression had higher peripheral blood myeloblast counts (p = 0.007), though the median was not different between the two groups. Between the high and low ANXA5 expression groups, no significant differences were observed in age, sex, ethnicity, white blood cell count, bone marrow blast, complex karyotype, or FLT3-ITD, NPM1, CEBPA, and c-KIT mutations in exons 8 and 17. In the GO treatment group, study participants with downregulated ANXA5 exhibited a higher frequency of CEBPA (p = 0.005) and FLT3-ITD (p = 0.022) mutations, whereas c-Kit mutations in exons 17 (p = 0.007) and white blood cell counts (p = 0.001) were lower. Overall, the clinical and molecular characteristics of the two treatment groups at diagnosis were similar (Supplemental Table S1), excluding the influence of ANXA5 expression. Interestingly, both treatment groups showed that patients with low ANXA5 expression were more often diagnosed with M1 or M2 compared with patients with high ANXA5 expression.
Table 1.
Comparison of clinical and molecular characteristics with ANXA5 expression in patients.
| Characteristic |
No gemtuzumab ozogamicin treatment
|
Gemtuzumab ozogamicin treatment
|
||||
|---|---|---|---|---|---|---|
| ANXA5high (n = 47) | ANXA5low (n = 48) | p-value | ANXA5high (n = 79) | ANXA5low (n = 79) | p-value | |
| Age/years, median (range) | 10.6 (0.1–20.3) | 10.3 (1.7–23.5) | 0.101* | 10.4 (0.4–22.5) | 10.3 (1.2–18.2) | 0.448* |
| Age group/n (%) | 0.612§ | 0.633§ | ||||
| <10 years | 23 (48.9) | 21 (43.8) | 40 (50.6) | 37 (46.8) | ||
| ⩾10 years | 24 (51.1) | 27 (56.2) | 39 (49.4) | 42 (53.2) | ||
| Sex/n (%) | 0.051§ | 0.076§ | ||||
| Male | 19 (40.4) | 30 (62.5) | 52 (65.8) | 40 (50.6) | ||
| Female | 28 (59.6) | 18 (37.5) | 27 (34.2) | 39 (49.4) | ||
| Ethnicity/n(%) | ||||||
| Hispanic or Latino | 11 (23.4) | 10 (20.8) | 0.763§ | 14 (17.7) | 16 (20.3) | 0.685§ |
| Not Hispanic or Latino | 32 (68.1) | 36 (75.0) | 0.455§ | 63 (79.7) | 61 (77.2) | 0.699§ |
| Unknown | 4 (8.5) | 2 (4.2) | 0.384§ | 2 (2.5) | 2 (2.5) | 1.000§ |
| WBC/×10 9 /l, median (range) | 33.5 (1.3–519) | 34.6 (0.9–439.2) | 0.183* | 53.5 (4.2–446) | 52.6 (1.5–263.1) | 0.001* |
| BM blast/%, median (range) | 71 (23–99) | 71 (20–99) | 0.961* | 77 (21–100) | 77 (21–99) | 0.885* |
| PB blast/%, median (range) | 62 (0–97) | 62 (2–97) | 0.007* | 61 (0–95) | 61 (0–97) | 0.792* |
| FAB subtypes/n (%) | ||||||
| M0 | 0 (0) | 2 (4.2) | 0.157§ | 2 (2.5) | 4 (5.1) | 0.405§ |
| M1 | 3 (6.4) | 10 (20.8) | 0.040§ | 3 (3.8) | 16 (20.3) | 0.001§ |
| M2 | 4 (8.5) | 19 (39.6) | 0.001§ | 8 (10.1) | 26 (32.9) | 0.001§ |
| M4 | 10 (21.3) | 4 (8.3) | 0.075§ | 35 (44.3) | 8 (10.1) | 0.001§ |
| M5 | 14 (29.8) | 2 (4.2) | 0.001§ | 23 (29.1) | 9 (11.4) | 0.006§ |
| M6 | 0 (0) | 1 (2.1) | 0.319§ | 0 (0) | 2 (2.5) | 0.155§ |
| M7 | 5 (10.6) | 1 (2.1) | 0.087§ | 1 (1.3) | 1 (1.3) | 1.000§ |
| Others | 11 (23.4) | 9 (18.6) | 0.578§ | 7 (8.9) | 13 (16.5) | 0.151§ |
| Cytogenetics/n (%) | ||||||
| Normal | 9 (19.1) | 11 (22.9) | 0.652§ | 17 (21.5) | 28 (35.4) | 0.056§ |
| Complex karyotype | 7 (14.9) | 7 (14.6) | 0.966§ | 12 (15.2) | 11 (13.9) | 0.822§ |
| inv(16)/CBFβ-MYH11 | 11 (23.4) | 1 (2.1) | 0.002§ | 23 (29.1) | 1 (1.3) | 0.001§ |
| 11q23/MLL | 14 (29.8) | 4 (8.3) | 0.008§ | 14 (17.7) | 9 (11.4) | 0.259§ |
| t(8;21)/RUNX1-RUNX1T1 | 3 (6.4) | 14 (29.2) | 0.004§ | 5 (6.3) | 16 (20.3) | 0.010§ |
| Others | 3 (6.4) | 11 (22.9) | 0.023§ | 8 (10.1) | 14 (17.7) | 0.168§ |
| Risk/n (%) | ||||||
| Good | 18 (38.2) | 21 (43.7) | 0.589§ | 33 (41.8) | 31 (39.2) | 0.746§ |
| Intermediate | 27 (57.4) | 19 (39.5) | 0.082§ | 35 (44.3) | 34 (43.0) | 0.873§ |
| Poor | 2 (4.2) | 8 (16.6) | 0.049§ | 7 (8.9) | 1 0 (12.7) | 0.441§ |
| Others | 0 (0) | 0 (0) | 1.000§ | 4 (5.1) | 4 (5.1) | 1.000§ |
| FLT3-ITD/n (%) | 0.473§ | 0.022§ | ||||
| Positive | 8 (17.0) | 11 (22.9) | 6 (7.6) | 16 (20.3) | ||
| Negative | 39 (83.0) | 37 (77.1) | 73 (92.4) | 63 (79.7) | ||
| NPM1/n (%) | 0.751§ | 0.548§ | ||||
| Mutation | 4 (8.5) | 5 (10.4) | 7 (8.9) | 5 (6.3) | ||
| Wild type | 43 (91.5) | 43 (89.6) | 72 (91.1) | 74 (93.7) | ||
| CEBPA/n (%) | 0.176§ | 0.005§ | ||||
| Mutation | 1 (2.1) | 4 (8.3) | 1 (1.3) | 10 (12.7) | ||
| Wild type | 46 (97.9) | 44 (91.7) | 78 (98.7) | 69 (87.3) | ||
| WT1/n (%) | 0.016§ | 0.148§ | ||||
| Mutation | 1 (2.1) | 8 (16.7) | 4 (5.1) | 9 (11.4) | ||
| Wild type | 46 (97.9) | 40 (83.3) | 75 (94.9) | 70 (88.6) | ||
| c-Kit mutation exon 8/n (%) | ||||||
| Yes | 4 (8.5) | 2 (4.2) | 0.384§ | 3 (3.8) | 5 (6.3) | 0.468§ |
| No | 10 (21.3) | 13 (27.1) | 0.509§ | 20 (25.3) | 10 (12.7) | 0.043§ |
| Not done | 33 (70.2) | 33 (68.8) | 0.878§ | 56 (70.9) | 64 (81.0) | 0.136§ |
| c-Kit mutation exon 17/n (%) | ||||||
| Yes | 2 (4.3) | 5 (10.4) | 0.250§ | 7 (8.9) | 0 (0) | 0.007§ |
| No | 12 (25.5) | 10 (20.8) | 0.587§ | 16 (20.3) | 15 (19.0) | 0.841§ |
| Not done | 33 (70.2) | 33 (68.8) | 0.878§ | 56 (70.9) | 64 (81.0) | 0.136§ |
| CNS disease/n (%) | 0.570§ | 0.230§ | ||||
| Yes | 1 (2.1) | 2 (4.2) | 4 (5.1) | 8 (10.1) | ||
| No | 46 (97.9) | 46 (95.8) | 75 (94.9) | 71 (89.9) | ||
| CR1 achieved/n (%) | ||||||
| Yes | 31 (66.0) | 31 (64.6) | 0.888§ | 70 (88.6) | 60 (75.9) | 0.037§ |
| No | 15 (31.9) | 17 (35.4) | 0.718§ | 9 (11.4) | 19 (24.1) | 0.037§ |
| Unevaluable | 1 (2.1) | 0 (0) | 0.310§ | 0 (0) | 0 (0) | 1.000§ |
BM, bone marrow; CNS, central nervous system; CR1, first complete remission; FAB, French American British; PB, peripheral blood; WBC, white blood cell.
denotes Mann–Whitney U test.
denotes chi-square test. Complex karyotype is defined as more than or equal to three chromosomal abnormalities.
Prognostic value of ANXA5 expression
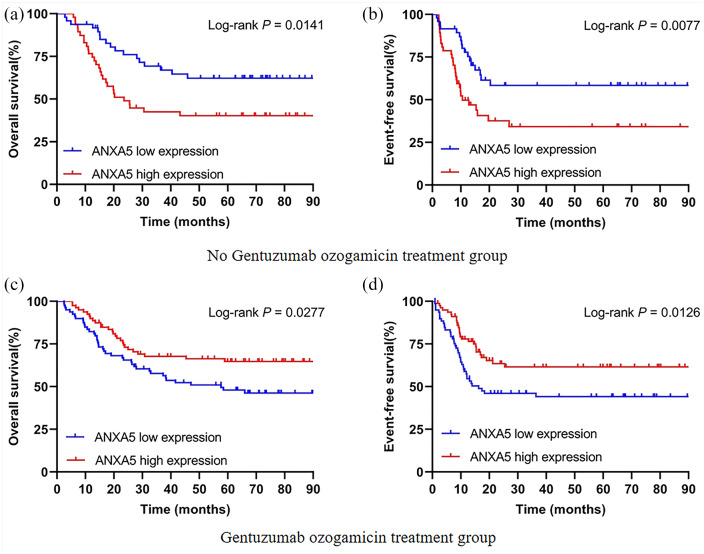
A log-rank test of Kaplan–Meier curves was used to describe the differences in survival to estimate clinical outcomes of ANXA5 in patients with different chemotherapy regimens. In the no-GO treatment group, survival distribution curves demonstrated that high ANXA5 expressers had shorter OS (HR = 2.086, 95% CI 1.158–3.758, p = 0.0141) and EFS (HR = 2.211, 95% CI 1.225–3.991, p = 0.0077) than low expressers [Figure 1(a and b)]. However, high expression of ANXA5 was considered a favorable prognostic factor for OS (HR = 0.583, 95% CI 0.352–0.939, p = 0.0277) and EFS (HR = 0.545, 95% CI 0.338–0.878, p = 0.0126) in patients with AML who received GO treatment [Figure 1(c and d)]. These results suggest that the curative effect of GO treatment may be closely related to the level of ANXA5 expression regulation in pediatric AML.
Figure 1.
Kaplan–Meier curves of survival in pediatric AML patients with respect to ANXA5 expression. (a, b) Effect of ANXA5 expression on OS and EFS in the no-GO treatment group (n = 95). (c, d) Patients with high ANXA5 expression had significantly prolonged OS and EFS in the GO treatment group (n = 158).
AML, acute myeloid leukemia; EFS, event-free survival; GO, gemtuzumab ozogamicin; OS, overall survival.
Univariate and multivariate analyses for prognostic factors
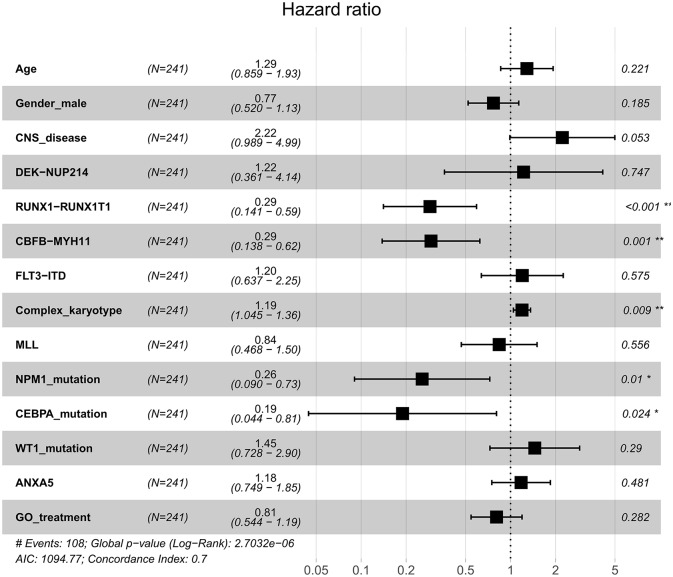
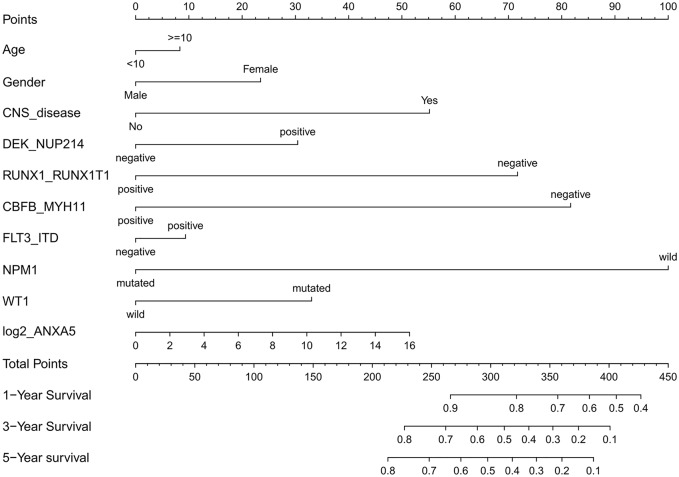
To evaluate the impact of clinical and molecular factors associated with ANXA5 expression level in pediatric AML, Cox proportional hazard models were constructed. The variables included expression levels of ANXA5 (high versus low), age (⩾10 versus <10 years), sex (male versus female), white blood cells (⩾50 versus <50 × 109/l), bone marrow blasts (⩾70% versus <70%), peripheral blood blasts (⩾50% versus <50%), NPM1 (mutated versus wild type), MLL (mutated versus wild type), FLT3-ITD (positive versus negative), CBFβ-MYH11 (positive versus negative), RUNX1-RUNX1T1 (positive versus negative), WT1 (mutated versus wild type), and risk (poor versus non-poor). A forest plot was constructed for OS for all patients according to prognostic factors (Figure 2), and the results showed that favorable factors in pediatric AML were found in terms of RUNX1-RUNX1T1, CBFβ-MYH11, NPM1 mutation, and CEBPA mutation, consistent with the results of the current risk stratification criteria.33 Meanwhile, we developed a nomogram to predict the probability of the 1- and 5-year OS including ANXA5 and clinical–biological prognostic index (see Figure 3).
Figure 2.
Forest plot of HR for OS according to prognostic factors with all patients with AML. Multivariate analyses of age, sex, CNS disease, DEK-NUP214, RUNX1-RUNX1T1, CBFB-MYH11, FLT3-ITD, complex karyotype, MLL, NPM1 mutation, CEBPA mutation, WT1 mutation, GO treatment and ANXA5 expression group for OS in all patients. The black squares on the transverse lines represent the HR, and the gray transverse lines represent 95% CI.
AML, acute myeloid leukemia; CI, confidence interval; CNS, central nervous system; GO, gemtuzumab ozogamicin; HR, hazard ratio; OS, overall survival.
*p < 0.05 and **p < 0.01.
Figure 3.
Nomogram for the prediction of OS at 1-, 3- and 5-years according to the clinical–biological prognostic index in patients with pediatric AML. By adding up the points assigned to each predictive variable, the total score on the bottom scale shows the probability of survival.
AML, acute myeloid leukemia; OS, overall survival.
In the no-GO treatment group, univariate analysis showed that high ANXA5 expression was associated with shorter OS (HR = 2.097, 95% CI 1.146–3.838, p = 0.016) and EFS (HR = 2.232, 95% CI 1.218–4.093, p = 0.009). Furthermore, multivariate analysis indicated that ANXA5 upregulation was an independent risk factor for OS (HR = 2.687, 95% CI 1.355–5.326, p = 0.005) and EFS (HR = 2.762, 95% CI 1.365–5.591, p = 0.005). Results are shown in Table 2.
Table 2.
Univariate and multivariate analysis for EFS and OS in patients without gemtuzumab ozogamicin treatment.
| Variables | OS |
EFS |
||
|---|---|---|---|---|
| HR (95% CI) | p-value | HR (95% CI) | p-value | |
| Univariate analyses | ||||
| ANXA5 (high versus low) | 2.097 (1.146–3.838) | 0.016 | 2.232 (1.218–4.093) | 0.009 |
| Age (⩾10 versus <10 years) | 1.017 (0.565–1.833) | 0.954 | 0.948 (0.526–1.709) | 0.859 |
| Sex (male versus female) | 1.288 (0.715–2.32) | 0.399 | 1.271 (0.706–2.289) | 0.425 |
| Ethnicity (Hispanic or Latino versus others) | 1.657 (0.854–3.213) | 0.135 | 1.507 (0.777–2.924) | 0.225 |
| WBC (⩾50 versus <50 × 109/l) | 1.003 (0.552–1.821) | 0.992 | 1.315 (0.723–2.393) | 0.37 |
| BM blasts (⩾70 versus <70%) | 0.957 (0.532–1.72) | 0.883 | 0.979 (0.545–1.76) | 0.943 |
| PM blasts (⩾50 versus <50%) | 0.718 (0.395–1.305) | 0.277 | 0.865 (0.476–1.571) | 0.634 |
| NPM1 (mutated versus wild type) | 0.578 (0.179–1.867) | 0.36 | 0.547 (0.169–1.767) | 0.313 |
| MLL (mutated versus wild type) | 1.807 (0.869–3.756) | 0.113 | 1.683 (0.809–3.5) | 0.163 |
| FLT3-ITD (positive versus negative) | 1.163 (0.576–2.351) | 0.673 | 1.288 (0.637–2.603) | 0.481 |
| CBFβ-MYH11 (positive versus negative) | 0.598 (0.214–1.669) | 0.326 | 0.687 (0.245–1.925) | 0.476 |
| RUNX1-RUNX1T1 (positive versus negative) | 0.507 (0.2–1.284) | 0.152 | 0.404 (0.159–1.026) | 0.057 |
| WT1 (mutated versus wild type) | 1.211 (0.433–3.385) | 0.715 | 1.4 (0.498–3.933) | 0.523 |
| Risk (poor versus non-poor) | 2.795 (1.415–5.521) | 0.003 | 3.259 (1.646–6.449) | 0.001 |
| Multivariate analyses | ||||
| ANXA5 (high versus low) | 2.687 (1.355–5.326) | 0.005 | 2.762 (1.365–5.591) | 0.005 |
| Ethnicity (Hispanic or Latino versus others) | 1.652 (0.845–3.231) | 0.142 | 1.408 (0.722–2.746) | 0.315 |
| MLL (mutated versus wild type) | 0.783 (0.335–1.828) | 0.572 | 0.709 (0.309–1.626) | 0.417 |
| FLT3-ITD (positive versus negative) | 1.343 (0.62–2.912) | 0.455 | 1.535 (0.696–3.383) | 0.288 |
| RUNX1-RUNX1T1 (positive versus negative) | 2.009 (0.572–7.06) | 0.277 | 1.884 (0.513–6.923) | 0.34 |
| Risk (poor versus non-poor) | 3.806 (1.518–9.542) | 0.004 | 4.492 (1.781–11.33) | 0.001 |
BM, bone marrow; CI, confidence interval; EFS, event-free survival; HR, hazard ratio; OS, overall survival; PB, peripheral blood; PM, peripheral blood; non-poor, intermediate and good; WBC, white blood cell.
In the GO treatment group, univariate analysis showed that high ANXA5 expression was associated with longer OS (HR = 0.583, 95% CI 0.359–0.948, p = 0.030) and EFS (HR = 0.544, 95% CI 0.334–0.884, p = 0.014), as well as CBFβ/MYH11-positive for OS (HR = 0.364, 95% CI 0.146–0.906, p = 0.030) and EFS (HR = 0.356, 95% CI 0.143–0.885, p = 0.026). In addition, multivariate analysis showed that high ANXA5 expression was an independent favorable factor for OS (HR = 0.629, 95% CI 0.372–1.064, p = 0.084) and EFS (HR = 0.544, 95% CI 0.321–0.922, p = 0.024) in combination with the curative effect of GO treatment. Results are shown in Table 3.
Table 3.
Univariate and multivariate analysis for EFS and OS in patients with gemtuzumab ozogamicin treatment.
| Variables | OS |
EFS |
||
|---|---|---|---|---|
| HR (95% CI) | p-value | HR (95% CI) | p-value | |
| Univariate analyses | ||||
| ANXA5 (high versus low) | 0.583 (0.359–0.948) | 0.03 | 0.544 (0.334–0.884) | 0.014 |
| Age (⩾10 versus <10 years) | 1.105 (0.687–1.778) | 0.68 | 0.956 (0.594–1.539) | 0.853 |
| Sex (male versus female) | 0.524 (0.325–0.845) | 0.008 | 0.538 (0.334–0.867) | 0.011 |
| Ethnicity (Hispanic or Latino versus others) | 1.19 (0.661–2.142) | 0.563 | 1.179 (0.655–2.124) | 0.582 |
| WBC (⩾50 versus <50 × 109/l) | 0.725 (0.449–1.17) | 0.188 | 0.711 (0.441–1.148) | 0.163 |
| BM blasts (⩾70 versus <70%) | 0.947 (0.584–1.536) | 0.827 | 0.906 (0.558–1.47) | 0.688 |
| PM blasts (⩾50 versus <50%) | 0.981 (0.601–1.6) | 0.938 | 0.905 (0.554–1.476) | 0.688 |
| NPM1 (mutated versus wild type) | 0.328 (0.08–1.34) | 0.121 | 0.303 (0.074–1.237) | 0.096 |
| MLL (mutated versus wild type) | 1.274 (0.632–2.57) | 0.499 | 1.291 (0.64–2.603) | 0.476 |
| FLT3-ITD (positive versus negative) | 1.546 (0.81–2.954) | 0.187 | 1.483 (0.777–2.829) | 0.232 |
| CBFβ-MYH11 (positive versus negative) | 0.364 (0.146–0.906) | 0.03 | 0.356 (0.143–0.885) | 0.026 |
| RUNX1-RUNX1T1 (positive versus negative) | 0.545 (0.236–1.261) | 0.156 | 0.528 (0.228–1.222) | 0.136 |
| WT1 (mutated versus wild type) | 1.935 (0.923–4.056) | 0.08 | 2.208 (1.052–4.632) | 0.036 |
| Risk (poor versus non-poor) | 3.115 (1.815–5.346) | 0.001 | 3.451 (2.009–5.928) | 0.001 |
| Multivariate analyses | ||||
| ANXA5 (high versus low) | 0.629 (0.372–1.064) | 0.084 | 0.544 (0.321–0.922) | 0.024 |
| NPM1 (mutated versus wild type) | 0.432 (0.103–1.811) | 0.251 | 0.409 (0.098–1.701) | 0.219 |
| CBFβ-MYH11 (positive versus negative) | 0.9 (0.297–2.729) | 0.852 | 1.034 (0.34–3.143) | 0.953 |
| WT1 (mutated versus wild type) | 1.569 (0.735–3.35) | 0.244 | 1.753 (0.822–3.738) | 0.146 |
| Risk (poor versus non-poor) | 2.778 (1.476–5.228) | 0.002 | 3.323 (1.764–6.26) | 0.001 |
BM, bone marrow; CI, confidence interval; EFS, event-free survival; HR, hazard ratio; OS, overall survival; PB, peripheral blood; PM, peripheral blood; non-poor, intermediate and good; WBC, white blood cell.
Patients overexpressing ANXA5 benefited from GO treatment
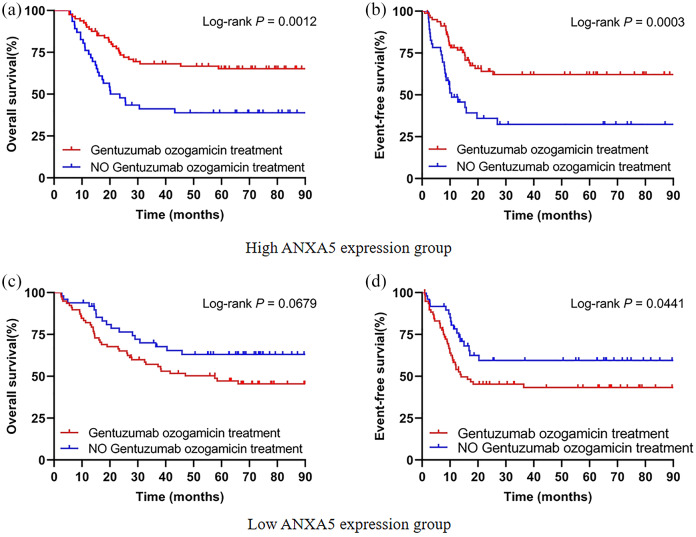
The 253 patients were divided into two groups based on median expression levels of ANXA5 in order to investigate whether GO treatment could overcome the unfavorable outcomes of high ANXA5 expression in pediatric AML. In the high ANXA5 expression group, the patients undergoing chemotherapy combined with GO had significantly better OS (p = 0.0012) and EFS (p = 0.0003) compared with patients treated with conventional chemotherapy alone [Figure 4(a and b)]. For the low ANXA5 expression group, there were no obvious differences in OS (p = 0.0679) regardless of whether or not the chemotherapy regimen was combined with GO [Figure 4(c)]. However, low expression of ANXA5 might be considered an unfavorable prognostic factor in EFS (p = 0.0441) under GO treatment [Figure 4(d)]. Pediatric AML patients with overexpression of ANXA5 may circumvent poor outcomes from chemotherapy combined with GO.
Figure 4.
GO treatment circumvents the unfavorable outcomes of high ANXA5 expression in pediatric AML patients. A total of 253 patients were divided into two groups based on the median expression levels of ANXA5. (a, b) Kaplan–Meier curves of OS and EFS in patients with GO treatment (n = 80) and without GO treatment (n = 46) in the high-ANXA5-expression group. (c, d) Kaplan–Meier curves of OS and EFS in patients with GO treatment (n = 78) and without GO treatment (n = 49) in the low-ANXA5-expression group.
AML, acute myeloid leukemia; EFS, event-free survival; GO, gemtuzumab ozogamicin; OS, overall survival.
Biological insights
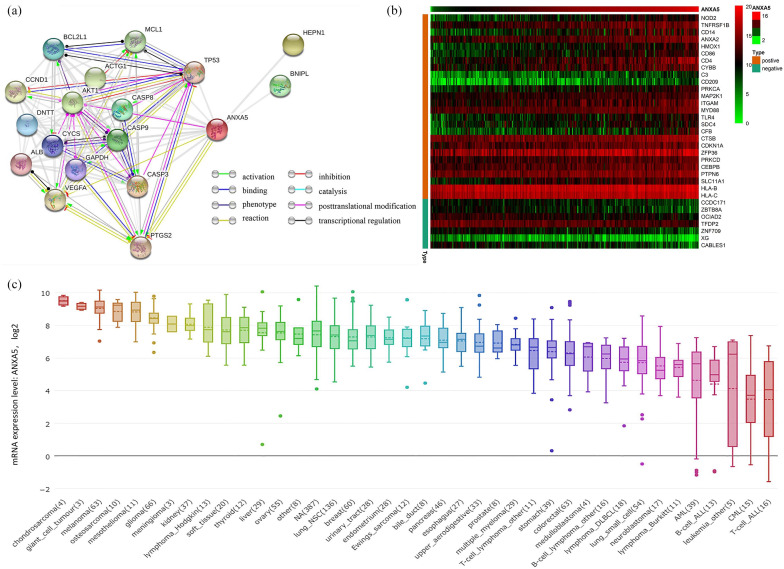
To generate insight into the biological functions of ANXA5 among pediatric patients with AML, we analyzed the features of gene expression connected with ANXA5 expression. An association between the expression of the most frequently altered neighbor genes and ANXA5 was observed; the network is shown in Figure 5(a). In addition, we extracted the gene matrix file including immune-related genes in pediatric AML, and a heat map of genes coexpressed with ANXA5 expression level was constructed. Among these genes, 7 were negatively correlated and 26 were positively correlated with the expression of ANXA5 [Figure 5(b)]. The results showed that ANXA5 expression was positively correlated with the expression of ANXA2, HMOX1, CD4, CD14, C3, and MAP2K1, as well as HLA-B and HLA-C. Notably, these molecular markers are crucial for the leukemogenesis and immune functions of regulation in AML.34,35 Figure 5(c) shows the expression of ANXA5 in different tumor cell lines according to the CCLE database.
Figure 5.
Analysis of ANXA5 gene expression. (a) The network for ANXA5 and the most frequently altered neighbor genes, analyzed by STRING. (b) Immune-related genes coexpressed of heat map with ANXA5 in pediatric AML. (c) The expression of ANXA5 in tumor cell lines, analyzed by CCLE.
AML, acute myeloid leukemia; CCLE, the Cancer Cell Line Encyclopedia; STRING, Search Tool for the Retrieval of Interacting Genes.
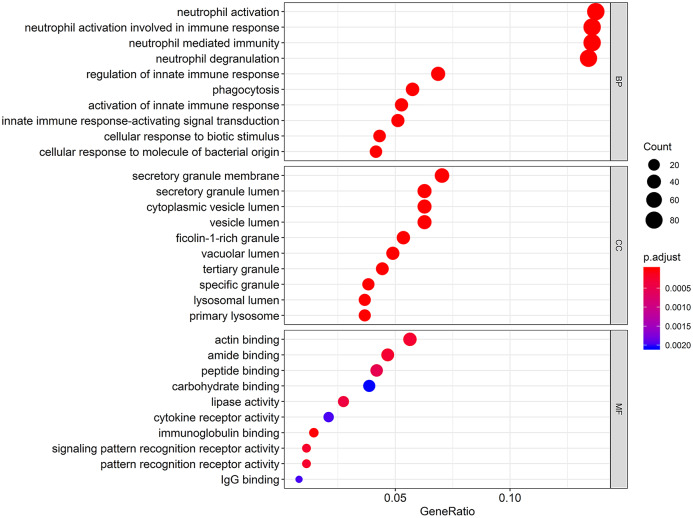
To understand the functional biological implications of aberrantly expressed ANXA5 in pediatric AML, a top 10 gene ontology enrichment analysis was conducted. Relevant biological processes included were neutrophil activation, neutrophil-mediated immunity, phagocytosis, and innate immune-response-activating signal transduction. Furthermore, the most enriched gene ontology terms in cellular component included lysosomal lumen and secretory granule membrane; in molecular function, the most enriched terms were immunoglobulin binding, cytokine receptor activity, and signaling pattern recognition receptor activity. The results of the bubble diagram are shown in Figure 6.
Figure 6.
Gene ontology terms of biological processes, cellular component, and molecular function in the ANXA5 associated expression profile with pediatric AML. The size of each dot represents the count of genes, the color represents the adjusted p-value.
AML, acute myeloid leukemia.
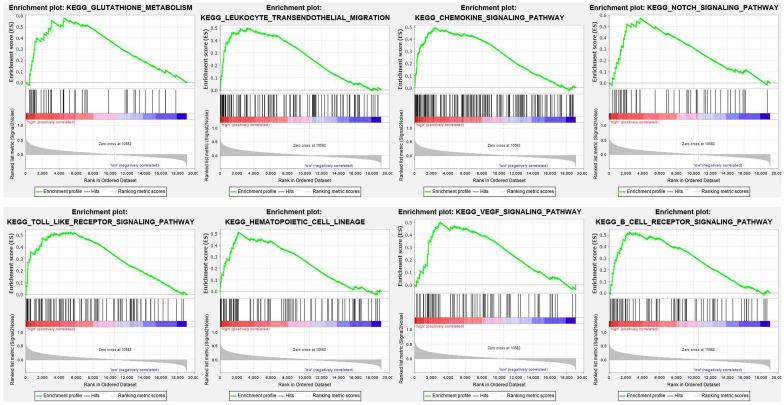
To explore signaling pathways in which ANXA5 is differentially activated in pediatric AML, we performed GSEA between high and low ANXA5 expression data sets. GSEA revealed significant differences (nominal < 0.05, false discovery rate p < 0.05) in the gene expression profile from the MSigDB collection (c2.cp.kegg.version 7.0.symbols). The significant pathways for gene sets are listed in order of significance in Table 4. Multiple pathways, including glutathione metabolism, leukocyte transendothelial migration, chemokine signaling pathway, NOTCH signaling pathway, Toll-like receptor signaling pathway, hematopoietic cell lineage, vascular endothelial growth factor (VEGF) signaling pathway, and B-cell receptor signaling pathway, were significant in ANXA5 high-expression phenotype. Enrichment plots were shown in Figure 7. Our results indicate that ANXA5 high-expression may be associated with AML progression and sensitivity to chemotherapeutic agents such as GO.
Table 4.
Gene set enrichment analysis demonstrated the correlation between ANXA5 expression phenotype.
| Name | ES | NES | NOM p-value | FDR q-value | Leading edge |
|---|---|---|---|---|---|
| KEGG_FC_GAMMA_R_MEDIATED_PHAGOCYTOSIS | 0.6027 | 2.0824 | 0 | 0.0069 | tags=39%, list=13%, signal=44% |
| KEGG_GLYCOSPHINGOLIPID_BIOSYNTHESIS_GANGLIO_SERIES | 0.706 | 2.0465 | 0.0021 | 0.0073 | tags=20%, list=2%, signal=20% |
| KEGG_GLYCOLYSIS_GLUCONEOGENESIS | 0.5859 | 1.984 | 0 | 0.0094 | tags=44%, list=19%, signal=54% |
| KEGG_LEUKOCYTE_TRANSENDOTHELIAL_MIGRATION | 0.5 | 1.9654 | 0 | 0.0096 | tags=32%, list=17%, signal=38% |
| KEGG_GLUTATHIONE_METABOLISM | 0.5781 | 1.9597 | 0.0021 | 0.0094 | tags=56%, list=24%, signal=74% |
| KEGG_NATURAL_KILLER_CELL_MEDIATED_CYTOTOXICITY | 0.5061 | 1.9264 | 0 | 0.0113 | tags=30%, list=13%, signal=34% |
| KEGG_CHEMOKINE_SIGNALING_PATHWAY | 0.4926 | 1.9236 | 0 | 0.0104 | tags=27%, list=13%, signal=31% |
| KEGG_PENTOSE_PHOSPHATE_PATHWAY | 0.6734 | 1.9087 | 0.0021 | 0.0128 | tags=65%, list=22%, signal=84% |
| KEGG_NOTCH_SIGNALING_PATHWAY | 0.5746 | 1.9063 | 0 | 0.0125 | tags=40%, list=19%, signal=50% |
| KEGG_TOLL_LIKE_RECEPTOR_SIGNALING_PATHWAY | 0.5263 | 1.8692 | 0.002 | 0.017 | tags=47%, list=28%, signal=65% |
| KEGG_HEMATOPOIETIC_CELL_LINEAGE | 0.5143 | 1.8585 | 0.004 | 0.0174 | tags=31%, list=11%, signal=34% |
| KEGG_ANTIGEN_PROCESSING_AND_PRESENTATION | 0.5053 | 1.8251 | 0.0084 | 0.0223 | tags=43%, list=27%, signal=58% |
| KEGG_VEGF_SIGNALING_PATHWAY | 0.5002 | 1.8199 | 0 | 0.0226 | tags=36%, list=16%, signal=42% |
| KEGG_ENDOCYTOSIS | 0.4643 | 1.8076 | 0 | 0.024 | tags=38%, list=20%, signal=47% |
| KEGG_CELL_ADHESION_MOLECULES_CAMS | 0.4348 | 1.7818 | 0.0021 | 0.0287 | tags=31%, list=22%, signal=40% |
| KEGG_BETA_ALANINE_METABOLISM | 0.5792 | 1.7665 | 0.0066 | 0.0304 | tags=64%, list=35%, signal=97% |
| KEGG_AMINO_SUGAR_AND_NUCLEOTIDE_SUGAR_METABOLISM | 0.5403 | 1.7504 | 0.0104 | 0.0336 | tags=26%, list=8%, signal=28% |
| KEGG_PYRUVATE_METABOLISM | 0.5485 | 1.7417 | 0.0085 | 0.0359 | tags=41%, list=20%, signal=51% |
| KEGG_TRYPTOPHAN_METABOLISM | 0.5075 | 1.7353 | 0.0086 | 0.0365 | tags=43%, list=26%, signal=57% |
| KEGG_B_CELL_RECEPTOR_SIGNALING_PATHWAY | 0.5245 | 1.7279 | 0.0085 | 0.0379 | tags=31%, list=13%, signal=35% |
ES, enrichment score; FDR, false discovery rate; NES, normalized enrichment score; NOM, nominal.
Figure 7.
GSEA results showing differential enrichment of genes related to glutathione metabolism, leukocyte transendothelial migration, chemokine signaling pathway, NOTCH signaling pathway, Toll-like receptor signaling pathway, hematopoietic cell lineage, VEGF signaling pathway, and B-cell receptor signaling pathway in pediatric AML with high ANXA5 expression.
AML, acute myeloid leukemia; GSEA, gene set enrichment analysis; VEGF, vascular endothelial growth factor.
Discussion
The prognosis of pediatric AML has improved tremendously over the last few decades, with the survival rate today reaching approximately 75%.36 Unfortunately, one third of cases eventually relapse.37 Cytarabine- and anthracycline-based induction chemotherapy for AML has been used for 30–40 years and needs to be optimized to prolong survival and increase CR rate without adopting dose escalation.38 Recent progress in the genetic and molecular etiology of leukemia has led to molecular-targeted therapies.39 The first targeted therapy was a combination of all-trans retinoic acid and arsenic trioxide for patients with AML with rearrangement of the retinoic acid receptor.40 Subsequently, fms-related tyrosine kinase 3 (FLT3) inhibitors targeting FLT3 mutations emerged, but the effective treatment period of any given drug as a single agent was only 2–6 months.41 Therefore, another more broad-spectrum FLT3 inhibitor, midostaurin, combined with chemotherapy, was approved by the US FDA for use in newly diagnosed patients with AML.42,43 However, patients treated with this regimen still had a high recurrence rate. More recently, inhibitors ivosidenib and enasidenib, targeting IDH1 and IDH2 mutations, as well as inhibitors targeting epigenetically related genes such as EZH2, KDM1A and DOT1L mutations were successively approved by the US FDA.44 These hypomethylated drugs showed good results in specific types of AML, such as patients with mutations in the TET2 or p53 genes.45,46 Venetoclax, an inhibitor of BCL2 gene mutation, has been used in elderly patients with AML.47 The effective rate of single-drug therapy in patients with recurrence was about 20%, and combined with hypomethylated drug therapy, 60%.48 Furthermore, GO can be used in AML induction chemotherapy to improve OS and relapse free survival (RFS) and reduce recurrence rates.26,38 Several clinical trials have evaluated the efficacy of GO in combination with conventional cytotoxic chemotherapy, but some may increase the toxic load. The cytotoxic effect of GO depends heavily on its intracellular trafficking and processing. Previous research has shown that activation of lysosomal functions in primary leukemia cells could enhance the cytotoxicity of GO.49 Therefore, it is necessary to integrate available data for evaluation of this drug.
Further research is required to guide clinical therapy for pediatric patients with AML, and to identify molecular predictive markers of efficacy. The present study analyzed the RNA-sequencing data from TARGET databases with a focus on pediatric AML. We evaluated the correlation among ANXA5 expression with clinical characteristics between GO and no-GO groups, and further described the differences in survival to estimate the clinical outcome of ANXA5 in patients with different chemotherapy regimens. Furthermore, the results suggest that high ANXA5 expression correlates with an adverse outcome in patients with pediatric AML treated with conventional chemotherapy. However, patients undergoing conventional chemotherapy combined with GO are able to overcome the adverse effect of high ANXA5 expression. Our findings suggest that ANXA5 can be considered a predictive molecule to design personalized therapy for individual patients. Patients with high expression of ANXA5 will benefit more from GO treatment in pediatric AML. Of note, we made an effort to study the effect of knockdown ANXA5 on AML cells lines using specific siRNA. ANXA5 was decreased upon siRNA knockdown; however, it did not significantly alter the expression of CD33. Therefore, ANXA5 may affect the efficacy of GO via an indirect mechanism, and high-throughput sequencing bioinformatics analysis helps to clarify the potential biological implications of this work. We noticed that the expression of ANXA5 was positively correlated with certain immune-related genes that were crucial for leukemogenesis and immune regulatory functions in AML. Functional enrichment analyses indicated that ANXA5 was enriched mainly in neutrophil activation, neutrophil-mediated immunity, phagocytosis, lysosomal lumen, immunoglobulin binding, cytokine receptor activity, and signaling pattern recognition receptor activity. GSEA further revealed that 20 pathways were enriched, and primarily involved the following: glutathione metabolism, leukocyte transendothelial migration, chemokine signaling pathway, and hematopoietic cell lineage. In addition, other pathways such as the NOTCH signaling pathway, Toll-like receptor signaling pathway, and VEGF signaling pathway were also related to leukemia.
Pediatric AML lacks an effective prognostic marker to guide the selection of appropriate treatment. In previous research, the expression level of annexin family members predicted clinical outcomes with regard to different variables in combination with clinical characteristics, many of which are frequently dysregulated in human cancers. Tyagi et al. demonstrated that patients with AML with high levels of annexin-V expression had significantly inferior OS in univariate analysis. It was also revealed that high apoptosis may be associated with a high-risk phenotype of disease.50 Several studies have reported that ANXA5 stimulates reduction of interleukin-6 production in bone-marrow-derived macrophages in vitro, reduces the infarcted area after ischemia-reperfusion injury, and improves cardiac function by inhibiting cardiac inflammatory response.51 ANXA5 improves diagnostic efficiency of conventional biomarkers in predicting mortality in patients with heart failure.52 ANXA5 increases the phosphorylation level of ERK, and ERK inhibitors reverse the activation of ANXA5 by the ERK/Nrf2 pathway.53 Linke B et al. suggested that manipulating annexin family members mediated immunosuppression may benefit patients with cancer or autoimmune diseases and chronic inflammation.54 ANXA5 was associated with monocyte differentiation, which correlates with favorable clinical outcome in AML.25,55 In addition, ANXA5 is highly expressed in cells with barrier functions, including vascular endothelium and placental trophoblast cells, and binds to lipopolysaccharide, reducing its endotoxin activity.56 Since ANXA5 has an influential biological role based on these findings, we propose ANXA5 as a predictive molecular marker to guide treatment choice in pediatric AML.
Taken together, the results of this study indicate that ANXA5 produces different prognostic outcomes in different treatment groups of pediatric AML, and patients with high ANXA5 expression benefit from GO therapy. Our analysis was based on information obtained from the TARGET database, and strengths of the trial included its strong eligibility criteria and a uniform treatment regimen according to standard guidelines. Despite the fact that our results provide a novel therapeutic option for pediatric AML, there are still certain limitations. Our analysis is a retrospective study design, and so the accuracy rate may drop in small-sample cases. This work is based on the results of an RNA-sequencing dataset, and more biological insights remain to be explored. Furthermore, whether this scheme is applicable to adult AML needs to be confirmed by research and practice.
In summary, our research results indicate that the curative effect of GO treatment may be closely related to the level of ANXA5 expression regulation in pediatric AML. This will improve risk stratification and decision-making regarding treatment options. Furthermore, overexpression of ANXA5 may circumvent poor outcomes from chemotherapy combined with GO. This discovery may further benefit investigations of the therapeutic direction to guide optimal treatment regimens for individual patients.
Supplemental Material
Supplemental material, Supplementary_Table_S1 for Overexpression of annexin A5 might guide the gemtuzumab ozogamicin treatment choice in patients with pediatric acute myeloid leukemia by Nan Zhang, Ying Zhang, Ping Zhang, Shifeng Lou, Ying Chen, Huan Li, Hanqing Zeng, Yan Shen and Jianchuan Deng in Therapeutic Advances in Medical Oncology
Footnotes
Conflict of interest: The authors declare that there is no conflict of interest.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: this study was supported by the Science and Technology Research Program of Chongqing Municipal Education Commission (grant no. KJQN201900412) and the Natural Science Foundation Project of CQ CSTC (grant no. cstc2017jcyjAX0239). In addition, we thank The National Cancer Institute Office of Cancer Genomics.
ORCID iDs: Nan Zhang  https://orcid.org/0000-0002-5877-1786
https://orcid.org/0000-0002-5877-1786
Jianchuan Deng  https://orcid.org/0000-0001-9927-579X
https://orcid.org/0000-0001-9927-579X
Supplemental material: Supplemental material for this article is available online.
Contributor Information
Nan Zhang, Department of Hematology, The Second Affiliated Hospital, Chongqing Medical University, Jiangnan, Chongqing, P.R. China.
Ying Zhang, Department of Hematology, The Second Affiliated Hospital, Chongqing Medical University, Jiangnan, Chongqing, P.R. China.
Ping Zhang, Hematology Laboratory, The Second Affiliated Hospital, Chongqing Medical University, Yuzhong, Chongqing, P.R. China.
Shifeng Lou, Department of Hematology, The Second Affiliated Hospital, Chongqing Medical University, Jiangnan, Chongqing, P.R. China.
Ying Chen, Department of Hematology, The Second Affiliated Hospital, Chongqing Medical University, Jiangnan, Chongqing, P.R. China.
Huan Li, Department of Hematology, The Second Affiliated Hospital, Chongqing Medical University, Jiangnan, Chongqing, P.R. China.
Hanqing Zeng, Department of Hematology, The Second Affiliated Hospital, Chongqing Medical University, Jiangnan, Chongqing, P.R. China.
Yan Shen, Department of Hematology, The Second Affiliated Hospital, Chongqing Medical University, Jiangnan, Chongqing, P.R. China.
Jianchuan Deng, Department of Hematology, The Second Affiliated Hospital, Chongqing Medical University, 76 Linjiang Road, Chongqing, 400010, P.R. China.
References
- 1. Miller KD, Nogueira L, Mariotto AB, et al. Cancer treatment and survivorship statistics, 2019. CA Cancer J Clin 2019; 69: 363–385. [DOI] [PubMed] [Google Scholar]
- 2. Weinberg OK, Sohani AR, Bhargava P, et al. Diagnostic work-up of acute myeloid leukemia. Am J Hematol 2017; 92: 317–321. [DOI] [PubMed] [Google Scholar]
- 3. de Rooij JD, Zwaan CM, van den Heuvel-Eibrink M. Pediatric AML: from biology to clinical management. J Clin Med 2015; 4: 127–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cornelissen JJ, Blaise D. Hematopoietic stem cell transplantation for patients with AML in first complete remission. Blood 2016; 127: 62–70. [DOI] [PubMed] [Google Scholar]
- 5. Wiernik PH. Optimal therapy for adult patients with acute myeloid leukemia in first complete remission. Curr Treat Options Oncol 2014; 15: 171–186. [DOI] [PubMed] [Google Scholar]
- 6. Laubach J, Rao AV. Current and emerging strategies for the management of acute myeloid leukemia in the elderly. Oncologist 2008; 13: 1097–1108. [DOI] [PubMed] [Google Scholar]
- 7. O’Dwyer K, Freyer DR, Horan JT. Treatment strategies for adolescent and young adult patients with acute myeloid leukemia. Blood 2018; 132: 362–368. [DOI] [PubMed] [Google Scholar]
- 8. Hofmann S, Schubert ML, Wang L, et al. Chimeric Antigen receptor (CAR) T cell therapy in acute myeloid leukemia (AML). J Clin Med 2019; 8: E200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang N, Chen Y, Lou S, et al. A six-gene-based prognostic model predicts complete remission and overall survival in childhood acute myeloid leukemia. Onco Targets Ther 2019; 12: 6591–6604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Godwin CD, Gale RP, Walter RB. Gemtuzumab ozogamicin in acute myeloid leukemia. Leukemia 2017; 31: 1855–1868. [DOI] [PubMed] [Google Scholar]
- 11. Bross PF, Beitz J, Chen G, et al. Approval summary: gemtuzumab ozogamicin in relapsed acute myeloid leukemia. Clin Cancer Res 2001; 7: 1490–1496. [PubMed] [Google Scholar]
- 12. Larson RA, Sievers EL, Stadtmauer EA, et al. Final report of the efficacy and safety of gemtuzumab ozogamicin (Mylotarg) in patients with CD33-positive acute myeloid leukemia in first recurrence. Cancer 2005; 104: 1442–1452. [DOI] [PubMed] [Google Scholar]
- 13. Jen EY, Ko CW, Lee JE, et al. FDA approval: gemtuzumab ozogamicin for the treatment of adults with newly diagnosed CD33-positive acute myeloid leukemia. Clin Cancer Res 2018; 24: 3242–3246. [DOI] [PubMed] [Google Scholar]
- 14. Balaian L, Ball ED. Cytotoxic activity of gemtuzumab ozogamicin (Mylotarg) in acute myeloid leukemia correlates with the expression of protein kinase syk. Leukemia 2006; 20: 2093–2101. [DOI] [PubMed] [Google Scholar]
- 15. Fostvedt LK, Hibma JE, Masters JC, et al. Pharmacokinetic/pharmacodynamic modeling to support the re-approval of gemtuzumab ozogamicin. Clin Pharmacol Ther 2019; 106: 1006–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Norsworthy KJ, Ko CW, Lee JE, et al. FDA approval summary: mylotarg for treatment of patients with relapsed or refractory CD33-positive acute myeloid leukemia. Oncologist 2018; 23: 1103–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bouter A, Carmeille R, Gounou C, et al. Review: annexin-A5 and cell membrane repair. Placenta 2015; 36(Suppl. 1): S43–S49. [DOI] [PubMed] [Google Scholar]
- 18. Boersma HH, Kietselaer BL, Stolk LM, et al. Past, present, and future of annexin A5: from protein discovery to clinical applications. J Nucl Med 2005; 46: 2035–2050. [PubMed] [Google Scholar]
- 19. van Genderen HO, Kenis H, Hofstra L, et al. Extracellular annexin A5: functions of phosphatidylserine-binding and two-dimensional crystallization. Biochim Biophys Acta 2008; 1783: 953–963. [DOI] [PubMed] [Google Scholar]
- 20. Xue G, Hao LQ, Ding FX, et al. Expression of annexin A5 is associated with higher tumor stage and poor prognosis in colorectal adenocarcinomas. J Clin Gastroenterol 2009; 43: 831–837. [DOI] [PubMed] [Google Scholar]
- 21. Tang S, Huang W, Zhong M, et al. Identification Keratin 1 as a cDDP-resistant protein in nasopharyngeal carcinoma cell lines. J Proteomics 2012; 75: 2352–2360. [DOI] [PubMed] [Google Scholar]
- 22. Lee EK, Cho H, Kim CW. Proteomic analysis of cancer stem cells in human prostate cancer cells. Biochem Biophys Res Commun 2011; 412: 279–285. [DOI] [PubMed] [Google Scholar]
- 23. Sofiadis A, Becker S, Hellman U, et al. Proteomic profiling of follicular and papillary thyroid tumors. Eur J Endocrinol 2012; 166: 657–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Han SH, Korm S, Han YG, et al. GCA links TRAF6-ULK1-dependent autophagy activation in resistant chronic myeloid leukemia. Autophagy 2019; 15: 2076–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Niu Y, Yang X, Chen Y, et al. Distinct prognostic values of Annexin family members expression in acute myeloid leukemia. Clin Transl Oncol 2019; 21: 1186–1196. [DOI] [PubMed] [Google Scholar]
- 26. Gamis AS, Alonzo TA, Meshinchi S, et al. Gemtuzumab ozogamicin in children and adolescents with de novo acute myeloid leukemia improves event-free survival by reducing relapse risk: results from the randomized phase III children’s oncology group trial AAML0531. J Clin Oncol 2014; 32: 3021–3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cooper TM, Franklin J, Gerbing RB, et al. AAML03P1, a pilot study of the safety of gemtuzumab ozogamicin in combination with chemotherapy for newly diagnosed childhood acute myeloid leukemia: a report from the Children’s Oncology Group. Cancer 2012; 118: 761–769. [DOI] [PubMed] [Google Scholar]
- 28. Oran B, Cortes J, Beitinjaneh A, et al. Allogeneic transplantation in first remission improves outcomes irrespective of FLT3-ITD allelic ratio in FLT3-ITD-positive acute myelogenous leukemia. Biol Blood Marrow Transplant 2016; 22: 1218–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Döhner H, Estey E, Grimwade D, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 2017; 129: 424–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res 2017; 45: D362–D368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Barretina J, Caponigro G, Stransky N, et al. Addendum: the cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2019; 565: E5–E6. [DOI] [PubMed] [Google Scholar]
- 32. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005; 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Swerdlow SH, Campo E, Pileri SA, et al. The 2016 revision of the world health organization classification of lymphoid neoplasms. Blood 2016; 127: 2375–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hobo W, Hutten T, Schaap N, et al. Immune checkpoint molecules in acute myeloid leukaemia: managing the double-edged sword. Br J Haematol 2018; 181: 38–53. [DOI] [PubMed] [Google Scholar]
- 35. Holmström MO, Hasselbalch HC. Cancer immune therapy for myeloid malignancies: present and future. Semin Immunopathol 2019; 41: 97–109. [DOI] [PubMed] [Google Scholar]
- 36. Kuhlen M, Klusmann JH, Hoell JI. Molecular approaches to treating pediatric leukemias. Front Pediatr 2019; 7: 368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang N, Chen Y, Shen Y, et al. Comprehensive analysis the potential biomarkers for the high-risk of childhood acute myeloid leukemia based on a competing endogenous RNA network. Blood Cells Mol Dis 2019; 79: 102352. [DOI] [PubMed] [Google Scholar]
- 38. Li X, Xu SN, Qin DB, et al. Effect of adding gemtuzumab ozogamicin to induction chemotherapy for newly diagnosed acute myeloid leukemia: a meta-analysis of prospective randomized phase III trials. Ann Oncol 2014; 25: 455–461. [DOI] [PubMed] [Google Scholar]
- 39. Shimada A. Hematological malignancies and molecular targeting therapy. Eur J Pharmacol 2019; 862: 172641. [DOI] [PubMed] [Google Scholar]
- 40. Raffoux E, Rousselot P, Poupon J, et al. Combined treatment with arsenic trioxide and all-trans-retinoic acid in patients with relapsed acute promyelocytic leukemia. J Clin Oncol 2003; 21: 2326–2334. [DOI] [PubMed] [Google Scholar]
- 41. Smith BD, Levis M, Beran M, et al. Single-agent CEP-701, a novel FLT3 inhibitor, shows biologic and clinical activity in patients with relapsed or refractory acute myeloid leukemia. Blood 2004; 103: 3669–3676. [DOI] [PubMed] [Google Scholar]
- 42. Stone RM, Mandrekar SJ, Sanford BL, et al. Midostaurin plus chemotherapy for acute myeloid leukemia with a FLT3 mutation. N Engl J Med 2017; 377: 454–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wei AH, Tiong IS. Midostaurin, enasidenib, CPX-351, gemtuzumab ozogamicin, and venetoclax bring new hope to AML. Blood 2017; 130: 2469–2474. [DOI] [PubMed] [Google Scholar]
- 44. Bohl SR, Bullinger L, Rücker FG. New targeted agents in acute myeloid leukemia: new hope on the rise. Int J Mol Sci 2019; 20: pii: E1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Itzykson R, Kosmider O, Cluzeau T, et al. Impact of TET2 mutations on response rate to azacitidine in myelodysplastic syndromes and low blast count acute myeloid leukemias. Leukemia 2011; 25: 1147–1152. [DOI] [PubMed] [Google Scholar]
- 46. Welch JS, Petti AA, Miller CA, et al. TP53 and decitabine in acute myeloid leukemia and myelodysplastic syndromes. N Engl J Med 2016; 375: 2023–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Agarwal S, Gopalakrishnan S, Mensing S, et al. Optimizing venetoclax dose in combination with low intensive therapies in elderly patients with newly diagnosed acute myeloid leukemia: an exposure-response analysis. Hematol Oncol 2019; 37: 464–473. [DOI] [PubMed] [Google Scholar]
- 48. DiNardo CD, Pratz KW, Letai A, et al. Safety and preliminary efficacy of venetoclax with decitabine or azacitidine in elderly patients with previously untreated acute myeloid leukaemia: a non-randomised, open-label, phase 1b study. Lancet Oncol 2018; 19: 216–228. [DOI] [PubMed] [Google Scholar]
- 49. Mizutani Y, Inase A, Maimaitili Y, et al. An mTORC1/2 dual inhibitor, AZD2014, acts as a lysosomal function activator and enhances gemtuzumab ozogamicin-induced apoptosis in primary human leukemia cells. Int J Hematol 2019; 110: 490–499. [DOI] [PubMed] [Google Scholar]
- 50. Tyagi A, Pramanik R, Bakhshi R, et al. Apoptosis: a biomarker of high-risk phenotype in pediatric acute myeloid leukemia. Int J Lab Hematol 2019; 41: 141–147. [DOI] [PubMed] [Google Scholar]
- 51. de Jong R, Pluijmert NJ, de Vries MR, et al. Annexin A5 reduces infarct size and improves cardiac function after myocardial ischemia-reperfusion injury by suppression of the cardiac inflammatory response. Sci Rep 2018; 8: 6753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Schurgers LJ, Burgmaier M, Ueland T, et al. Circulating annexin A5 predicts mortality in patients with heart failure. J Intern Med 2016; 279: 89–97. [DOI] [PubMed] [Google Scholar]
- 53. Zhang L, Qin Z, Li R, et al. The role of ANXA5 in DBP-induced oxidative stress through ERK/Nrf2 pathway. Environ Toxicol Pharmacol 2019; 72: 103236. [DOI] [PubMed] [Google Scholar]
- 54. Linke B, Abeler-Dörner L, Jahndel V, et al. The tolerogenic function of annexins on apoptotic cells is mediated by the annexin core domain. J Immunol 2015; 194: 5233–5242. [DOI] [PubMed] [Google Scholar]
- 55. Forthun RB, Aasebø E, Rasinger JD, et al. Phosphoprotein DIGE profiles reflect blast differentiation, cytogenetic risk stratification, FLT3/NPM1 mutations and therapy response in acute myeloid leukaemia. J Proteomics 2018; 173: 32–41. [DOI] [PubMed] [Google Scholar]
- 56. Rand JH, Wu XX, Lin EY, et al. Annexin A5 binds to lipopolysaccharide and reduces its endotoxin activity. mBio 2012; 3: e00292-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, Supplementary_Table_S1 for Overexpression of annexin A5 might guide the gemtuzumab ozogamicin treatment choice in patients with pediatric acute myeloid leukemia by Nan Zhang, Ying Zhang, Ping Zhang, Shifeng Lou, Ying Chen, Huan Li, Hanqing Zeng, Yan Shen and Jianchuan Deng in Therapeutic Advances in Medical Oncology