Abstract
Coronavirus related infectious diseases seems to be biggest challenge of 21 century that have been constantly emerging and threating public health around the globe. Coronavirus disease-19 (COVID-19) that was detected as cause of respiratory tract infection in China by end the December 2019 impelled World Health Organization to declare in January 2020 public health emergency of international concern and consequently pandemic in March 2020. Over a past six months COVID-19 pandemic has wrapped up all continents except Antarctica. Scientists around the globe are finding way to tackle and reduce the ultimate risk and size of pandemic with lower morbidity and mortality rates. In this context, technologies such as sequencing, Crispr and artificial intelligence are playing vital role in diagnosis and management of infectious disease in contrast to conventional methods. Despite of this, there is a need to have rapid and early diagnostic tools and systems that recognize infectious disease in asymptotic condition. Here we provide an overview on the recent CoV outbreak and contribution of technologies with the emphasis on the future management for detection of such infectious diseases.
Keywords: Infectious diseases, Coronavirus, COVID-19, Sequencing, CRISPR, Artificial intelligence
Introduction
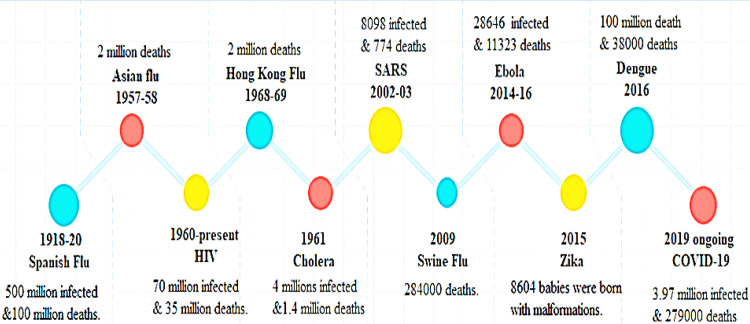
Infectious diseases, from Spanish flu to COVID-19 are frequently emerging and posing greater global health threats than terrorism resulting enormous losses to individual health, economies and social wellbeing [1]. Several pathogens comprising of bacteria, viruses, fungi, and parasites are principal source of infectious diseases [2]. Due to their miniscule architecture and hasty mode of circulation, there is always possibility that an identified or unidentified pathogen could develop and spread speedily to damage any population at any time [3]. Despite of safety measures and surveillance system infectious diseases are arising with more lethal effects globally (Fig. 1). Evidence shown that such infectious diseases threating both physical and mental health of suspected people, their family and healthcare staff [4]. Over past 20 years, infectious diseases related to coronavirus (CoV) including severe acute respiratory syndrome (SARS- 2002; 8000 infected cases and 774 deaths), Middle East respiratory syndrome (MERS-2012; 858 casualties and 2494 infected cases) and ongoing COVID-19 pandemic are repeatedly evolving from zoonotic reservoirs to brandishes severe health calamity on global inhabitants [5]. Few months back in late December 2019, pneumonia of unknown etiology was reported in Wuhan China which later named as COVID-19 due to immersion of new strain of CoV namely SARS CoV-2 as causative agent of respiratory tract infection. Due to its vibrant outreach around the globe, World Health Organization (WHO) declared in January 2020 public health emergency of international concern and consequently pandemic in March 2020 [6]. As of 9th May 2020, more than 3.97 million infected cases and more than 276,000 death have been reported in over 187 countries and territories. Worldwide spreading of COVID-19 highlighted masses to control this pandemic at earliest possible otherwise it could cross figures of Spanish flu, one of the deadliest infectious disease of 20th century that killed million people than world war [3]. Though technologies are available to detect causing agent rapidly with targeting therapeutic agents but still there is need to have diagnostic tools that provide aid in early detection. Here we provide an overview on the CoV outbreaks and contribution of technologies with the emphasis on the future management for detection of such infectious diseases.
Fig. 1.
History of infectious diseases occurred around the Globe between 1918 and 2019
Coronavirus classification, origin, epidemiology and genetics
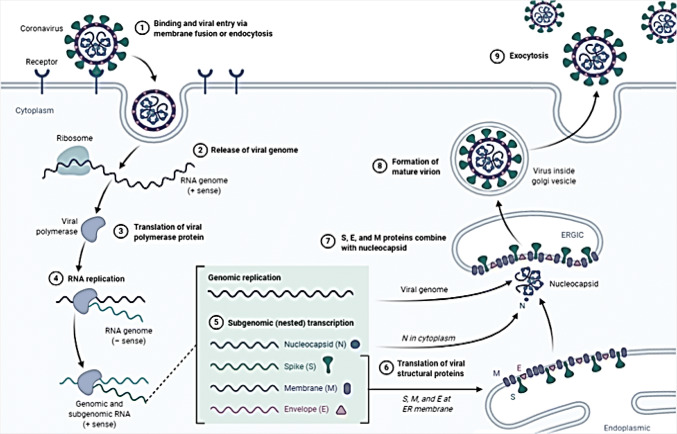
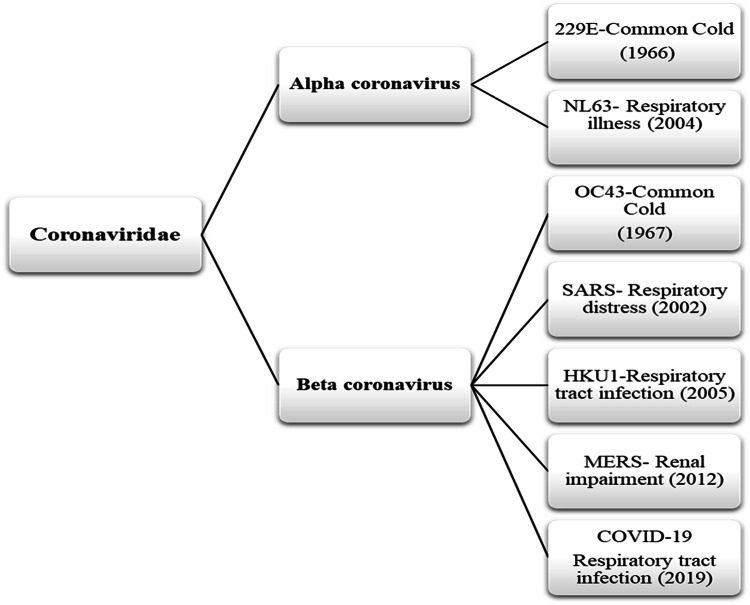
Coronavirus (CoV) are circulating in nature since creation of world but their origin is unclear although they exists in animals and human as well. Scientific efforts disclosed their hierarchy and include them in order Nidovirales, family Coronaviridae, subfamily Orthocoronavirinae, subgenus Sarbecovirus and genus and genus beta Coronavirus [7]. Under microscopic observation, these viruses look like tennis ball with prickle like projection on their outer covering or membrane which give them crown like shape as well as helps them to attach and enter in the host (animals or human) system [4]. After entering into the host cell, they releases their RNA genome and using host cell machinery produce their progeny in massive quantity which not only infect host organ/cells to make them ill but on expulsion transmits to another host to cause infections thus known to be infection causing agents [8] (Fig. 2). CoV infections including respiratory, bronchitis, pneumonia, renal impairment, gastrointestinal, and neurological diseases are prevalent in both animals and human. Prevalence of CoV infection in human was marked with mild common cold (B814) in 1965. Since then to 2019, seven new strains of CoV belong to genus alpha and beta coronaviruses are being identified to cause human infectious diseases globally (Fig. 3). Out of seven, four strains including 229E, OC43, NL63, and HKU1 are maintained and circulating within human population with mild cold/infection. However, three strains containing SARS, MERS and COVID-19 are circulating in susceptible human population through intermediate carrier/host infected from primary zoonotic reservoirs [4, 9].
Fig. 2.
Life cycle of CoV in host cells. Infection with CoV starts upon entry into the host cell via spike protein which attaches to cell membrane due to cell specific enzyme activation (1). After attachment, CoV via spike protein enters in host cell cytoplasm and releases their genome RNA (positive sense RNA) for the replication and transcription of mRNA (messenger RNA) (2). Viral genome (replicase genes) then translated into poly proteins pp1a and 1ab which later cleave and convert into small products for replication (3). In the next stage, viral RNA replicates to produce negative stranded RNA (sub genomic RNA) for the synthesis of genomic positive sense RNA through discontinuous transcription (4–5). Genomic RNA and N protein assembled into nucleocapsids followed by budding in the intercellular membrane of the endoplasmic reticulum (ER), Golgi intermediate compartment (6–7–8). Once they complete their life cycle, viruses are then ready to exit from the cell through exocytosis for transmission (9).
Fig. 3.
Time line of human corona virus diseases that traced back from 1960 to 2019.
On the basis of host vulnerability epidemiological investigation of CoV has always been doubtful. For instance, outbreak of SARS-CoV epidemic was connected with palm civet from animal market of Guangdong, China. However, follow-up studies exposed wild horseshoe bats (sold and served in Guangdong market and restaurants) as cradle of SARS-CoV outbreak [10]. Similarly, dromedary camels were thought to be potential source of MERS-CoV outbreak but studies on bat originated HKU4-CoV assumed that MERS-CoV might be circulated from bats to camels on the assumption of CD26 receptor which was common point in both CoV strains for host entry [11]. Likewise, recent COVID-2019 outbreak reported to be traced from seafood market located in Wuhan with assumption of wildlife animal involvement in disease prognosis. It has been figured out that SARS-CoV-2 cause of COVID-19 belong to bat strains that was involved in SARS-CoV epidemic thus pointing towards an unknown animal being a facilitator of crossing over between bats and human [6]. In addition, minks and pangolins are also connected with outbreak as intermediate hosts. Such evidence on host variability and susceptibility confirms that CoV has high mutation and recombination capacity to modify genes with high frequency nearly 25% of the entire genome thus stimulating to cross species transmission (new host), pathogenesis and genetic diversity [12].
Genetic diversity or recombination ability in CoV is frequent due to large genome. As we know 26–32 kilobases sized genome of all CoV is centered on single stranded, non-segmented, positive sense RNA that having 5′–3′ end with 7–9 genes containing nonstructural (replicase gene) and structural (Spike, Membrane, Envelope and Nucleocapsid) genes. Among nonstructural genes/protein, four protein including chymotrypsin like protease, papain like protease, helicase and RNA-dependent RNA polymerase are indispensable for the replication and transcription of viral genome. However, structural protein such as Spike protein with 2 sub units S1 and S2 assists in cell binding and fusion between the viral and host cell membranes resulting CoV genome release into the host cell for infection [5, 9].
Studies have figured out mutation in SARS and COVID-19 spike protein residue have likely to triggered process of pathogenesis via angiotensin converting enzyme 2 (ACE2) which is mainly found in human lungs (alveolar epithelial cells) and enterocytes of small intestine. It is evident that SARS-CoV-2 uses receptor binding domain ACE2 for transmission and host tropism that also have identified in SARS pathogenesis [6, 13]. In addition to this, samples of COVID patients from different parts of china have shown difference in mutated SARS-CoV-2 sequence which indicate that there is need to closely monitor the SARS-CoV-2 in order to scrutinize the virulence and pandemic.
CoV transmission, clinical observation and treatment
CoV infection often goes asymptomatic and most of patients present with mild symptoms of flu with cough fatigue, and headache. An asymptomatic incubation period varying from is 0–14 days, occasionally be 21 days long cause of fueling up CoVID-19 infection across the borders and wrapping up almost people of all ages including new born to elderly via direct or indirect contacts [14]. Direct contact (blood, slaiva, semens and nasal/body secretions, animal meat or products) or in direct contact (with bite of infected organism, contaminated food, water, bedding or cloths) are equally detrimental in CoV outbreaks. In comparison to SARS and MERS, COVID-19 is spreading much faster because SARS-CoV-2 replicates in upper respiratory tract producing immense number of pathogenic progeny that release and transport from one person to another on sneezing and coughing via respiratory dissemination. Including this, during coughing and sneezing mucus droplets suspend in the air for hours and can travel with air flows to infect passersby via airborne dissemination or infected droplets when landed on any object could allow SARS-CoV-2 to transmit via contact with contaminated surfaces thus providing contact, aerosols and fecal oral transmission routes [15, 16]. Earlier studies shown COVID-19 transmitted and infected more likely to male patients rather female but recent studies on ICU and non ICU patients declined this with justification that male could be victim more than females due to their work at Hunan wet animal market [17, 18]. Likewise, Chen and colleagues in clinical data finding from COVID-19 infected nine pregnant showed that none of pregnant women developed severe pneumonia in neonates and fetal infection that might be caused by intrauterine vertical transmission [19]. However another study on 10 neonates born to mothers showed neonatal infection through mother to child transmission, suggesting that COVID-19 have no age, gender and health status discrimination for transmission [20].
According to clinical observation of COVID-19 patients, asymptomatic condition includes fever, fatigue, dry cough, nasal obstruction, runny nose, sore throat, abdominal distress, nausea, vomiting, stomachache and diarrhea. In severe cases these symptoms rapidly evolve into a severe condition leading to hypotension, multi organ failure, shock and death [19, 21]. In addition to this, cellular immune deficit, coagulation instigation and myocardia hepatic kidney injury were also observed in COVID-19 patients as previously diagnosed in SARS and MERS [20]. Although several studies on clinical observation are contributing but due to certain limitation it is difficult to assess ultimate risk factors regarding gender, age or health status so continuous observations of the natural history of the disease is needed for pathogenesis and treatment.
Treatment of COVID-19 is a major challenge for health care settings because no antiviral therapies of proven efficacy are available yet due to insufficient data on disease pathogenesis. Due to lack of proper medication, several options including vaccines, monoclonal antibodies, peptides, interferon and oligonucleotide based therapies have been used to cure patients [18]. In the beginning of epidemics, antiviral treatments such as lopinavir and ritonavir oseltamivir and ganciclovir along with corticosteroids were used that already were practiced in SARS and MERS cases but as per WHO interim guideline corticosteroid treatment was discontinue due to prompted lung injury or shock [22]. In addition, antibacterial including moxifloxacin, ceftriaxone, azithromycin and glucocorticoid therapy have been used because vaccine preparation will take more than year as per WHO thus supportive and personal care options are in practice to minimize effects of deadly COVID-19.
CoV detection through technologies
Exponential growth and advancement in technologies have provided rapid methods for the identification of infectious agents along with their genomes, microbial traits similarity and influence of host and viral factor on diseases chronicity [23]. Globally, RT-PCR (real time polymerase chain reaction) technology have been used as standard diagnostic tool to detect nucleic acid (RNA) of SARS-CoV-2 from the nasal, throat swab or other samplings of COVID-19 patients [24]. Furthermore, COVID-19 diagnosis through computed tomography (CT) scans using artificial intelligence tools help in differentiating community acquired pneumonia, other lung diseases and COVID-19 related lungs complications in rapid and accurate manner [25]. Further verification could be done through sequencing technology.
Sequencing technology
In recent COVID-19 outbreak, DNA sequencing technology as molecular diagnostic tool has shown a remarkable paradigm in the early disease diagnosis, medication and management of infections via identifying mutated or drug resistance genes. MinION, (nanopore sequencing) is one of portable DNA sequencing gadget which detect causative SARS-CoV-2 sequence within one hour through extraction of nucleic acid from samples followed by amplification and bioinformatics analysis [15]. Such molecular blueprints details via sequencing provided foundation on which other technologies built their strategies for the diagnosis, prevention and cure of such infectious diseases.
CRISPR technology
Clustered regularly interspaced short palindromic repeats (Crispr) associated protein (Cas) system adopted from natural immune system of prokaryotes is powerful tool against communicable and non-communicable infections. In order to work against infections host immune system operates through three stages namely, an adaptation, an expression and an interference [26]. At first stage, Cas protein cut genetic material of invader phage or virus into short pieces which then deposited in bacterial genome or Crispr array as spacers so that bacteria can reminisce the viruses when they attack them again. In expression stage, spacers transcribed into small crRNA containing an invader spacer sequence and complementary tracrRNA, hybridize together to form single guide RNA (sgRNA) for the recruitment of Cas protein. In last step, sgRNA and Cas protein fuse to form complex whereby sgRNA identify target through sequence similarity and let Cas protein to scan and cut target sequence at particular point [27]. Using customizable RNA and various Cas protein Crispr technology can edit, alter and diagnose gene of interest involved in any infections [28]. Crispr as a gene editing technique has facilitated gene modifications in both pathogen and host cells for the molecular analysis of infection. On the other side, screening of protein in host and pathogen that activate expression of infection and the development of vaccine and antimicrobial drugs are also possible through crispr technology [29].
Since, its discovery Crispr technology provide cutting edge routes for the disease diagnosis, prevention and cure using indispensable tools including sgRNA and Cas enzyme. Currently specific high sensitivity enzymatic reporter unlocking (SHERLOCK) relied on Cas13 enzyme has been used for detection, monitoring and genotyping of COVID-19 viral signatures from samples [30]. Conversely, DNA endonuclease targeted CRISPR transreporter (DETECTR) also used to sense target RNA sequence from nasopharyngeal or oropharyngeal swabs from COVID-19 patients with help of sgRNA and Cas12. Notably, the future development of portable microfluidic based cartridges and reagents to run the assay could enable point-of-care testing outside of the clinical diagnostic laboratory [31].
CoV outbreaks and future perspectives
After CoVID-19 outbreak, several sources are constantly sharing and recording epidemiological, clinical evidence with public in order to understand risk factor, routes of transmission and geographic spread of infections. In addition to this, such data could be vital for policy makers and international health organizations to improve surveillance system for ongoing and emerging epidemics. Around the globe researcher are finding way to tackle and reduce the ultimate size of such outbreaks with lower morbidity and mortality rates at local and international level [32]. Alleviating global infectious disease demands for rapid and early diagnostic tools and systems to control outbreak. Notable improved diagnostic approaches and monitoring technologies usually relied on blood, swab or other body fluids that could increase risk of infectious agents in other and health care staff as reported in COVID-19 pandemic [24]. Therefore, there is need of noninvasive diagnostics tools that help in early diagnosis particularly in asymptomatic condition, as majority of viral infections used to appear after incubation time ranging from 1 to 20 days and with more or less communal clinical observation/symptoms. Incubation period with no sign or symptoms is proved to be lethal in terms of disease spreading that pointing towards the dearth of diagnostics tools which work promptly in obscure state. No doubt, several breakthrough techniques have been launched for disease diagnosis but still there is a lot to do.
One of the technique that supposed to be helpful in disease diagnosis, particularly in asymptomatic condition is breath analysis. In past two decades, researchers are working on the breath analysis techniques using artificial intelligence (AI) techniques for the diagnosis of cancer, respiratory, gastrointestinal diseases and infectious diseases [33, 34]. Exhaled breath which contain distinctive breath print or volatile organic compound (VOC) is crucial in differentiating healthy and unhealthy individuals which produced in the body during metabolic reactions. Sometime oxidative stress, inflammation, smoking, pollution or microbe’s intrusion triggers alteration in biochemical pathways resulting production of voc with variable composition and concentration. Schnabel et al. and Nakhleh et al. figured out exceptional voc signatures in patients suffering from different diseases including cancer, pulmonary, Parkinson, ventilator associated pneumonia and kidney diseases [35, 36]. These data suggested that voc contain disease specific unique pattern or biomarkers (produce due to stress or microbes and also representing pathogen signatures) that could be reconnoitered for diagnosis of infectious diseases [37]. Therefore on the basis of voc disease specific pattern it could be hypothesized that patients with viral infection might possess shifts in voc profile and could be detectable upon the establishment of infections in the body. Upon recognition of such voc pattern using algorithms could benefit in early diagnosis and control of infectious diseases.
In addition to breath analysis technique, disease detection through speech or voice using artificial intelligence technologies is attracting increasing attention. Study with enough evidence suggested that speech signals (such as certain vowel, words and numbers) contain vocal biomarkers that could be great aid to pathologist for the disease diagnosis. Several machine learning algorithms showed superior accuracy in disease prediction including Parkinson, depression, cardio and other diseases [38–40]. In the light of finding, it can be hypothesized that whether viral infections contain disease specific vocal biomarkers for disease analysis. Both breath and voice analysis techniques could thought to be beneficial in early diagnosis of infectious disease through noninvasive device. Non-invasive diagnostic tools based on voice or voc could hold great promise in the control of infectious diseases. Nevertheless, the challenges to identify microbial voc discerning between human and pathogens are still missing [37].
On the other side, 70% of emerging infectious diseases of viral origin are derive from livestock and poultry which carry global public health risks of infrequent human zoonotic infections [41]. Due to recent CoVID-19 pandemic, china has band transaction of wild animal in accordance with previous strategy that was adopted in SARS epidemics [42]. Rather imposing legislation on wild life, there should be rapid, improved and efficient early detection tools for pathogen that prevalent in animals. In era of the technology, chemical sensors, microelectronic designs, and artificial intelligence are contributing towards prevention, diagnosis and management of animals ailments. Technological advancements such as wearable technologies and biosensors make it possible to manage the health status of livestock and birds [43–45]. In addition to this precision livestock forming technology could also help to manage and monitor animal’s status health through providing a continuous picture of health in real time and enabling fast mediations that benefit the safety of both animals and consumers [46, 47].
Concluding remarks
Spread of zoonotic infectious diseases require an integrated tactics for disease surveillance. It could be assumed that in future CoV host versatility such as insects, mosquitos, bits and flies can pose serious consequences on health and economy due to their countless generation and limited source of detection and elimination, thus there is a dire need of precision diagnosis approaches for classification, identification and cure of emerging infectious diseases. Infectious disease outbreaks are always challenging in terms of diagnosis and cure so diagnostic strategy like voice or breathe analysis (particularly focusing on asymptotic period) might be helpful in exploiting disease incidence and prognosis. This could be hypothesized that either patient voice or breathe might have significant role in early viral disease detection. Success in getting biomarker could lead to an alternative, noninvasive, portable devices that could be used for personalized screening tool in a nonclinical setting reducing risk of ultimate outbreak. Breathe or vocal signals based diagnostic apps or devices could be viable solution for the timely diagnosis of infectious disease that likely to reduce disease risk intensity and spread.
Funding
No funding received/not applicable
Availability of data and materials
All the data used to support the findings of this study are available from the corresponding author upon reasonable request.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Shireen Akhter, Email: shireen.soomro@iba-suk.edu.pk.
Shahzeen Akhtar, Email: Shahzeen.akhtar@boltonft.nhs.uk.
References
- 1.Butler CD. Infectious disease emergence and global change: thinking systemically in a shrinking world. Infect Dis Poverty. 2012;1:5. doi: 10.1186/2049-9957-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Furuse Y. Analysis of research intensity on infectious disease by disease burden reveals which infectious diseases are neglected by researchers. PNAS. 2019;116(2):478–483. doi: 10.1073/pnas.1814484116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bloom DE, Cadarette D. Infectious disease threats in the twenty-first century: strengthening the global response. Front Immunol. 2019;10:549. doi: 10.3389/fimmu.2019.00549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qiu W, Chu C, Mao A, Wu J. The impacts on health, society, and economy of SARS and H7N9 outbreaks in China: a case comparison study. J Env and pub health. 2018;2018(7):1–7. doi: 10.1155/2018/2710185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Su S, Wong G, Shi W, Liu J, Lai ACK, Zhou J, et al. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbial. 2016;24(6):490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yi-Chia W, Ching-Sunga C, Yu-Jiuna C. The outbreak of COVID-19. An overview. J Chi Med Asso. 2020;83(3):217–220. doi: 10.1097/jcma.0000000000000270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lau SKP, Chan JFW. Coronaviruses: emerging and re-emerging pathogens in humans and animals. Virol J. 2015;12:209. doi: 10.1186/s12985-015-0432-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sicard A, Pirolles E, Gallet R, Vernerey MS, Yvon M, Urbino C, et al. A multicellular way of life for a multipartite virus. Elife. 2019;8:e43599. doi: 10.7554/elife.43599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fehr AR, Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kan B, Wang M, Jing H, Xu H, Jiang X, Yan M, et al. Molecular evolution analysis and geographic investigation of severe acute respiratory syndrome coronavirus-like virus in palm civets at an animal market and on farms. J Virol. 2005;79:11892–11900. doi: 10.1128/JVI.79.18.11892-11900.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haagmans BL, Al Dhahiry SH, Reusken CB, Raj VS, Galiano M, Myers R, et al. Middle east respiratory syndrome coronavirus in dromedary camels: an outbreak investigation. Lancet Infect Dis. 2014;14(2):140–145. doi: 10.1016/S1473-3099(13)70690-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liao CL, Lai MM. RNA recombination in a coronavirus: recombination between viral genomic RNA and transfected RNA fragments. J Virol. 1992;66(10):6117–6124. doi: 10.1128/JVI.66.10.6117-6124.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, et al. Genomic characterization and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cupertino MC, Resende MB, Mayer NA, Carvalho LM, Siqueira-Batista R. Emerging and re-emerging human infectious diseases: a systematic review of the role of wild animals with a focus on public health impact. Asian Pac J Trop Med. 2020;13:99–106. doi: 10.4103/1995-7645.277535. [DOI] [Google Scholar]
- 15.Chan JFW, Yuan S, Kok KH, To KKW, Chu H, Yang J, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang B, Wang X, Li Q, Bragazzi NL, Tang S, Xiao Y, et al. Estimation of the transmission risk of 2019-nCov and its implication for public health interventions. J Clin Med. 2020;9:462. doi: 10.3390/jcm9020462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus infected pneumonia in Wuhan, China. JAMA. 2020 doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen H, Guo J, Wang C, Lau F, Yu X, Zhang W, et al. Clinical characteristics and intrauterine vertical transmission potential of COVID-19 infection in nine pregnant women: a retrospective review of medical records. Lancet. 2020;395(10226):809–815. doi: 10.1016/S0140-6736(20)30360-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu H, Wang L, Fang C, Peng S, Zhang L, Chang G, et al. Clinical analysis of 10 neonates born to mothers with 2019-nCoV pneumonia. Trans Pediatr. 2020;9:51–60. doi: 10.21037/tp.2020.02.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan. China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Russell D, Millar DE, Baillie JK. Clinical evidence does not support corticosteroid treatment for 2019-nCoV lung injury. Lancet. 2020;395:473–475. doi: 10.1016/S0140-6736(20)30317-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kiselev D, Matsvay A, Abramov I, Dedkov V, Shipulin G, Khafizov K, et al. Current trends in diagnostics of viral infections of unknown etiology. Viruses. 2020;12(2):211. doi: 10.3390/v12020211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Islam A, Ahmed A, Naqvi IH, et al. Emergence of deadly severe acute respiratory syndrome coronavirus-2 during 2019–2020. VirusDis. 2020 doi: 10.1007/s13337-020-00575-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang Lu, Han Rui, Ai Tao, Pengxin Yu, Kang Han, Tao Qian, Xia Liming. Serial quantitative chest CT assessment of COVID-19: deep-learning approach. Radiology. 2020 doi: 10.1148/ryct.2020200075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hille F, Richter H, Wong SP, Bratovic M, Ressel S, Charpentier E. The biology of CRISPR-Cas: backward and forward. Cell. 2018;172:1239–1259. doi: 10.1016/j.cell.2017.11.032. [DOI] [PubMed] [Google Scholar]
- 27.Ratan ZA, Son YJ, Haidere MF, Mahtab Uddin B, Yusuf MA, Zaman SB, et al. CRISPR-Cas9: a promising genetic engineering approach in cancer research. Ther Adv Med Oncol. 2018 doi: 10.1177/1758834018755089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H, Yang Y, Hong W, Huang M, Wu M, Zhao X. Applications of genome editing technology in the targeted therapy of human diseases: mechanisms, advances and prospects. Sig Transduct Target Ther. 2020;5:1. doi: 10.1038/s41392-019-0089-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Doerflinger M, Forsyth W, Ebert G, Pellegrini M, Herold MJ. CRISPR/Cas9-The ultimate weapon to battle infectious diseases? Cell Microbio. 2016 doi: 10.1111/cmi.12693. [DOI] [PubMed] [Google Scholar]
- 30.Gootenberg JS, Abudayyeh OO, Kellner MJ, Joung J, Collins JJ, Zhang F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science. 2018;360(6387):439–444. doi: 10.1126/science.aaq0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Broughton JP, Deng X, Yu G, Fasching CL, Servellita V, Singh J, et al. CRISPR–Cas12-based detection of SARS-CoV-2. Nat Biotechnol. 2020 doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steele L, Orefuwa E, Dickmann P. Drivers of earlier infectious disease outbreak detection. Int J of infec diseas. 2016;53:15–20. doi: 10.1016/j.ijid.2016.10.005. [DOI] [PubMed] [Google Scholar]
- 33.Sethi S, Nanda R, Chakraborty T. Clinical application of volatile organic compound analysis for detecting infectious diseases. Clin Microbiol Rev. 2013;26(3):462–475. doi: 10.1128/CMR.00020-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang C, Dong R, Wang X, Lian A, Chi C, Ke C, et al. Exhaled volatile organic compounds as lung cancer biomarkers during one-lung ventilation. Sci Rep. 2014;4:7312. doi: 10.1038/srep07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schnabel R, Fijten R, Smolinska A, Dallinga J, Boumans ML, Stobberinghet E, et al. Analysis of volatile organic compounds in exhaled breath to diagnose ventilator-associated pneumonia. Sci Rep. 2015;5:17179. doi: 10.1038/srep17179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakhleh MK, Amal H, Jeries R, Yoav Y, Broza YY, Aboud M, et al. Diagnosis and classification of 17 diseases from 1404 subjects via pattern analysis of exhaled molecules. ACS Nano. 2017;11(1):112–125. doi: 10.1021/acsnano.6b04930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Palma SICJ, Traguedo AP, Porteira AR, Frias MJ, Gamboa H, Roque ACA. Machine learning for the meta-analyses of microbial pathogens’ volatile signatures. Sci Rep. 2018;8(1):3360. doi: 10.1038/s41598-018-21544-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Erdogdu Sakar B, Serbes G, Sakar CO. Analyzing the effectiveness of vocal features in early telediagnosis of Parkinson’s disease. PLoS ONE. 2017;12(8):e0182428. doi: 10.1371/journal.pone.0182428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uddin S, Khan A, Hossain M, Moni MA. Comparing different supervised machine learning algorithms for disease prediction. BMC Med Inform Decis Mak. 2019;19:281. doi: 10.1186/s12911-019-1004-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pan W, Flint J, Shenhav L, Liu T, Liu M, Hu B, et al. Re-examining the robustness of voice features in predicting depression: compared with baseline of confounders. PLoS ONE. 2019;14(6):e0218172. doi: 10.1371/journal.pone.0218172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuiken T, Leighton FA, Fouchier RA, LeDuc JW, Peiris JS, Schudel A, et al. Public health. Pathogen surveillance in animals. Science. 2005;309:1680–1681. doi: 10.1126/science.1113310. [DOI] [PubMed] [Google Scholar]
- 42.Li J, Li J, Xie X, Cai X, Huang J, Tian X, et al. Game consumption and the 2019 novel coronavirus. Lancet. 2020;20(3):275–276. doi: 10.1016/S1473-3099(20)30063-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Astill J, Dara RA, Fraser EDG, Sharif S. Detecting and predicting emerging disease in poultry with the implementation of new technologies and big data: a focus on avian influenza virus [published correction appears in Front Vet Sci 2019; 5:337] Front Vet Sci. 2018;5:263. doi: 10.3389/fvets.2018.00263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neethirajan S. Recent advances in wearable sensors for animal health management. Sens and Bio Sens Res. 2017;12:15–29. doi: 10.1016/j.sbsr.2016.11.004. [DOI] [Google Scholar]
- 45.Halachmi I, Guarino M, Bewley J, Pastell M. Smart animal agriculture: application of real-time sensors to improve animal well-being and production. Annu Rev Anim Biosci. 2019;7:403–425. doi: 10.1146/annurev-animal-020518-114851. [DOI] [PubMed] [Google Scholar]
- 46.Rowe E, Dawkins MS, Gebhardt-Henrich SG. A systematic review of precision livestock farming in the poultry sector: Is technology focussed on improving bird welfare? Animals. 2019;9(9):614. doi: 10.3390/ani9090614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Benjamin M, Yik S. Precision livestock farming in swine welfare: a review for swine practitioners. Animals. 2019;9(4):133. doi: 10.3390/ani9040133. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the data used to support the findings of this study are available from the corresponding author upon reasonable request.