Abstract
Skull-stripping is an essential pre-processing step in computational neuro-imaging directly impacting subsequent analyses. Existing skull-stripping methods have primarily targeted non-pathologicallyaffected brains. Accordingly, they may perform suboptimally when applied on brain Magnetic Resonance Imaging (MRI) scans that have clearly discernible pathologies, such as brain tumors. Furthermore, existing methods focus on using only T1-weighted MRI scans, even though multi-parametric MRI (mpMRI) scans are routinely acquired for patients with suspected brain tumors. Here we present a performance evaluation of publicly available implementations of established 3D Deep Learning architectures for semantic segmentation (namely DeepMedic, 3D U-Net, FCN), with a particular focus on identifying a skull-stripping approach that performs well on brain tumor scans, and also has a low computational footprint. We have identified a retrospective dataset of 1,796 mpMRI brain tumor scans, with corresponding manually-inspected and verified gold-standard brain tissue segmentations, acquired during standard clinical practice under varying acquisition protocols at the Hospital of the University of Pennsylvania. Our quantitative evaluation identified DeepMedic as the best performing method (Dice = 97.9, Hausdorf f95 = 2.68). We release this pre-trained model through the Cancer Imaging Phenomics Toolkit (CaPTk) platform.
Keywords: Skull-stripping, Brain extraction, Glioblastoma, GBM, Brain tumor, Deep learning, DeepMedic, U-Net, FCN, CaPTk
1. Introduction
Glioblastoma (GBM) is the most aggressive type of brain tumors, with a grim prognosis in spite of current treatment protocols [1,2]. Recent clinical advancements in the treatment of GBMs have not increased the overall survival rate of patients with this disease by any substantial amount. The recurrence of GBM is virtually guaranteed and its management is often indefinite and highly case-dependent. Any assistance that can be gleaned from the computational imaging and machine learning communities could go a long way towards making better treatment plans for patients suffering from GBMs [3–11]. One of the first steps towards the goal of a good treatment plan is to ensure that the physician is observing only the areas that are of immediate interest, i.e., the brain and the tumor tissues, which would ensure better visualization and quantitative analyses.
Skull-stripping is the process of removing the skull and non-brain tissues from brain magnetic resonance imaging (MRI) scans. It is an indispensable pre-processing operation in neuro-imaging analyses that directly affects the efficacy of subsequent analyses. The effects of skull-stripping on subsequent analyses have been reported in the literature, including studies on brain tumor segmentation [12–14] and neuro-degeneration [15]. Manual removal of the non-brain tissues is a very involved and grueling process [16], which often results in inter- and intra-rater discrepancies affecting reproducibility in large scale studies.
In recent years, with theoretical advances in the field and with the proliferation of inexpensive computing power, including consumer-grade graphical processing units [17], there has been an explosion of deep learning (DL) algorithms that use heavily parallelized learning techniques for solving major semantic segmentation problems in computer vision. These methods have the added advantage of being easy to implement by virtue of the multitude of mature tools available, most notable of these being TensorFlow [18] and PyTorch [19]. Importantly, DL based segmentation techniques, which were initially adopted from generic applications in computer vision, have promoted the development of novel methods and architectures that were specifically designed for segmenting 3-dimensional (3D) MRI images [20–23]. DL, specifically convolutional neural networks, have been applied for segmentation problems in neuroimaging (including skull-stripping), obtaining promising results [16]. Unfortunately, most of these DL algorithms either require a long time to train or have unrealistic run-time inference requirements.
In this paper, we evaluate the performance of 3 established and validated DL architectures for semantic segmentation, which have out-of-the-box publicly-available implementations. Our evaluation is focusing on skull-stripping of scans that have clearly discernible pathologies, such as scans from subjects diagnosed with GBM. We also perform extensive comparisons using models trained on various combinations of different MRI modalities, to evaluate the benefit of utilizing multi-parametric MRI (mpMRI) data that are typically acquired in routine clinical practice for patients with suspected brain tumors on the final segmentation.
2. Materials and Methods
2.1. Data
We retrospectively collected 1,796 mpMRI brain tumor scans, from 449 glioblastoma patients, acquired during standard clinical practice under varying acquisition protocols at the Hospital of the University of Pennsylvania. Corresponding brain tissue annotations were manually-approved by an expert and used as the gold-standard labels to quantitatively evaluate the performance of the algorithms considered in this study.
In this study, we have chosen to take advantage of the richness of the mpMRI protocol that is routinely acquired in the cases of subjects with suspected tumors. Specifically, four structural modalities are included at baseline pre-operative time-point: native (T1) and post-contrast T1-weighted (T1Gd), native T2-weighted (T2), and T2-weighted Fluid Attenuated Inversion Recovery (FLAIR) MRI scans (Fig. 1). To conduct our quantitative performance evaluation we split the available data, based on an 80/20 ratio, in a training and testing subset of 1,432 and 364 mpMRI brain tumor scans, from 358 and 91 patients, respectively.
Fig. 1.
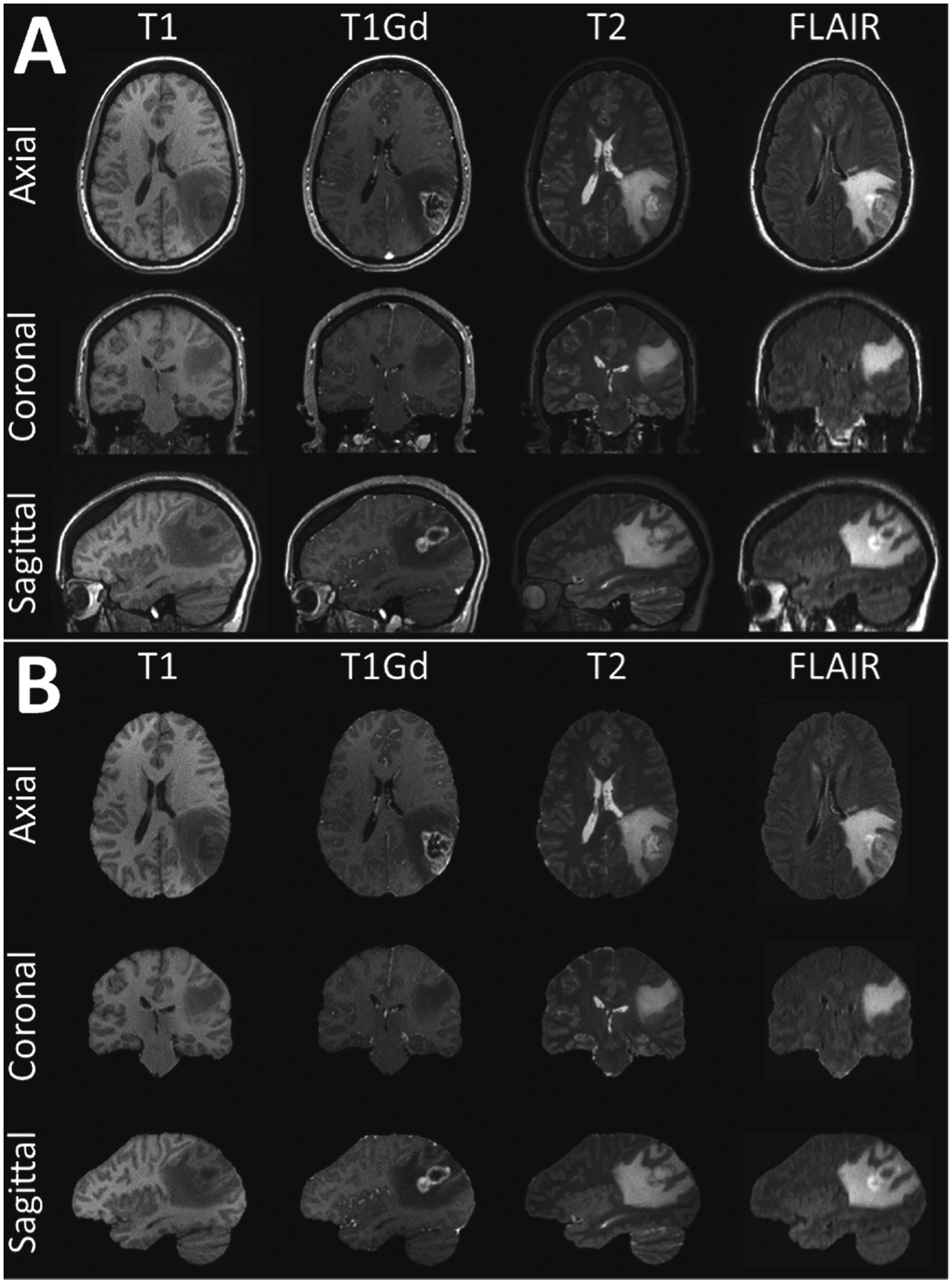
Example mpMRI brain tumor scans from a single subject. The original scans including the non-brain-tissues are illustrated in A, whereas the same scans after applying the manually-inspected and verified gold-standard brain tissue segmentations are illustrated in B.
2.2. Pre-processing
To guarantee the homogeneity of the dataset, we applied the same pre-processing pipeline across all the mpMRI scans. Specifically, all the raw DICOM scans obtained from the scanner were initially converted to the NIfTI [24] file format and then followed the protocol for pre-processing, as defined in the International Brain Tumor Segmentation (BraTS) challenge [12–14,25,26]. Specifically, each patient’s T1Gd scan was rigidly registered to a common anatomical atlas of 240 × 240 × 155 image size and resampled to its isotropic resolution of 1 mm3 [27]. The remaining scans of each patient (namely, T1, T2, FLAIR) were then rigidly co-registered to the same patient’s resampled T1Gd scan. All the registrations were done using “Greedy” (github.com/pyushkevich/greedy) [28], which is a CPU-based C++ implementation of the greedy diffeomorphic registration algorithm [29]. “Greedy” is integrated into the ITK-SNAP (itksnap.org) segmentation software [30,31], as well as the Cancer Imaging Phenomics Toolkit (CaPTk - www.cbica.upenn.edu/captk) [32,33]. After registration, all scans were down-sampled from a resolution of 240 × 240 × 155 to a resolution of 128 × 128 × 128, with anisotropic spacing of 1.875 × 1 875 × 1.25 mm3 with proper padding measures to ensure the anisotropic spacing is attained. Finally, the intensities found on each scan below the 2nd percentile and above the 95th percentile were capped, to ensure suppression of spurious intensity changes due to the scanner acquisition parameters.
2.3. Network Topologies
For our comparative performance evaluation, we focused on the most well-established DL network topologies for 3D semantic segmentation. The selection was done after taking into consideration their wide application in related literature, their state-of-the-art performance on other segmentation tasks, as established by various challenges [12,13,34,35], as well as their out-of-the-box applicability of their publicly available implementation, in low resource settings. The specific architectures included in this evaluation comprise the a) DeepMedic [20,21], b) 3D U-Net [22], and c) Fully Convolutional Neural network (FCN) [23].
DeepMedic [20,21] is a novel architecture, which came into the foreground after winning the 2015 ISchemic LEsion Segmentation (ISLES) challenge [34]. DeepMedic is essentially a 3D convolutional neural network with a depth of 11-layers, along with a double pathway to provide sufficient context and detail in resolution, simultaneously. In our study, we have applied DeepMedic using its default parameters, as provided in its GitHub repository github.com/deepmedic/deepmedic. As a post-processing step, we also include a hole filling algorithm.
As a second method, we have applied a 3D U-Net [22], an architecture that is widely used in neuroimaging. We used 3D U-Net with an input image size of 128 × 128 × 128 voxels. Taking into consideration our requirement for a low computational footprint, we reduced the initial number of “base” filters from 64 (as was originally proposed) to 16.
The third method selected for our comparisons was a 3D version of an FCN [23]. Similarly to the 3D U-Net, we used an input image size of 128 × 128 × 128 voxels. For both 3D U-Net and FCN, we used ‘Leaky ReLU’ instead of ‘ReLU’ for back-propagation with leakiness defined as α = 0.01. Furthermore, we used instance normalization instead of batch normalization due to batch size being equal to 1, due to the high memory consumption.
2.4. Experimental Design
Current state of the art methods typically use only the T1 modality for skull-stripping [36–41]. Here, we followed a different approach, by performing a set of experiments using various input image modality combinations for training DL models. Our main goal was to investigate potential contribution of different modalities, which are obtained as part of routine mpMRI acquisitions in patients with suspected brain tumors, beyond using T1 alone for skull-stripping. Accordingly, we first trained and inferred each topology on each individual modality separately to measure segmentation performance using different independent modalities, resulting in 4 models for each topology (“T1”, “T1Gd”, “T2”, “Flair”). Additionally, we trained and inferred models on a combination of modalities; namely, using a) both T1 and T2 modalities (“Multi-2”), and b) all 4 structural modalities together (“Multi-4”). The first combination was chosen as it has been shown that addition of the T2 modality improves the skull-stripping performance [14] and can also be used in cases where contrast medium is not used and hence the T1Gd modality is not available, i.e., in brain scans without any tumors. The second combination approach (i.e., “Multi-4”) was chosen to evaluate a model that uses all available scans. Finally, we utilized an ensembling approach (i.e., “Ens-4”) where the majority voting of the 4 models trained and inferred on individual modalities was used to produce the final label for skull-stripping.
We ensured that the learning parameters stayed consistent across each experiment. Each of the applied topologies needed different time for convergence, based on their individual parameters. For Deep-Medic, we trained with default parameters (as provided at the original github repository - 0.7.1 [commit dbdc1f1]) and it trained for 44 h. 3D U-Net and FCN were trained with Adam optimizer with a learning rate of 0.01 over 25 epochs. The number of epochs was determined according to the amount of improvement observed. Each of them trained for 6 h.
The average inference time for DeepMedic including the pre-processing and post-processing for a single brain tumor scan was 10.72 s, while for 3D U-Net and FCN was 1.06 s. These times were estimated based on the average time taken to infer on 300 patients. The hardware we used to train and infer were NVIDIA P100 GPUs with 12 GB VRAM utilizing only a single CPU core with 32 GB of RAM from nodes of the CBICA’s high performance computing (HPC) cluster.
2.5. Evaluation Metrics
Following the literature on semantic segmentation we use the following metrics to quantitatively evaluate the performance of the trained methods.
Dice Similarity Coefficient.
The Dice Similarity Coefficient (Dice) is typically used to evaluate and report on the performance of semantic segmentation. Dice measures the extent of spatial overlap between the predicted masks (PM) and the provided ground truth (GT ), and is mathematically defined as:
| (1) |
where it would range between 0–100, with 0 describing no overlap and 100 perfect agreement.
Hausdorf f95.
Evaluating volumetric segmentations with spatial overlap agreement metrics alone can be insensitive to differences in the slotted edges. For our stated problem of brain extraction, changes in edges might lead to minuscule differences in spatial overlap, but major differences in areas close to the brain boundaries resulting in inclusion of skull or exclusion of a tumor region. To robustly evaluate such differences, we used the 95th percentile of the Hausdorff95 distance to measure the maximal contour distance d, on a radial assessment, between the PM and GT masks.
| (2) |
3. Results
The median and inter-quartile range for Dice and Hausdorff95 scores for each of the constructed models using 3 topologies and 7 input image combinations, are shown in Tables 1 and 2 and Fig. 2 and 3. DeepMedic showed a consistent superior performance to 3D U-Net and FCN, for all input combinations.
Table 1.
Median and inter-quartile range for Dice scores of all trained models and input image combinations. DM:DeepMedic, 3dU:3D U-Net.
| T1 | T2 | T1Gd | Flair | Multi-4 | Multi-2 | Ens-4 | |
|---|---|---|---|---|---|---|---|
| DM | 98.09 ± 1.18 | 97.88 ± 1.07 | 97.86 ± 1.08 | 97.88 ± 1.03 | 98.19 ± 1.08 | 98.13 ± 1.08 | 97.94 ± 1.10 |
| 3dU | 94.77 ± 2.30 | 96.01 ± 1.58 | 97.15 ± 1.66 | 96.08 ± 1.92 | 98.20 ± 1.19 | 98.13 ± 1.02 | 98.05 ± 0.76 |
| FCN | 97.65 ± 0.74 | 96.34 ± 1.32 | 97.16 ± 0.99 | 96.74 ± 1.11 | 93.34 ± 1.13 | 97.82 ± 1.18 | 97.46 ± 1.07 |
Table 2.
Median and inter-quartile range for Hausdorf f95 scores for all trained models and input image combinations. DM:DeepMedic, 3dU:3D U-Net.
| T1 | T2 | T1Gd | Flair | Multi-4 | Multi-2 | Ens-4 | |
|---|---|---|---|---|---|---|---|
| DM | 2.24 ± 1.41 | 2.24 ± 1.00 | 2.24 ± 1.27 | 2.24 ± 1.00 | 2.00 ± 1.41 | 2.24 ± 1.41 | 2.24 ± 1.00 |
| 3dU | 11.45 ± 2.98 | 19.21 ± 2.85 | 3.00 ± 2.01 | 5.20 ± 5.85 | 1.73 ± 1.41 | 2.00 ± 1.50 | 2.00 ± 0.61 |
| FCN | 2.24 ± 1.00 | 3.61 ± 1.18 | 2.83 ± 0.93 | 3.00 ± 1.55 | 5.92 ± 0.93 | 2.24 ± 1.00 | 2.24 ± 0.76 |
Fig. 2.
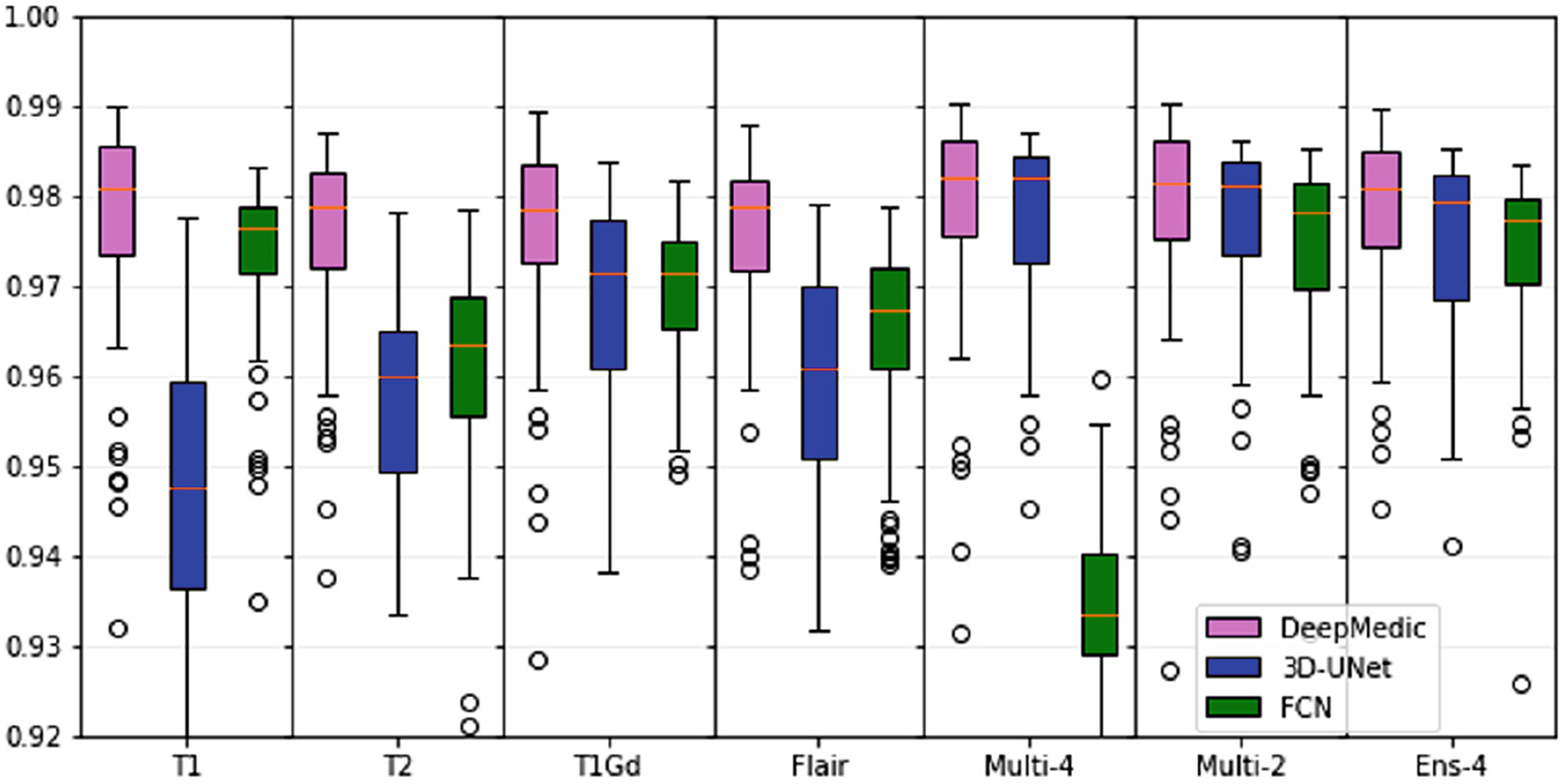
Median and inter-quartile range for Dice scores of all trained models and input image combinations.
Fig. 3.
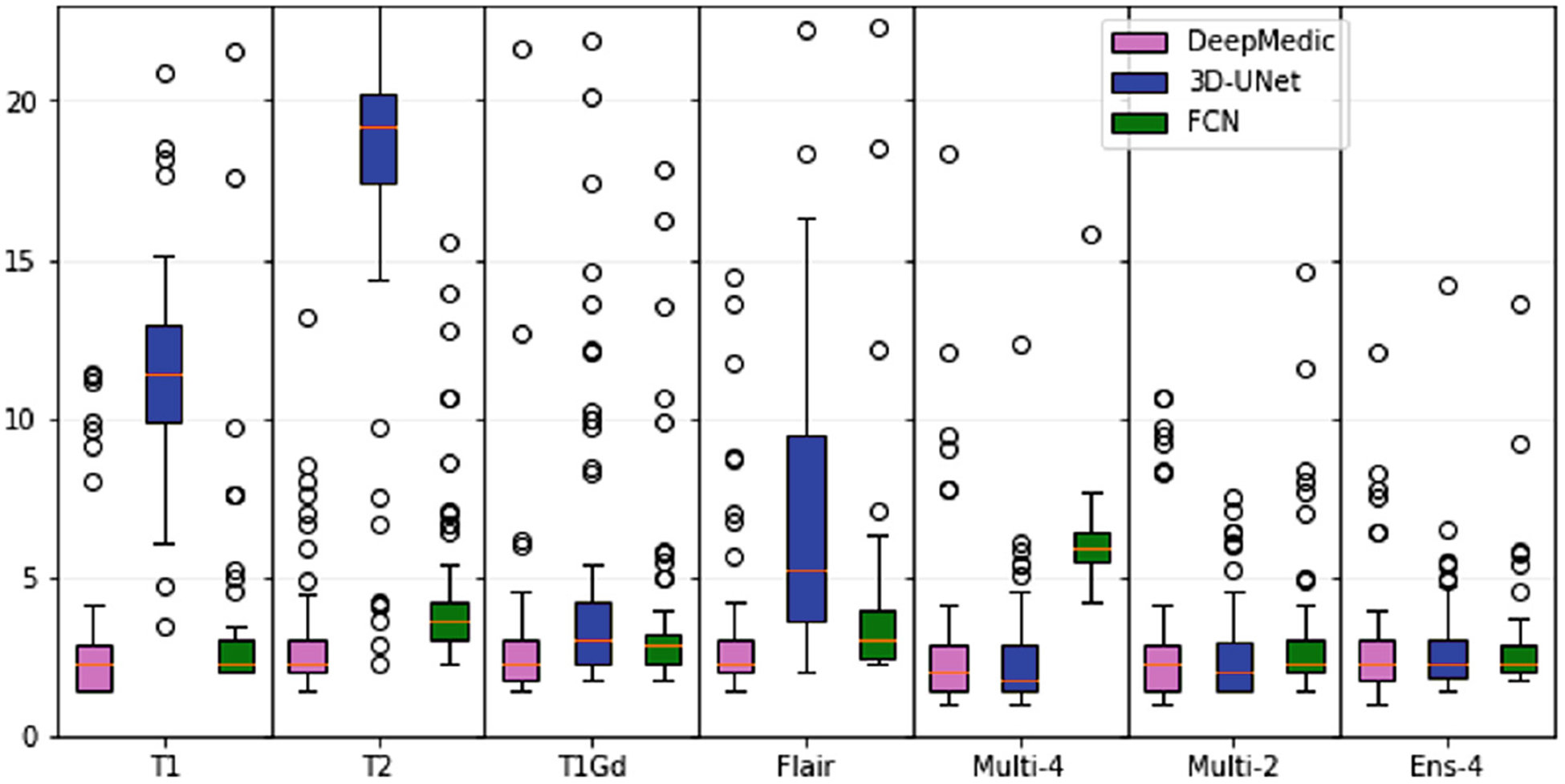
Median and inter-quartile range for Hausdorf f95 scores of all trained models and input image combinations.
Overall, best performance was obtained for the DeepMedic-Multi-4 model (Dice = 97.9, Hausdorf f95 = 2.68). However, for the model trained using DeepMedic and only the T1 modality obtained comparable (statistically insignificant, p > 0.05 - Wilcoxon signed-rank test) performance (Dice = 97.8, Hausdorf f95 = 3.01). This result reaffirms the use of T1 in current state of the art methods for skull-stripping.
Performance of 3D U-Net was consistently lower when the network was trained on single modalities. However, the 3D U-Net-Multi-4 model obtained performance comparable to DeepMedic. Despite previous literature reporting a clear benefit of the ensemble approach [12,13], in our validations we found that the ensemble of models trained and inferred on individual modalities did not offer a noticeable improvement.
Illustrative examples of the segmentations for the best performing model (DeepMedic-Multi-4) are shown in Fig. 4. We showcase the best and the worst segmentation results, selected based on the Dice scores.
Fig. 4.
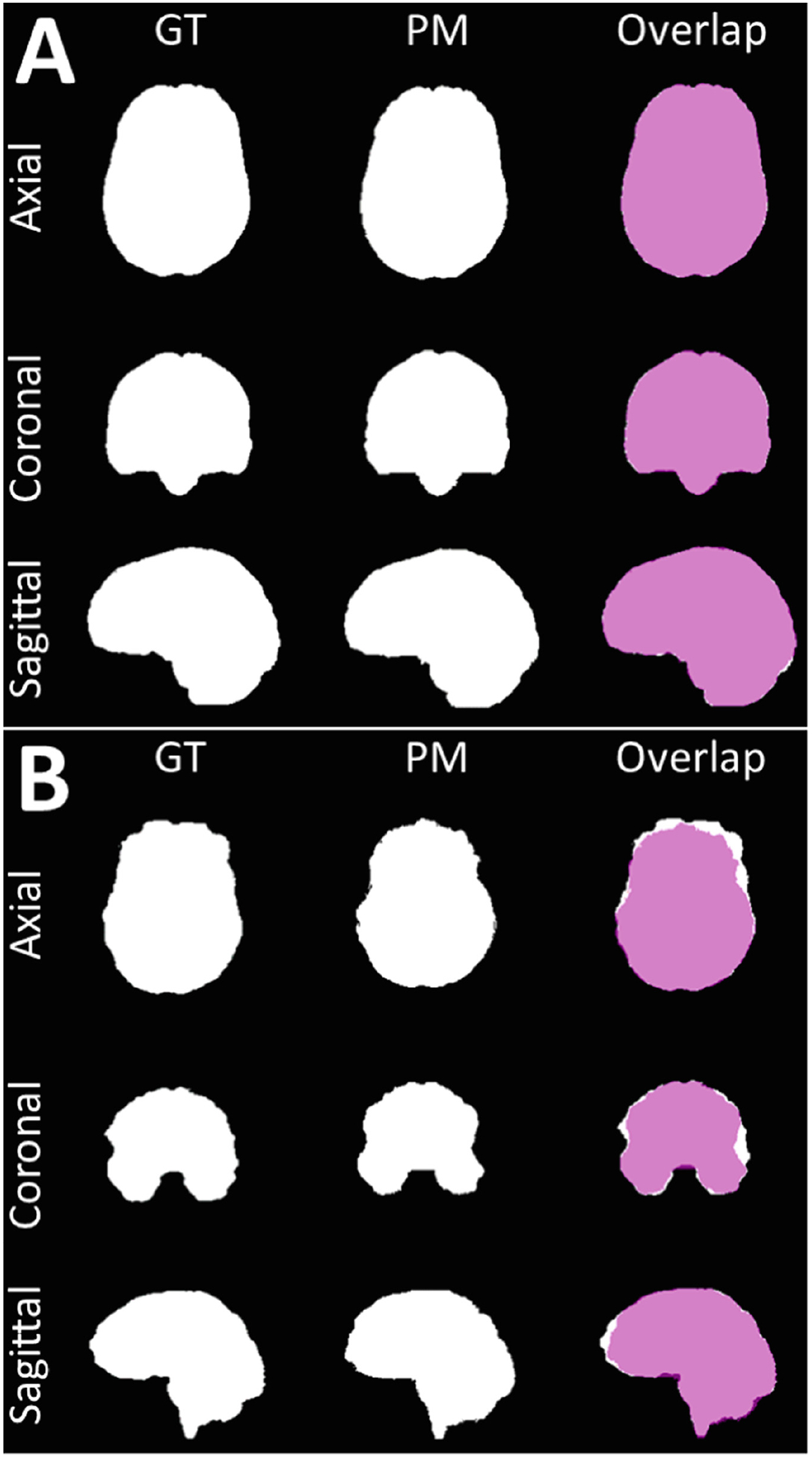
Visual example of the best (A) and the worst (B) output results.
4. Discussion
We compared three of the most widely used DL architectures for semantic segmentation in the specific problem of skull-stripping of images with brain tumors. Importantly, we trained models using different combinations of input image modalities that are typically acquired as part of routine clinical evaluations of patients with suspected brain tumors, to investigate contribution of these different modalities to overall segmentation performance.
DeepMedic consistently outperformed the other 2 methods with all input combinations, suggesting that it is more robust. In contrast, 3D U-Net and FCN had highly variable performance with different image combinations. With the addition of mpMRI input data, the 3D U-Net models (“Multi-4” and “Multi-2”) performed comparably with DeepMedic.
We have made the pre-trained DeepMedic model, including all pre-processing and post-processing steps, available for inference to others through our cancer image processing toolkit, namely the CaPTk [32,33], which provides readily deployable advanced computational algorithms to facilitate clinical research.
In future work, we intend to extend application of these topologies to multi-institutional data along with other topologies for comparison.
Acknowledgement.
Research reported in this publication was partly supported by the National Institutes of Health (NIH) under award numbers NIH/NINDS: R01NS042645, NIH/NCI:U24CA189523, NIH/NCI:U01CA242871. The content of this publication is solely the responsibility of the authors and does not represent the official views of the NIH.
References
- 1.Ostrom Q, et al. : Females have the survival advantage in glioblastoma. Neuro-Oncology 20, 576–577 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Herrlinger U, et al. : Lomustine-temozolomide combination therapy versus standard temozolomide therapy in patients with newly diagnosed glioblastoma with methylated MGMT promoter (CeTeG/NOA 2009): a randomised, open-label, phase 3 trial. Lancet 393, 678–688 (2019) [DOI] [PubMed] [Google Scholar]
- 3.Gutman DA, et al. : MR imaging predictors of molecular profile and survival: multi-institutional study of the TCGA glioblastoma data set. Radiology 267(2), 560–569 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gevaert O: Glioblastoma multiforme: exploratory radiogenomic analysis by using quantitative image features. Radiology 273(1), 168–174 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jain R: Outcome prediction in patients with glioblastoma by using imaging, clinical, and genomic biomarkers: focus on the nonenhancing component of the tumor. Radiology 272(2), 484–93 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aerts HJWL: The potential of radiomic-based phenotyping in precision medicine: a review. JAMA Oncol. 2(12), 1636–1642 (2016) [DOI] [PubMed] [Google Scholar]
- 7.Bilello M, et al. : Population-based MRI atlases of spatial distribution are specific to patient and tumor characteristics in glioblastoma. NeuroImage: Clin. 12, 34–40 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zwanenburg A, et al. : Image biomarker standardisation initiative. Radiology, arXiv:1612.07003 (2016). 10.1148/radiol.2020191145 [DOI] [Google Scholar]
- 9.Bakas S, et al. : In vivo detection of EGFRvIII in glioblastoma via perfusion magnetic resonance imaging signature consistent with deep peritumoral infiltration: the phi-index. Clin. Cancer Res 23, 4724–4734 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Binder Z, et al. : Epidermal growth factor receptor extracellular domain mutations in glioblastoma present opportunities for clinical imaging and therapeutic development. Cancer Cell 34, 163–177 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Akbari H, et al. : In vivo evaluation of EGFRvIII mutation in primary glioblastoma patients via complex multiparametric MRI signature. Neuro-Oncology 20(8), 1068–1079 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Menze BH, et al. : The multimodal Brain Tumor Image Segmentation Benchmark (BRATS). IEEE Trans. Med. Imaging 34(10), 1993–2024 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bakas S, et al. : Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the BRATS challenge. arXiv e-prints. arxiv:1811.02629 (2018) [Google Scholar]
- 14.Bakas S, et al. : Advancing the cancer genome atlas glioma MRI collections with expert segmentation labels and radiomic features. Nat. Sci. Data 4, 170117 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gitler AD, et al. : Neurodegenerative disease: models, mechanisms, and a new hope. Dis. Models Mech 10(5), 499–502 (2017). https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5451177/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Souza R, et al. : An open, multi-vendor, multi-field-strength brain MR dataset and analysis of publicly available skull stripping methods agreement. NeuroImage 170, 1053–8119 (2018) [DOI] [PubMed] [Google Scholar]
- 17.Lin HW, et al. : Why does deep and cheap learning work so well? J. Stat. Phys 168, 1223–1247 (2017) [Google Scholar]
- 18.Abadi M, et al. : TensorFlow: large-scale machine learning on heterogeneous systems, Software. tensorflow.org (2015) [Google Scholar]
- 19.Paszke A, et al. : Automatic differentiation in PyTorch. In: NIPS Autodiff Workshop (2017) [Google Scholar]
- 20.Kamnitsas K, et al. : Multi-scale 3D convolutional neural networks for lesion segmentation in brain MRI. Ischemic Stroke Lesion Segment. 13, 46 (2015) [Google Scholar]
- 21.Kamnitsas K, et al. : Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med. Image Anal 36, 61–78 (2017) [DOI] [PubMed] [Google Scholar]
- 22.Çiçek Ö, Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O: 3D U-Net: learning dense volumetric segmentation from sparse annotation In: Ourselin S, Joskowicz L, Sabuncu MR, Unal G, Wells W (eds.) MICCAI 2016. LNCS, vol. 9901, pp. 424–432. Springer, Cham; (2016). 10.1007/978-3-319-46723-8_49 [DOI] [Google Scholar]
- 23.Long J et al. : Fully convolutional networks for semantic segmentation. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 3431–3440 (2015) [Google Scholar]
- 24.Cox R, et al. : A (Sort of) new image data format standard: NIfTI-1: WE 150. Neuroimage 22 (2004) [Google Scholar]
- 25.Bakas S, et al. : Segmentation labels and radiomic features for the pre-operative scans of the TCGA-GBM collection. Cancer Imaging Arch. 286 (2017) [Google Scholar]
- 26.Bakas S, et al. : Segmentation labels and radiomic features for the pre-operative scans of the TCGA-LGG collection. Cancer Imaging Arch. (2017) [Google Scholar]
- 27.Rohlfing T, et al. : The SRI24 multichannel atlas of normal adult human brain structure. Hum. Brain Mapp 31, 798–819 (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yushkevich PA, et al. : Fast automatic segmentation of hippocampal subfields and medial temporal lobe subregions in 3 Tesla and 7 Tesla T2-weighted MRI. Alzheimer’s Dement. J. Alzheimer’s Assoc 12(7), P126–P127 (2016) [Google Scholar]
- 29.Joshi S, et al. : Unbiased diffeomorphic atlas construction for computational anatomy. NeuroImage 23, S151–S160 (2004) [DOI] [PubMed] [Google Scholar]
- 30.Yushkevich PA, et al. : User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. NeuroImage 31(3), 1116–1128 (2006) [DOI] [PubMed] [Google Scholar]
- 31.Yushkevich PA, et al. : User-guided segmentation of multi-modality medical imaging datasets with ITK-SNAP. Neuroinformatics 17(1), 83–102 (2018). 10.1007/s12021-018-9385-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davatzikos C, et al. : Cancer imaging phenomics toolkit: quantitative imaging analytics for precision diagnostics and predictive modeling of clinical outcome. J. Med. Imaging 5(1), 011018 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rathore S, et al. Brain cancer imaging phenomics toolkit (brain-CaPTk): an interactive platform for quantitative analysis of glioblastoma In: Brainlesion : Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries: Third International Brain-Les Workshop Held in Conjunction with MICCAI 2017, Quebec City, QC, Canada, vol. 10670, pp. 133–145 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maier O, et al. : ISLES 2015 - a public evaluation benchmark for ischemic stroke lesion segmentation from multispectral MRI. Med. Image Anal 35, 250–269 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simpson AL, et al. : A large annotated medical image dataset for the development and evaluation of segmentation algorithms, arXiv e-prints, arXiv:1902.09063 (2019) [Google Scholar]
- 36.Shattuck DW, et al. : Magnetic resonance image tissue classification using a partial volume model. NeuroImage 13(5), 856–876 (2001) [DOI] [PubMed] [Google Scholar]
- 37.Smith SM: Fast robust automated brain extraction. Hum. Brain Mapp 17(3), 143–155 (2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iglesias JE, et al. : Robust brain extraction across datasets and comparison with publicly available methods. IEEE Trans. Med. Imaging 30(9), 1617–1634 (2011) [DOI] [PubMed] [Google Scholar]
- 39.Eskildsen SF: BEaST: brain extraction based on nonlocal segmentation technique. NeuroImage 59(3), 2362–2373 (2012) [DOI] [PubMed] [Google Scholar]
- 40.Doshi J, et al. : Multi-atlas skull-stripping. Acad. Radiol 20(12), 1566–1576 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Doshi J, et al. : MUSE: MUlti-atlas region segmentation utilizing ensembles of registration algorithms and parameters, and locally optimal atlas selection. Neuroimage 127, 186–195 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]


