Abstract
Brain lesions composed of pathological tau help to drive neurodegeneration in Alzheimer’s disease (AD) and related tauopathies. Here, we identified the mammalian suppressor of tauopathy 2 (MSUT2) gene as a modifier of susceptibility to tau toxicity in two mouse models of tauopathy. Transgenic PS19 mice overexpressing tau, a model of AD, and lacking the Msut2 gene exhibited decreased learning and memory deficits, reduced neurodegeneration, and reduced accumulation of pathological tau compared to PS19 tau transgenic mice expressing Msut2. Conversely, Msut2 overexpression in 4RTauTg2652 tau transgenic mice increased pathological tau deposition and promoted the neuroinflammatory response to pathological tau. MSUT2 is a poly(A) RNA binding protein that antagonizes the canonical nuclear poly(A) binding protein PABPN1. In individuals with AD, MSUT2 abundance in postmortem brain tissue predicted an earlier age of disease onset. Postmortem AD brain tissue samples with normal amounts of MSUT2 showed elevated neuroinflammation associated with tau pathology. We observed co-depletion of MSUT2 and PABPN1 in postmortem brain samples from a subset of AD cases with higher tau burden and increased neuronal loss. This suggested that MSUT2 and PABPN1 may act together in a macromolecular complex bound to poly(A) RNA. Although MSUT2 and PABPN1 had opposing effects on both tau aggregation and poly(A) RNA tail length, we found that increased poly(A) tail length did not ameliorate tauopathy, implicating other functions of the MSUT2/PABPN1 complex in tau proteostasis. Our findings implicate poly(A) RNA binding proteins both as modulators of pathological tau toxicity in AD and as potential molecular targets for interventions to slow neurodegeneration in tauopathies.
INTRODUCTION
The molecular mechanisms underpinning neurodegenerative diseases include the cellular disruption of proteostasis. In Alzheimer’s disease (AD), this disruption manifests as the deposition of amyloid plaques and neurofibrillary tangles (NFTs), the diagnostic pathological lesions of the disorder. Whereas the mechanistic relationship between plaques and tangles remains unclear, abnormal tau and Aβ act synergistically to drive neurodegeneration in AD. A large body of evidence supports the idea of Aβ amyloid pathology initiating the disease process in AD. However, the discovery of tau mutations in frontotemporal lobar degeneration with tau inclusions (FTLD-tau) (1–4) demonstrates that tau pathology can cause neurodegeneration independent of amyloid plaques. Furthermore, tau pathology, not amyloid deposition, correlates with the severity of dementia in AD (5). Thus, findings to date justify active investigation of the mechanistic underpinnings of both amyloid- and tau-mediated neurodegeneration in AD. Despite a diverse array of highly powered AD clinical trials targeting amyloid production, clearance, or deposition, none have been successful. Together, these observations suggest that tau-targeted therapies in conjunction with removal of amyloid may be required to achieve cognitive preservation when treating AD (6, 7).
Abnormally aggregated highly phosphorylated tau becomes deposited as tangles or other lesions in tauopathy disorders. For AD and many other tauopathies, the molecular role tau plays in disease initiation and progression remains unknown. However, in FTLD-tau, mutations in the gene encoding tau cause the disease by reducing tau’s affinity for microtubules and increasing the propensity of tau to aggregate (8, 9). Because tau binds to microtubules, one hypothesis suggests that abnormal tau impairs the function of the cytoskeleton. The reduced affinity of tau for microtubules caused by FTLD mutations may disrupt microtubule stability and axonal transport (10). An alternate hypothesis is that tau aggregation reduces the amount of tau available for binding to microtubules (6, 11). Evidence suggests that toxic tau aggregates or oligomers can spread by a seeding mechanism following neuronal connectivity pathways (12, 13). The critical neurotoxic species remain poorly defined, and dimers, low-level tau oligomers, higher-order assemblies of tau, and end-stage NFTs are all candidate triggers of neurotoxicity. The phosphorylation state of tau likely contributes to toxicity as tau phosphorylation can drive tau from microtubules and promote aggregation [reviewed in (14, 15)]. Together, recent evidence suggests that a diversity of related and varying neurotoxic species likely contributes to both the spreading of tau pathology and tau-mediated neurodegeneration [reviewed in (16, 17)].
How abnormal tau kills neurons remains unknown. The identification of genes mediating susceptibility or resistance to pathological tau may inform disease mechanisms in AD and related disorders. To date, genomic studies in AD patients implicate many genes in susceptibility, but only the APOE2 allele is strongly protective against AD. Among the risk-causing genetic variants, genes involved in innate immune responses and expressed in microglia are unusually common [reviewed in (18)]. Whereas tau pathology in glial cells does not commonly occur in AD, it is a feature of some pure tauopathy disorders (19), occurring in astrocytes in progressive supranuclear palsy (PSP) and in oligodendrocytes in some forms of FTLD. Furthermore, reactive gliosis is a common feature of tauopathy disorders including AD. Neuroinflammation and tau pathology appear to be mutually reinforcing features of AD and related disorders (20–22).
To identify genes controlling tau toxicity, we previously generated a tauopathy model by expressing human tau in the nematode Caenorhabditis elegans using a promoter that drives expression in all neurons. The phenotype of this model includes uncoordinated locomotion, accumulation of insoluble tau, neurodegeneration, and a shortened life span (23). We used this model to identify loss-of-function mutations suppressing tau-induced neurodegenerative phenotypes (24, 25). In this model, loss-of-function mutations in the suppressor of tauopathy 2 gene (sut-2) decreased tau aggregation and protected against neurodegeneration (26). The sut-2 gene encodes a CCCH finger protein with conserved homologs in all species from yeast to humans. MSUT2 (also known as ZC3H14) is the mammalian homolog of the C. elegans sut-2 gene. We hypothesized that loss of function of MSUT2 in mammals would ameliorate neurodegenerative tauopathy by a previously unknown mechanism. Here, we examine how loss of MSUT2 provides protection against tau pathology and gliosis in mice and humans.
RESULTS
Loss of MSUT2 protects neurons in mouse brain from pathological tau
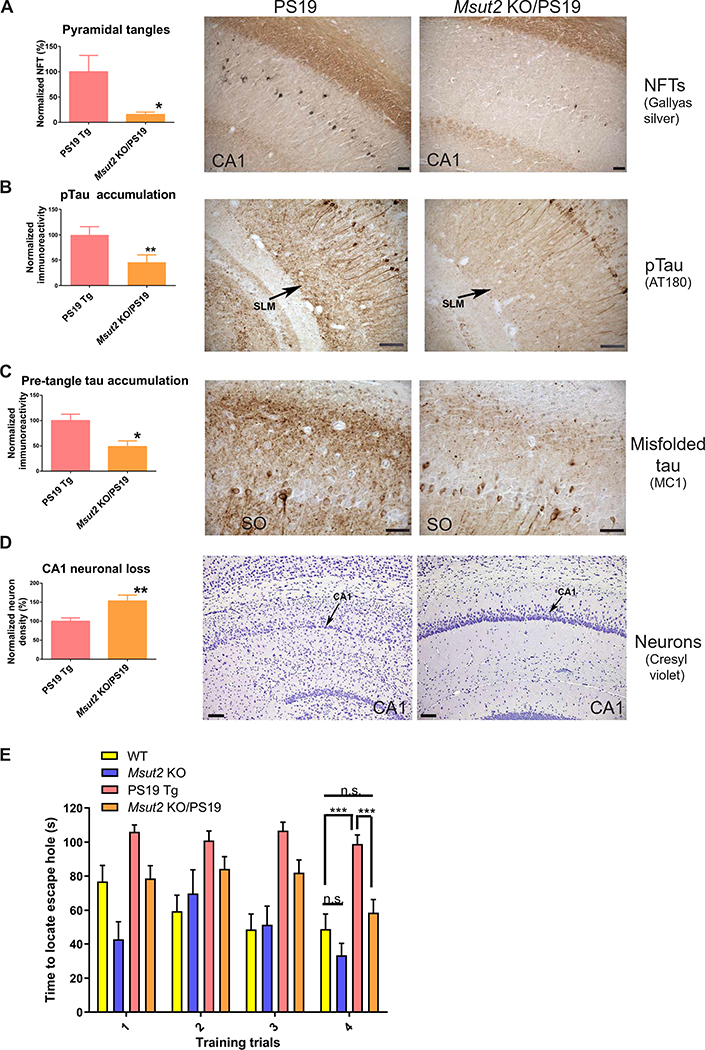
To test whether MSUT2 function affects the effects of pathological tau on neurons in the intact mammalian brain, we generated mice lacking the MSUT2 CCCH finger domains (fig. S1, A to E), which have been shown to be critical for SUT-2 function in C. elegans (24). Mice homozygous for this disruption (henceforth called Msut2 KO mice) were fully viable, exhibited no visible phenotypes, and appeared neurologically intact. We crossed these mice with the well-characterized PS19 mouse model of tauopathy that overexpresses human tau carrying the P301S mutation that causes the tauopathy frontotemporal lobar degeneration (FTLD) (27). We generated PS19 mice with a homozygous deletion of Msut2 and compared the abundance of tau-positive pathological lesions in PS19 mice with or without functional MSUT2. Given that PS19 mice exhibit deficits in hippocampal-dependent memory tasks (28), we focused our neuropathological analysis on the hippocampal formation and projecting regions. PS19 mice develop age-dependent neurodegeneration that correlates with the progression of pathological tau deposition in NFTs similar to that seen in human patients with AD or other tauopathy disorders (27). PS19 mice have an abundance of NFT-like inclusions in hippocampal CA1 pyramidal neurons and the entorhinal cortex. In contrast, PS19 mice with a homozygous deletion of Msut2 had reduced numbers of hippocampal NFTs (85% reduction; P = 0.0129 by two-tailed t test; Fig. 1A and fig. S1F). Formation of pathological tau progresses from monomeric normal tau, to phosphorylated tau (pTau), to oligomeric and pre-tangle conformations of tau, to fibrillary tau culminating in deposition of mature NFTs. Both pTau and misfolded tau lesions positive for MC1 (a pre-tangle tau-specific monoclonal antibody) were reduced by over 50% in Msut2 KO/PS19 mice relative to PS19 animals, especially in neuronal processes of hippocampal subfields (stratum lacunosum moleculare and stratum oriens) as well as in the entorhinal cortex [Fig. 1B (55% reduction; P = 0.016 by two-tailed t test), Fig. 1C (50% reduction; P = 0.0062 by two-tailed t test), and fig. S2, A and B]. When MSUT2 was absent, the overall decrease in pTau- and MC1-positive tau lesions paralleled the decrease of NFTs in the hippocampus and entorhinal cortex (Fig. 1A). Likewise, the Msut2 KO/PS19 mice did not show the neuronal loss observed in the CA1 region of the hippocampus of PS19 mice with intact Msut2 (53% reduction; P = 0.0062 by two-tailed t test; Fig. 1D). However, loss of MSUT2 alone did not obviously change neuronal abundance in the mouse hippocampus (fig. S2C). Our findings suggest that loss of MSUT2 reduced accumulation of pathologic tau and protected against neuronal loss in mouse brain.
Fig. 1. Genetic ablation of Msut2 protects against tauopathy in mice.
(A) Deletion of Msut2 decreases accumulation of neurofibrillary tangles (NFTs) in the CA1 region of the hippocampus of PS19 mice with tauopathy. Representative brain sections from 9-month-old PS19 mice with tauopathy are shown. NFTs were detected by Gallyas silver stain. Left: Number of Gallyas-positive NFTs in brain sections from Msut2 KO/PS19 transgenic (Tg) mice (n = 24) compared to PS19 Tg mice (n = 24) (P = 0.0129 by two-tailed t test). Scale bars, 100 μm.
(B) Deletion of Msut2 decreases accumulation of pTau in the stratum lacunosum moleculare (SLM) of brains of PS19 mice with tauopathy. Representative brain sections from 9-month-old PS19 mice with tauopathy stained with AT180 monoclonal antibody to reveal pTau phosphorylated on Thr231 are shown. Left: Densitometry analysis of AT180-positive pathological tau deposits in Msut2 KO/PS19 Tg brain sections (n = 24) compared to PS19 Tg brain sections (n = 24) (P=0.0156 by two-tailed t test). Scale bars, 100 μm.
(C) Deletion of Msut2 decreases accumulation of pre-tangle tau species in the stratum oriens (SO) region of the hippocampus of PS19 mice with tauopathy. Pre-tangle pathological tau species were detected by the conformation-dependent anti-tau monoclonal antibody MC1. Representative brain sections from 9-month-old PS19 mice with tauopathy stained with MC1 antibody are shown. Left: Densitometry analysis of MC1-positive tau lesions in Msut2 KO/PS19 Tg mouse brain (n = 24) compared to PS19 Tg mouse brain (n = 24) (P = 0.0042 by two-tailed t test). Scale bars, 50 μm.
(D) Deletion of Msut2 decreases loss of CA1 hippocampal neurons in PS19 mice with tauopathy. Representative brain sections from 9-month-old PS19 mice with tauopathy stained with cresyl violet to identify neuronal cell bodies are shown (black arrows indicate the pyramidal cell layer). Left: Densitometry analysis of pyramidal neuron density in Msut2 KO/PS19 Tg mouse brains (n = 21) compared to PS19 Tg mouse brains (n = 19) (P = 0.0062 by two-tailed t test). Photomicrographs in (A) to (D) were modified by equally adjusting brightness and contrast to optimize visualization of staining.
(E) Deletion of Msut2 enhances performance of 8-month-old PS19 mice with tauopathy in the Barnes maze training test. PS19 Tg animals performed poorly on the Barnes maze test, taking longer to locate the escape hole compared to either wild-type (WT) or Msut2 KO/PS19 animals (***P < 0.001 by ANOVA). n.s., not significant.
Loss of MSUT2 improves cognitive function in mice with tauopathy
Msut2 KO mice exhibited typical motor functions and appeared cognitively similar to wild-type mice (fig. S3 and Fig. 1E). To assess the effect of Msut2 loss on PS19 mouse motor activity and behavior, we characterized PS19 mice with or without a functional Msut2 gene (PS19 mice versus Msut2 KO/PS19 mice) at 8 months of age (n = 32 to 33 mice per genotype). As with the wild-type background, loss of MSUT2 did not alter gross physical characteristics, motor function, or spontaneous activity of PS19 mice (fig. S3, B to G). However, MSUT2 loss did protect against age-related weight loss in PS19 male mice (fig. S3D). We examined the cognitive performance of these PS19 mice using the Barnes maze test of hippocampal-dependent learning and memory (29), a task at which PS19 animals have a reproducible impairment (Fig. 1E) (30, 31). Msut2 KO/PS19 mice exhibited better performance during the training trials (as measured by time to locate the escape hole) and learnt the Barnes maze task faster than did their PS19 siblings (P < 0.001; Fig. 1E). These data show that loss of MSUT2 function slowed cognitive decline caused by the mutant tau transgene in PS19 mice.
MSUT2 binds to the poly(A) binding protein PABPN1 through multifunctional CCCH domains
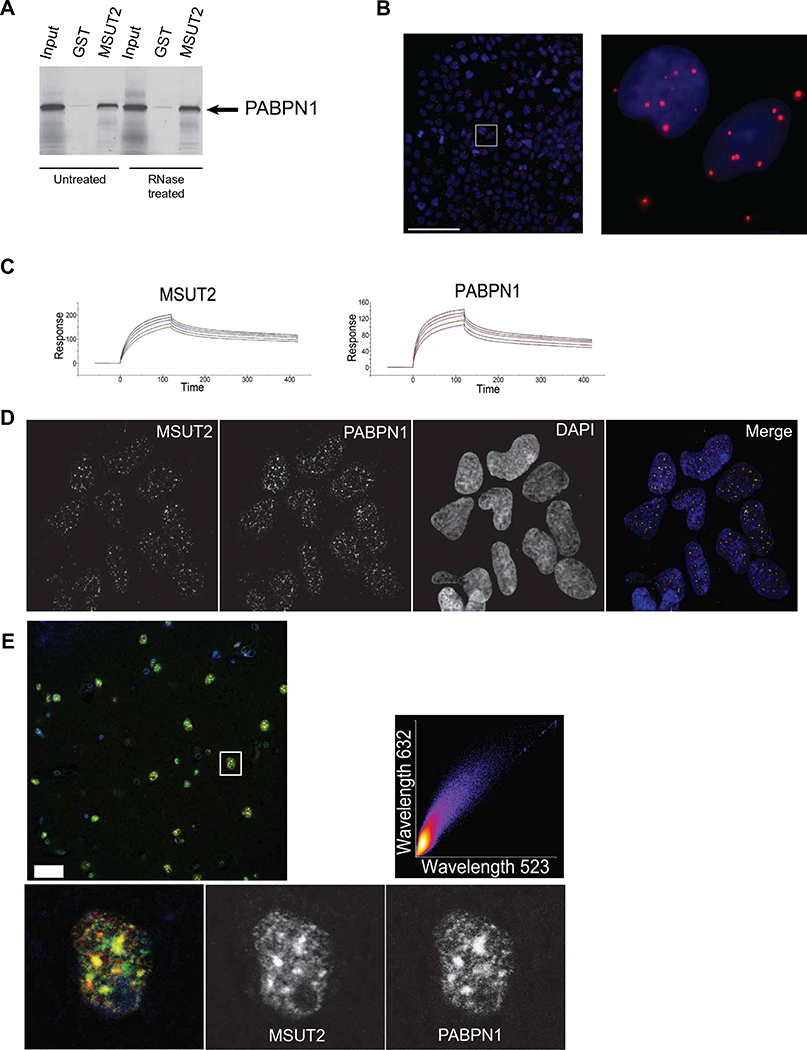
To explore the mechanism of MSUT2 function, we focused on the C terminus of MSUT2 and SUT-2, which appears to be the critical functional domain necessary for suppressing tauopathy phenotypes (24, 32). To explore the functional partners of the SUT-2 protein, we conducted a yeast two-hybrid screen using the sut-2 CCCH domains as bait. The major SUT-2 binding protein identified, representing over 90% of the cDNAs recovered, was the homolog of nuclear polyadenylate binding protein (PABPN1). PABPN1 binds to the poly(A) tail of mRNAs through its RNA recognition motif (RRM) domain and regulates poly(A) tail length in both yeast and mammals (33, 34). We demonstrated a conserved protein-protein interaction between human PABPN1 and MSUT2 proteins using purified recombinant human proteins in an in vitro glutathione S-transferase (GST) pulldown assay and showed that the interaction could occur in the absence of detectable RNA (Fig. 2A). We validated the binding of MSUT2 to PABPN1 in a cellular context in situ using a proximity ligation assay of human HEK293 cells (35) and found that MSUT2 bound to PABPN1 predominantly in the nuclear compartment and, to a lesser extent, in the cytoplasm (Fig. 2B). Given that CCCH domains can bind to both RNA and protein, we measured MSUT2 association with poly(A) RNA using a surface plasmon resonance assay. MSUT2 affinity for poly(A) RNA was about fourfold greater than for PABPN1 for a single poly(A) binding site [MSUT2 KD = 60 ± 15 nM versus PABPN1 KD = 237 ± 21 nM for poly(A)15] (Fig. 2C).
Fig. 2. MSUT2 binds independently to both poly(A) RNA tails and PABPN1.
(A) In vitro confirmation of the yeast two-hybrid interaction between the MSUT2 CCCH finger domain and PABPN1.35S radiolabeled PABPN1 was tested against immobilized recombinant glutathione S-transferase (GST)-MSUT2 (ZF) fusion protein or recombinant GST alone using GST pulldown assays. RNase pretreatment of samples did not prevent MSUT2 and PABPN1 protein-protein interactions.
(B) The proximity ligation assay detects MSUT2/PABPN1 interactions within intact HEK293 cell nuclei. Blue, DAPI nuclear stain; red, interactions detected by proximity ligation assay. Left: High-magnification stitched panel image of a large field of immunostained HEK293 cells. Scale bar, 100 μm. The white square in the left represents the magnified area shown in the right.
(C) Surface plasmon resonance detection of MSUT2 and PABPN1 binding to poly(A) RNA. Dissociation constants were calculated for biotinylated poly(A)15 from the binding kinetics of each protein at five concentrations (1, 0.9, 0.8, 0.7, and 0.6 μM). MSUT2 KD = 60 ± 15 nM compared to PABPN1 KD = 237 ± 21 nM for poly(A)15.
(D) MSUT2 and PABPN1 colocalization in SC35-positive nuclear speckles in HEK293 cells [Pearson coefficient of correlation (PCC) = 0.7976]. A representative single-channel image of nuclear MSUT2, PABPN1, and DAPI stain is depicted. Colocalization analysis of MSUT2 with SC35 (PCC = 0.7578) and PABPN1 with SC35 (PCC = 0.4347) is shown in fig. S4 (C and D).
(E) MSUT2 and PABPN1 colocalization in nuclear speckles in neuronal nuclei from human frontal cortex brain tissue. Representative confocal image shows immunofluorescent staining for MSUT2 (green), PABPN1 (red), and DAPI (blue). Inset: Digital magnification of a single nucleus displayed as split channels for MSUT2 and PABPN1 and merged channels. MSUT2 and PABPN1 colocalized with a Pearson coefficient of correlation (PCC) = 0.88 in this image (see also fig. S4). Scale bar, 25 μm. Where necessary, image adjustments applied linear brightness and contrast changes.
MSUT2 colocalizes with PABPN1 and poly(A) RNA within nuclear speckles
Given that MSUT2 bound to PABPN1 and poly(A) RNA independently in vitro, we examined the relationship between MSUT2, PABPN1, and poly(A) RNA in cultured human cells. HEK293 cells were immunostained for MSUT2 and PABPN1 and hybridized with poly(A) RNA-specific fluorescent probes. Analysis showed that MSUT2 protein colocalized with PABPN1 and poly(A) RNA in the nucleus of HEK293 cells [Pearson coefficient of correlation (PCC) = 0.7976; Fig. 2D and fig. S4, A and B]. To explore whether the speckle-like appearance of MSUT2 and PABPN1 immunostaining represented authentic nuclear speckle localization, we costained with antibodies against SC35, a nuclear splicing factor, and observed labeling of nuclear speckles. We saw colocalization of both MSUT2 and PABPN1 together with SC35 within nuclear speckles (fig. S4, C and D). We also investigated the localization of MSUT2 and PABPN1 in postmortem normal human brain tissue. Coimmunostaining for MSUT2 and PABPN1 revealed clear nuclear colocalization of PABPN1 and MSUT2 in frontal cortex neurons of postmortem human brain tissue (PCC = 0.88; Fig. 2E). Together, these data support the cellular colocalization of MSUT2 and PABPN1 with poly(A) RNA in nuclear speckles, consistent with their demonstrated physical interaction with each other and with RNA.
MSUT2 and PABPN1 reciprocally regulate tau aggregation
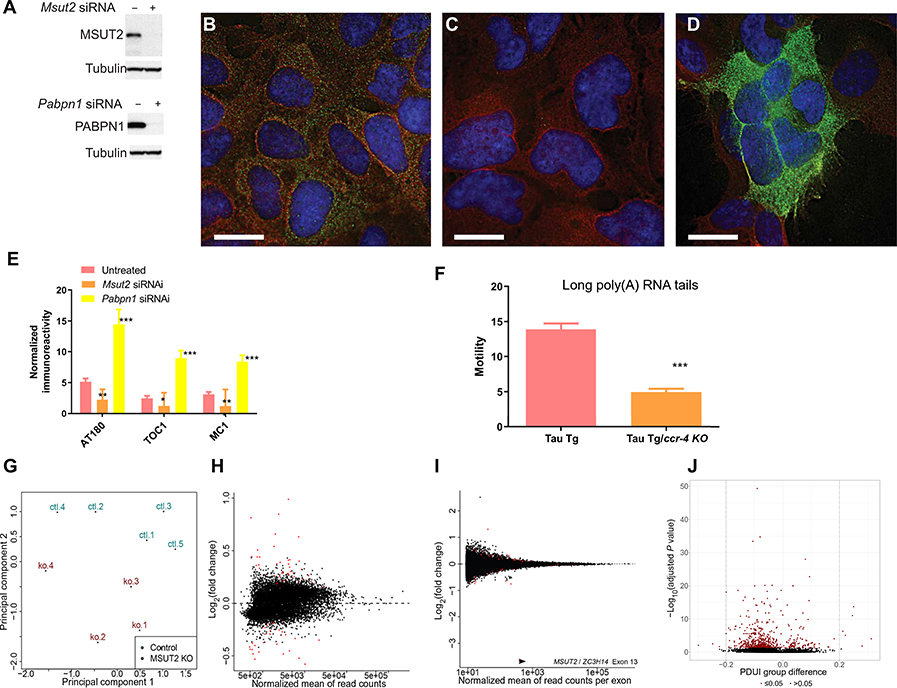
Recent work demonstrated that siRNA knockdown of PABPN1 decreased the poly(A) tail length on bulk mRNAs, whereas knockdown of MSUT2 increased poly(A) tail length, indicating opposing functions of PABPN1 and MSUT2 in polyadenylation (36, 37). To explore whether the PABPN1/MSUT2 interaction mediates changes in accumulation of pathological tau, we analyzed tau aggregation in cultured human HEK293 cells constitutively overexpressing human tau in which MSUT2 or PABPN1 was knocked down by gene-specific siRNA treatment (Fig. 3A). We observed decreased pTau species in cells where MSUT2 was knocked down, as assessed by immunostaining with AT180 monoclonal antibody, which detects tau phosphorylated on Thr231 (Fig. 3B, mock siRNA treatment; Fig. 3C, MSUT2 siRNA treatment). Similar quantitative measures showed that siRNA-mediated knockdown of MSUT2 decreased both pTau and tau oligomeric species accumulation as detected by quantitative immunofluorescence using the conformation-specific monoclonal antibodies, tau oligomeric complex 1 (TOC1) (38) and MC1 (39) (Fig. 3E). In contrast, PABPN1 knockdown exacerbated accumulation of aggregated tau in both assays (Fig. 3, D and E). Thus, MSUT2 and PABPN1 seemed to have reciprocal effects on tau aggregation in cultured cells, with MSUT2 promoting tau aggregation and PABPN1 inhibiting tau aggregation. Likewise, knockdown of MSUT2 and PABPN1 together resulted in an intermediate tau aggregation phenotype in the cultured cells (fig. S5A). However, knockdown of MSUT2 did not obviously alter PABPN1 protein expression (fig. S5B) or expression pattern (fig. S5C). In contrast, knockdown of PABPN1 led to decreased MSUT2 expression (fig. S5D). To investigate whether the effects of PABPN1 and MSUT2 on tau-related phenotypes could be attributed to effects on mRNA poly(A) tail length, we directly manipulated mRNA poly(A) tail length using mutations in the gene encoding CCR-4, the major mRNA poly(A) deadenylase in C. elegans. Mutations in ccr-4 are known to cause lengthening of bulk mRNA poly(A) tails in C. elegans (40), similar to what is seen for human MSUT2 knockdown in human cells (36). Analysis of tau transgenic C. elegans carrying a loss-of-function mutation in ccr-4 demonstrated enhancement of a tauopathy-related locomotion behavioral deficit (2.7 fold decrease in locomotion; P < 0.0001 by two-tailed t test; Fig. 3F). In contrast, previous work has shown that loss-of-function mutations in sut-2 suppressed accumulation of pathological tau and tau-mediated neurodegeneration in C. elegans (24, 32). Together, these data suggest that changes in bulk polyadenylation did not underpin the molecular mechanisms by which MSUT2 and PABPN1 modulated tauopathy phenotypes.
Fig. 3. MSUT2 mediates effects on tauopathy through poly(A) tail length but not gene expression.
(A) Western blot shows that synthetic siRNA treatment eliminated MSUT2 and PABPN1 protein in HEK293 cells.
(B) Immunofluorescence staining of HEK293 cells overexpressing human tau. DAPI nuclear stain is blue, AT180 monoclonal antibody staining of pTau phosphorylated on Thr231 is green (86), and the positive staining control for total tau is red. Scale bar, 15 μm.
(C) Immunofluorescence image of HEK293 cells overexpressing human tau shows MSUT2 knockdown by siRNA and a decrease in pTau accumulation. DAPI nuclear stain is blue, AT180 monoclonal antibody staining of pTau is green (86), and the positive staining control for total tau is red. Scale bar, 15 μm.
(D) Immunofluorescence image of HEK293 cells overexpressing human tau shows PABPN1 knockdown by siRNA and an increase in pTau accumulation. DAPI nuclear stain is blue, AT180 monoclonal antibody staining of pTau is green (86), and the positive staining control for total tau is red. Scale bar, 15 μm. Image adjustments applied linear contrast and brightness changes.
(E) Quantitation of immunostaining for pTau detected by AT180 monoclonal antibody, conformation-dependent tau detected by MC1 monoclonal antibody (87), and tau oligomeric complex-1 detected by TOC1 monoclonal antibody (38, 88). Error bars are SEM. Comparison of MSUT2 siRNA-treated to untreated HEK293 cells overexpressing human tau stained with AT180 antibody, P < 0.005; comparison of PABPN1 siRNA-treated to untreated HEK293 cells overexpressing human tau stained with AT180 antibody, P < 0.001.
(F) Effect on swimming behavior of modulating poly(A) RNA tail length through ccr-4 mutation in tau transgenic C. elegans. Four-day-old tau transgenic (Tg) worms with or without the loss-of-function ccr-4 mutation were placed in liquid, and their swimming activity was recorded. Tau transgenic worms exhibited better locomotion relative to tau transgenic ccr-4 mutant animals (P < 0.0001 by two-tailed Student’s t test).
(G) Total brain RNA from Msut2 KO and C57BL/6J mice was analyzed by RNA-seq. Multidimensional scaling and principal components analysis of RNA sequencing data are shown.
(H) Transcriptomic changes in RNA sequencing data from total brain RNA of Msut2 KO and C57BL/6J mice (normalized mean read count by gene). Differentially regulated genes are depicted as red spots. See table S1 for listing of all differentially expressed genes.
(I) Alternative splicing changes in RNA sequencing data from total brain RNA of Msut2 KO and C57BL/6J mice (normalized mean read count by exon). Two hundred one differentially spliced exons were detected (indicated by red spots). See table S2 for listing of all differentially spliced exons.
(J) Volcano plot of alternative polyadenylation site selection changes in RNA sequencing data from total brain RNA of Msut2 KO and C57BL/6J mice. Analysis of polyadenylation site selection was conducted using Dynamic Analysis of Alternative Polyadenylation from RNA-seq (DaPars). Ten genes exhibited a change in percentage of distal poly(A) site usage (PDUI) of greater than 20% (see red dots). See table S3 for listing of all genes with significant alterations in polyadenylation site selection.
Given that poly(A) tails may influence the stability of specific mRNAs, we examined the consequences of knocking out Msut2 on the mouse brain transcriptome by sequencing total RNA isolated from the brains of 2.5-month-old C57BL/6J mice. Multidimensional scaling and principal components analysis revealed clustering of control and Msut2 KO transcriptome profiles (Fig. 3G). Overall, very few transcripts exhibited robust changes in abundance, with few mRNAs exhibiting significant changes in expression (n < 70) and none greater than twofold except for Msut2 (Fig. 3H and table S1). Likewise, surprisingly few genes exhibited differential exon inclusion, with only 201 differentially spliced exons detected (Fig. 3I and table S2). These data suggest that MSUT2 may mediate its effects on tau through mechanisms other than regulation of mRNA and not through global changes in mRNA processing/stability as might have been expected for a bulk poly(A) binding protein. Because PABPN1 has been shown to affect alternative polyadenylation site selection during mRNA processing (41), we analyzed alternative polyadenylation site selection in our Msut2 KO RNA-seq dataset using the Dynamic Analysis of Polyadenylation site by RNA-seq (DaPars) method (Fig. 3J and table S3) (42). Unlike PABPN1 depletion, depletion of MSUT2 had only modest consequences for alternative polyadenylation site utilization, with 10 mRNAs showing a change greater than the 20% threshold. By contrast, PABPN1 knockdown caused marked changes in alternative polyadenylation site utilization for 500 mRNAs over the 20% threshold, with more than 200 mRNAs exhibiting greater than a twofold change in alternative polyadenylation site utilization (41).
MSUT2 overexpression synergizes with pathological tau causing premature neuronal death
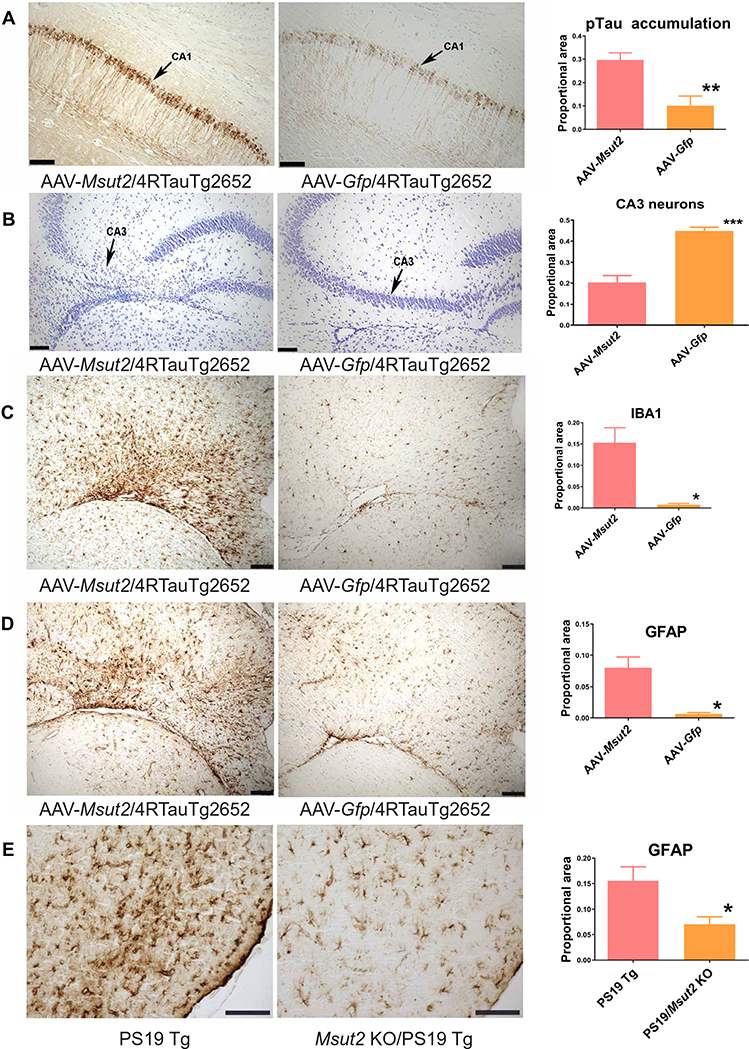
To test whether MSUT2 overexpression could provoke tau pathology in the mammalian brain, we expressed human MSUT2 in the brains of tau transgenic mice using an adeno-associated virus (AAV) vector. We used an existing transgenic mouse line with mild and nonprogressive tauopathy driven by high pan-neuronal expression of wild-type human tau (43). We stereotactically injected the AAV serotype 9 vector carrying the gene encoding human MSUT2 into the hippocampus of Tau4RTg2652 mice. The AAV9 vector drove strong expression of human MSUT2 throughout the mouse hippocampus with a normal nuclear distribution as well as an abnormal distribution in the cytoplasm and within the neuronal processes (fig. S6, A to D). By 1 month after injection, hippocampal sites with MSUT2 overexpression exhibited exacerbation of tau neuropathology in the form of pTau accumulation, whereas injection of AAV9 carrying the gene encoding green fluorescent protein (GFP) did not result in exacerbation of tauopathy (a 2.9 fold increase in tau accumulation for MSUT2 overexpression compared to GFP overexpression; P = 0.004 by two-tailed t test; Fig. 4A). Accumulation of pTau-positive lesions in the CA1 hippocampal region of mice injected with AAV9-Msut2 was accompanied by evidence of neuronal loss in associated projection regions, which was not seen in AAV9-Gfp injected control mice (a 2.2 fold decrease in neurons; P < 0.0001 by two-tailed t test; Fig. 4B). In contrast to pTau, MSUT2 overexpression did not affect total tau abundance (fig. S6E).
Fig. 4. MSUT2 potentiates neuroinflammation, tau pathology, and neurodegeneration in mouse brain.
(A) Representative brain sections from Tau4RTg2652 mice injected with AAV-Msut2 or AAV-Gfp as control. Brain sections of 4-month-old Tau4RTg2652 mice with tauopathy were stained with the phospho-dependent anti-tau monoclonal antibody AT180 to detect pTau phosphorylated on Thr231 (arrows indicate the CA1 pyramidal cell layer). MSUT2 overexpression exacerbated pTau accumulation in the ipsilateral CA1 region of mouse brain compared to mice receiving an AAV-Gfp injection. Right: Densitometry analysis of AT180-positive pathological tau deposits in Tau4RTg2652 mice overexpressing Msut2 (n = 15) compared to GFP (n = 5) (P = 0.0044 by two-tailed t test). Scale bars, 100 μm.
(B) Representative brain sections of the ipsilateral CA3 region of Tau4RTg2652 mice injected with AAV-Msut2 or AAV-Gfp as control. MSUT2 overexpression exacerbated neuronal loss in the CA3 region compared to GFP overexpression. Neuronal cell bodies were detected by cresyl violet staining (purple; arrows indicate CA3 pyramidal cell layer). Right: Densitometry analysis of pyramidal layer staining in Tau4RTg2652 mice overexpressing MSUT2 (n = 15) compared to GFP (n = 5) (P = 0.0001 by two-tailed t test). Scale bars, 100 μm.
(C) Representative brain sections of the ipsilateral CA3 region of Tau4RTg2652 mice injected with AAV-Msut2 or AAV-Gfp. Microgliosis was detected by immunohistochemical staining of glia with the IBA1 monoclonal antibody in CA3 brain sections from 4-month-old Tau4RTg2652 mice. MSUT2 overexpression exacerbated microgliosis compared to GFP overexpression. Right: Densitometry analysis of IBA1 immunoreactivity in Tau4RTg2652 mice injected with AAV-Msut2 (n = 13) compared to AAV-Gfp (n = 5) (P = 0.024 by two-tailed t test). Scale bars, 100 μm.
(D) Representative brain sections of the ipsilateral CA3 region of Tau4RTg2652 mice injected with AAV-Msut2 or AAV-Gfp. Astrocytosis was detected by immunostaining of brain sections from 4-month-old Tau4RTg2652 mice with anti-GFAP monoclonal antibody. MSUT2 overexpression exacerbated astrocyte reactivity compared to GFP overexpression. Right: Densitometry analysis of GFAP immunoreactivity in Tau4RTg2652 mice overexpressing MSUT2 (n = 14) compared to GFP (n = 5) (P = 0.022 by two-tailed t test). Scale bars, 100 μm.
(E) Representative brain sections of the entorhinal cortex of PS19 transgenic (Tg) mice analyzed for astrocytosis by immunohistochemical staining with anti-GFAP monoclonal antibody. Right: Densitometry analysis of GFAP reactivity in the entorhinal cortex of Msut2 KO/PS19 Tg mice (n = 23) compared to PS19 Tg mice (n = 23) (P = 0.0104 by two-tailed t test). Scale bars, 100 μm. Photomicrographs in (A) to (E) were modified by adjusting brightness and contrast to optimize visualization of staining.
Neuroinflammation, as evidenced by activated microglia and reactive astrocytes, occurs in association with pathological tau in tauopathies including AD. We examined whether MSUT2 overexpression exacerbated gliosis in response to pathological tau in the mild nonprogressive Tau4RTg2652 mouse model of tauopathy. We observed marked neuroinflammatory changes in the hippocampus of MSUT2-overexpressing Tau4RTg2652 mice, including increased microgliosis as detected by immunostaining with IBA1 antibody (a 19-fold increase in microgliosis with MSUT2 overexpression; P = 0.024 by two-tailed t test; Fig. 4C and fig. S6F). Likewise, increased astrocytosis as detected by GFAP immunostaining occurred in MSUT2-overexpressing Tau4RTg2652 mice (a 12-fold increase in astrocytosis; P < 0.02 by two-tailed t test; Fig. 4D and fig. S7A). To explore whether deletion of Msut2 could prevent the neuroinflammatory responses to tauopathy in the more severe PS19 tau transgenic mouse model of tauopathy, we immunostained mouse brain tissue for markers of astrocytosis and microgliosis. We saw decreased astrocytosis in Msut2 KO/PS19 mice compared to PS19 mice with normal MSUT2 expression (an about twofold decrease; P < 0.010 by two-tailed t test; Fig. 4E and fig. S7B). Furthermore, we observed a trend toward decreased microgliosis as detected by IBA1 staining (fig. S8, A to C). However, Msut2 KO/PS19 mice were not obviously protected from neuroinflammatory changes induced by an exogenous chemical trigger such as kainic acid (fig. S9).
Neuronal depletion of the MSUT2/PABPN1 complex occurs in AD and predicts age at disease onset
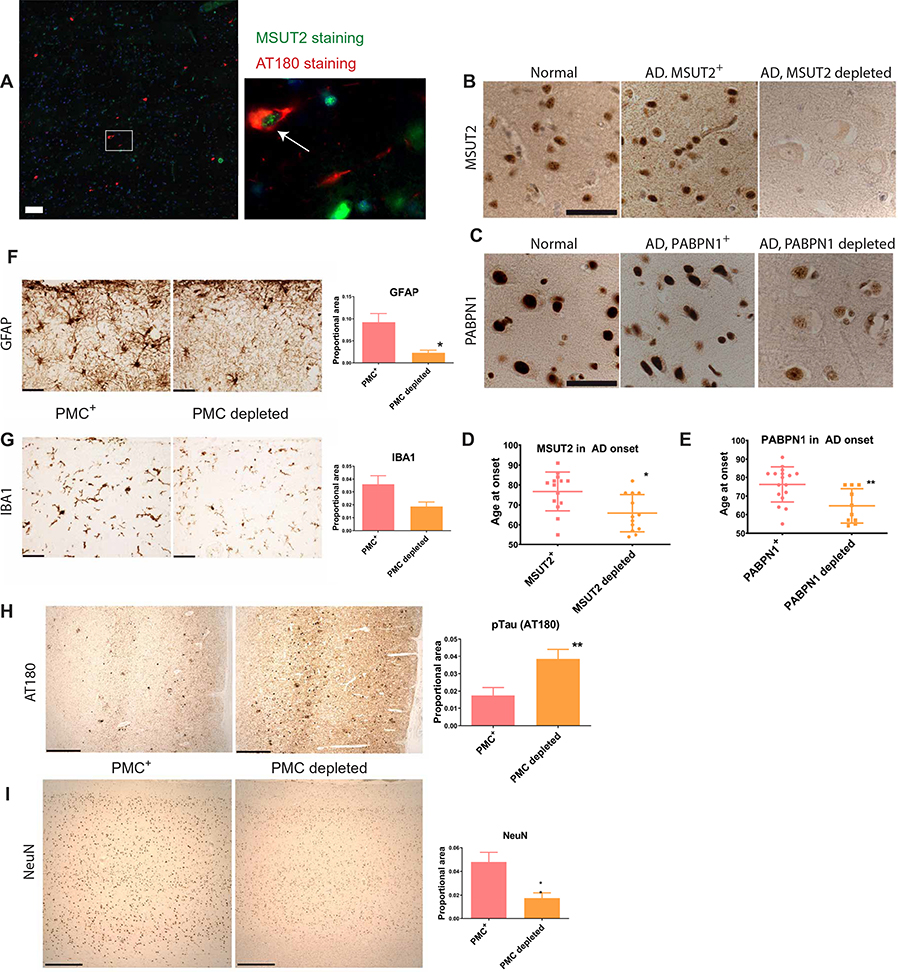
To investigate the involvement of MSUT2 in a human disease, we examined MSUT2 protein expression in postmortem human brain tissue from normal control individuals and AD cases. We observed that in the frontal cortex of postmortem brain tissue from AD subjects, MSUT2-positive neurons harbored pathological tau species without obvious colocalization of nuclear MSUT2 protein and cytoplasmic tau deposits (Fig. 5A). To investigate how MSUT2 expression might influence tau pathology in AD, we characterized MSUT2 protein expression by immunostaining of postmortem brain tissue sections from a cohort of 32 AD cases (table S4). In about half of the AD cases, we observed diminished MSUT2 expression compared to control brain tissue, whereas the remainder exhibited similar amounts of MSUT2 compared to control brain tissue (Fig. 5B). This difference was not due to variability in the postmortem interval between death and brain tissue collection (fig. S8D and table S4). We then classified our AD cases into two groups, MSUT2 normal and MSUT2 depleted, on the basis of MSUT2 immunostaining. In the subset of AD cases with depleted MSUT2 protein, the number of MSUT2-positive neurons was much lower (Fig. 5B and table S4).
Fig. 5. MSUT2 activity potentiates neuroinflammation induced by tau-opathy in human brain.
(A) Representative brain section of postmortem brain frontal cortex from an AD case that was coimmunostained for pathological tau (red) and MSUT2 (green). White arrow indicates an MSUT2-positive neuron harboring tau tangles. Pathological tau was detected by immunofluorescence using the AT180 monoclonal antibody (86), and MSUT2 was detected using a polyclonal anti-MSUT2 antibody (32). Scale bar, 100 μm.
(B) Representative brain sections of postmortem brain cortex from two AD cases and a neurologically normal control immunostained for MSUT2 (brown). Middle: AD cortex brain section with abundant MSUT2 protein. Right: AD cortex brain section with undetectable MSUT2 protein (see table S4 for characteristics of AD cases). Scale bar, 50 μm.
(C) Representative brain sections of postmortem brain cortex from two AD cases and a neurologically normal control immunostained for PABPN1 (brown). PABPN1 protein was abundant in the normal control and one AD case (middle), whereas the other AD case had greatly reduced PABPN1. Scale bar, 50 μm.
(D) Different amounts of MSUT2 in AD postmortem brain tissue depicted as a function at age at disease onset. AD cases with normal amounts of MSUT2 in postmortem brain (n = 14) had a later age of disease onset compared to those with depleted MSUT2 (n = 13) (P = 0.0306 by Student’s t test).
(E) Different amounts of PABPN1 in AD postmortem brain tissue depicted as a function of age at disease onset. AD cases with normal amounts of PABPN1 in postmortem brain (n = 16) had a later age of disease onset compared to those with depleted PABPN1 (n = 11) (P = 0.0084 by Student’s t test).
(F) Representative sections from postmortem brain cortex of an AD case with a normal amount of PABPN1/MSUT2 complexes (PMC+) compared to an AD case depleted of PABPN1/MSUT2 complexes (PMC depleted). Brain sections were stained with anti-GFAP monoclonal antibody to detect activated astrocytes. Right: Densitometry analysis of GFAP-positive astrocyte reactivity in PMC-normal AD cases (n = 9) compared to PMC-depleted AD cases (n = 6) (P = 0.0139 by two-tailed t test). Scale bars, 50 μm.
(G) Representative brain sections from postmortem brain frontal cortex of an AD case with a normal amount of PABPN1/MSUT2 complexes (PMC+) compared to an AD case depleted of PABPN1/MSUT2 complexes (PMC-depleted). Brain sections were stained with anti-IBA1 monoclonal antibody to detect microgliosis. Right: Densitometry analysis of IBA1 reactivity in PMC-positive AD cases (n = 9) compared to PMC-depleted AD cases (n = 6) (P = 0.069 by two-tailed t test). Scale bars, 50 μm.
(H) Representative brain sections from postmortem brain cortex of an AD case with a normal amount of PABPN1/MSUT2 complexes in the brain (PMC+) compared to an AD case depleted of PABPN1/MSUT2 complexes in the brain (PMC-depleted). Brain tissue from PMC-depleted AD cases exhibited more pathological tau as shown by immunostaining with the anti-phosphorylated tau antibody AT180. Right: Densitometry analysis of AT180-positive reactivity in PMC-positive AD cases (n = 14) compared to PMC-depleted AD cases (n = 13) (P = 0.0089 by two-tailed t test). Scale bars, 500 μm.
(I) Representative brain sections from postmortem brain cortex of an AD case with a normal amount of PABPN1/MSUT2 complexes in the brain (PMC+) compared to an AD case depleted of PABPN1/MSUT2 complexes in the brain (PMC-depleted). Brain tissue from PMC-depleted AD cases exhibited decreased NeuN immunoreactivity (indicative of more neuronal loss) than did brain tissue from PMC-positive AD cases. Right: Densitometry analysis of NeuN reactivity in PMC-positive AD cases (n = 14) compared with PMC-depleted AD cases (n = 13) (P = 0.0039 by two-tailed t test). Scale bars, 500 μm. Photomicrographs in (B), (C), and (F) to (I) were modified by adjusting brightness and contrast to optimize visualization of staining.
To explore whether PABPN1 also varied in AD, we examined PABPN1 expression and observed depleted PABPN1 in neuronal nuclei in postmortem brain tissue from a subset of AD cases (Fig. 5C and table S4). Depletion of PABPN1 correlated with depletion of MSUT2 (Spearman correlation coefficient = 0.71, P = 0.0003; table S4). These findings suggested that there was depletion of the molecular complex containing PABPN1 and MSUT2 proteins bound to poly(A) RNA in a subset of AD cases. To explore whether the co-depletion of PABPN1 and MSUT2 might influence clinical AD, we examined the relationship between age at onset and either MSUT2 or PABPN1 depletion in neuronal nuclei of AD postmortem brain tissue. We observed that AD cases with depleted brain MSUT2 protein exhibited a relatively younger age of disease onset (P = 0.031; Fig. 5D). Likewise, AD cases exhibiting nuclear depletion of PABPN1 also exhibited an earlier age of disease onset (P = 0.008; Fig. 5E).
Depletion of the PABPN1/MSUT2 complex exacerbates disease severity in AD
Previous investigations by others have demonstrated a clear relationship between pathological tau burden and severity of dementia (44, 45). However, it is unknown whether depletion of the PABPN1/MSUT2 complex correlates with pathological neurodegenerative changes in AD. To further explore the relationship between the PABPN1/MSUT2 complex and severity of neuroinflammation, we examined frontal cortex tissue in our AD postmortem brain tissue collection for markers of astrocytosis and microgliosis. We observed a significant decrease in astrocytosis in the frontal cortex of AD brain tissue showing depletion of the PABPN1/MSUT2 complex (P < 0.0139 by two-tailed t test; Fig. 5F). This was consistent with our observations in Msut2 KO/PS19 mice (Fig. 4E and fig. S7B). A parallel trend for decreased microgliosis was also evident in AD brain tissue depleted of PABPN1/MSUT2 complexes (~52% decrease; P = 0.069 by two-tailed t test; Fig. 5G). This decrease in microgliosis was also observed in Msut2 KO/PS19 mice (fig. S8, A to C). Like-wise, we observed a significant increase in pTau burden (P = 0.0089 by two-tailed t test; Fig. 5H) and increased neuronal loss detected by NeuN staining (P = 0.0039 by two-tailed t test; Fig. 5I) in AD brain tissue depleted of PABPN1/MSUT2 complexes. Loss of Msut2 alone did not provoke neurodegeneration in the mouse brain (fig. S2C). It seems that loss of PABPN1/MSUT2 complexes associated with poly(A) RNA may have exacerbated disease severity in AD brain tissue, as pathological tau species increased when PABPN1 was depleted in cultured cells, resulting in a concomitant reduction in MSUT2 (Fig. 3 and fig. S5).
DISCUSSION
Pathological accumulation of abnormal tau occurs in normal aging and, to a much greater extent, in AD and other related tauopathies. Abnormal assembly of tau and other aggregating proteins represents a toxic gain of function disrupting neuronal proteostasis in neurodegenerative disease. In AD specifically, accumulation of tau-positive lesions drives cognitive decline and neuronal loss (44, 45). We hypothesized that MSUT2, the mammalian homolog of sut-2, plays a central role in tauopathy disorders by determining neuronal susceptibility to pathological tau accumulation.
We demonstrated that MSUT2 influences tauopathy-related phenotypes in mammals. Mice lacking MSUT2 exhibited reduced accumulation of phosphorylated and aggregated human tau driven by a human tau transgene. Likewise, Msut2 KO mice were protected from the cognitive impairment and neuronal loss caused by neuropathological tau species. Msut2 KO mice were also protected from neuroinflammatory changes in response to tauopathy as indicated by reduced astrocytosis. In contrast, introduction of excess MSUT2 exacerbated normally mild and nonprogressive tauopathy phenotypes caused by a wild-type human tau transgene. MSUT2 overexpression in both nucleus and cytoplasm of neurons caused elevation of pTau accumulation leading to loss of neurons. MSUT2 has a nuclear export signal and is not normally seen in the cytoplasm, suggesting that MSUT2 mislocalization may drive pathological tau accumulation through a direct but possibly supraphysiological mechanism. Regardless, MSUT2 overexpression also exacerbated neuroinflammation, as markers of both microgliosis and astrocytosis were elevated in brain regions with increased MSUT2 protein. Our findings in tauopathy mouse models reveal that neuronal MSUT2 determines both accumulation of pathological tau species and neuronal vulnerability (fig. S10A).
In our model systems, the mechanism of MSUT2 modulation of tauopathy involved the nuclear RNA binding functions of MSUT2. We demonstrated that MSUT2 bound both to poly(A) RNA and to another nuclear poly(A) binding protein, PABPN1. We showed that MSUT2 and PABPN1 colocalized together with poly(A) RNA in nuclear speckles, forming a macromolecular complex. Furthermore, the constituents of this complex were co-depleted in postmortem brain tissue from AD cases with earlier disease onset or more extensive tau pathology. Others showed that PABPN1 and MSUT2 have opposing effects on poly(A) tail length (36, 46). We found that MSUT2 and PABPN1 also functioned together to influence tauopathy. MSUT2 normally promotes tau aggregation, whereas PABPN1 normally promotes clearance of aggregated tau. To test the linkage between RNA polyadenylation state and susceptibility to tauopathy, we inhibited the predominant poly(A) nuclease in our C. elegans model of tauopathy and found that increasing poly(A) tail length did not ameliorate tauopathy. These findings implicate functions of MSUT2 and PABPN1 other than mediation of poly(A) tail length as the critical control point in tau proteostasis.
How does the RNA binding function of MSUT2 affect tau pathology? We initially hypothesized that loss of MSUT2 function ameliorated tauopathy through changes in gene expression. However, RNA-seq analysis of brain tissue from Msut2 KO mice revealed only limited changes in gene expression, alternative splicing, or alternative polyadenylation site selection. Another hypothesis implicates RNA stress granules in the process of pathological protein aggregation whereby poly(A)+ mRNA becomes recruited to stress granules containing poly(A) binding proteins (47). In AD, RNA stress granule assembly factors TIA1 and G3BP overlap with tau-positive lesions in neurons (48, 49). TIA1 participates in RNA stress granule assembly, binds to poly(A) RNA, and has similarity to other poly(A) binding proteins (50, 51). Thus, one possible mechanism of MSUT2 action could be through promotion of stress granule formation leading to increased tau aggregation. Formation of tau/RNA coacervates, the liquid-liquid phase separation of tau/RNA complexes, has recently been suggested as a mechanism for initiating tau fibrillization (52, 53). Another possible mechanism could involve a direct tau-RNA interaction whereby RNA, as a polyanion, can both sequester tau from its function as a tubulin-binding protein and potentially promote pathological aggregation (53, 54). An alternative hypothesis incorporates the relationship between microtubule dynamics and polyadenylation. Previous work demonstrated that in tau transgenic mice, pathological tau causes microtubule hyperdynamicity (extensive microtubule growth and recurrent microtubule collapse) (55). Tau pathology and consequent neurocognitive deficits could be ameliorated by stabilization of microtubules (30, 31, 55). Previous investigation of the role of mRNA polyadenylation showed that defects in any one of several polyadenylation factors limits polyadenylation and causes microtubule destabilization (56). Thus, we hypothesize that the effects of Msut2 loss on polyadenylation may restore microtubule stability, thereby modulating tauopathy phenotypes (fig. S10B).
Whereas investigation of the molecular and cellular functions of MSUT2 has received attention in the context of RNA metabolism, investigation of its overall importance in brain function remains underdeveloped. However, its abundant expression in neurons suggests that it may play a role in brain function. Recent reports implicate MSUT2 in learning and cognitive function in flies and mice (36, 46, 57). Furthermore, our data suggest that MSUT2 may play an important role in mediating tauopathy-induced cognitive dysfunction. Nevertheless, the precise relationship between MSUT2 function, poly(A) RNA, and cognition remains unclear. The nuclear polyadenylation machinery and regulators of cytoplasmic mRNA polyadenylation also regulate synaptic function and memory (58, 59). For instance, the cytoplasmic polyadenylation element RNA binding protein (CPEB) has prion-like protein aggregation properties and influences poly(A) abundance through synaptic modification (60). Likewise, the RNA binding protein Cst-64 binds to RNA and can influence learning and behavior in mice (61). Collectively, poly(A) binding proteins and regulatory factors affect cognition, and the co-depletion of MSUT2 and PABPN1 in AD suggests that poly(A) binding protein homeostasis becomes disrupted in a subset of AD cases.
In the human brain, MSUT2 protein expression occurs primarily in neurons. In postmortem brain tissue from AD cases, we observed many tangle-bearing neurons that were MSUT2 positive. Extensive characterization of MSUT2 expression revealed that in about half of AD cases examined, MSUT2 protein was markedly reduced. MSUT2 depletion occurred more often in AD cases with earlier disease onset and more extensive tau pathology, suggesting the possible loss of MSUT2-positive neurons. In the same AD cases with depleted MSUT2, PABPN1 was also reduced, suggesting that depletion of the MSUT2/PABPN1 complex occurred in earlier onset AD. Whereas it is difficult to discern functional relationships between postmortem examination of pathological tau and underlying disease mechanisms, we hypothesize that MSUT2-positive neurons may die when challenged with pathological tau. Thus, AD cases with earlier disease onset could exhibit more extensive pathological tau and loss of MSUT2-positive neurons, whereas cases with later disease onset and milder tauopathy could show a sparing of MSUT2-positive neurons. Our experiments overexpressing MSUT2 in mouse neurons demonstrated a marked exacerbation of tau pathology, consistent with our hypothesis that MSUT2 activity determines sensitivity to neuronal challenge by pathological tau.
Although the relationship between tau pathology and neuroinflammation remains poorly understood, pathological tau provokes neuroinflammation. Likewise, increasing neuroinflammation frequently exacerbates tauopathy. It remains less clear whether an adaptive form of neuroinflammation can mediate clearance of pathological tau and promote neuronal survival. Here, we show that MSUT2 function promotes the brain’s neuroinflammatory response to pathological tau. Deletion of Msut2 leads to a decrease in astrocytosis and microgliosis in mice with tauopathy. Overexpression of MSUT2 in neurons in mouse brain provokes additional astrocytosis and microgliosis in mice with tauopathy. Previous work showed that poly(A) RNA can modulate inflammatory pathways (62). Thus, MSUT2 function may drive susceptibility to tauopathy and gliosis by changing the neuroinflammatory response to tau pathology. Postmortem AD brain tissue with a higher number of PABPN1/MSUT2 complexes came from AD cases with a later disease onset in the face of a more robust neuroinflammatory response. Conversely, postmortem AD brain tissue with a reduced number of PABPN1/MSUT2 complexes came from AD cases with an earlier disease onset and a weak neuroinflammatory response. Together, these observations suggest that MSUT2 through the PABPN1/MSUT2 complex could promote an adaptive neuroinflammatory response to AD neuropathology.
Our study has a number of limitations. The primary limitation was the use of human tau protein expression to drive pathological tau accumulation and neurodegeneration in mouse brain. An additional potential limitation is the genomic divergence of species used to study tauopathy, as the function of sut-2 in tauopathy may not be fully conserved between C. elegans and mammals. To overcome these limitations, we examined human brain autopsy tissue from AD cases to study the relationship between MSUT2, PABPN1, and neuropathology in AD. Nonetheless, studies of human postmortem brain tissue also have their limitations, as inference of cause and consequence in disease pathogenesis is not feasible from examination of a single endpoint in AD neuropathology. Despite advancing our understanding of tauopathy, our study leaves many open questions about tau toxic species and precise molecular mechanisms of tau-mediated neurodegeneration.
Our work with MSUT2 demonstrates that molecular disease mechanisms of tauopathy remain conserved from C. elegans to humans. In mice, reduced activity of MSUT2 protects neurons against abnormal tau, whereas increased MSUT2 activity exacerbates tauopathy. Analysis of MSUT2 function implicates RNA polyadenylation machinery as a critical node of cellular function in tauopathy. Likewise, we present evidence that MSUT2 activity potentiates the neuroinflammatory response to pathological tau. Given the current lack of treatments for tauopathies, strategies for therapeutically targeting abnormal tau and approaches targeting RNA processing warrant further investigation. Reducing MSUT2 function may have potential as a therapeutic approach for treating tauopathies because complete loss of this protein appears to have little effect on mouse development or function. Given that tau accumulation in AD appears to occur concomitant with amyloid plaque deposition, it may be possible to stop the progression of neurodegeneration even after the Aβ-initiated AD process has begun by targeting tau through MSUT2.
MATERIALS AND METHODS
Study design
Our objective in conducting this study was to investigate the genetic and molecular involvement of MSUT2 in tauopathy. Previous work has suggested that MSUT2 expression modulates susceptibility to pathological tau in simple model organisms (24, 32, 63). This study evaluated the impact of MSUT2 on pathological tau deposition in rodents and in postmortem brain tissue from AD cases. All mice (131 in total) had the C57BL/6J genetic background. Msut2 KO mice were generated by the Knockout Mouse Project (KOMP) using standard knockout mouse technologies (64). The PS19 tau transgenic mouse model expressing human P301S mutant human tau was used for its well-characterized and highly progressive tauopathy-related phenotypes (27). A second, less severe tauopathy model was also chosen on the basis of its mild, consistent, and nonprogressive tauopathy phenotype driven by wild-type human tau (43). Animals were assigned to experimental (Msut2 KO) and control (Msut2 +/+) groups based on genotype, with no animals of the appropriate genotype excluded from the study. Experimental group sample sizes for neuropathology were based on previous pathological findings (27, 43). Formalin-fixed paraffin-embedded (FFPE) tissue sections from each mouse brain were collected using standard histological techniques (see below). For each experiment shown for mice, representative sections from each experimental animal were immunostained and visualized by standard light microscopy. Histology was analyzed under identical conditions for both experimental and control brain tissue sections. Except where indicated, analyses were performed unblinded using identical experimental procedures for both imaging and analysis. Where necessary, image adjustments applied linear contrast and brightness changes. For behavioral analysis, group sizes were based on previously published findings with these models (28, 43). For RNA-seq analysis, RNA isolated from brains of Msut2 KO mice was compared to wild-type C57BL/6 control mice. All animals were studied in groups assembled based on genotype. All animal experiments were conducted under the supervision of the VA Puget Sound Health Care System Institutional Animal Use and Care Committee and used approved protocols and standard operating procedures.
We obtained samples of postmortem brain tissue from the University of Washington Alzheimer’s Disease Research Center (ADRC) Neuropathology Core (PI, C. Dirk Keene) after receiving human subjects approval (University of Washington Human Subjects Division approval: HSD no. 06–0492-E/A 01). Human postmortem brain samples were also obtained from the brain bank at the Center for Neurodegenerative Disease Research, University of Pennsylvania (www.med.upenn.edu/cndr/biosamples-brainbank.html). AD cases were selected on the basis of having an autopsy-confirmed diagnosis of AD (Braak stage V or VI with a CERAD score of moderate or frequent) (65). Control postmortem brain samples were from neurologically healthy participants, who were of a similar age but with a low AD pathology score (Braak stage III or less and CERAD score of none or sparse). Postmortem brain specimens from a total of 31 AD cases were subjected to histological and neuropathological analysis. Evaluation of tissue specimens was conducted using the same workflows as outlined for the rodent experiments.
Construction of knockout mice
B6-Zc3h14tm1a(KOMP)Wtsi mice were generated from a KOMP ES cell line as described (64). B6-Zc3h14tm1a animals were crossed with strains carrying the Flp and Cre recombinases [129S4/SvJaeSor-Gt (ROSA)26Sortm1(FLP1)Dym/J and B6.129S4-Meox2tm1(cre)Sor/J, respectively] to remove exon 13 of the Msut2 gene as well as all inserted transgene sequences. PS19 animals [B6-Tg(Prnp-MAPT*P301S)PS19Vle] were backcrossed to C57BL/6 for >10 generations. Tau4RTg2652 tauopathy model mice were generated as described (43). Briefly, the cDNA encoding the most abundant brain isoform (1N4R) of tau was cloned into a mouse neuron-specific expression vector, pThy1.2 (66). Transgenic mice were generated by pronuclear microinjection of the Thy1.2::Tau (1N4R) transgene at the University of Washington Nathan Shock Center Transgenic Animal Model Core (Warren Ladiges, PI). Founder mice were intercrossed with C57BL/6J mice to establish lines. The Tau4RTg2652 line was the focus of characterization due to its high-level tau expression and robust phenotype. Mice from the Tau4RTg2652 line used in these studies were backcrossed 10 generations (incipient congenic) with the C57BL/6J strain. This mouse strain has been deposited with the Mutant Mouse Regional Resource Centers (MMRRC) and is available under the stock number MMRRC:036717 and the strain name of B6.Cg-Tg(Thy1-MAPT*)2652Gds.
Immunohistochemistry and histological staining
Immunohistochemistry was performed on paraffin-embedded frontal cortex sections from over 31 AD cases. In addition, the following transgenic mouse lines were examined by immunohistochemistry: 9-month-old PS19 Tg mice (n = 24), Msut2 KO/PS19 Tg mice (n = 24), 4-month-old Tau4RTg2652 mice injected with AAV-Msut2 (n = 15), or AAV-Gfp (n = 5). Mice were anesthetized and fixed by transcardial perfusion with 4% paraformaldehyde. Brains were removed and paraffin-embedded for sectioning. Coronal sections from the hippocampus were cut and stored at 4°C until use. Human and mouse brain sections were deparaffinized and rehydrated through alcohols, and an antigen retrieval step consisting of heat pretreatment by microwave or autoclave in citrate buffer was used when necessary. Sections were treated for endogenous peroxidases with 3% hydrogen peroxide in PBS (pH 7.4), blocked in 5% nonfat milk in PBS, and incubated with one of the following primary antibodies overnight at 4°C: MC1 (67), AT180 (Thermo Fisher Scientific, catalog no. MN1040), GFAP (Dako Cytomation, Z0334), IBA1 (Wako, 019–19741), MSUT2 (24), PABPN1 (Abcam, ab81224), tau [Rb17025, (68)], or NeuN (Millipore, MAB377). Biotinylated secondary antibody was applied for 45 min at room temperature. Last, sections were incubated in an avidin-biotin complex (Vector’s Vectastain Elite ABC kit, Burlingame, CA), and the reaction product was visualized with 0.05% diaminobenzidine (DAB)/0.01% hydrogen peroxide in PBS. Negative controls with secondary antibody alone did not immunostain tissue sections. The presence of NFTs was assessed by Gallyas silver staining using standard methods (69). In addition, cresyl violet staining was performed to assess neuronal loss.
For double-label immunofluorescence of AT180 and MSUT2 in human AD brain, Alexa Fluor 488 goat anti-rabbit and Alexa Fluor 594 goat anti-mouse secondary antibodies (Molecular Probes) were used and autofluorescence was quenched with 0.1% Sudan Black.
For quantitation, stained sections were analyzed using the computerized image analysis system, MicroComputer Imaging Device (MCID; Imaging Research, St. Catherines, Ontario, Canada). Blinded assessment of optical density measurements was obtained relative to the proportional area for AT180, MC1, GFAP, IBA1, NeuN, and cresyl violet staining in frontal cortex and hippocampus. Data were averaged and are represented as means ± SEM. A two-tailed Student’s t test was used to assess differences in staining intensity between experimental groups.
Immunohistochemistry photomicrographs were taken with a digital camera and imported into Adobe Photoshop. To optimize visualization of staining, photomicrographs were modified when necessary by adjusting brightness and contrast. Fluorescent and immunofluorescent microscopy was performed on a Delta Vision microscope (GE Inc.) using a 60× oil immersion objective, a sCMOS camera, and 2 × 2 binning. Image analysis was performed using softWoRx 6.0 Beta software (GE Inc.).
Rodent behavioral analysis
All mice were bred and housed at the VAPSHCS animal facility, and all experiments were approved by the IACUC committee. Mice were housed on a 12:12 light cycle in static microisolator cages, with rodent chow and water available ad libitum. Environmental enrichment was added at every cage change in accordance with VAPSHCS IACUC mouse care guidelines. All behavioral experiments were conducted during the light phase.
Open field testing was conducted in a 30-inch-diameter circular plastic arena. All animals were moved into the testing room about 1 hour before start of testing. The arena was cleaned with 70% ethanol between animals. Total locomotor activity was measured for 10 min using SMART behavioral tracking system (San Diego Instruments).
The Barnes maze is a circular white plastic platform 36 inches in diameter with 20 holes around the perimeter. One hole leads to a dark target escape box containing clean bedding material. Latencies to locate and to enter the escape box were recorded. Spatial cues (blue circle, yellow triangle, and black/white striped square) were placed around the maze to be used as landmarks to find the escape box. Mice are placed in a clear box in the center of the maze for 30 s at the beginning of each trial, so the direction each mouse faces at the trial start is random. When the start box is removed, each animal has a maximum of 2 min to locate the escape box. If the animal locates the hole, it is allowed to shelter there for 1 min before being returned to the home cage. If the animal fails the task, it is gently guided to the escape box by the investigator and allowed to shelter there for 1 min. All animals were moved into the testing room about 1 hour before start of testing, and the maze was cleaned with 70% ethanol between animals. Training trials were conducted twice daily on two consecutive days.
Rotarod testing was conducted using an accelerating UGO Basile RotaRod. The rod surface is covered with ridged plastic, located 18 inches above a padded floor. All mice were transferred to the testing room about 1 hour before the start of testing and were tested on four consecutive days. On the first 3 days, all mice received three training trials separated by a minimum of 30-min rest time. Animals were placed onto a stationary rotarod and allowed to acclimate for a minimum of 10 s before accelerating the rod up to 8 rpm over 2 min on all training trials. On the final test day, all animals were tested in four trials with acceleration up to 8, 16, 24, and 32 rpm.
Cell culture, RNA interference, and transient transfections
HEK293 and HEK/tau cells (Zeocin, 100 μg/ml) were cultured under standard tissue culture conditions [DMEM, 10% defined fetal bovine serum, penicillin (1000 IU/ml), and streptomycin (1000 μg/ml)] as previously described (26, 32). RNA interference and plasmid transient transfections were conducted following the manufacturer’s protocol (RNAiMAX, Invitrogen).
Immunofluorescent staining and fluorescent in situ hybridization
Cells were grown on poly-D-lysine-coated 12-mm round coverslips and fixed in 4% formaldehyde solution. Cells were washed three times for 5 min in PBS/Ca2+/Mg2+ and then blocked in antibody buffer (PBS, 0.5% Triton X-100, 1 mM EDTA, 0.1% BSA, and 0.05% NaN3) with 10% normal goat serum. Primary antibodies were applied and incubated for 1 hour at room temperature. Primary antibodies used were as follows: PABPN1 (catalog no. 10782–2-AP, Proteintech Group), MSUT2 (Rabbit 9857–2-AP) (32), oligomeric tau (TOC1) (70), MC1 (67), AT180 (Thermo Fisher Scientific, catalog no. MN1040), and SC35 (catalog no. S4045, Sigma-Aldrich). Cells were washed three times for 5 min in PBS/Ca2+/Mg2+ and then reblocked for 10 min. Appropriate Alexa dye-labeled secondary antibodies (Invitrogen) were applied and incubated for 20 min at room temperature. Cells were again washed three times for 5 min in PBS/Ca2+/Mg2+, counterstained with 300 nM DAPI, and mounted with ProLong Gold antifade (Molecular Probes). TOC1 staining was conducted with 10% rabbit serum in antibody buffer. Rabbit anti-mouse secondary antibody used for TOC1 was FITC-labeled (Invitrogen). Fluorescence in situ hybridization was conducted before counterstaining with DAPI as per the following: After secondary antibody staining, cells were washed three times for 5 min in PBS/Ca2+/Mg2+ and 5′ Alexa 647-labeled poly(T) oligonucleotide (100 pg/μl) (sequence = TTTTT TTTTT TTTTT TTTTT TTTTT TTTTT TTTTT TTTTT TTT; IDT) and incubated in the dark for 2 hours at 37°C. After hybridization, the samples were washed once in PBS/Ca2+/Mg2+ in the dark 30 min at 37°C. Proximity ligation assays were performed using Duolink PLA technology as recommended by the manufacturer (catalog no. DUO92101, Sigma-Aldrich). Human brain samples stained for MSUT2 and PABPN1 were imaged on a Leica TCS SP5 II confocal microscope using a 63× oil immersion objective. Image normalization and confocal image colocalization (Coloc 2 plugin) was conducted in ImageJ 1.51n (71).
Recombinant protein purification and in vitro binding assays
The MSUT2 ZF and PABPN1 protein expression constructs were prepared by inserting the MSUT2 ZF or PABPN1 cDNA into the pGEX6P-1 expression vector (Pharmacia) to generate a construct encoding a glutathione S-transferase (GST)-MSUT2 ZF or GST-PABPN1 fusion protein. The GST moiety allows one-step affinity purification of recombinant protein on glutathione-coupled Sepharose beads. A log phase culture of BL21 (DE3) cells carrying the pGEX-MSUT2 or PABPN1 vector was induced for 3 hours at 37°C with shaking. Escherichia coli lysates were treated with benzonase nuclease, which degrades both RNA and DNA before clearing and purification. Glutathione Sepharose (Pharmacia) was used as the affinity resin and purified via the method of Frangioni and Neel (72). In vitro protein-binding assays were performed essentially as described using 35S-labeled PABPN1 protein generated with TNT reticulocyte lysates (Promega Inc.); labeled PABPN1 was pretreated with RNase (73). Briefly, GST beads coupled to MSUT2 were blocked and then incubated with labeled PABPN1, both of which had been pretreated with nucleases to remove RNA. Unbound protein was repeatedly washed away, and then labeled proteins were eluted by boiling and analysis on SDS-PAGE.
Behavioral assay in C. elegans
Cultures were maintained at 20°C on standard 60-mm NGM plates containing OP50 E. coli as previously described (74). A timed egg lay using 10 to 20 gravid adults was performed, and worms were grown for 4 days at 20°C. To assess swimming behavior, a plate containing a synchronized population of worms was flooded with 1 ml of M9 buffer. The M9 solution containing C. elegans was pipetted onto an unseeded 35-mm NGM plate. One minute after the addition of M9 buffer, swimming worms were recorded using WormLab (MBF Bioscience) for 1 min at 14 frames per second. Videos of swimming worms were analyzed using WormLab 2017 software. Swimming behavior was quantified by measuring the number of turns made by a worm during the video. A turn was calculated using Bending Angles-Multiple and setting the sample angles to 3, the amplitude threshold to 20°, and the duration threshold to 2 frames (0.14 s). Data from the angle 2 (the angle made from the body quarterpoints and midpoint) were used. The number of turns a worm made was divided by the time a worm was tracked by the software to calculate turns per minute. Only worms that were analyzed for at least 20 s were kept in the analysis. At least 100 worms were analyzed per strain.
RNA-seq analysis
Raw reads from RNA-seq experiments had their 3′ Illumina TruSeq Indexed adaptors (AGATCGGAAGAGCACACGTCTGAACTCCAGTCAC) trimmed using cutadapt v1.8.3 with parameters “-e 0.06 -O 6 -m 14 -n 2 --length-tag ‘length’” (75). Reads were then mapped to the mm10 Mus musculus genome build using STAR version 2.4.1d with parameters “--outFilterMultimapNmax 1 --out-FilterMismatchNmax 2 --outFilterMismatchNoverReadLmax 0.06” (76) and were further filtered to uniquely mapping reads with a maximum mismatch rate of 0.06. The mapped reads from the RNA sequencing pipeline were analyzed for differential expression by using HTSeq with parameters “-s reverse -t exon -i gene_id” (77) to compute the number of reads mapping to each gene using Ensembl mm10 gene annotations (78). Gene counts were then analyzed for differential expression with the DESeq2 R package v1.0.19 (79). Splicing analysis was performed using the DEXSeq R package (80) following the protocol described in the paper and vignette. Briefly, exon reads were counted using the dexseq_count.py script and read into R v3.0.1 (81), where the DEXSeq analysis pipeline was followed using the GENCODE mm10 genome annotation. For both the differential expression and the differential splicing analyses, significantly differentially expressed genes/exons were defined as having an adjusted P value of less than 0.05 using the Benjamini-Hochberg multiple testing correction (82).
Alternative polyadenylation analysis
To perform alternative polyadenylation analysis, the DaPars tool was used (42). Ensembl gene annotations for the mm10 M. musculus genome were downloaded from the UCSC Table Browser (83), and the DaPars_Extract_Anno.py script was used to generate 3′ UTR annotations. Then, the RNA-seq data were converted to BedGraph format using the bedtools genomecov command (84). Last, the DaPars_main. py script was used to perform the alternative polyadenylation analysis.
Surface plasmon resonance
Surface plasmon resonance (SPR) experiments were conducted on a Biacore T200 using the Biotin Capture Kit Series S following the manufacturer’s instructions (GE). Biotin capture reagent was applied to the sensor chip at a flow rate of 2 μl/min for 300 s. Biotinylated RNA (1 nM) (BiotinTEG-AAAAAAAAAAAAAAA) was captured at a flow rate of 10 μl/min for 180 s. Samples were flowed across the sensor chip at 50 μl/min for 120 s, and the chip was regenerated for 120 s at a flow rate of 10 μl/min. KD values for binding and disassociation were calculated from the binding kinetics of each protein at five concentrations using the Biacore T200 Software v3.1 application package (1, 0.9, 0.8, 0.7, and 0.6 μM; GE).
Immunoblotting
Tau fractions were obtained as described (23). Protein samples were brought to 10 mM tris (pH 6.8), 1 mM EDTA, 40 mM DTT, 1% SDS, and 10% sucrose by addition of 5× sample buffer boiled for 5 min and loaded onto 4 to 15% precast criterion SDS-PAGE gradient gels (Bio-Rad). For immunoblotting, we detected human tau using antibody 17025 at a dilution of 1:6000 as described previously (24). We used anti-tubulin antibody at a dilution of 1:1000 (Developmental Studies Hybridoma Bank, catalog E7). MSUT2 antibody Rbt9857 was prepared as described above and used at a dilution of 1:1000. Secondary goat anti-mouse or goat anti-rabbit IgGs were the secondary antibody reagents used at a dilution of 1:1000 (GE Lifesciences). Signals were measured by densitometry using Adobe Photoshop.
Stereotaxic injections of AAV viral vector
AAV viral vector preparations expressing either MSUT2 or GFP under control of the chicken β-actin promoter were obtained from R. Klein at Louisiana State University. Intracerebral injections were performed using a stereotaxic apparatus (Parkland Scientific) as described in detail previously (85). Three-month-old male and female Tau4RTg2652 mice were anesthetized with isoflurane and received a single injection of buprenorphine SR (0.3 mg/kg) during surgery for extended pain relief support. All animals received a single unilateral injection of 1.0 μl of AAV9 (containing about 5 × 1011 viral vectors) into the right ventral hippocampus delivered over an 8-min period at the following coordinates: AP, −2.92 mm from bregma; ML, +2.80 mm; DV, −3.50 mm from the surface of the skull. The needle was left in place for an additional 4 min after the injection, and then the needle was slowly withdrawn. Animals were sutured, removed from the stereotaxic apparatus, and allowed to recover. Animals were sacrificed 1 month later for analysis of neuropathological changes.
Statistical analysis
Standard ANOVA or t tests were used (two-tailed) for statistical analysis. Statistical analysis was performed using the Statistica software package (StatSoft). For all measures, sex was included as a factor in the ANOVA. If sex did not have a significant effect on a particular measure, then the analysis was repeated without that factor. For repeated measures (rotarod and Barnes tests), ANOVA was followed up with planned comparisons of the two genotypes on each testing day.
Supplementary Material
Acknowledgments
We thank E. Loomis, A. Franklin, K. Chickering, S. Danner, T. Martin, and Samantha Rice for technical assistance. We thank A. Beller and N. Czajkiewicz for administrative support. The Msut2 KO mouse strain used for this research project was generated by KOMP and was obtained from the KOMP Repository (www.komp.org). We thank P. Davies for anti-tau antibodies. We thank J. Sumida and H. Won Kown at the Analytical Biopharmacy Core within the School of Pharmacy and the Molecular Engineering and Sciences Institute at the University of Washington for their assistance with surface plasmon resonance experiments. We thank A. Corbett for antibodies against mouse ZC3H14 and the Developmental Studies Hybridoma Bank (NICHD) for the anti-β-tubulin antibody E7.
Funding: This work was supported by grants from the Department of Veterans Affairs (Merit Review grant no. I01BX002619 to B.K. and Career Development Award 2 no. I01BX007080 to N.F.L.); NIH grant nos. RF1AG055474 and R01NS064131 (to B.K.), P01AG017856 [to project 2 (G.D.S. as PI) and project 4 (J.Q.T. as PI and V.M.Y.L. as program director)], and P50AG05136 (to C.D.K. for the UW ADRC); and the Nancy and Buster Alvord endowment (to C.D.K.).
Footnotes
Competing interests: G.D.S. has consulted for Biogen, Texas Alzheimer’s Disease Research and Care Consortium, and the Oxford Parkinson Disease Center. E.P. serves on the advisory boards of Avanir and Acadia. N.M.K. serves as a consultant and is a member of the Scientific Advisory Board for Tau biologic corporation. The other authors declare that they have no competing interests.
Data and materials availability: All data associated with this study are in the main text or the Supplementary Materials. Animal strains and cell lines will be made available upon request to B.K. through a materials transfer agreement.
REFERENCES AND NOTES
- 1.Poorkaj P, Bird TD, Wijsman E, Nemens E, Garruto RM, Anderson L, Andreadis A, Wiederholt WC, Raskind M, Schellenberg GD, Tau is a candidate gene for chromosome 17 frontotemporal dementia. Ann. Neurol 43, 815–825 (1998). [DOI] [PubMed] [Google Scholar]
- 2.Spillantini MG, Murrell JR, Goedert M, Farlow MR, Klug A, Ghetti B, Mutation in the tau gene in familial multiple system tauopathy with presenile dementia. Proc. Natl. Acad. Sci. U.S.A 95, 7737–7741 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clark LN, Poorkaj P, Wszolek Z, Geschwind DH, Nasreddine ZS, Miller B, Li D, Payami H, Awert F, Markopoulou K, Andreadis A, D’Souza I, Lee VM-Y, Reed L, Trojanowski JQ, Zhukareva V, Bird T, Schellenberg GD, Wilhelmsen KC, Pathogenic implications of mutations in the tau gene in pallido-ponto-nigral degeneration and related neurodegenerative disorders linked to chromosome 17. Proc. Natl. Acad. Sci. U.S.A 95, 13103–13107 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hutton M, Lendon CL, Rizzu P, Baker M, Froelich S, Houlden H, Pickering-Brown S, Chakraverty S, Isaacs A, Grover A, Hackett J, Adamson J, Lincoln S, Dickson D, Davies P, Petersen RC, Stevens M, de Graaff E, Wauters E, van Baren J, Hillebrand M, Joosse M, Kwon JM, Nowotny P, Che LK, Norton J, Morris JC, Reed LA, Trojanowski J, Basun H, Lannfelt L, Neystat M, Fahn S, Dark F, Tannenberg T, Dodd PR, Hayward N, Kwok JBJ, Schofield PR, Andreadis A, Snowden J, Craufurd D, Neary D, Owen F, Oostra BA, Hardy J, Goate A, van Swieten J, Mann D, Lynch T, Heutink P, Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature 393, 702–705 (1998). [DOI] [PubMed] [Google Scholar]
- 5.Bierer LM, Hof PR, Purohit DP, Carlin L, Schmeidler J, Davis KL, Perl DP, Neocortical neurofibrillary tangles correlate with dementia severity in Alzheimer’s disease. Arch. Neurol 52, 81–88 (1995). [DOI] [PubMed] [Google Scholar]
- 6.Khanna MR, Kovalevich J, Lee VM-Y, Trojanowski JQ, Brunden KR, Therapeutic strategies for the treatment of tauopathies: Hopes and challenges. Alzheimers Dement. 12, 1051–1065 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ballatore C, Lee VM-Y, Trojanowski JQ, Tau-mediated neurodegeneration in Alzheimer’s disease and related disorders. Nat. Rev. Neurosci 8, 663–672 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Hong M, Zhukareva V, Vogelsberg-Ragaglia V, Wszolek Z, Reed L, Miller BI, Geschwind DH, Bird TD, McKeel D, Goate A, Morris JC, Wilhelmsen KC, Schellenberg GD, Trojanowski JQ, Lee VM-Y, Mutation-specific functional impairments in distinct tau isoforms of hereditary FTDP-17. Science 282, 1914–1917 (1998). [DOI] [PubMed] [Google Scholar]
- 9.Barghorn S, Zheng-Fischhöfer Q, Ackmann M, Biernat J, von Bergen M, Mandelkow E-M, Mandelkow E, Structure, microtubule interactions, and paired helical filament aggregation by tau mutants of frontotemporal dementias. Biochemistry 39, 11714–11721 (2000). [DOI] [PubMed] [Google Scholar]
- 10.Hong M, Zhukareva V, Vogelsberg-Ragaglia V, Wszolek Z, Reed L, Miller BI, Geschwind DH, Bird TD, McKeel D, Goate A, Morris JC, Wilhelmsen KC, Schellenberg GD, Trojanowski JQ, Lee VM-Y, Mutation-specific functional impairments in distinct Tau isoforms of hereditary FTDP-17. Science 282, 1914–1917 (1998). [DOI] [PubMed] [Google Scholar]
- 11.Lee VM-Y, Daughenbaugh R, Trojanowski JQ, Microtubule stabilizing drugs for the treatment of Alzheimer’s disease. Neurobiol. Aging 15 (suppl. 2), S87–S89 (1994). [DOI] [PubMed] [Google Scholar]
- 12.Braak H, Braak E, Staging of Alzheimer’s disease-related neurofibrillary changes. Neurobiol. Aging 16, 271–278; discussion 278–284 (1995). [DOI] [PubMed] [Google Scholar]
- 13.Clavaguera F, Bolmont T, Crowther RA, Abramowski D, Frank S, Probst A, Fraser G, Stalder AK, Beibel M, Staufenbiel M, Jucker M, Goedert M, Tolnay M, Transmission and spreading of tauopathy in transgenic mouse brain. Nat. Cell Biol 11, 909–913 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y, Mandelkow E, Tau in physiology and pathology. Nat. Rev. Neurosci 17, 5–21 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Ballatore C, Brunden KR, Trojanowski JQ, Lee VM-Y, Smith III AB, Huryn DM, Modulation of protein-protein interactions as a therapeutic strategy for the treatment of neurodegenerative tauopathies. Curr. Top. Med. Chem. 11, 317–330 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sahara N, Maeda S, Takashima A, Tau oligomerization: A role for tau aggregation intermediates linked to neurodegeneration. Curr. Alzheimer Res 5, 591–598 (2008). [DOI] [PubMed] [Google Scholar]
- 17.Goedert M, The ordered assembly of tau is the gain-of-toxic function that causes human tauopathies. Alzheimers Dement. 12, 1040–1050 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Naj AC, Schellenberg GD; Alzheimer’s Disease Genetics Consortium (ADGC), Genomic variants, genes, and pathways of Alzheimer’s disease: An overview. Am. J. Med. Genet. B Neuropsychiatr. Genet 174, 5–26 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Komori T, Tau-positive glial inclusions in progressive supranuclear palsy, corticobasal degeneration and Pick’s disease. Brain Pathol. 9, 663–679 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maphis N, Jiang S, Xu G, Kokiko-Cochran ON, Roy SM, Van Eldik LJ, Watterson DM, Lamb BT, Bhaskar K, Selective suppression of the α isoform of p38 MAPK rescues late-stage tau pathology. Alzheimers Res. Ther 8, 54 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maphis N, Xu G, Kokiko-Cochran ON, Cardona AE, Ransohoff RM, Lamb BT, Bhaskar K, Loss of tau rescues inflammation-mediated neurodegeneration. Front. Neurosci 9, 196 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhaskar K, Konerth M, Kokiko-Cochran ON, Cardona A, Ransohoff RM, Lamb BT, Regulation of tau pathology by the microglial fractalkine receptor. Neuron 68, 19–31 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kraemer BC, Zhang B, Leverenz JB, Thomas JH, Trojanowski JQ, Schellenberg GD, Neurodegeneration and defective neurotransmission in a Caenorhabditis elegans model of tauopathy. Proc. Natl. Acad. Sci. U.S.A 100, 9980–9985 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guthrie CR, Schellenberg GD, Kraemer BC, SUT-2 potentiates tau-induced neurotoxicity in Caenorhabditis elegans. Hum. Mol. Genet 18, 1825–1838 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kraemer BC, Schellenberg GD, SUT-1 enables tau-induced neurotoxicity in C. elegans. Hum. Mol. Genet 16, 1959–1971 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Guthrie CR, Kraemer BC, Proteasome inhibition drives HDAC6-dependent recruitment of tau to aggresomes. J. Mol. Neurosci 45, 32–41 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoshiyama Y, Higuchi M, Zhang B, Huang S-M, Iwata N, Saido TC, Maeda J, Suhara T, Trojanowski JQ, Lee VM-Y, Synapse loss and microglial activation precede tangles in a P301S tauopathy mouse model. Neuron 53, 337–351 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Takeuchi H, Iba M, Inoue H, Higuchi M, Takao K, Tsukita K, Karatsu Y, Iwamoto Y, Miyakawa T, Suhara T, Trojanowski JQ, Lee VM-Y, Takahashi R, P301S mutant human tau transgenic mice manifest early symptoms of human tauopathies with dementia and altered sensorimotor gating. PLOS ONE 6, e21050 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barnes CA, Memory deficits associated with senescence: A neurophysiological and behavioral study in the rat. J. Comp. Physiol. Psychol 93, 74–104 (1979). [DOI] [PubMed] [Google Scholar]
- 30.Brunden KR, Zhang B, Carroll J, Yao Y, Potuzak JS, Hogan A-M, Iba M, James MJ, Xie SX, Ballatore C, Smith AB III, Lee VM-Y, Trojanowski JQ, Epothilone D improves microtubule density, axonal integrity, and cognition in a transgenic mouse model of tauopathy. J. Neurosci 30, 13861–13866 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang B, Carroll J, Trojanowski JQ, Yao Y, Iba M, Potuzak JS, Hogan A-M, Xie SX, Ballatore C, Smith AB III, Lee VM-Y, Brunden KR, The microtubule-stabilizing agent, epothilone D, reduces axonal dysfunction, neurotoxicity, cognitive deficits, and Alzheimer-like pathology in an interventional study with aged tau transgenic mice. J. Neurosci 32, 3601–3611 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guthrie CR, Greenup L, Leverenz JB, Kraemer BC, MSUT2 is a determinant of susceptibility to tau neurotoxicity. Hum. Mol. Genet 20, 1989–1999 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kühn U, Nemeth A, Meyer S, Wahle E, The RNA binding domains of the nuclear poly(A)-binding protein. J. Biol. Chem 278, 16916–16925 (2003). [DOI] [PubMed] [Google Scholar]
- 34.Banerjee A, Apponi LH, Pavlath GK, Corbett AH, PABPN1: Molecular function and muscle disease. FEBS J. 280, 4230–4250 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Söderberg O, Leuchowius KJ, Gullberg M, Jarvius M, Weibrecht I, Larsson L-G, Landegren U, Characterizing proteins and their interactions in cells and tissues using the in situ proximity ligation assay. Methods 45, 227–232 (2008). [DOI] [PubMed] [Google Scholar]
- 36.Kelly SM, Leung SW, Pak C, Banerjee A, Moberg KH, Corbett AH, A conserved role for the zinc finger polyadenosine RNA binding protein, ZC3H14, in control of poly(A) tail length. RNA 20, 681–688 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Soucek S, Corbett AH, Fasken MB, The long and the short of it: The role of the zinc finger polyadenosine RNA binding protein, Nab2, in control of poly(A) tail length. Biochim. Biophys. Acta 1819, 546–554 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ward SM, Himmelstein DS, Lancia JK, Fu Y, Patterson KR, Binder LI, TOC1: Characterization of a selective oligomeric tau antibody. J. Alzheimers Dis 37, 593–602 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jicha GA, Bowser R, Kazam IG, Davies P, Alz-50 and MC-1, a new monoclonal antibody raised to paired helical filaments, recognize conformational epitopes on recombinant tau. J. Neurosci. Res 48, 128–132 (1997). [DOI] [PubMed] [Google Scholar]
- 40.Nousch M, Techritz N, Hampel D, Millonigg S, Eckmann CR, The Ccr4-Not deadenylase complex constitutes the main poly(A) removal activity in C. elegans. J. Cell Sci 126, 4274–4285 (2013). [DOI] [PubMed] [Google Scholar]
- 41.Jenal M, Elkon R, Loayza-Puch F, van Haaften G, Kühn U, Menzies FM, Oude Vrielink JAF, Bos AJ, Drost J, Rooijers K, Rubinsztein DC, Agami R, The poly(A)-binding protein nuclear 1 suppresses alternative cleavage and polyadenylation sites. Cell 149, 538–553 (2012). [DOI] [PubMed] [Google Scholar]
- 42.Xia Z, Donehower LA, Cooper TA, Neilson JR, Wheeler DA, Wagner EJ, Li W, Dynamic analyses of alternative polyadenylation from RNA-seq reveal a 3′-UTR landscape across seven tumour types. Nat. Commun 5, 5274 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wheeler JM, McMillan PJ, Hawk M, Iba M, Robinson L, Xu GJ, Dombroski BA, Jeong D, Dichter MA, Juul H, Loomis E, Raskind M, Leverenz JB, Trojanowski JQ, Lee VM-Y, Schellenberg GD, Kraemer BC, High copy wildtype human 1N4R tau expression promotes early pathological tauopathy accompanied by cognitive deficits without progressive neurofibrillary degeneration. Acta Neuropathol. Commun 3, 33 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gómez-Isla T, Hollister R, West H, Mui S, Growdon JH, Petersen RC, Parisi JE, Hyman BT, Neuronal loss correlates with but exceeds neurofibrillary tangles in Alzheimer’s disease. Ann. Neurol 41, 17–24 (1997). [DOI] [PubMed] [Google Scholar]
- 45.Nelson PT, Alafuzoff I, Bigio EH, Bouras C, Braak H, Cairns NJ, Castellani RJ, Crain BJ, Davies P, Del Tredici K, Duyckaerts C, Frosch MP, Haroutunian V, Hof PR, Hulette CM, Hyman BT, Iwatsubo T, Jellinger KA, Jicha GA, Kövari E, Kukull WA, Leverenz JB, Love S, Mackenzie IR, Mann DM, Masliah E, McKee AC, Montine TJ, Morris JC, Schneider JA, Sonnen JA, Thal DR, Trojanowski JQ, Troncoso JC, Wisniewski T, Woltjer RL, Beach TG, Correlation of Alzheimer disease neuropathologic changes with cognitive status: A review of the literature. J. Neuropathol. Exp. Neurol 71, 362–381 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kelly SM, Bienkowski R, Banerjee A, Melicharek DJ, Brewer ZA, Marenda DR, Corbett AH, Moberg KH, The Drosophila ortholog of the Zc3h14 RNA binding protein acts within neurons to pattern axon projection in the developing brain. Dev. Neurobiol 76, 93–106 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, Scheuner D, Kaufman RJ, Golan DE, Anderson P, Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol 169, 871–884 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vanderweyde T, Apicco DJ, Youmans-Kidder K, Ash PEA, Cook C, Lummertz da Rocha E, Jansen-West K, Frame AA, Citro A, Leszyk JD, Ivanov P, Abisambra JF, Steffen M, Li H, Petrucelli L, Wolozin B, Interaction of tau with the RNA-binding protein TIA1 regulates tau pathophysiology and toxicity. Cell Rep 15, 1455–1466 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vanderweyde T, Yu H, Varnum M, Liu-Yesucevitz L, Citro A, Ikezu T, Duff K, Wolozin B, Contrasting pathology of the stress granule proteins TIA-1 and G3BP in tauopathies. J. Neurosci 32, 8270–8283 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tian Q, Streuli M, Saito H, Schlossman SF, Anderson P, A polyadenylate binding protein localized to the granules of cytolytic lymphocytes induces DNA fragmentation in target cells. Cell 67, 629–639 (1991). [DOI] [PubMed] [Google Scholar]
- 51.Kawakami A, Tian Q, Duan X, Streuli M, Schlossman SF, Anderson P, Identification and functional characterization of a TIA-1-related nucleolysin. Proc. Natl. Acad. Sci. U.S.A 89, 8681–8685 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wegmann S, Eftekharzadeh B, Tepper K, Zoltowska KM, Bennett RE, Dujardin S, Laskowski PR, MacKenzie D, Kamath T, Commins C, Vanderburg C, Roe AD, Fan Z, Molliex AM, Hernandez-Vega A, Muller D, Hyman AA, Mandelkow E, Taylor JP, Hyman BT, Tau protein liquid-liquid phase separation can initiate tau aggregation. EMBO J. 37, e98049 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang X, Lin Y, Eschmann NA, Zhou H, Rauch JN, Hernandez I, Guzman E, Kosik KS, Han S, RNA stores tau reversibly in complex coacervates. PLOS Biol. 15, e2002183 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bryan JB, Nagle BW, Doenges KH, Inhibition of tubulin assembly by RNA and other polyanions: Evidence for a required protein. Proc. Natl. Acad. Sci. U.S.A 72, 3570–3574 (1975). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barten DM, Fanara P, Andorfer C, Hoque N, Wong PYA, Husted KH, Cadelina GW, Decarr LB, Yang L, Liu V, Fessler C, Protassio J, Riff T, Turner H, Janus CG, Sankaranarayanan S, Polson C, Meredith JE, Gray G, Hanna A, Olson RE, Kim S-H, Vite GD, Lee FY, Albright CF, Hyperdynamic microtubules, cognitive deficits, and pathology are improved in tau transgenic mice with low doses of the microtubule-stabilizing agent BMS-241027. J. Neurosci 32, 7137–7145 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cappell KM, Larson B, Sciaky N, Whitehurst AW, Symplekin specifies mitotic fidelity by supporting microtubule dynamics. Mol. Cell. Biol 30, 5135–5144 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rha J, Jones SK, Fidler J, Banerjee A, Leung SW, Morris KJ, Wong JC, Inglis GAS, Shapiro L, Deng Q, Cutler AA, Hanif AM, Pardue MT, Schaffer A, Seyfried NT, Moberg KH, Bassell GJ, Escayg A, Garcia PS, Corbett AH, The RNA-binding protein, ZC3H14, is required for proper poly(A) tail length control, expression of synaptic proteins, and brain function in mice. Hum.Mol. Genet 26, 3663–3681 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Du L, Richter JD, Activity-dependent polyadenylation in neurons. RNA 11, 1340–1347 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kwak JE, Drier E, Barbee SA, Ramaswami M, Yin JCP, Wickens M, GLD2 poly(A) polymerase is required for long-term memory. Proc. Natl. Acad. Sci. U.S.A 105, 14644–14649 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Si K, Giustetto M, Etkin A, Hsu R, Janisiewicz AM, Miniaci MC, Kim J-H, Zhu H, Kandel ER, A neuronal isoform of CPEB regulates local protein synthesis and stabilizes synapse-specific long-term facilitation in aplysia. Cell 115, 893–904 (2003). [DOI] [PubMed] [Google Scholar]
- 61.Harris JC, Martinez JM, Grozdanov PN, Bergeson SE, Grammas P, MacDonald CC, The Cstf2t polyadenylation gene plays a sex-specific role in learning behaviors in mice. PLOS ONE 11, e0165976 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rose KM, Bell LE, Jacob ST, Specific inhibition of chromatin-associated poly(A) synthesis in vitro by cordycepin 5′-triphosphate. Nature 267, 178–180 (1977). [DOI] [PubMed] [Google Scholar]
- 63.Wheeler JM, Guthrie CR, Kraemer BC, The role of MSUT-2 in tau neurotoxicity: A target for neuroprotection in tauopathy? Biochem. Soc. Trans 38, 973–976 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lloyd KCK, A knockout mouse resource for the biomedical research community. Ann. N. Y. Acad. Sci 1245, 24–26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Montine TJ, Phelps CH, Beach TG, Bigio EH, Cairns NJ, Dickson DW, Duyckaerts C, Frosch MP, Masliah E, Mirra SS, Nelson PT, Schneider JA, Thal DR, Trojanowski JQ, Vinters HV, Hyman BT; National Institute on Aging; Alzheimer’s Association, National Institute on Aging-Alzheimer’s Association guidelines for the neuropathologic assessment of Alzheimer’s disease: A practical approach. Acta Neuropathol. 123, 1–11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Seki T, Spurr N, Obata F, Goyert S, Goodfellow P, Silver J, The human Thy-1 gene: Structure and chromosomal location. Proc. Natl. Acad. Sci. U.S.A 82, 6657–6661 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Carmel G, Mager EM, Binder LI, Kuret J, The structural basis of monoclonal antibody Alz50’s selectivity for Alzheimer’s disease pathology. J. Biol. Chem 271, 32789–32795 (1996). [DOI] [PubMed] [Google Scholar]
- 68.Ishihara T, Zhang B, Higuchi M, Yoshiyama Y, Trojanowski JQ, Lee VM-Y, Age-dependent induction of congophilic neurofibrillary tau inclusions in tau transgenic mice. Am. J. Pathol 158, 555–562 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sun A, Nguyen XV, Bing G, Comparative analysis of an improved thioflavin-S stain, Gallyas silver stain, and immunohistochemistry for neurofibrillary tangle demonstration on the same sections. J. Histochem. Cytochem 50, 463–472 (2002). [DOI] [PubMed] [Google Scholar]
- 70.Ward SM, Himmelstein DS, Lancia JK, Binder LI, Tau oligomers and tau toxicity in neurodegenerative disease. Biochem. Soc. Trans 40, 667–671 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A, Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Frangioni JV, Neel BG, Solubilization and purification of enzymatically active glutathione S-transferase (pGEX) fusion proteins. Anal. Biochem 210, 179–187 (1993). [DOI] [PubMed] [Google Scholar]
- 73.Kraemer B, Crittenden S, Gallegos M, Moulder G, Barstead R, Kimble J, Wickens M, NANOS-3 and FBF proteins physically interact to control the sperm-oocyte switch in Caenorhabditis elegans. Curr. Biol 9, 1009–1018 (1999). [DOI] [PubMed] [Google Scholar]
- 74.Brenner S, The genetics of Caenorhabditis elegans. Genetics 77, 71–94 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Martin M, Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12 (2011). [Google Scholar]
- 76.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR, STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Anders S, Pyl PT, Huber W, HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cunningham F, Amode MR, Barrell D, Beal K, Billis K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fitzgerald S, Gil L, Girón CG, Gordon L, Hourlier T, Hunt SE, Janacek SH, Johnson N, Juettemann T, Kähäri AK, Keenan S, Martin FJ, Maurel T, McLaren W, Murphy DN, Nag R, Overduin B, Parker A, Patricio M, Perry E, Pignatelli M, Riat HS, Sheppard D, Taylor K, Thormann A, Vullo A, Wilder SP, Zadissa A, Aken BL, Birney E, Harrow J, Kinsella R, Muffato M, Ruffier M, Searle SMJ, Spudich G, Trevanion SJ, Yates A, Zerbino DR, Flicek P, Ensembl 2015. Nucleic Acids Res. 43, D662–D669 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Anders S, Reyes A, Huber W, Detecting differential usage of exons from RNA-seq data. Genome Res. 22, 2008–2017 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.R Development Core Team, R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2011). [Google Scholar]
- 82.Benjamini Y, Hochberg Y, Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B. Methodol 57, 289–300 (1995). [Google Scholar]
- 83.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D, The human genome browser at UCSC. Genome Res. 12, 996–1006 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Quinlan AR, Hall IM, BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Szot P, Franklin A, Sikkema C, Wilkinson CW, Raskind MA, Sequential loss of LC noradrenergic and dopaminergic neurons results in a correlation of dopaminergic neuronal number to striatal dopamine concentration. Front. Pharmacol 3, 184 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Goedert M, Jakes R, Crowther RA, Cohen P, Vanmechelen E, Vandermeeren M, Cras P, Epitope mapping of monoclonal antibodies to the paired helical filaments of Alzheimer’s disease: Identification of phosphorylation sites in tau protein. Biochem. J 301 (Pt 3), 871–877 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Weaver CL, Espinoza M, Kress Y, Davies P, Conformational change as one of the earliest alterations of tau in Alzheimer’s disease. Neurobiol. Aging 21, 719–727 (2000). [DOI] [PubMed] [Google Scholar]
- 88.Patterson KR, Ward SM, Combs B, Voss K, Kanaan NM, Morfini G, Brady ST, Gamblin TC, Binder LI, Heat shock protein 70 prevents both tau aggregation and the inhibitory effects of preexisting tau aggregates on fast axonal transport. Biochemistry 50, 10300–10310 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.