Abstract
Modular self-assembly of biomolecules in two dimensions (2D) is straightforward with DNA but has been difficult to realize with proteins, due to the lack of modular specificity similar to Watson-Crick base pairing. Here we describe a general approach to design 2D arrays using de novo designed pseudosymmetric protein building blocks. A homodimeric helical bundle was reconnected into a monomeric building block, and the surface was redesigned in Rosetta to enable self-assembly into a 2D array in the C12 layer symmetry group. Two out of ten designed arrays assembled to micrometer scale under negative stain electron microscopy, and displayed the designed lattice geometry with assembly size up to 100 nm under atomic force microscopy. The design of 2D arrays with pseudosymmetric building blocks is an important step toward the design of programmable protein self-assembly via pseudosymmetric patterning of orthogonal binding interfaces.
Graphical Abstract
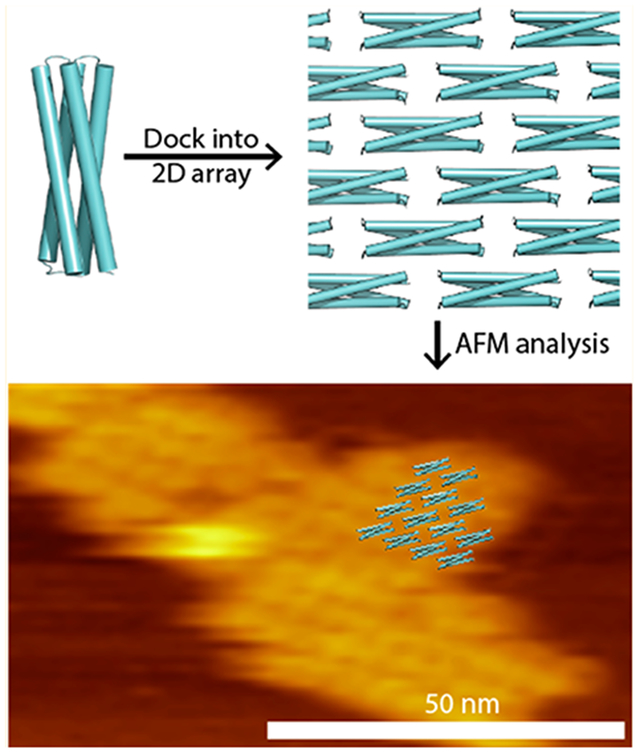
INTRODUCTION
Programmatic molecular self-assembly—the ability of complex assemblies to be built from simple building blocks—is ubiquitous in Nature. In particular, naturally occurring protein assemblies combine symmetry with pseudosymmetry,1 use conformational flexibility to break symmetry,2 or bind scaffolding molecules such as nucleic acids3 to construct large but bounded assemblies from simple protein building blocks. Mimicking native assemblies, we aim to design a modular scheme for programmatically controlling protein assemblies. Due to the simplicity of design, we focus on the use of pseudosymmetry—through sequence variations on a fixed backbone—as a preferred route of engineering programmable protein self-assemblies.
Several prior efforts have attempted to design such assemblies. Drawing off the specificity of Watson−Crick base pairing, a series of methods have been developed to accurately pattern DNA in two dimensions.4–6 For many applications, protein is a more attractive building material due to its inherent bioreactivity. Previous efforts in making patterned 2D materials with native proteins have resulted in some success.7–12 These previous efforts all made use of native proteins as building blocks, which is practically limiting: the geometry and overall symmetry of the final assembly is limited by the oligomeric state and symmetry of the native building block. De novo protein building blocks can be custom designed to have the desired symmetry13–15 and high stability,16 properties that should result in more diverse and robust building blocks for designing 2D self-assembling materials.
Here we describe a general approach for generating pseudosymmetric 2D assemblies based on a C12 symmetric layer group. Starting from a de novo designed homodimer, we first design a new loop to monomerize the backbone of our building block, then identify configurations of this backbone capable of forming 2D arrays with pseudo-C12 symmetry, and finally redesign the interface so that the building block will be programmatically assembled into 2D arrays with the prescribed unit cell dimensions and subunit configuration. The resulting layer group symmetry is pseudo-C12 because the addition of a loop breaks the C2 symmetry of the building block. The use of pseudosymmetric building blocks only requires the redesign of a single interface to form a 2D array. The monomerization of the multimeric protein building block allows unique sequences to be designed on each of the 4 binding interfaces, ultimately enabling the modular assembly of higher order interactions through the design of mutually orthogonal interfaces with the same subunit placement and unit cell dimensions. This study experimentally characterizes a key step toward this overall design goal, which will enable various applications including patterned enzymatic reactions.
RESULTS
Building off Gonen et al.,7 we developed a general strategy for the design of pseudosymmetric 2D protein assemblies using de novo designed proteins as building blocks, fully described in Methods. Figure 1 presents a high-level overview of the approach. Briefly, a previously designed helical bundle homodimer 2L4HC2_23 (PDB ID 5J0K,14 Figure 1A) was connected into a single chain monomer via a designed loop, resulting in a pseudo-C2 symmetric building block (SC_2L4HC2_23, Figure 1B). This building block has lower-order symmetry than any of the native building blocks used in a prior study.7 We solved the X-ray crystal structure of the building block, revealing a backbone nearly identical to the design model and the original 2L4HC2_23 homodimer structure, with a Cα root-mean-square deviation (RMSD) of1.08 Å between the design and crystal structure (Figure 1F).
Figure 1.
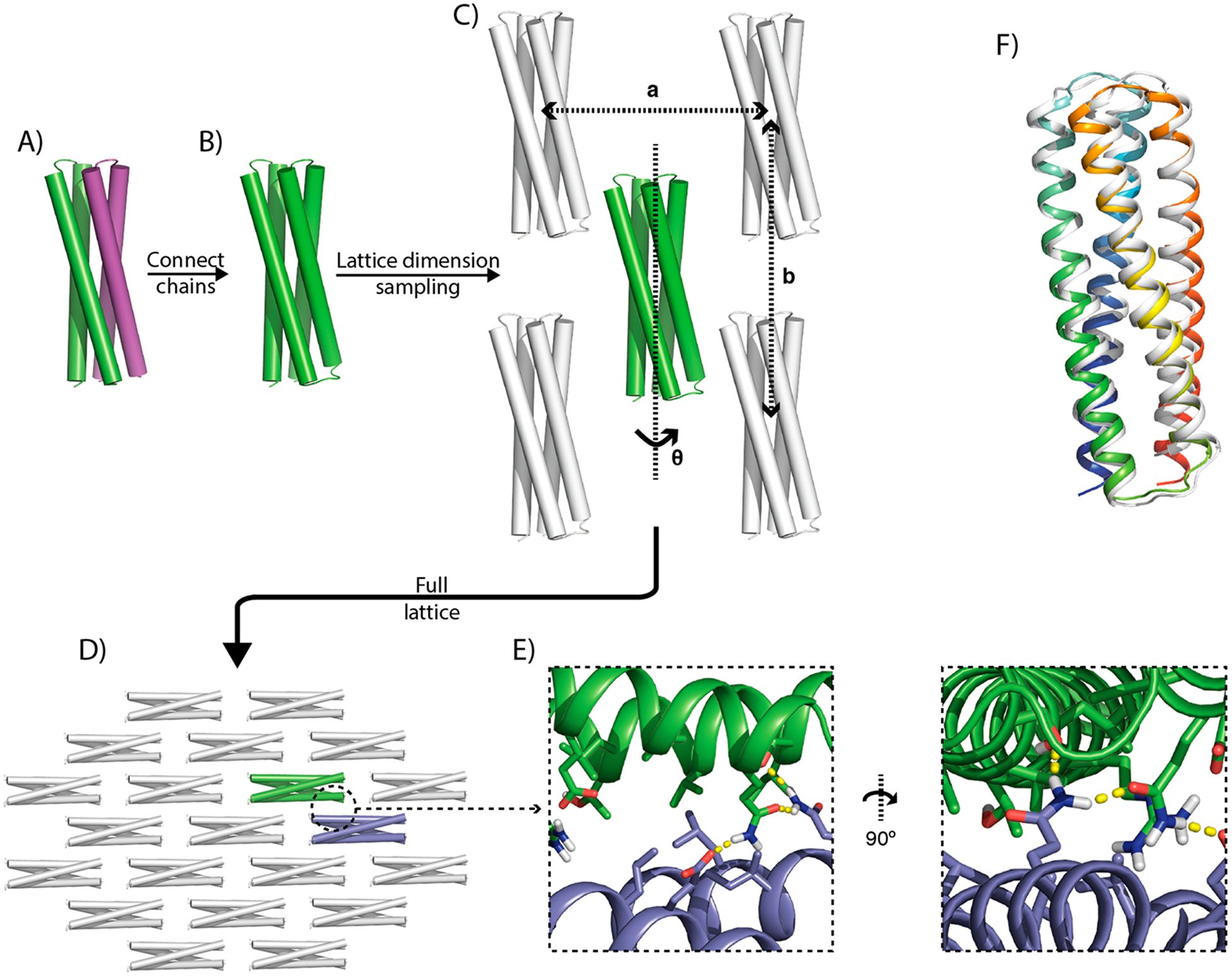
Overview of the design process. A de novo designed homodimer, with its two monomers colored in green and purple (A), is connected into a single chain (B), and docked in a C12 layer group symmetry with three parameters a, b, and θ (C), resulting in a 2D lattice (D). Intersubunit binding interfaces are designed with hydrogen bond networks to confer specificity, show here as an example between the green and blue building blocks (E). (F) A 1.74 Å resolution crystal structure of the design SC_2L4HC2_23 (PDB ID 6EGC, white) superimposed onto the design model (rainbow); the design model deviates from the crystal by 1.08 Å RMSD.
Using this monomerized building block as a starting point for pseudosymmetric assembly, we subsequently enumerated all possible pseudo-C12 symmetric layer assemblies compatible with this design, exhaustively sampling three degrees of freedom: two parameters (a, b) describing the lattice dimensions, and one parameter (θ) controlling rotation of the building block around its central axis (Figure 1C). We sampled 576,000 settings of these three parameters, and removed those which were not capable of forming a connected, nonclashing 2D assembly (Figure 1D). The remaining ~1000 designs had their surfaces redesigned to self-assemble into the corresponding lattice arrangement using standard Rosetta fixed backbone design.17 Using computationally predicted interface energies as well as visual inspection, seven designs were selected for experimental characterization, out of which two did not express, two purified solubly, and three formed higher-order assemblies that could be purified from the insoluble fraction.
Examination by negative-stain electron microscopy (EM) and atomic force microscopy (AFM) revealed regular arrays on the micrometer scale for one of the insoluble designs with exclusively hydrophobic residues at the binding interfaces (2DHP, Figure 2A,B), with 84% sequence identity to SC_2L4HC2_23 (22 mutations). The design was found to be α-helical and stable up to 95 °C (Supporting Information Figure S1C,D) as measured by circular dichroism. Negative-stain EM revealed the clustering of 2D arrays into bundle-like structures that are sensitive to different staining molecules (Figure 2C, Supporting Information Figure S1A,B). AFM analysis showed the formation of highly ordered assemblies on mica surfaces, with clearly evident rectangular packing (Figure 2D). Higher magnification revealed lattice dimensions of 6.3 ±0.5 nm by 4.7 ± 0.4 nm, in close agreement with the designed lattice dimensions of 6.1 nm by 4.7 nm. The measured single layer thickness of the 2D assemblies was 2.4 nm (Figure 2E), closely consistent with the thickness of 2.3 nm from the design model (Figure 2F).
Figure 2.
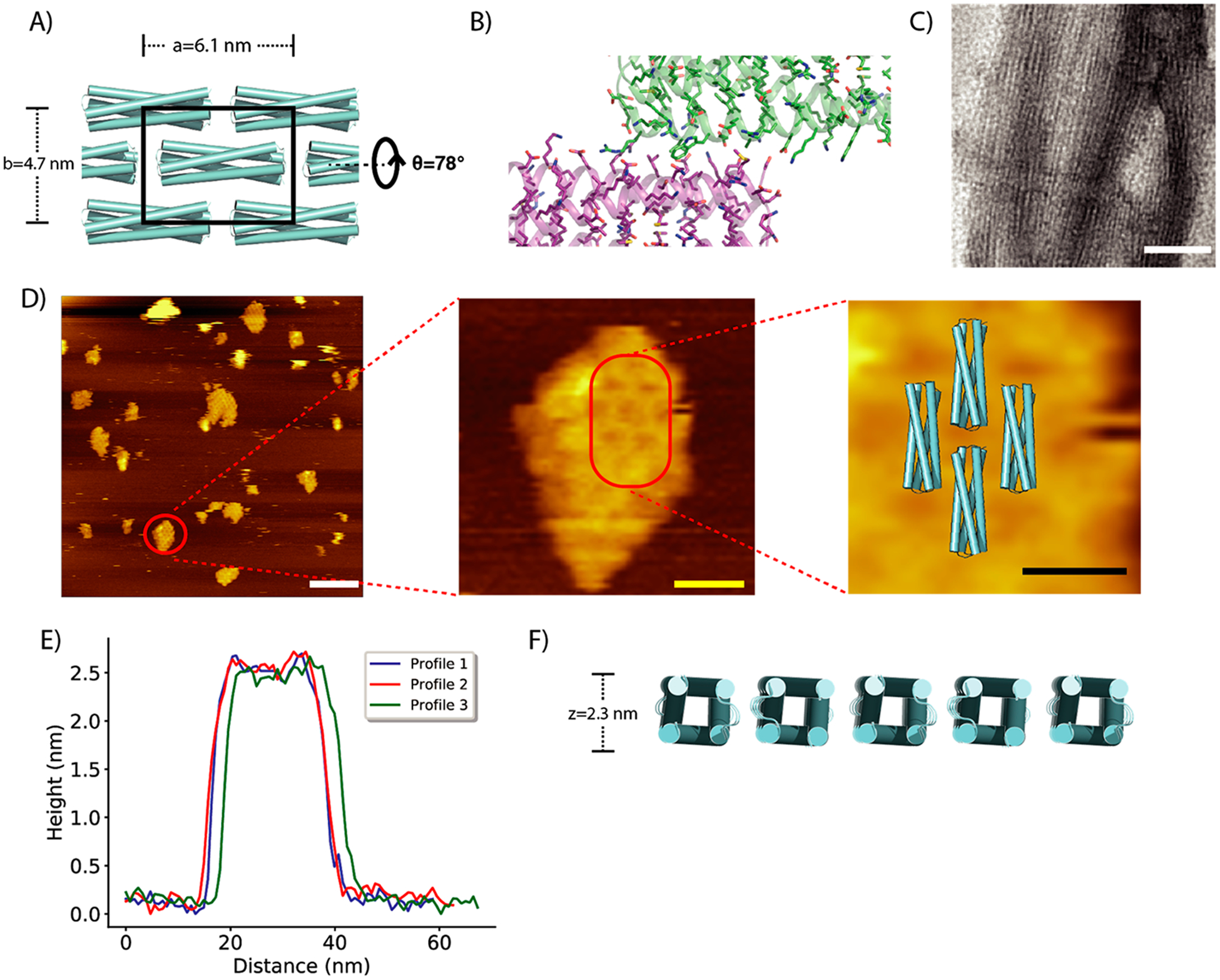
Structural analysis of the designed 2D assembly 2D-HP. (A) Lattice design of 2D-HP, with the black box showing unit cell. (B) Designed interface of 2D-HP with exclusive hydrophobic packing across the interface. (C) Negatively stained array of 2D-HP under electron microscopy.(D) AFM image of 2D-HP assemblies, with zoomed-in views of one of the assemblies superimposed onto the design model. (E) AFM height profiles of 2D-HP assemblies. (F) Thickness of the design model is 2.3 nm based on Cβ distances. All scale bars: white, 50 nm; yellow, 10 nm; black, 5 nm.
Given the nonspecific clustering of 2D-HP assemblies under EM, we sought to further improve the binding specificity among building blocks by using the Rosetta HBNet algorithm14 to design buried hydrogen bonds at the interface (Figure 1E). A systematic search of interfacial hydrogen bond networks on 576,000 lattice dimensions resulted in 24 designs with no buried unsatisfied polar heavy atoms and good interfacial binding energy. After a round of in silico selection with Rosetta docking,18 three such designs were ordered, with one of the designs (2D-HBN, Figure 3A,B) forming more extended and regular assemblies compared to that of 2D-HP (Figure 3C), likely due to better binding specificity conferred by hydrogen bond networks. The design 2D-HBN has 52% (68 mutations) and 55% (62 mutations) sequence identity to SC_2L4HC2_23 and 2D-HP, respectively. To rule out the possibility of domain swapping from the single chain building block contributing to the final assembly, we additionally expressed the building block protein of 2D-HBN as individual homodimers of helix hairpins, which similarly assembled into 2D arrays of the same morphology under the same condition (Supporting Information Figure S2).
Figure 3.
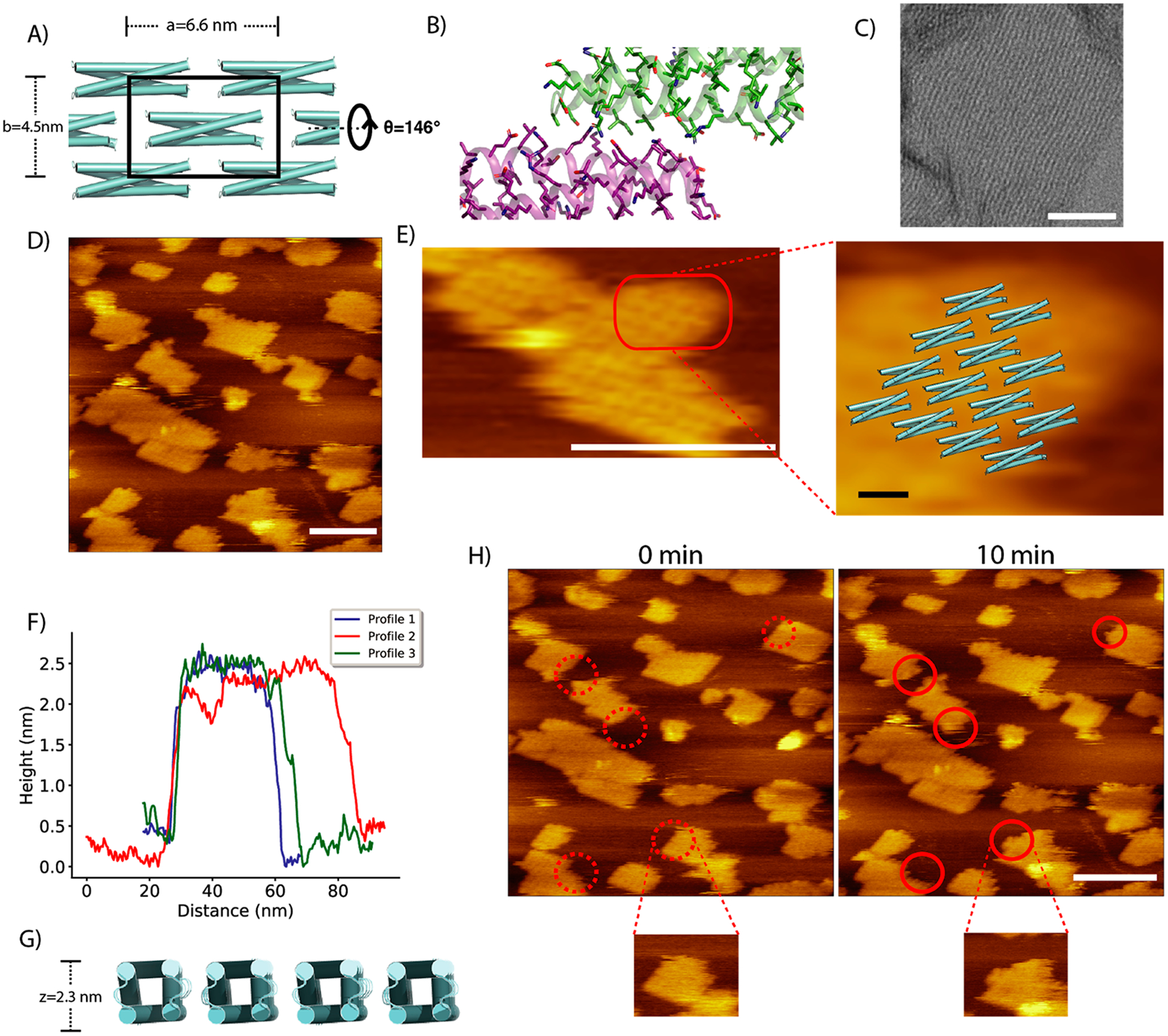
Structural analysis of the designed 2D assembly 2D-HBN. (A) Lattice design of 2D-HBN, and its designed interface with a hydrogen bond network (B). (C) Negatively stained array of 2D-HBN showing an extensive and flexible 2D assembly. (D) AFM image of 2D-HBN assemblies, with a zoomed-in view of one of the assemblies superimposed onto the design model (E). (F) AFM height profiles of 2D-HBN assemblies. (G) Thickness of the design model is 2.3 nm based on Cβ distances. (H) In situ self-assembly of 2D-HBN, red circles show regions of growth at t = 0 min (dotted) and t = 10 min (solid). Inset, zoomed-in view of one of the growing assemblies. All scale bars: white, 50 nm; black, 5 nm.
To verify that the array was forming a regular 2D grid, we collected a larger negative stain data set of the best-behaved arrays (Supporting Information Figure S3A). Subsequent 2D classification and averaging of 1893 boxed ~20 nm regions yielded an image showing an ordered two-dimensional assembly with a power spectrum indicating first-order spots (Supporting Information Figure S3B,C). While the resulting images were consistent with a C12-symmetric complex, the unit-cell dimensions were different than designed: while the design had a 6.6 by 4.5 nm unit cell, the experimental images indicated approximately a 6.8 by 2.2 nm unit cell. Given the inability to pack the designed model into this observed spacegroup (Figure S4), we believe that this apparent difference is due to either an artifact of image averaging or stacking of individual layers (Figure S5).
We further validated the 2D-HBN design with AFM. Direct measurement on single-layered assemblies revealed highly ordered rectangular packing, with individual monomers clearly resolved (Figure 3D,E). Lattice dimensions of 6.7 ± 0.3 nm by4.7 ± 0.3 nm with a thickness of 2.4 nm were observed (Figure 3F), in close agreement with the designed lattice dimensions of6.6 nm by 4.5 nm with a thickness of 2.3 nm (Figure 3G). Furthermore, we observed the in situ growth and dissolution of 2D-HBN under AFM, showing protein building blocks being dynamically and reversibly added onto already assembled 2D arrays (Figure 3H).
DISCUSSION
We showed that by systematically sampling lattice dimensions followed by computational interface design, the same de novo designed helical bundle building block can be modularly self-assembled into two arrays with unique cell dimensions. Limited by available de novo designed protein building blocks, the designed interfaces are much smaller than those in previously designed 2D protein assemblies, resulting in a high degree of structural flexibility under EM. Future work could address this in two ways: (1) Using bigger de novo building blocks, more rigid arrays should be possible to realize with bigger binding interfaces; (2) designing in nonpolar layer groups, which have a rotation about the layer plane (e.g., P321 and P4212), effectively canceling out any “curvature” errors in binding along the z axis, further flattening out the 2D assembly.7 As more de novo building blocks are designed, particularly with higher-order symmetry, a variety of 2D assemblies with unique layer group symmetries should be achievable with the same design protocol.
The monomerization of the homodimer building block coupled with designed hydrogen bond networks should allow orthogonal interfaces to be designed at each intermolecular binding site, paving the way for the programmatic self-assembly of proteins into finite shapes, which requires the design of multiple such interfaces on a single pseudosymmetric building block (Figure 4A,B). Such interfaces can be applied modularly, by plugging the designed sequence onto the corresponding helical bundle. Our work represents a key step toward this goal and shows that de novo designed proteins can be engineered to self-assemble into regular 2D arrays.
Figure 4.
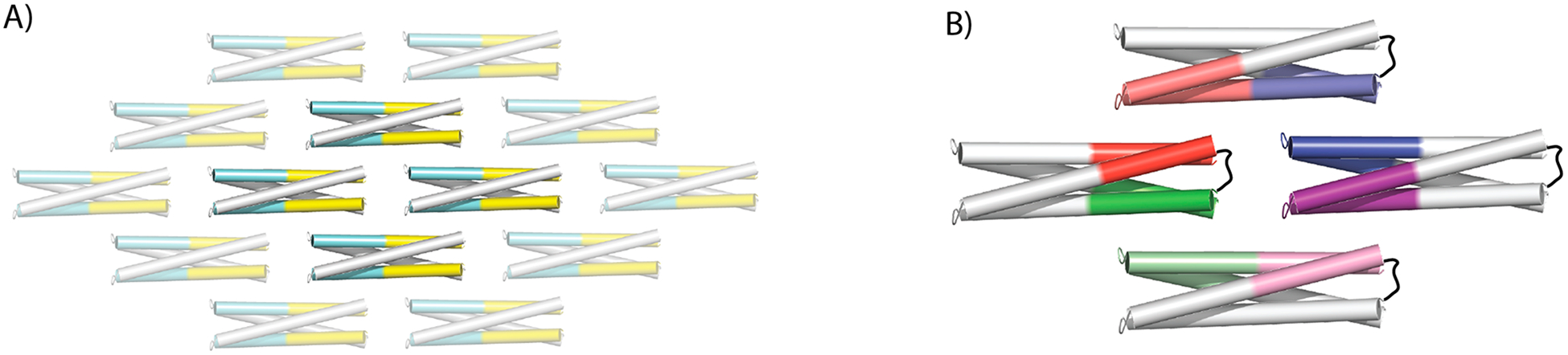
Pseudosymmetry enables the design of programmatic assemblies. (A) 2D self-assembly using homodimers as building blocks. Designed interbuilding block binding interfaces are highlighted in cyan and yellow, indicating heterotypic interactions. In this scenario the assembly process will result in an infinite 2D lattice. (B) By using designed loops to monomerize the building block, and modularly mixing orthogonal binding interfaces (e.g., dark red exclusively binds light red), programmatic assembly design is enabled (in this case, a heterotetramer).
Supplementary Material
ACKNOWLEDGMENTS
We thank P. Lu for help with X-ray crystallography, F. Praetorius for helpful discussions. F.D. was supported by NIH GM123089. J.M.K. was supported by NIH R01 GM127648. S.E.B. was supported by the Burroughs Wellcome Fund Career Award at the Scientific Interface. AFM data collection and analysis was supported by the US Department of Energy (DOE), Office of Basic Energy Sciences, Division of Materials Science and Engineering, Biomolecular Materials Program under contract DE-SC0018940 and Grant 12569199 and was carried out at Pacific Northwest National Laboratory (PNNL). PNNL is a multiprogram national laboratory operated for DOE by Battelle under Contract No. DE-AC05-76RL01830. Electron microscopy data was collected at the Arnold and Mabel Beckman Center for CryoEM at the University of Washington. This work was facilitated through the use of advanced computational, storage, and networking infrastructure provided by the Hyak supercomputer system and in part funded by the STF at the University of Washington.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.9b01978.
Computational protocols, experimental procedures, and design sequences (PDF)
Data and results (ZIP)
The authors declare the following competing financial interest(s): Z.C., D.B., and F.D. are inventors on on US provisional patent application no. 62/833,902 describing this work.
REFERENCES
- (1).Takenoya M; Nikolakakis K; Sagermann M Crystallographic Insights into the Pore Structures and Mechanisms of the EutL and EutM Shell Proteins of the Ethanolamine-Utilizing Microcompartment of Escherichia Coli. J. Bacteriol 2010, 192 (22), 6056–6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Guo F; Liu Z; Fang P-A; Zhang Q; Wright ET; Wu W; Zhang C; Vago F; Ren Y; Jakana J; Chiu W; Serwer P; Jiang W Capsid Expansion Mechanism of Bacteriophage T7 Revealed by Multistate Atomic Models Derived from Cryo-EM Reconstructions. Proc. Natl. Acad. Sci. U. S. A 2014, 111 (43), E4606–E4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Klug A The Tobacco Mosaic Virus Particle: Structure and Assembly. Philos. Trans. R. Soc. London B Biol. Sci 1999, 354 (1383), 531–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Rothemund PWK Folding DNA to Create Nanoscale Shapes and Patterns. Nature 2006, 440 (7082), 297–302. [DOI] [PubMed] [Google Scholar]
- (5).Wei B; Dai M; Yin P Complex Shapes Self-Assembled from Single-Stranded DNA Tiles. Nature 2012, 485 (7400), 623–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Tikhomirov G; Petersen P; Qian L Programmable Disorder in Random DNA Tilings. Nat. Nanotechnol 2017, 12 (3), 251–259. [DOI] [PubMed] [Google Scholar]
- (7).Gonen S; DiMaio F; Gonen T; Baker D Design of Ordered Two-Dimensional Arrays Mediated by Noncovalent Protein-Protein Interfaces. Science 2015, 348 (6241), 1365–1368. [DOI] [PubMed] [Google Scholar]
- (8).Suzuki Y; Cardone G; Restrepo D; Zavattieri PD; Baker TS; Tezcan FA Self-Assembly of Coherently Dynamic, Auxetic, Two-Dimensional Protein Crystals. Nature 2016, 533 (7603), 369–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Liu Y; Gonen S; Gonen T; Yeates TO Near-Atomic Cryo-EM Imaging of a Small Protein Displayed on a Designed Scaffolding System. Proc. Natl. Acad. Sci. U. S. A 2018, 115 (13), 3362–3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Jiang T; Xu C; Zuo X; Conticello VP Structurally Homogeneous Nanosheets from Self-Assembly of a Collagen-Mimetic Peptide. Angew. Chem., Int. Ed 2014, 53 (32), 8367–8371. [DOI] [PubMed] [Google Scholar]
- (11).Bai Y; Luo Q; Liu J Protein Self-Assembly via Supra-molecular Strategies. Chem. Soc. Rev 2016, 45 (10), 2756–2767. [DOI] [PubMed] [Google Scholar]
- (12).Luo Q; Hou C; Bai Y; Wang R; Liu J Protein Assembly: Versatile Approaches to Construct Highly Ordered Nanostructures. Chem. Rev 2016, 116 (22), 13571–13632. [DOI] [PubMed] [Google Scholar]
- (13).Fletcher JM; Boyle AL; Bruning M; Bartlett GJ; Vincent TL; Zaccai NR; Armstrong CT; Bromley EHC; Booth PJ; Brady RL; Thomson AR; Woolfson DN A Basis Set of de Novo Coiled-Coil Peptide Oligomers for Rational Protein Design and Synthetic Biology. ACS Synth. Biol 2012, 1 (6), 240–250. [DOI] [PubMed] [Google Scholar]
- (14).Boyken SE; Chen Z; Groves B; Langan RA; Oberdorfer G; Ford A; Gilmore JM; Xu C; DiMaio F; Pereira JH; Sankaran B; Seelig G; Zwart PH; Baker D De Novo Design of Protein Homo-Oligomers with Modular Hydrogen-Bond Network-Mediated Specificity. Science 2016, 352 (6286), 680–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Fallas JA; Ueda G; Sheffler W; Nguyen V; McNamara DE; Sankaran B; Pereira JH; Parmeggiani F; Brunette TJ; Cascio D; Yeates TR; Zwart P; Baker D Computational Design of Self-Assembling Cyclic Protein Homo-Oligomers. Nat. Chem 2017, 9 (4), 353–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Huang P-S; Oberdorfer G; Xu C; Pei XY; Nannenga BL; Rogers JM; DiMaio F; Gonen T; Luisi B; Baker D High Thermodynamic Stability of Parametrically Designed Helical Bundles. Science 2014, 346 (6208), 481–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Leaver-Fay A; Tyka M; Lewis SM; Lange OF; Thompson J; Jacak R; Kaufman K; Renfrew PD; Smith CA; Sheffler W; Davis IW; Cooper S; Treuille A; Mandell DJ; Richter F; Ban YA; Fleishman SJ; Corn LE; Kim DE; Lyskov S; Berrondo M; Mentzer S; Popovic Z; Havranek JJ; Karanicolas J; Das R; Meiler J; Kortemme T; Gray JJ; Kuhlman B; Baker D; Bradley P ROSETTA3: An Object-Oriented Software Suite for the Simulation and Design of Macromolecules. Methods Enzymol. 2011, 487, 545–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Gray JJ; Moughon S; Wang C; Schueler-Furman O; Kuhlman B; Rohl CA; Baker D Protein-Protein Docking with Simultaneous Optimization of Rigid-Body Displacement and Side-Chain Conformations. J. Mol. Biol 2003, 331 (1), 281–299. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


