Abstract
Purpose
Accumulating studies have explored the potential diagnostic value of lncRNA MALAT1 in various cancers. However, there are still inconsistent results in diagnostic accuracy and reliability in individual studies. The aim of this pooled study was to summarize the overall diagnostic capacity of lncRNA MALAT1 in cancer detection and diagnosis.
Methods
Eligible studies satisfying the inclusion criteria were screened and selected from the online database. All statistical analyses were performed using Stata 14.0.
Results
A total of 17 eligible studies were included in this pooled analysis, with 1777 cases and 1478 controls. The overall results were shown as follows: sensitivity, 0.74 (95% CI=0.65–0.81), specificity, 0.79 (95% CI=0.73–0.84), positive likelihood ratio (PLR), 3.48 (95% CI=2.79–4.32), negative likelihood, 0.33 (95% CI=0.25–0.44), diagnostic score, 2.34 (95% CI=1.99–2.69), diagnostic odds ratio, 10.41 (95% CI=7.33–14.78) and area under the curve, 0.83 (95% CI=0.80–0.86). Deeks’ funnel plot asymmetry test (p = 0.66) suggested no potential publication bias.
Conclusion
All these results indicate that lncRNA MALAT1 achieves a relatively moderate accuracy in cancer detection and diagnosis, and could serve as a diagnostic biomarker for cancers.
Keywords: lncRNA, MALAT1, cancer, pooled analysis, detection, diagnosis
Introduction
With a leading cause of morbidity and mortality in recent years, cancer has become a significant public health problem that threatening peoples’ life quality all over the world.1,2 Despite the rapid development of cancer treatments, such as surgery, chemotherapy, radiotherapy, and targeted therapy, the prognosis of patients with cancers still remains unsatisfactory.3–5 The lack of early diagnostic techniques and Delay in the diagnosis of cancer are the major obstacles in the current situation, contributing to missing the optimal opportunity for treatment, less chance of surviving, and more expensive costs.6–8 Thus, it is urgent to identify a novel diagnostic marker with good sensitivity and specificity for the early detection of cancer.
LncRNAs are often described as non-coding transcripts more than 200 nucleotides in length, lacking functional open reading frames (ORFs) and protein-coding capability.9,10 Previous studies have shown that lncRNAs perform multiple physiological and pathological biological functions.11 Aberrant expression of lncRNAs is widely observed in many cancers, involving in cancer initiation, progression, and metastasis.12–14 Furthermore, increasing evidence indicates that lncRNAs could serve as potential biomarkers in cancer detection and diagnosis, with high sensitivity and specificity.15,16
MALAT1, located at 11q13, was firstly characterized by Ji et al17 in a study of early-stage non-small-cell lung cancer, with a total 8.5 kb full length. A subsequent research study has demonstrated that MALAT1 is widely expressed in normal tissues, involving in cell viability and normal development.18 While MALAT1 was initially described as a prognostic marker of lung cancer metastasis, emerging evidence has shown that this lncRNA contributes greatly to other types of cancer’s development and progression.19–22 Furthermore, an increasing number of studies suggest that the aberrant expression of MALAT1 in tumor tissues or body fluids may serve as a biomarker for tumor diagnosis and prognosis.
However, the diagnostic accuracy of MALAT1 in individual studies is still inconsistent and controversial. For instance, Liu et al23 revealed that MALAT1 achieve a moderate-high sensitivity and specificity of 86.0% and 75.0% in the diagnosis of gastric cancer, respectively, whereas Yazarlou et al24 demonstrated that a relatively low sensitivity and specificity of 62.7% and 69.4%, respectively, in bladder cancer detection. The agreement is hard to reach among these results because of ethnicity, study design, types of tumors, stage of cancer, and the small sample size. Previously, Mei et al performed a meta-analysis of MALAT1 expression in cancer detection for the first time, revealing that MALAT1 might be an effective cancer diagnostic marker. However, a limited number of cancer cases and small sample size in single tumor type make it unpersuasive enough.25 Thus, we conducted this pooled analysis to summarize the overall diagnostic performance of MALAT1 in cancer detection and diagnosis and further explored its clinical value.
Methods
Search Strategy and Study Eligibility Criteria
Literature research was performed in the database including PubMed, Cochrane library, CNKI and Wanfang library, up to December 20, 2019, by the following searching strategy: “cancer” or “tumor” or “carcinoma” or “neoplasm” or “malignancy” or “neoplasm” and “MALAT1” and “sensitivity” or “specificity” or “ROC curve” or “accuracy”. Two investigators (YZ and YQY) check the titles and abstract information of the studies, and then scanned the full articles to delete irrelevant studies by using the following inclusion criteria: (1) human clinical studies; (2) the diagnostic value of lncRNA MALAT1 for detecting cancer evaluated in studies; (3) the number of true positive (TP), false positive (FP), false negative (FN) and true negative (TN) in cancer patients and controls could be drawn or calculated from the original included studies and (4) being published in English or Chinese. Accordingly, the articles were excluded on the basis of the following exclusion criteria: (1) laboratory studies on animal models or cell lines; (2) reviews, meta-analysis, case reports, commentaries and (3) lack of sufficient data to calculate TP, FP, FN and TN.
Data Extraction and Quality Assessment
The data and primary information from each included study, including first author, year of publication, country, ethnicity, cancer type, Normalizer, sample type, test method, cutoff value, sample size, true positive (TP), false positive (FP), true negative (TN), and false negative (FN) were extracted by two reviewers (YZ and YQY) independently and cross-checked. Divergences will come to an agreement by another two authors (SY and CMZ).
Quality Assessment
The QUADAS-2 was applied to evaluate the quality of the studies included in this pooled analysis systematically. With the maximum QUADAS-2 score of 7, we can judge the quality of the included studies based on the score. Studies with four or more scores were defined as moderate-high quality. All of these were carried out independently by two authors (YZ and SY); Any disagreement was resolved in the conference by two authors (LW and WXZ).
Statistical Analysis
All statistical analysis were performed using Stata 14.0 (Stata Corporation, College Station, TX, USA). The pooled sensitivity, specificity, diagnostic score (DS), diagnostic odds ratio (DOR), positive likelihood ratio (PLR) and negative likelihood ratio (NLR) and other parameters were calculated by the bivariate model. Then, summary receiver operator characteristic (SROC) curves are applied to analyze and calculated the area under the ROC curves (AUC), to assess the overall diagnostic value of lncRNA MALAT1 in cancer detection and diagnosis. These data were confirmed by a hierarchical summary receiver operating characteristics (HSROC) model. Cochran-Q and Inconsistency index (I2) test were applied in order to evaluate the statistical heterogeneity across the included publications. A P value less than 0.10 for the Q test or I2 value higher than 50% indicated obvious heterogeneity between the studies.26 In addition, Fagan’s Nomogram was used to confirm relationships between prior-test probability, likelihood ratio, and posttest probability. Deeks’ funnel plot was applied for publication bias evaluation.
Results
Studies Selection and General Features of Included Studies
A total of 17 eligible studies23,24,27–41 including 1777 cases and 1478 controls were finally included in the pooled analysis after a systematic search of PubMed, Cochrane library, CNKI and Wanfang library database from 2013 to 2019, according to inclusion and exclusion criteria (Figure 1). The main characteristics of the included studies were displayed in Table 1. In total, there were studies on breast cancer (N=4), lung cancer (N=4), bladder cancer (N=3), gastric cancer (N=1), oral squamous cell carcinoma (N=1), osteosarcoma (N=1), endometrial cancer (N=1), nasopharyngeal carcinoma (N=1) and prostate cancer (N=1). The expression of lncRNA MALAT1 was measured by qRT-PCR methods based on serum (N=8), plasma (N=4), tissue (N=2), urine (N=2) and Pleural effusion (N=1). In all included studies, the expression of MALAT1 showed an up-regulation trend in cancer samples when compared with controls.
Figure 1.
The flow diagram of relevant studies from the electronic databases.
Abbreviations: CNKI, China National Knowledge Infrastructure; MALAT1, metastasis-associated lung adenocarcinoma transcript 1.
Table 1.
Characteristics of the Included Studies
| Author | Year | Country | Ethnicity | Cancer Type | Normalizer | Sample Type | Test Method | Cutoff | Case/Control | TP | FP | FN | TN |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Liu23 | 2019 | China | Asian | GC | GAPDH | Serum | qRT-PCR | N/A | 100/100 | 86 | 25 | 14 | 75 |
| Wu27 | 2018 | China | Asian | BRC | GAPDH | Plasma | qRT-PCR | 1.69 | 102/50 | 55 | 7 | 47 | 43 |
| Wang.J28 | 2018 | China | Asian | OSCC | GAPDH | Plasma | qRT-PCR | 1.18 | 70/50 | 61 | 14 | 9 | 36 |
| Li29 | 2018 | China | Asian | BRC | GAPDH | Tissue | qRT-PCR | N/A | 64/64 | 50 | 22 | 14 | 42 |
| Yazarlou24 | 2018 | Iran | Asian | BC | 5s rRNA | Urine | qRT-PCR | 6.86 | 59/49 | 37 | 15 | 22 | 34 |
| Huang30 | 2018 | China | Asian | BRC | GAPDH | Serum | qRT-PCR | 0.6200 (Training set) | 158/107 | 140 | 25 | 18 | 82 |
| 2.1345 (Validation set) | |||||||||||||
| Zhan31 | 2018 | China | Asian | BC | GAPDH | Urine | qRT-PCR | N/A | 184/184 | 138 | 42 | 46 | 142 |
| Wang.W32 | 2018 | China | Asian | LC | GAPDH | Pleural effusion | qRT-PCR | 6.975 | 217/132 | 161 | 12 | 56 | 120 |
| He33 | 2017 | China | Asian | NPC | GAPDH | Serum | qRT-PCR | N/A | 101/101 | 67 | 11 | 34 | 90 |
| Huo34 | 2017 | China | Asian | OS | GAPDH | Serum | qRT-PCR | 3.68 | 46/40 | 37 | 11 | 9 | 29 |
| Zidan35 | 2017 | Egypt | African | BRC | β-actin | Serum | qRT-PCR | 2.6 | 80/80 | 67 | 15 | 13 | 65 |
| Han36 | 2016 | China | Asian | EC | GAPDH | Tissue | qRT-PCR | 6.445 | 104/104 | 47 | 18 | 57 | 86 |
| Wen37 | 2016 | China | Asian | LC | GAPDH | Serum | qRT-PCR | 0.03 | 84/60 | 49 | 11 | 35 | 49 |
| Duan38 | 2016 | China | Asian | BC | GAPDH | Serum | qRT-PCR | N/A | 120/120 | 68 | 39 | 52 | 81 |
| Peng39 | 2016 | China | Asian | LC | GAPDH | Serum | qRT-PCR | 1.096 (Training set) | 156/107 | 151 | 63 | 5 | 44 |
| 2.0845 (Validation set) | |||||||||||||
| Ren40 | 2013 | China | Asian | PC | GAPDH | Plasma | qRT-PCR | 867.8 copies/mL | 87/105 | 51 | 16 | 36 | 89 |
| Weber41 | 2013 | Germany | Caucasian | LC | GAPDH | Plasma | qRT-PCR | −0.41 | 45/25 | 21 | 0 | 24 | 25 |
Abbreviations: GC, gastric cancer; BRC, breast cancer; OSCC, oral squamous cell carcinoma; BC, bladder cancer; LC, lung cancer; NPC, nasopharyngeal carcinoma; OS, osteosarcoma; EC, endometrial cancer; PC, prostate cancer; NA, not available; TP, true positive; FP, false positive; FN, false negative; TN, true negative.
Quality Assessment
The scores of the Quality Assessment of Diagnostic Accuracy Studies-2 (QUADAS-2) study quality assessment were shown in Table 2. Among the 17 studies, seven studies achieved 6 stars, four studies achieved 5 stars, and six studies achieved 4 stars, indicating a moderate-high quality for most of the studies.
Table 2.
Study Quality of the Diagnostic Studies Judged by the QUADAS II Checklist
| Author | Year | Risk of Bias | Concerns Regarding Applicability | Total Stars | |||||
|---|---|---|---|---|---|---|---|---|---|
| Patient Selection | Index Test | Reference Standard | Flow and Timing | Patient Selection | Index Test | Reference Standard | |||
| Liu23 | 2019 | Low | Low | Low | Unclear | Low | Low | Low | 6 |
| Wu27 | 2018 | Low | Low | Low | Unclear | Low | Low | Low | 6 |
| Wang.J28 | 2018 | Low | Low | Low | Unclear | High | Low | Low | 5 |
| Li29 | 2018 | Low | High | Low | Unclear | Unclear | Low | Low | 4 |
| Yazarlou24 | 2018 | Unclear | Unclear | Low | Low | Unclear | Low | Low | 4 |
| Huang30 | 2018 | Low | Low | Low | Low | Unclear | Low | Low | 6 |
| Zhan31 | 2018 | Low | Low | Low | Unclear | Unclear | Low | Low | 5 |
| Wang.W32 | 2018 | Low | Unclear | Low | Unclear | Low | Low | Low | 5 |
| He33 | 2017 | Low | Unclear | Low | Low | Unclear | Low | Low | 5 |
| Huo34 | 2017 | Unclear | Low | Low | Unclear | Unclear | Low | Low | 4 |
| Zidan35 | 2017 | Unclear | Low | Low | Unclear | Unclear | Low | Low | 4 |
| Han36 | 2016 | Low | High | Low | Unclear | Unclear | Low | Low | 4 |
| Wen37 | 2016 | Low | Low | Low | Unclear | Low | Low | Low | 6 |
| Duan38 | 2016 | Low | Unclear | Low | Unclear | Unclear | Low | Low | 4 |
| Peng39 | 2016 | Low | Low | Low | Low | High | Low | Low | 6 |
| Ren40 | 2013 | Low | Low | Low | Low | Unclear | Low | Low | 6 |
| Weber41 | 2013 | Unclear | Low | Low | Low | Low | Low | Low | 6 |
Pooled Results
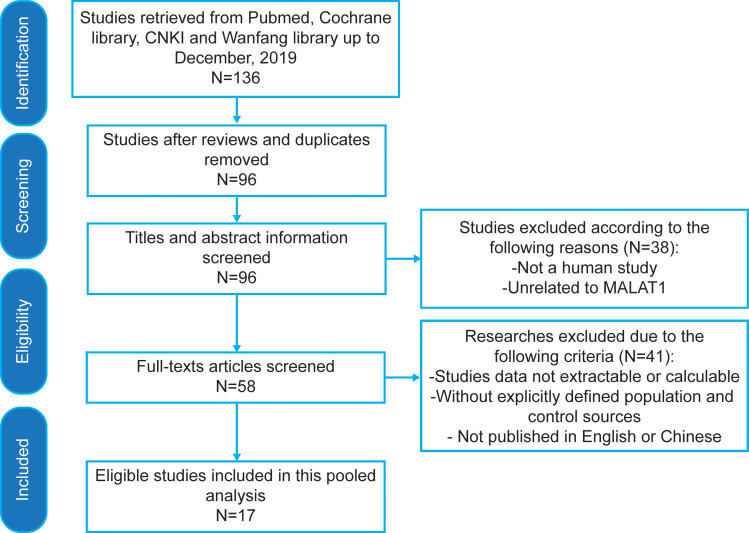
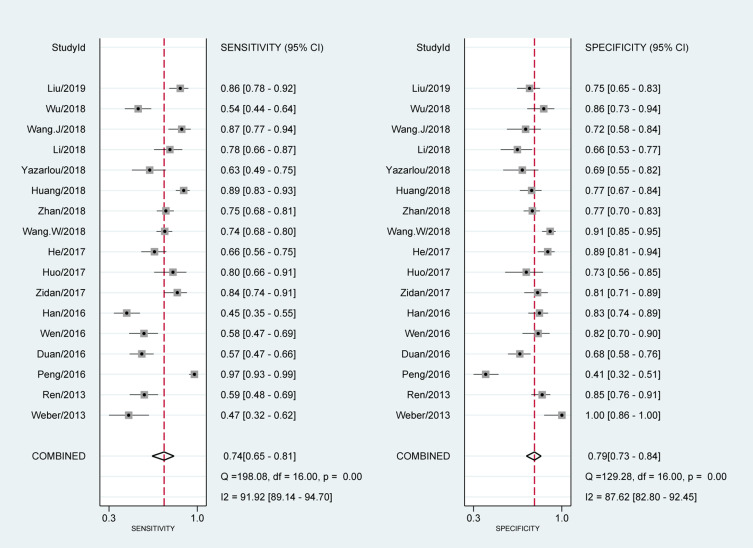
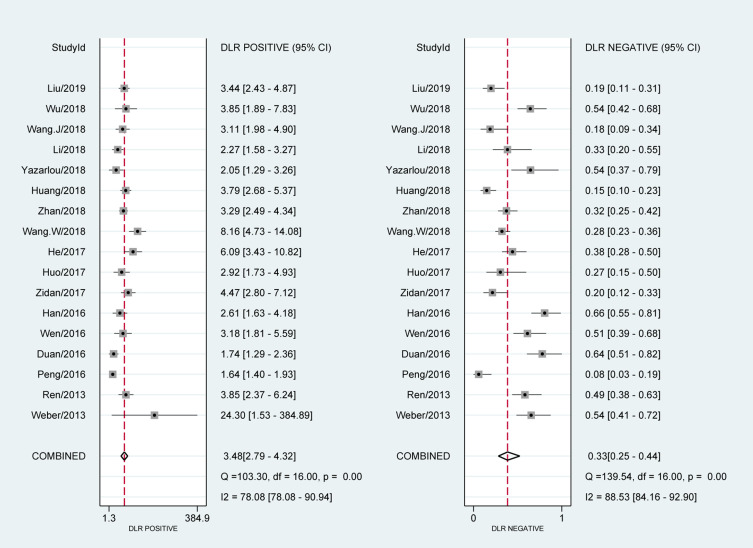
The forest plot of data from 17 included articles on sensitivity and specificity for MALAT1 in diagnosing cancer was shown in Figure 2. The sensitivity and specificity for the pooled data were 0.74 [95% CI=0.65–0.81] and 0.79 [95% CI=0.73–0.84], respectively. Significant heterogeneity was found for both sensitivity [I2=91.92%, 95% CI=89.14–94.70%] and specificity [I2=87.62%, 95% CI=82.80–92.45%]. Meanwhile, we found that the pooled PLR was 3.48 [95% CI=2.79–4.32], the NLR was 0.33 [95% CI=0.25–0.44] (Figure 3). The DS and DOR were 2.34 [95% CI=1.99–2.69] and 10.41 [95% CI=7.33–14.78], respectively (Figure 4).
Figure 2.
Forest plots of pooled sensitivity and specificity of 17 included studies.
Figure 3.
Forest plots of positive likelihood ratio (PLR) and negative likelihood ratio (NLR) for MALAT1 in the diagnosis of cancer.
Abbreviation: DLR, diagnostic likelihood ratio.
Figure 4.
Forest plots of pooled diagnostic score (DS) and diagnostic odds ratio (DOR) for MALAT1 in the diagnosis of cancer.
Pooled SROC and HSROC
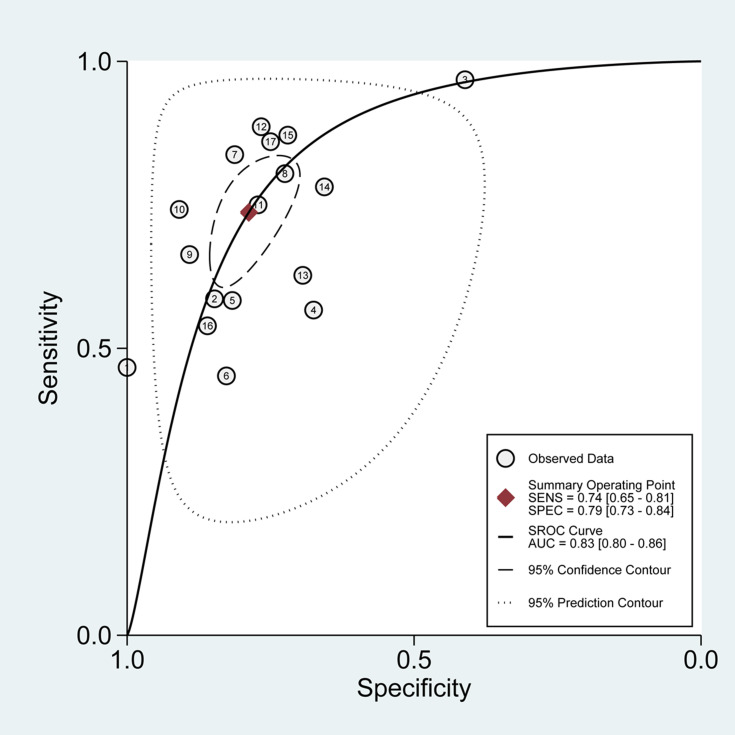
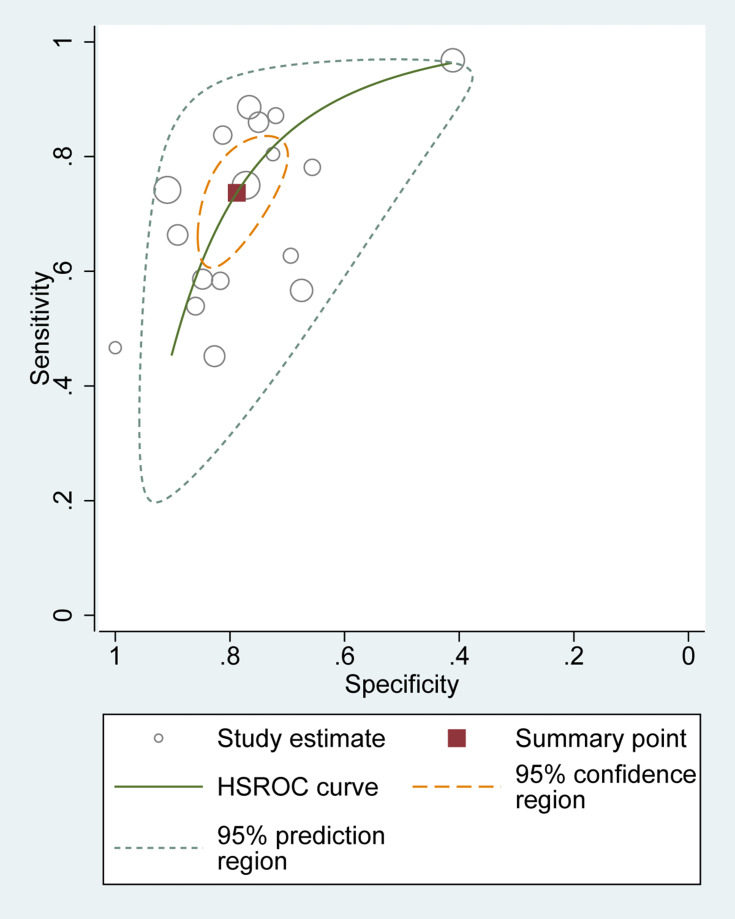
The SROC curve for the 17 included studies was 0.83 [95% CI=0.80–0.86], as shown in Figure 5, indicating a relatively moderate-high diagnostic value. Besides, The HSROC curve of these included studies was consistent with the results of the bivariate model. The calculated β value was −0.29 (95% CI=−0.76–0.17), and the P value was 0.210, implying that the HSROC was symmetrical (Figure 6).
Figure 5.
Summary receiver operating characteristic (SROC) graph of 17 included studies.
Abbreviations: SENS, sensitivity; SPEC, specificity; AUC, area under curve.
Figure 6.
Hierarchical summary receiver operating characteristics (HSROC) curve for MALAT1 in the diagnosis of cancer.
Clinical Utility of the Index Test
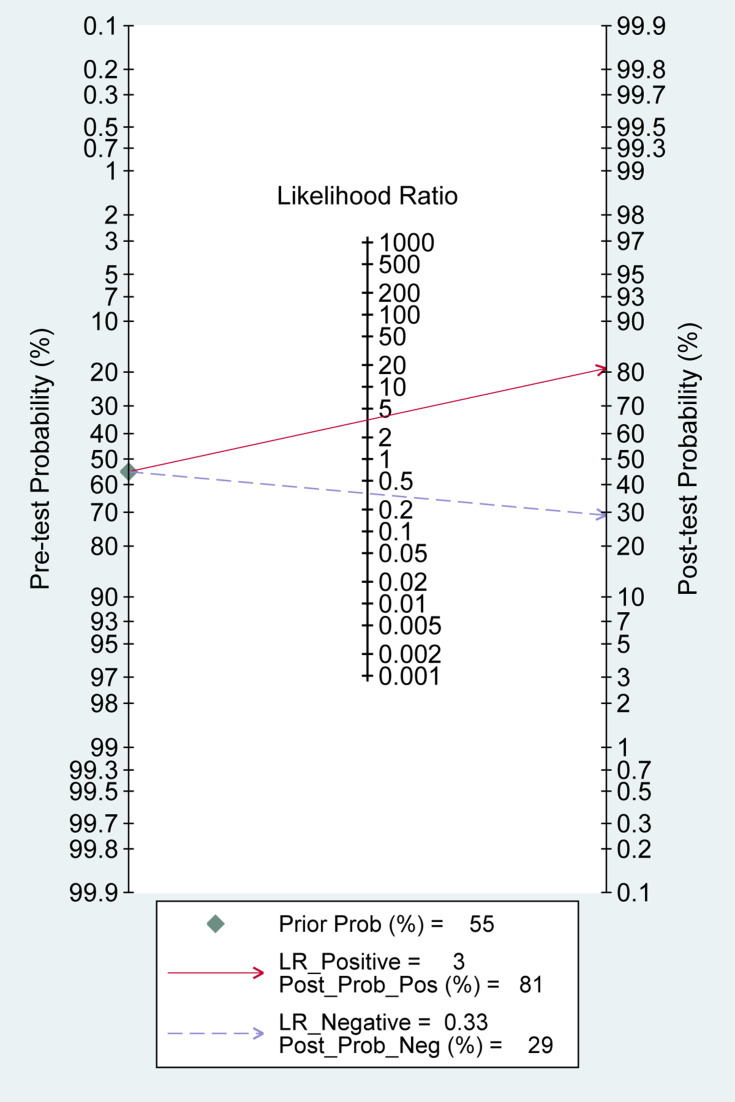
We performed Fagan’s Nomogram to predict the increasing inerrability about a positive diagnosis by using the value of the test, and it is used for estimating posttest probabilities. When MALAT1 assays were tested for all individuals with a pretest probability of 55% to have cancer, a positive result would improve posttest probability having cancer to 81%, whereas a negative result would drop the posttest probability to 29% (Figure 7).
Figure 7.
Fagan’s Nomogram for calculation of posttest probabilities.
Abbreviation: LR, likelihood ratio.
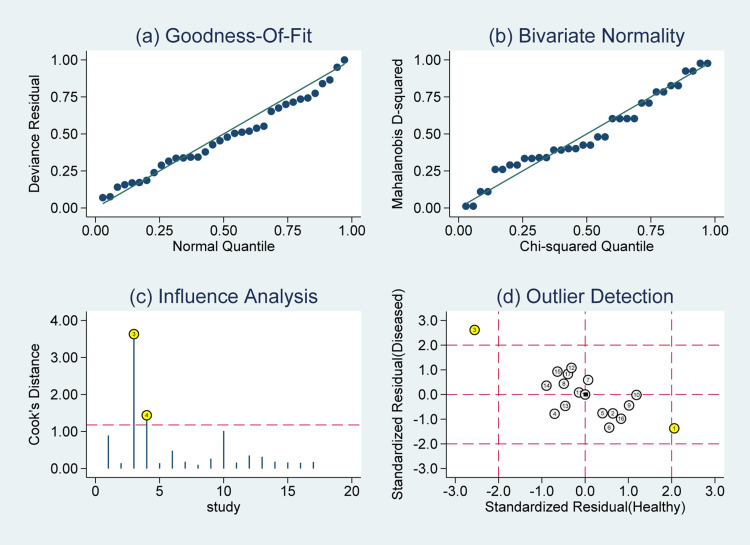
Influence Analysis and Robustness Tests
God-of-ft and bivariate normality analysis (Figure 8A and B) showed that the bivariate model was moderately robust. And then, we performed sensitivity analysis after further excluded 3 outliers found by influence analysis and outlier detection in Figure 8C and D. The sensitivity dropped from 0.74 to 0.73, the specificity increased from 0.79 to 0.80, the PLR increased from 3.48 to 3.69, the NLR remain the same as 0.33, the DS increased from 2.34 to 2.40, the DOR increased from 10.41 to 11.04, and the AUC increased from 0.83 to 0.84, showing no significant change after the exclusion of the outliers (Supplementary Figures 1–3).
Figure 8.
Graphs for sensitivity analysis: (A) goodness of fit, (B) bivariate normality, (C) influence analysis, and (D) outlier detection.
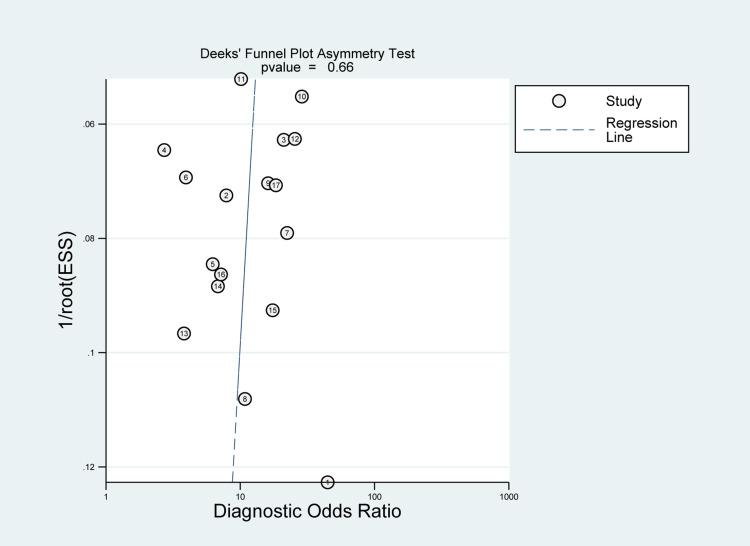
Publication Bias Evaluation
The publication bias was evaluated by Deeks’ funnel plot in this pooled analysis and indicated no significant publication bias for MALAT1 (P=0.66) (Figure 9).
Figure 9.
Graph of Deeks’ funnel plot asymmetry test.
Subgroup Analysis
According to the cancer type, we performed the subgroup analysis for lung cancer and breast cancer (Supplementary Table 1 and Supplementary Figure 4). The pooled sensitivity, specificity, PLR, DLR, DS, DOR and AUC for lung cancer were 0.76, 0.85, 5.02, 0.28, 2.88, 17.73 and 0.88, respectively. Whereas for breast cancer, the pooled sensitivity, specificity, PLR, DLR, DS, DOR and AUC were 0.78, 0.78, 3.56, 0.28, 2.55, 12.86 and 0.84, respectively. Meanwhile, subgroup analysis, according to different sample types, were also performed (Supplementary Table 2 and Supplementary Figure 5). The pooled results from studies based on serum sample detection were as follows: sensitivity, 0.81; specificity, 0.75; PLR, 3.18; NLR, 0.26; DS 2.52; DOR 12.43 and AUC, 0.84. As for studies based on plasma sample detection, the pooled results were as follows: sensitivity, 0.63; specificity, 0.85; PLR, 4.29; NLR, 0.43; DS 2.29; DOR 9.90 and AUC, 0.84.
Discussion
In recent years, lncRNA MALAT1 has been reported to participate in the prognosis of tumors by regulating a variety of biological processes, such as proliferation, apoptosis, invasion and angiogenesis.22,42–44 Furthermore, several studies demonstrated that aberrant expression of MALAT1 in tumor tissues or body fluids might serve as a biomarker for tumor diagnosis and prognosis.28,29,31,32 An ideal biomarker should be easily obtained with minimal risk and discomfort to patients, as well as detectable and reproducible in standard clinical laboratories. Luckily, lncRNA MALAT1 possesses all these features as a biomarker.45 MALAT1 is not only expressed in tissues but also detectable in body fluids, such as blood and urine,24,38 which are easily obtainable with minimal damage to the patient. However, the diagnostic accuracy of MALAT1 still remains inconsistent and controversial. Thus, in our analysis, 17 studies with 3255 subjects were pooled to evaluate the diagnostic value of lncRNA MALAT1 in human cancers detection.
In the present pooled analysis, the results showed that the sensitivity, specificity and AUC of MALAT1 were 0.74, 0.79 and 0.83, respectively, indicating a capacity to distinguish cancer patients from normal people. Meanwhile, a pooled PLR of 3.48 and NLR of 0.33 implied that patients with cancer have a 3.48-fold higher possibility of being MALAT1 positive for patients with cancer compared with controls, and 33% of all individuals have negative results, suggesting that the diagnostic value of MALAT1 is relatively moderate. Moreover, the pooled DS and DOR were 2.34 and 10.41, reflecting a moderate level of diagnostic accuracy. In addition, Fagan’s nomogram showed MALAT1 could raise the probability of cancer detection by 26% (post-test probability 81%, pre-test probability 55%).
Similar analysis were also performed in lung cancer and breast cancer subgroup. The pooled AUC, DS and DOR were 0.88, 2.88 and 17.73 in lung cancer, and 0.84, 2.55 and 12.86 in breast cancer, indicating that MALAT1 could act as an effective diagnostic biomarker in both types of these two cancers. Meanwhile, MALAT1 achieved a high diagnostic value in circulating blood for cancer detection, with a pooled AUC of 0.84, DS of 2.52 and DOR of 12.43 in serum sample detection, and an AUC of 0.84, DS of 2.29 and DOR of 9.90 in plasma sample detection. Furthermore, a pooled sensitivity of 0.81 and specificity of 0.75 in serum, and a pooled sensitivity of 0.63 and specificity of 0.85 in plasma, demonstrated that circulating MALAT1 had relatively moderate accuracy in human cancer detection.
The pooled analysis in our study also had its limitations. First, a very high ratio of data in Chinese populations was included in our analysis, which might contribute to inevitable publication bias. Second, not all of the studies reported the cutoff values of lncRNA MALAT1. Third, only studies published in English or Chinese were screened and included in the pooled analysis, studies published in other languages should not be ignored.
Conclusion
In summary, these pooled results demonstrated that lncRNA MALAT1 could serve as a detection and diagnosis biomarker in various human cancers, especially in serum/plasma, with a relatively moderate accuracy in distinguishing cancer patients from all individuals. We also proved that the moderate diagnostic value of MALAT1 in lung cancer and breast cancer subgroup; However, more cancer types studies with a large sample size are needed to strengthen our conclusion.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grant numbers 81871963, 81572335) and the China Scholarship Council (No:201906310088 to Yue Zhao).
Disclosure
All authors declare that there are no conflicts of interest.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. doi: 10.3322/caac.21590 [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi: 10.3322/caac.21551 [DOI] [PubMed] [Google Scholar]
- 3.Poon RT. Prevention of recurrence after resection of hepatocellular carcinoma: a daunting challenge. Hepatology. 2011;54(3):757–759. doi: 10.1002/hep.24569 [DOI] [PubMed] [Google Scholar]
- 4.Cree IA, Kurbacher CM, Lamont A, Hindley AC, Love S. A prospective randomized controlled trial of tumour chemosensitivity assay directed chemotherapy versus physicians? Choice in patients with recurrent platinum-resistant ovarian cancer. Anticancer Drugs. 2007;18(9):1093–1101. doi: 10.1097/CAD.0b013e3281de727e [DOI] [PubMed] [Google Scholar]
- 5.Yang K, Park W, Huh SJ, Bae DS, Kim BG, Lee JW. Clinical outcomes in patients treated with radiotherapy after surgery for cervical cancer. Radiat Oncol J. 2017;35(1):39–47. doi: 10.3857/roj.2016.01893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Car LT, Papachristou N, Urch C, et al. Preventing delayed diagnosis of cancer: clinicians’ views on main problems and solutions. J Glob Health. 2016;6(2):020901. doi: 10.7189/jogh.06.020901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gildea TR, DaCosta Byfield S, Hogarth DK, Wilson DS, Quinn CC. A retrospective analysis of delays in the diagnosis of lung cancer and associated costs. Clinicoecon Outcomes Res. 2017;9:261–269. doi: 10.2147/CEOR.S132259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bradley CJ. Cancer, Financial Burden, and Medicare Beneficiaries. J Clin Oncol. 2017;35(22):2461–2462. doi: 10.1200/JCO.2017.73.1877 [DOI] [PubMed] [Google Scholar]
- 9.Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77(15):3965–3981. doi: 10.1158/0008-5472.CAN-16-2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. 2013;20(3):300–307. doi: 10.1038/nsmb.2480 [DOI] [PubMed] [Google Scholar]
- 11.Beermann J, Piccoli MT, Viereck J, Thum T. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev. 2016;96(4):1297–1325. doi: 10.1152/physrev.00041.2015 [DOI] [PubMed] [Google Scholar]
- 12.Xie CR, Wang F, Zhang S, et al. Long noncoding RNA HCAL facilitates the growth and metastasis of hepatocellular carcinoma by acting as a ceRNA of LAPTM4B. Mol Ther Nucleic Acids. 2017;9:440–451. doi: 10.1016/j.omtn.2017.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Terashima M, Tange S, Ishimura A, Suzuki T. MEG3 long noncoding RNA contributes to the epigenetic regulation of epithelial-mesenchymal transition in lung cancer cell lines. J Biol Chem. 2017;292(1):82–99. doi: 10.1074/jbc.M116.750950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsumura K, Kawasaki Y, Miyamoto M, et al. The novel G-quadruplex-containing long non-coding RNA GSEC antagonizes DHX36 and modulates colon cancer cell migration. Oncogene. 2017;36(9):1191–1199. doi: 10.1038/onc.2016.282 [DOI] [PubMed] [Google Scholar]
- 15.Wang M, Dong X, Feng Y, Sun H, Shan N, Lu T. Prognostic role of the long non-coding RNA, SPRY4 intronic transcript 1, in patients with cancer: a meta-analysis. Oncotarget. 2017;8(20):33713–33724. doi: 10.18632/oncotarget.16735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tian GW, Li N, Xin Y. Prognostic and clinicopathological significance of CCAT2 in Chinese patients with various tumors. Int J Biol Markers. 2017;32(3):e344–e351. doi: 10.5301/ijbm.5000281 [DOI] [PubMed] [Google Scholar]
- 17.Ji P, Diederichs S, Wang W, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22(39):8031–8041. doi: 10.1038/sj.onc.1206928 [DOI] [PubMed] [Google Scholar]
- 18.Eissmann M, Gutschner T, Hammerle M, et al. Loss of the abundant nuclear non-coding RNA MALAT1 is compatible with life and development. RNA Biol. 2012;9(8):1076–1087. doi: 10.4161/rna.21089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Z, Katsaros D, Biglia N, et al. High expression of long non-coding RNA MALAT1 in breast cancer is associated with poor relapse-free survival. Breast Cancer Res Treat. 2018;171(2):261–271. doi: 10.1007/s10549-018-4839-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li C, Cui Y, Liu LF, et al. High expression of long noncoding RNA MALAT1 indicates a poor prognosis and promotes clinical progression and metastasis in bladder cancer. Clin Genitourin Cancer. 2017;15(5):570–576. doi: 10.1016/j.clgc.2017.05.001 [DOI] [PubMed] [Google Scholar]
- 21.Malakar P, Shilo A, Mogilevsky A, et al. Long noncoding RNA MALAT1 promotes hepatocellular carcinoma development by SRSF1 upregulation and mTOR activation. Cancer Res. 2017;77(5):1155–1167. doi: 10.1158/0008-5472.CAN-16-1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhuang M, Zhao S, Jiang Z, et al. MALAT1 sponges miR-106b-5p to promote the invasion and metastasis of colorectal cancer via SLAIN2 enhanced microtubules mobility. EBioMedicine. 2019;41:286–298. doi: 10.1016/j.ebiom.2018.12.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Y, Jiang M, Ni L, Xu Z, Xu J. Diagnostic value of serum carcinoembryonic antigen and carbohydrate antigen 724 combined with long non-coding RNA MALAT1 detection in gastric cancer. Cancer Res Clin. 2019;31:88–92. [Google Scholar]
- 24.Yazarlou F, Modarressi MH, Mowla SJ, et al. Urinary exosomal expression of long non-coding RNAs as diagnostic marker in bladder cancer. Cancer Manag Res. 2018;10:6357–6365. doi: 10.2147/CMAR.S186108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mei H, Liu Y, Zhou Q, Hu K, Liu Y. Long noncoding RNA MALAT1 acts as a potential biomarker in cancer diagnosis and detection: a meta-analysis. Biomark Med. 2019;13(1):45–54. doi: 10.2217/bmm-2018-0128 [DOI] [PubMed] [Google Scholar]
- 26.Rutter CM, Gatsonis CA. A hierarchical regression approach to meta-analysis of diagnostic test accuracy evaluations. Stat Med. 2001;20(19):2865–2884. doi: 10.1002/sim.942 [DOI] [PubMed] [Google Scholar]
- 27.Wu J, Yu L, Wang X, Li J, Zhu M, Yan F. Expression and clinical significance of plasma MALAT1 in patients with breast cancer. Chin J Lab Med. 2018;41:92–96. [Google Scholar]
- 28.Wang J, Zhao S, Ouyang S, Liao L. Expression and clinical significance of plasma long non-coding RNA MALAT1 in patients with oral squamous cell carcinoma. J Pract Stomatol. 2018;34:188–192. [Google Scholar]
- 29.Li Z, Zhou J, Zhou L, Yang H, Li M. Expression and clinical significance of long non-coding RNA MALAT1 in breast cancer. J Med Res. 2018;47:79–81. [Google Scholar]
- 30.Huang SK, Luo Q, Peng H, et al. A panel of serum noncoding RNAs for the diagnosis and monitoring of response to therapy in patients with breast cancer. Med Sci Monit. 2018;24:2476–2488. doi: 10.12659/MSM.909453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhan Y, Du L, Wang L, et al. Expression signatures of exosomal long non-coding RNAs in urine serve as novel non-invasive biomarkers for diagnosis and recurrence prediction of bladder cancer. Mol Cancer. 2018;17(1):142. doi: 10.1186/s12943-018-0893-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang WW, Zhou XL, Song YJ, Yu CH, Zhu WG, Tong YS. Combination of long noncoding RNA MALAT1 and carcinoembryonic antigen for the diagnosis of malignant pleural effusion caused by lung cancer. Onco Targets Ther. 2018;11:2333–2344. doi: 10.2147/OTT.S157551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He B, Zeng J, Chao W, et al. Serum long non-coding RNAs MALAT1, AFAP1-AS1 and AL359062 as diagnostic and prognostic biomarkers for nasopharyngeal carcinoma. Oncotarget. 2017;8(25):41166–41177. doi: 10.18632/oncotarget.17083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huo Y, Li Q, Wang X, et al. MALAT1 predicts poor survival in osteosarcoma patients and promotes cell metastasis through associating with EZH2. Oncotarget. 2017;8(29):46993–47006. doi: 10.18632/oncotarget.16551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zidan HE, Karam RA, El-Seifi OS, Abd Elrahman TM. Circulating long non-coding RNA MALAT1 expression as molecular biomarker in Egyptian patients with breast cancer. Cancer Genet. 2018;220:32–37. doi: 10.1016/j.cancergen.2017.11.005 [DOI] [PubMed] [Google Scholar]
- 36.Han C. The expression and clinical significance of long non-coding RNA MALAT1 in endometrial cancer. Chin J Family Plann Gynecotokology. 2016;8:51–55. [Google Scholar]
- 37.Wen Z, Li X. Diagnostic value of long non-coding RNA metastasis-associated lung adenocarcinoma transcript 1 in serum for clinical diagnosis of lung cancer. J Clin Pulm Med. 2016;21:2154–2158. [Google Scholar]
- 38.Duan W, Du L, Jiang X, et al. Identification of a serum circulating lncRNA panel for the diagnosis and recurrence prediction of bladder cancer. Oncotarget. 2016;7(48):78850–78858. doi: 10.18632/oncotarget.12880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peng H, Wang J, Li J, et al. A circulating non-coding RNA panel as an early detection predictor of non-small cell lung cancer. Life Sci. 2016;151:235–242. doi: 10.1016/j.lfs.2016.03.002 [DOI] [PubMed] [Google Scholar]
- 40.Ren S, Wang F, Shen J, et al. Long non-coding RNA metastasis associated in lung adenocarcinoma transcript 1 derived miniRNA as a novel plasma-based biomarker for diagnosing prostate cancer. Eur J Cancer. 2013;49(13):2949–2959. doi: 10.1016/j.ejca.2013.04.026 [DOI] [PubMed] [Google Scholar]
- 41.Weber DG, Johnen G, Casjens S, et al. Evaluation of long noncoding RNA MALAT1 as a candidate blood-based biomarker for the diagnosis of non-small cell lung cancer. BMC Res Notes. 2013;6(1):518. doi: 10.1186/1756-0500-6-518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liao K, Lin Y, Gao W, et al. Blocking lncRNA MALAT1/miR-199a/ZHX1 axis inhibits glioblastoma proliferation and progression. Mol Ther Nucleic Acids. 2019;18:388–399. doi: 10.1016/j.omtn.2019.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu Y, Lin J, Fang H, et al. Targeting the MALAT1/PARP1/LIG3 complex induces DNA damage and apoptosis in multiple myeloma. Leukemia. 2018;32(10):2250–2262. doi: 10.1038/s41375-018-0104-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qiu JJ, Lin XJ, Tang XY, Zheng TT, Lin YY, Hua KQ. Exosomal metastasis associated lung adenocarcinoma transcript 1 promotes angiogenesis and predicts poor prognosis in epithelial ovarian cancer. Int J Biol Sci. 2018;14(14):1960–1973. doi: 10.7150/ijbs.28048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li ZX, Zhu QN, Zhang HB, Hu Y, Wang G, Zhu YS. MALAT1: a potential biomarker in cancer. Cancer Manag Res. 2018;10:6757–6768. doi: 10.2147/CMAR.S169406 [DOI] [PMC free article] [PubMed] [Google Scholar]