Abstract
Purpose
miR-205 is a tumor suppressor and plays an important role in tumor invasiveness. However, the role of miR-205 in human gastric cancer (GC) epithelial-mesenchymal transition (EMT) remains unclear. The aim of this study was to investigate the molecular mechanism of miR-205 in the regulation of EMT in GC invasion.
Materials and Methods
Quantitative polymerase chain reaction (qPCR) was used to detect the expression of miR-205 in GC. Further, the correlation between the pathological parameters and prognosis of GC was statistically analyzed. A transwell model was used to evaluate the effect of miR-205-3p on the invasion and migration of GC cells. qPCR, western blotting, and luciferase assay were performed to analyze the relationship and target effects between miR-205-3p and the expression of zinc finger electron box binding homologous box 1 (ZEB1) and 2 (ZEB2).
Results
We found that the levels of miR-205-3p were significantly lower (P<0.05) in GC tissues than in matched normal tissues. Additionally, the expression of miR-205-3p was related to the tumor invasion depth, lymph node metastasis, lymph node invasion, and tumor, node, metastasis stage. Patients with lower miR-205-3p expression levels in the tumors had a poorer prognosis. The in vitro assays indicated that miR-205-3p could affect the invasion ability and EMT of GC cells by targeting the expression of both ZEB1 and ZEB2.
Conclusions
miR-205-3p promotes GC progression and affects the prognosis of patients by targeting both ZEB1 and ZEB2 to directly influence EMT.
Keywords: miR-205, Stomach neoplasms, Epithelial-mesenchymal transition, Prognosis
INTRODUCTION
Gastric cancer (GC) is one of the most common malignancies of the digestive system in China [1,2]. Most GC deaths are caused by cancer cell invasion and metastasis [2]. Thus, it is important to study the underlying mechanisms of invasion and migration of GC. MicroRNAs (miRNAs) are small noncoding RNAs that act as post-transcriptional regulators of gene expression during tumor development and carcinogenesis [3,4,5]. A series of studies has shown that numerous miRNAs influence the capacity for invasiveness, migration, and proliferation of cancer cells in GC [5]. miR-205 has been reported to be a tumor suppressor in several types of cancer, such as prostate cancer [6], breast cancer [7], and colon cancer [8]. Furthermore, previous studies have shown that miR-205 is downregulated in GC and the inhibition of miR-205 significantly promotes GC cell proliferation via cell-cycle progression [9]. However, the mechanisms by which miR-205-3p promotes GC cell invasion, especially through epithelial-mesenchymal transition (EMT) induction, have not yet been reported.
EMT plays a key role in cancer cell invasion during the progression of cancer metastasis. Downregulation of adhesion molecules, such as E-cadherin, and upregulation of mesenchymal markers, such as N-cadherin, are core events during EMT. After EMT, the cancer cells lose their connections to epithelial cells and acquire an interstitial cell phenotype under the influence of a number of factors; finally, they gain the ability to migrate [10,11]. Accumulating evidence indicates that zinc finger electron box binding homologous box 1 (ZEB1) and 2 (ZEB2) act as EMT-regulating transcription factors by negatively regulating the expression of E-cadherin and promoting invasiveness and migration in many tumors [12,13,14]. Upon searching miRBase, TargetScan release 5.0 (http://www.targetscan.org/), and PicTar databases, ZEB1 and ZEB2 were identified as candidate target genes for miR-205-3p. Thus, we speculated that miR-205-3p possibly promotes the progression of GC through its EMT-inducing effect, which is influenced by targeting the expression of both ZEB1 and ZEB2. In the present study, we investigated the correlation between miR-205-3p expression levels in GC tissues and clinicopathological parameters. The association between miR-205-3p, ZEB1 and ZEB2 expression, GC cell invasion, and EMT markers was also investigated in vitro. Finally, we hypothesized that miR-205-3p promotes GC cell invasion and poor prognosis by directly targeting ZEB1 and ZEB2, which then induce EMT.
MATERIALS AND METHODS
Samples and patients
All fresh GC tissue samples were collected with the written informed consent of 70 patients who underwent GC resection at the Department of General Surgery of Tongde Hospital of Zhejiang Province (People's Republic of China) from 2012 to 2013. The histological tumor type was diagnosed by 3 independent pathologists and all cases were classified according to the American Joint Committee on Cancer classification (7th and 8th versions) of GC tumors [15,16]. The matched normal gastric epithelial tissues, which were collected from an area more than 5 cm away from the tumors, were also verified at the same time. None of the patients received chemotherapy prior to surgery. The study designs and methods were approved by the Ethics Committee of Tongde Hospital of Zhejiang Province (No. 2019074). Upon admission, all patients or their relatives provided informed consent within the written treatment contract prior to their inclusion in the study. All patients were followed-up for over 5 years or until December 2018. The survival time was calculated from the date of surgery to the end of the follow-up period and/or the date of death. The age of the GC patients ranged from 17 to 80 years (with a median age of 59.3 years). The clinicopathological characteristics of the GC patients are summarized in Table 1.
Table 1. Association between miR-205-3p expression and clinicopathological factors.
| Clinical parameters | miR-205-3p expression | ||||
|---|---|---|---|---|---|
| Low | High | t/χ2 | P | ||
| Age (yr) | 58.56±11.51 | 63.06±8.02 | 1.461 | 0.149 | |
| Gender | 0.308 | 0.579 | |||
| Man | 33 (61.1) | 11 (68.8) | |||
| Woman | 21 (38.9) | 5 (31.3) | |||
| Location | 2.245 | 0.326 | |||
| Proximal | 10 (18.5) | 5 (31.3) | |||
| Middle | 24 (44.4) | 4 (25.0) | |||
| Distal | 20 (37.0) | 7 (43.8) | |||
| Size (cm) | 0.054 | 0.816 | |||
| ≥5 | 32 (59.3) | 10 (62.5) | |||
| <5 | 22 (40.7) | 6 (37.5) | |||
| Histology type | 7.616 | 0.055 | |||
| Papillary adenocarcinoma | 2 (3.7) | 3 (18.8) | |||
| Tubular adenocarcinoma | 36 (66.7) | 12 (75.0) | |||
| Mucinous adenocarcinoma | 4 (7.4) | 1 (6.3) | |||
| Signet-ring cell carcinoma | 12 (22.2) | 0 (0.0) | |||
| Lauren classification | 0.235 | 0.628 | |||
| Diffuse type | 34 (63.0) | 9 (56.3) | |||
| Intestinal type | 20 (37.0) | 7 (43.8) | |||
| Differentiation | 10.952 | 0.004 | |||
| Well | 0 (0.0) | 3 (18.8) | |||
| Moderately | 12 (22.2) | 4 (25.0) | |||
| Poorly | 42 (77.8) | 9 (56.3) | |||
| Invasion depth (T grade) | 37.926 | 0.000 | |||
| T1 | 0 (0.0) | 5 (31.3) | |||
| T2 | 3 (5.6) | 7 (43.8) | |||
| T3 | 32 (59.3) | 4 (25.0) | |||
| T4 | 19 (35.2) | 0 (0.0) | |||
| Lymphatic metastasis (N grade) | 50.087 | 0.000 | |||
| N0 | 1 (1.9) | 9 (56.3) | |||
| N1 | 1 (1.9) | 5 (31.3) | |||
| N2 | 16 (29.6) | 2 (12.5) | |||
| N3 | 36 (66.7) | 0 (0.0) | |||
| Distant metastasis (M grade) | 3.457 | 0.063 | |||
| M0 | 44 (81.5) | 16 (100) | |||
| M1 | 10 (18.5) | 0 (1.0) | |||
| TNM stages | 64.959 | 0.000 | |||
| I | 0 (0.0) | 8 (50.0) | |||
| II | 1 (1.9) | 8 (50.0) | |||
| III | 43 (79.6) | 0 (0.0) | |||
| IV | 10 (18.5) | 0 (0.0) | |||
| Lymphatic invasion | 34.277 | 0.000 | |||
| Yes | 53 (98.1) | 6 (37.5) | |||
| No | 1 (1.9) | 10 (62.5) | |||
| Vascular invasion | 13.547 | 0.000 | |||
| No | 5 (9.3) | 8 (50.0) | |||
| Yes | 49 (90.7) | 8 (50.0) | |||
All cases were classified according to the American Joint Committee on Cancer (2016, 8th version) pathological classification of gastric cancer. Invasion depth (T grade) grade T4 includes T4a and T4b. Lymphatic metastasis (N grade) grade N3 includes N3a and N3b. TNM grade I includes Ia and Ib, TNM grade II includes IIa and IIb, and TNM grade III includes IIIa, IIIb, and IIIc.
TNM = tumor, node, metastasis.
Cell culture
Human GC cell lines (7901, MKN-45, AGS, and GES-1) were purchased from the Cell Bank of Shanghai Institute of Cell Biology (Shanghai, People's Republic of China) and cultured in RPMI-1640 (HyClone, Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS; Sigma-Aldrich Co., St. Louis, MO, USA) at 37°C in a humidified atmosphere containing 5% CO2.
miRNA inhibitor and mimic transfection
AGS and 7901 cells were seeded (at a density of 1×105 cells per well) into 6-well plates. After 24 hours, the cells were transfected with a micrON miR-205-3p mimic and inhibitor (miR10009197-1-5 or miR20009197-1-5; RiboBio Co. Ltd., Guangzhou, China) or the corresponding negative controls at a final concentration of 10 nM using Lipofectamine 2000 transfection reagent (Thermo Fisher Scientific, Waltham, MA, USA), according to the manufacturer's protocol. After transfection, the cells were collected for future assays, such as quantitative polymerase chain reaction (qPCR), an invasion assay, and western blotting (WB).
Luciferase reporter assays
The pYr-mirTarget-ZEB1-3′UTR-wild-type (WT), pYr-mirTarget-ZEB2-3′UTR-WT, and corresponding mutated luciferase vectors, which contained the putative binding site or mutated site of miR-205-3p, were purchased from Yinrun Biotechnology (Changsha, China). HEK293 cells were seeded into 96-well plates. The cells were then co-transfected with the WT or mutated (Mut) reporter plasmids using Lipofectamine 2000 reagent along with the miR-205-3p mimic (100 nM). Forty-eight hours after transfection, the luciferase activity was assessed using the DualGlo Luciferase Assay System (Promega, Tokyo, Japan), according to the manufacturer's instructions. All transfection experiments were conducted in triplicate and were repeated 3 times independently. To further confirm whether ZEB1 and ZEB2 are target genes for miR-205-3p in GC cells, following transfection with an miR-205-3p inhibitor or mimic, the mRNA and protein expression levels of ZEB1 and ZEB2 in GC cells were assessed using qPCR and WB, respectively.
RNA isolation and qPCR
Total RNA was isolated from the tissue samples and GC cells, according to the protocol of the RNAsimple Total RNA kit (DP419; Tiangen Biotech Co. Ltd., Beijing, China). Reverse transcription was performed using a One-step PrimeScript miRNA cDNA synthesis kit (D350A; TaKaRa Biotechnology Co. Ltd., Dalian, China) and a PrimeScript™ RT reagent kit with gDNA Eraser (RR047A; TaKaRa Biotechnology Co. Ltd.). To detect the expression levels of miR-205-3p, ZEB1, ZEB2, E-cadherin, and N-cadherin, qPCR was carried out in an MX3000P system (Stratagene, La Jolla, CA, USA) using gene-specific primers with a SYBR Premix ExTaq kit (DRR081A; TaKaRa Biotechnology Co. Ltd.). All reactions were performed in triplicate. U6 (RNU6B) and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) were used as internal standards for normalization of miR-205-3p expression levels and ZEB1, ZEB2, E-cadherin, and N-cadherin expression levels, respectively. The primers for the candidate genes were selected from PrimerBank (https://pga.mgh.harvard.edu/primerbank/) and are listed in Table 2. The qPCR reaction conditions used were as follows: initial denaturation (4 minutes at 95°C) and then 40 cycles of denaturation at 95°C for 20 seconds, annealing at 58°C for 20 seconds, and extension at 72°C for 20 seconds. The relative expression levels were calculated using the 2−∆∆ct method.
Table 2. Sequence of primers used in this study.
| Primer name | Sequence (5′–3′) | Annealing temperatures (°C) |
|---|---|---|
| miRNA-205-3p | GATTTCAGTGGAGTGAAGTTC | 58 |
| U6B | CGCTTCACGAATTTGCGTGTCAT | 58 |
| GAPDH-F | ACAACTTTGGTATCGTGGAAGG | 50–60 |
| GAPDH-F | GCCATCACGCCACAGTTTC | 50–60 |
| ZEB1-F | GATGATGAATGCGAGTCAGATGC | 55 |
| ZEB1-R | ACAGCAGTGTCTTGTTGTTGT | 55 |
| ZEB2-F | CAAGAGGCGCAAACAAGCC | 58 |
| ZEB2-R | GGTTGGCAATACCGTCATCC | 58 |
| E-cadherin-F | CGAGAGCTACACGTTCACGG | 56 |
| E-cadherin-R | GGGTGTCGAGGGAAAAATAGG | 56 |
| N-cadherin-F | TCAGGCGTCTGTAGAGGCTT | 58 |
| N-cadherin-R | ATGCACATCCTTCGATAAGACTG | 58 |
GAPDH = glyceraldehyde 3-phosphate dehydrogenase.
Migration and invasion assays
The migration assay was performed using transwell plates containing membranes with 8 μm pores (3422; Corning Incorporated, Corning, NY, USA). Cell invasion assays were performed using invasion chambers pre-coated with Matrigel (354480; BD, Franklin Lakes, NJ, USA). The cells were resuspended (at a density of 2×105 cells for invasion assays and 5×104 cells for migration assays) in serum-free medium and seeded into the upper chamber; RPMI 1640 medium supplemented with 20% FBS was added to the lower chamber as a chemoattractant. After 24 or 48 hours, the cells were fixed and stained. Non-invading cells in the upper chambers were removed with cotton swabs. Then, the number of migrating or invading cells that had attached to the lower surface were counted in 5 random fields under a microscope (×200).
WB
Briefly, protein was extracted from cells using radio immunoprecipitation assay lysis buffer (Beyotime, Shanghai, China). Samples were separated using 10% sodium dodecyl sulphate-polyacrylamide gel electrophoresis and then transferred onto polyvinylidene fluoride membranes. The membranes were blocked using 5% bovine serum albumin for 2 hours and then incubated with primary anti-human antibodies for detection of N-cadherin (#4061 at 1:1,000 dilution; Cell Signaling Technology, Inc., Danvers, MA, USA), E-cadherin (#14472 at 1:2,000 dilution; Cell Signaling Technology, Inc.), ZEB1 (ab203829, at 1:2,000 dilution; Abcam, San Francisco, CA, USA), ZEB2 (ab138222, at 1:4,000 dilution; Abcam) and GAPDH (ab181602, at 1:3,000 dilution; Abcam) overnight at 4°C. The membranes were incubated with the corresponding horseradish peroxidase-labeled goat anti-rabbit or anti-mouse immunoglobulin G antibody for 1 hour. After washing, the WB signal was detected using an enhanced chemiluminescence system (Bio-Rad, Hercules, CA, USA).
Statistical analyses
All statistical analyses were performed using Statistical Package for the Social Sciences version 13.0 (SPSS Inc., Chicago, IL, USA) and Prism software. Statistical differences in miR-205-3p expression between cancer tissues and normal tissues were determined using a 2-tailed paired student's t-test. Data from the cell migration and cell invasion assays, WB, and qPCR were expressed as mean±standard error and the significant differences were determined using independent samples t-test. The receiver operating characteristic (ROC) curve was used to analyze the cut-off value of miR-205-3p expression in GC samples. The relationship between miR-205-3p expression and clinicopathological characteristics was tested using the χ2 test. Survival curves were plotted using the Kaplan-Meier method and were compared by log-rank test. The significance of various survival-related variables in this GC cohort was assessed using a Cox regression model in a multivariate analysis. A P-value of less than 0.05 (P<0.05) was considered statistically significant.
RESULTS
miR-205-3p is downregulated in GC tissues and GC cell lines
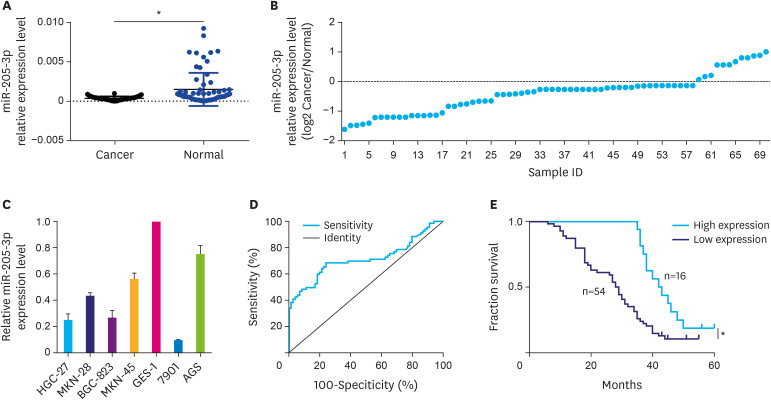
The qPCR results indicated that miR-205-3p expression in GC tissues was significantly lower (P<0.05) than that in matched normal tissues (Fig. 1A and B). At the same time, miR-205-3p expression was found to be lower in all the 4 GC cell lines compared to the normal gastric epithelial cell line GES-1. Among them, the highest and lowest expression levels of miR-205-3p were detected in AGS and 7901 cells, respectively (Fig. 1C). These 2 cell lines were, therefore, selected for the in vitro experiments.
Fig. 1. Expression of miR-205-3p in GC tissues and Kaplan-Meier survival curves of GC patients. (A, B) The expression of miR-205-3p in 70 paired GC tissues. (C) The expression of miR-205-3p in GC cell lines. (D) Receiver operating characteristic curve was employed to establish a cut-off value for the expression levels of miR-205-3p in GC tissues. (E) Kaplan-Meier survival curves of miR-205-3p expression in GC.
GC = gastric cancer.
*P<0.05.
Clinical significance of miR-205-3p in GC
A significant difference was observed in miR-205-3p expression levels between GC tissues and matched normal tissues (0.000363±0.000027 vs. 0.001485±0.000250, t=4.437, P<0.05) (Fig. 1A). The ROC curve was employed to establish a cut-off value for the miR-205-3p expression levels in GC tissues; the area under the ROC cure was 0.7141 (Fig. 1D). Based on the Youden Index (sensitivity+specificity−1) calculation, the relative expression value of miRNA-205-3p in tissues that provided the best accuracy (cut-off value, 0.0004745) was identified and the tumor specimens were classified based on this cut-off value into low-expression and high-expression groups. Finally, among the 70 cases of GC, 54 cases had low expression of miR-205-3p and 16 cases had high expression of miR-205-3p. The decrease in miR-205-3p expression level was found to be associated with tumor differentiation, invasion depth, lymphatic metastasis, tumor, node, metastasis (TNM) stage, lymphatic invasion, and vascular invasion (P<0.05, Table 1) but was unrelated to gender, Lauren classification, histological type, and distant metastasis (P>0.05, Table 2).
Low miR-205-3p expression in GC is associated with poor prognosis
We also analyzed the relationship between the miR-205-3p expression level and the prognosis of GC. In the present cohort of patients (n=70), the overall survival time was 31.46±13.16 months. Kaplan-Meier analysis indicated a significantly worse (P<0.05) survival in patients with low miR-205-3p expression (Fig. 1E). The 5-year survival time of patients with low miR-205-3p expression was significantly shorter than that of patients with high expression (28.46±1.81 months vs. 42.50±2.57 months, χ2=9.798, P<0.01). The 5-year survival rate of patients with low miR-205-3p expression (11.11%) was also significantly lower than that of patients with high miR-205-3p expression (18.75%, P<0.05). However, the Cox multivariate analysis showed that TNM stage, Lauren classification, vascular invasion, and miR-205-3p expression level were not independent prognostic factors in this cohort of GC patients (P>0.05).
miR-205-3p expression affects GC cell invasion and migration
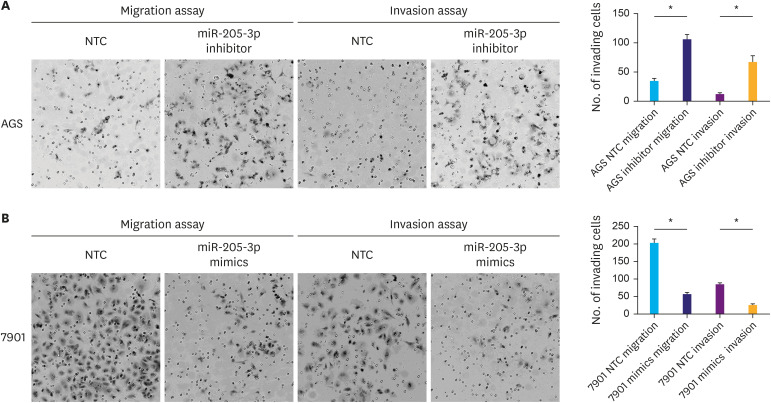
The AGS cell line, which expresses relatively high levels of miR-205-3p, was transfected with an miR-205-3p inhibitor or a negative control. As expected, decreased miR-205-3p expression significantly increased AGS cell migration and invasion abilities compared with corresponding negative control (P<0.05, Fig. 2A). In contrast, 7901 cells express relatively low levels of miR-205-3p. To increase the miR-205-3p expression in 7901 cells, the cells were transfected with miR-205-3p mimics, which resulted in significantly reduced invasion and migration capacity compared to the corresponding negative control (P<0.05, Fig. 2B). These results confirmed that miR-205-3p could affect the invasion and migration ability of GC cells.
Fig. 2. miR-205-3p affects the invasiveness of human gastric cancer cells. (A) Transwell migration and invasion assays showed that a decrease in the miR-205-3p expression in AGS cells led to a significant increase in the number of invading cells. (B) Transwell migration and invasion assays showed that upregulation of miR-205-3p expression in 7901 cells led to a significant decrease in the number of invading cells (magnification: ×200).
*P<0.05.
miR-205-3p targets ZEB1 and ZEB2 and is involved in EMT in vitro
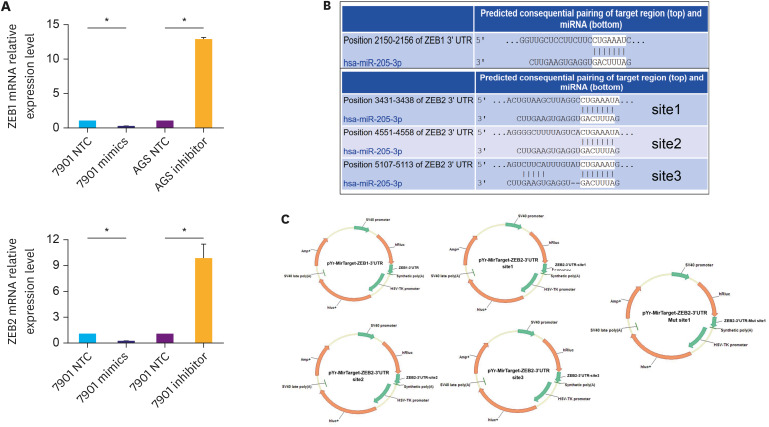
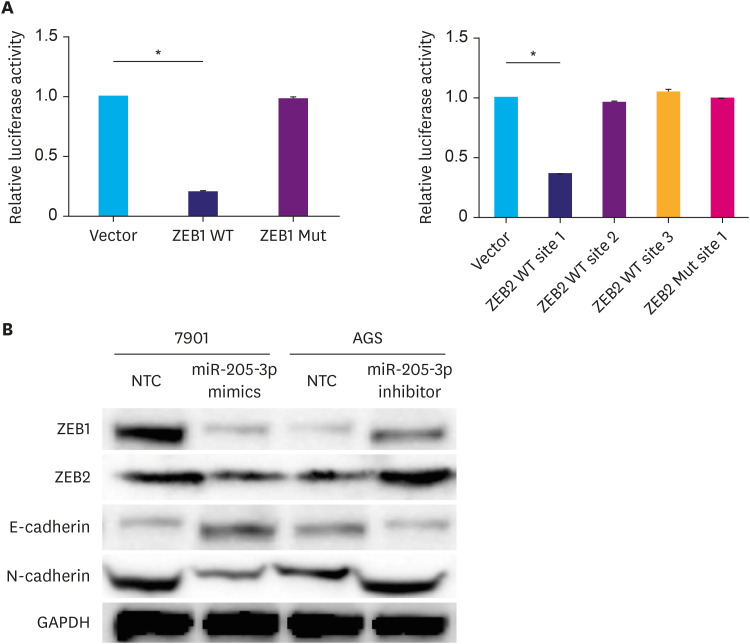
To investigate the possible mechanisms by which miR-205-3p affects GC cell invasiveness, we searched miRBase, TargetScan release 5.0, and PicTar databases and identified ZEB1 and ZEB2 as possible targets of miR-205-3p. The relationship between ZEB1, ZEB2, and miR-205-3p expression levels was first assayed using qPCR in GC cells transfected with an miR-205-3p mimic or inhibitor. We found that the ZEB1 and ZEB2 expression levels were inversely correlated with miR-205-3p expression in GC cells (Fig. 3A). To clarify whether miR-205-3p interacts directly with the 3′-UTR regions of ZEB1 and ZEB2, a binding site investigation was performed, which revealed that while the 3′-UTR of ZEB1 mRNA contained one target site for miR-205-3p, 3′-UTR of ZEB2 mRNA contained 3 target regions (Fig. 3B). Thus, we inserted these 4 wild-type sequences of the human ZEB1 and ZEB2 3′-UTR regions into a pYr-mirTarget-3′UTR luciferase reporter vector and co-transfected them with a miR-205-3p mimic into HEK293 cells (Fig. 3C). The results of the luciferase assay indicated that vectors containing ZEB1 3′-UTR and only site 1 of ZEB2 3′-UTR displayed significantly lower luciferase activity, in the presence of the miR-205-3p mimic, compared to the negative control (both P<0.05, Fig. 4A). These results indicated that miR-205-3p could directly affect ZEB1 expression by binding to its predicted site, whereas site 1 of the ZEB2 3′-UTR region was the true binding site for miR-205-3p. To confirm this, the corresponding mutated binding regions of the ZEB1 and ZEB2 3′-UTRs were cloned into the pYr-mirTarget-3′UTR reporter vector. These were subsequently co-transfected along with miR-205-3p mimics as 4 groups, 2 groups were transfected with the wild-type (ZEB1-wt, ZEB2-site1-wt) 3′-UTR targeting regions and the others were transfected with the mutant (ZEB1-Mut, ZEB2-site1-Mut) 3′-UTR targeting regions. While miR-205-3p was found to significantly reduce the luciferase activity in the WT groups, compared to the negative control (P<0.05, Fig. 4D), it did not alter the activity of the mutated ZEB1 and ZEB2 luciferase reporter groups. This indicated that miR-205-3p specifically and directly targets ZEB1 and ZEB2 by binding to the predicted 3′-UTR region of wild-type ZEB1 and site 1 of the ZEB2 3′-UTR region (P<0.05, Fig. 4A).
Fig. 3. ZEB1 and ZEB2 are both targets of miR-205-3p. (A) Changes in the mRNA levels of ZEB1 and ZEB2 after transfection of gastric cancer cells with miR-205-3p inhibitor (AGS) and mimic (7901). (B) The positions of the miR-205-3p target sites in ZEB1 and ZEB2 3′-UTRs, showing sequence alignment of miR-205-3p with the ZEB1 and ZEB2 3′-UTRs. (C) Profiles of pYr-MirTarget-ZEB1-3′UTR and pYr-MirTarget-ZEB1-3′UTR luciferase reporter plasmids.
ZEB1 = zinc finger electron box binding homologous box 1; ZEB2 = zinc finger electron box binding homologous box 2.
*P<0.05.
Fig. 4. Luciferase assay and WB. (A) HEK293 cells were co-transfected with luciferase reporter plasmids containing either wild-type or mutant ZEB1 and ZEB2 3′-UTRs (indicated as WT or Mut) along with the miR-205-3p mimic (100 nM). The luciferase activity of wild-type ZEB1 and ZEB2 site 1 was found to be much lower than that of mutant type ZEB1, ZEB2, and negative control. This indicated that miR-205-3p direct targets ZEB1 and the binding site 1 of ZEB2. (B) Changes in protein expression levels of ZEB1, ZEB2, and EMT markers (E-cadherin and N-cadherin) after transfection of gastric cancer cells with miR-205-3p inhibitor (AGS) and mimic (7901).
ZEB1 = zinc finger electron box binding homologous box 1; ZEB2 = zinc finger electron box binding homologous box 2; WT = wild-type; Mut = mutated; GAPDH = glyceraldehyde 3-phosphate dehydrogenase.
*P<0.05.
Furthermore, qPCR and WB analyses also showed that ZEB1 and ZEB2 were significantly downregulated in 7901 cells transfected with the miR-205-3p mimic, while the 2 proteins were significantly upregulated in AGS cells transfected with the miR-205-3p inhibitor, compared to the corresponding negative controls (P<0.05, Figs. 3A and 4B). Previous studies have indicated that ZEB1 and ZEB2 are inhibitors of E-cadherin, which is involved in tumor EMT [17,18]. Therefore, EMT markers were also investigated in the present study. We confirmed that N-cadherin expression was significantly increased (P<0.05), while E-cadherin expression was significantly decreased (P<0.05) in AGS cells transfected with the miR-205-3p inhibitor (Fig. 4B). Opposite patterns of N-cadherin and E-cadherin expression were detected in 7901 cells transfected with the miR-205-3p mimic (Fig. 4B). These data indicate that miR-205-3p is an important regulator of the candidate target genes ZEB1 and ZEB2, which participate in EMT by regulating E-cadherin expression.
DISCUSSION
Many miRNAs have been reported to function as oncogenes or tumor suppressors in cancer via the epigenetic regulation of target gene expression. Each miRNA can act on hundreds of target genes by partially complementing the 3′-UTR of those mRNAs, so that it forms a complex regulatory network that influences cancer invasion, metastasis, drug resistance, stemness, EMT, and signaling pathways, among other factors [3,19,20]. Many previous studies have reported the importance of miRNAs in the initiation and progression of GC [3]. Therefore, improvements in our knowledge regarding alterations in miRNA expression during GC progression or metastasis may provide novel options for the treatment of GC.
miR-205 has been reported to be a tumor suppressor in several types of cancer, such as colorectal cancer [8], glioblastoma [21], prostate cancer [6], and adrenal cortical carcinoma [22]. However, some reports have also indicated that miRNA-205 acts as an onco-miRNA in esophageal cancer [23], ovarian cancer [24], and lung cancer [25,26]. A previous study reported that the expression levels of miR-205 were significantly downregulated in GC tissues compared to normal gastric tissues and that the inhibition of miR-205 significantly promoted the proliferation and invasion of GC cells by targeting the expression of Yin Yang 1 and ICT1 oncoproteins [9,27]. However, the effects of miR-205 on EMT and cancer progression, as well as the underlying molecular mechanisms, remain largely unknown. In this present study, we compared miR-205-3p expression in 70 paired GC tissues and normal gastric tissues and provided important evidence in support of downregulation of miR-205-3p in GC tissues. Further analysis involving clinical studies revealed that a low level of miRNA-205-3p is associated with tumor differentiation, invasion depth, lymphatic metastasis, TNM stage, lymphatic invasion, vascular invasion, and poor prognosis, which serves as evidence for the tumor suppressor-like role of miR-205-3p in GC. The in vitro transwell assay also indicated that decreased miR-205-3p levels in GC cells could promote cancer invasion and migration abilities and that upregulation of its expression abolishes this facilitating effect.
EMT is a fundamental process during embryonic development and is also considered an important step that leads to tumor invasion and metastasis [10,11]. In previous studies, miRNAs have been found to induce gene silencing of various target mRNAs by partially complementing the 3′-UTR regions of those mRNAs. As such, miRNAs are important components of the cancer-signaling network and have been reported to be novel modulators of EMT regulation. To further identify the mechanisms underlying the suppressive effects of miR-205-3p, putative targets of miR-205-3p were explored. Among the targets identified, ZEB1 and ZEB2 genes were selected for further study.
Both ZEB1 and ZEB2 are members of the ZEB family and serve as transcriptional repressors of E-cadherin by binding to its CAGGTA/G E-box-like promoter elements [28]. Previous studies have demonstrated that these proteins suppress the expression of E-cadherin, which can induce EMT and contribute to the progression of many malignant tumors [29,30,31,32]. The results of the present study demonstrate that alterations in miR-205-3p expression in GC cells cause a negative regulatory effect on ZEB1 and ZEB2 expression. Furthermore, a mutagenesis assay of the miR-205-3p binding site in a luciferase reporter vector confirmed that the 3′-UTRs of both ZEB1 and ZEB2 mRNAs contain binding sites for miR-205-3p. At the same time, transfection with miR-205-3p resulted in the suppression of the mRNA and protein expression levels of ZEB1 and ZEB2. Since ZEB1 and ZEB2 are suppressors of E-cadherin, we also investigated changes in the expression levels of EMT markers, such as E-cadherin and N-cadherin. Consistent with the inhibitory effects of ZEB1 and ZEB2, we found that when miR-205-3p expression was downregulated in GC, the expression levels of ZEB1, ZEB2, and N-cadherin increased, whereas that of E-cadherin decreased. Taken together, these findings suggest that miR-205-3p induced the invasiveness and EMT of GC cells by negatively regulating the effects of ZEB1 and ZEB2. In order to increase the opportunities for the suppression of GC progression, additional research is needed to determine the exact mechanisms underlying the miR-205-3p-mediated functions in GC cells.
The results of the present study indicate that dysregulation of miR-205-3p may have a role in the tumor progression and prognosis of patients with GC. Furthermore, the data presented in this study suggest that miR-205-3p may modulate GC cell invasion and EMT progression by directly and negatively regulating ZEB1 and ZEB2. Therefore, restoration of miR-205-3p expression may be considered as a potential therapeutic strategy for GC.
Footnotes
Funding: This work was supported by the Medicine and Health Research Foundation of Zhejiang Province (2013KYB022 and 2020357780), National Natural Science Foundation of China (81502090), the Zhejiang Provincial Natural Science Foundation (LY14H160039, LY18H160043, Y18H030007, Y18H160037, and 2013RCA002) and the Science and Technology Plan of Zhejiang Province (2017F30045 and 2017C33130).
- Conceptualization: W.F., S.Y.
- Data curation: M.J., Y.Y.
- Funding acquisition: W.F., H.X.
- Investigation: Z.Z., X.J., Z.G., Y.Y., M.J.
- Methodology: W.F., S.Y., H.X.
- Project administration: W.F., S.Y.
- Validation: N.H.
- Writing - original draft: Z.Z., X.J.
- Writing - review & editing: Z.Z., W.F., H.X.
Conflict of Interest: No potential conflict of interest relevant to this article was reported.
References
- 1.Rawla P, Barsouk A. Epidemiology of gastric cancer: global trends, risk factors and prevention. Prz Gastroenterol. 2019;14:26–38. doi: 10.5114/pg.2018.80001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feng RM, Zong YN, Cao SM, Xu RH. Current cancer situation in China: good or bad news from the 2018 Global Cancer Statistics? Cancer Commun (Lond) 2019;39:22. doi: 10.1186/s40880-019-0368-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu X, Yang X, Xing C, Zhang S, Cao J. miRNA: The nemesis of gastric cancer (Review) Oncol Lett. 2013;6:631–641. doi: 10.3892/ol.2013.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ventura A, Jacks T. MicroRNAs and cancer: short RNAs go a long way. Cell. 2009;136:586–591. doi: 10.1016/j.cell.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shukla GC, Singh J, Barik S. MicroRNAs: processing, maturation, target recognition and regulatory functions. Mol Cell Pharmacol. 2011;3:83–92. [PMC free article] [PubMed] [Google Scholar]
- 6.Verdoodt B, Neid M, Vogt M, Kuhn V, Liffers ST, Palisaar RJ, et al. MicroRNA-205, a novel regulator of the anti-apoptotic protein Bcl2, is downregulated in prostate cancer. Int J Oncol. 2013;43:307–314. doi: 10.3892/ijo.2013.1915. [DOI] [PubMed] [Google Scholar]
- 7.Adachi R, Horiuchi S, Sakurazawa Y, Hasegawa T, Sato K, Sakamaki T. ErbB2 down-regulates microRNA-205 in breast cancer. Biochem Biophys Res Commun. 2011;411:804–808. doi: 10.1016/j.bbrc.2011.07.033. [DOI] [PubMed] [Google Scholar]
- 8.Li P, Xue WJ, Feng Y, Mao QS. MicroRNA-205 functions as a tumor suppressor in colorectal cancer by targeting cAMP responsive element binding protein 1 (CREB1) Am J Transl Res. 2015;7:2053–2059. [PMC free article] [PubMed] [Google Scholar]
- 9.Yin WZ, Li F, Zhang L, Ren XP, Zhang N, Wen JF. Down-regulation of microRNA-205 promotes gastric cancer cell proliferation. Eur Rev Med Pharmacol Sci. 2014;18:1027–1032. [PubMed] [Google Scholar]
- 10.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 11.Lyons JG, Lobo E, Martorana AM, Myerscough MR. Clonal diversity in carcinomas: its implications for tumour progression and the contribution made to it by epithelial-mesenchymal transitions. Clin Exp Metastasis. 2008;25:665–677. doi: 10.1007/s10585-007-9134-2. [DOI] [PubMed] [Google Scholar]
- 12.Sánchez-Tilló E, Lázaro A, Torrent R, Cuatrecasas M, Vaquero EC, Castells A, et al. ZEB1 represses E-cadherin and induces an EMT by recruiting the SWI/SNF chromatin-remodeling protein BRG1. Oncogene. 2010;29:3490–3500. doi: 10.1038/onc.2010.102. [DOI] [PubMed] [Google Scholar]
- 13.Schmalhofer O, Brabletz S, Brabletz T. E-cadherin, beta-catenin, and ZEB1 in malignant progression of cancer. Cancer Metastasis Rev. 2009;28:151–166. doi: 10.1007/s10555-008-9179-y. [DOI] [PubMed] [Google Scholar]
- 14.Koopmansch B, Berx G, Foidart JM, Gilles C, Winkler R. Interplay between KLF4 and ZEB2/SIP1 in the regulation of E-cadherin expression. Biochem Biophys Res Commun. 2013;431:652–657. doi: 10.1016/j.bbrc.2013.01.070. [DOI] [PubMed] [Google Scholar]
- 15.Amin MB, Greene FL, Edge SB, Compton CC, Gershenwald JE, Brookland RK, et al. The eighth edition AJCC cancer staging manual: continuing to build a bridge from a population-based to a more "personalized" approach to cancer staging. CA Cancer J Clin. 2017;67:93–99. doi: 10.3322/caac.21388. [DOI] [PubMed] [Google Scholar]
- 16.Washington K. 7th edition of the AJCC cancer staging manual: stomach. Ann Surg Oncol. 2010;17:3077–3079. doi: 10.1245/s10434-010-1362-z. [DOI] [PubMed] [Google Scholar]
- 17.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He Y, Northey JJ, Pelletier A, Kos Z, Meunier L, Haibe-Kains B, et al. The Cdc42/Rac1 regulator CdGAP is a novel E-cadherin transcriptional co-repressor with ZEB2 in breast cancer. Oncogene. 2017;36:3490–3503. doi: 10.1038/onc.2016.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Acunzo M, Croce CM. MicroRNA in cancer and cachexia--a mini-review. J Infect Dis. 2015;212(Suppl 1):S74–S77. doi: 10.1093/infdis/jiv197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiu D, Wang D, Wang J, Ji F, Zhang W. MicroRNA-543 suppresses liver cancer growth and induces apoptosis via the JAK2/STAT3 signaling pathway. Oncol Lett. 2019;17:2451–2456. doi: 10.3892/ol.2018.9838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yue X, Wang P, Xu J, Zhu Y, Sun G, Pang Q, et al. MicroRNA-205 functions as a tumor suppressor in human glioblastoma cells by targeting VEGF-A. Oncol Rep. 2012;27:1200–1206. doi: 10.3892/or.2011.1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu Y, Wang W, Hu W, Xu W, Xiao G, Nie Q, et al. MicroRNA-205 suppresses the growth of adrenocortical carcinoma SW-13 cells via targeting Bcl-2. Oncol Rep. 2015;34:3104–3110. doi: 10.3892/or.2015.4295. [DOI] [PubMed] [Google Scholar]
- 23.Dong Y, Si JW, Li WT, Liang L, Zhao J, Zhou M, et al. miR-200a/miR-141 and miR-205 upregulation might be associated with hormone receptor status and prognosis in endometrial carcinomas. Int J Clin Exp Pathol. 2015;8:2864–2875. [PMC free article] [PubMed] [Google Scholar]
- 24.Chu P, Liang A, Jiang A, Zong L. miR-205 regulates the proliferation and invasion of ovarian cancer cells via suppressing PTEN/SMAD4 expression. Oncol Lett. 2018;15:7571–7578. doi: 10.3892/ol.2018.8313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lei L, Huang Y, Gong W. miR-205 promotes the growth, metastasis and chemoresistance of NSCLC cells by targeting PTEN. Oncol Rep. 2013;30:2897–2902. doi: 10.3892/or.2013.2755. [DOI] [PubMed] [Google Scholar]
- 26.Bai J, Zhu X, Ma J, Wang W. miR-205 regulates A549 cells proliferation by targeting PTEN. Int J Clin Exp Pathol. 2015;8:1175–1183. [PMC free article] [PubMed] [Google Scholar]
- 27.Tao Y, Song Y, Han T, Wang C, Zhao T, Gu Y. miR-205 regulation of ICT1 has an oncogenic potential via promoting the migration and invasion of gastric cancer cells. Biomed Pharmacother. 2017;96:191–197. doi: 10.1016/j.biopha.2017.09.147. [DOI] [PubMed] [Google Scholar]
- 28.Wong TS, Gao W, Chan JY. Transcription regulation of E-cadherin by zinc finger E-box binding homeobox proteins in solid tumors. BioMed Res Int. 2014;2014:921564. doi: 10.1155/2014/921564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagathihalli NS, Merchant NB. Src-mediated regulation of E-cadherin and EMT in pancreatic cancer. Front Biosci (Landmark Ed) 2012;17:2059–2069. doi: 10.2741/4037. [DOI] [PubMed] [Google Scholar]
- 30.Yao X, Ireland SK, Pham T, Temple B, Chen R, Raj MH, et al. TLE1 promotes EMT in A549 lung cancer cells through suppression of E-cadherin. Biochem Biophys Res Commun. 2014;455:277–284. doi: 10.1016/j.bbrc.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qin Y, Tang B, Hu CJ, Xiao YF, Xie R, Yong X, et al. An hTERT/ZEB1 complex directly regulates E-cadherin to promote epithelial-to-mesenchymal transition (EMT) in colorectal cancer. Oncotarget. 2016;7:351–361. doi: 10.18632/oncotarget.5968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Voutsadakis IA. Epithelial-mesenchymal transition (EMT) and regulation of EMT factors by steroid nuclear receptors in breast cancer: a review and in silico investigation. J Clin Med. 2016;5:E11. doi: 10.3390/jcm5010011. [DOI] [PMC free article] [PubMed] [Google Scholar]