Abstract
Predicting regulatory potential from primary DNA sequences or transcription factor binding patterns is not possible. However, the annotation of the genome by chromatin proteins, histone modifications, and differential compaction is largely sufficient to reveal the locations of genes and their differential activity states. The Polycomb Group (PcG) and Trithorax Group (TrxG) proteins are the central players in this cell type–specific chromatin organization. PcG function was originally viewed as being solely repressive and irreversible, as observed at the homeotic loci in flies and mammals. However, it is now clear that modular and reversible PcG function is essential at most developmental genes. Focusing mainly on recent advances, we review evidence for how PcG and TrxG patterns change dynamically during cell type transitions. The ability to implement cell type–specific transcriptional programming with exquisite fidelity is essential for normal development.
Keywords: Polycomb, Trithorax, bivalent chromatin, gene regulation, epigenetics
INTRODUCTION
Chromatin proteins, histone modifications, and differential accessibility specifically mark the small fraction of DNA that comprises genes and their regulatory elements within the vast genomes of higher organisms. Genes are annotated on their 5′ ends and gene bodies in distinct ways, depending on their active or silent state. This annotation needs to be both stable and reversible, as gene expression programs differ in each cell type.
A set of conserved chromatin regulatory factors, collectively known as the Polycomb Group (PcG), play a central role in developmental patterning and cell type–specific transcriptional programs. The binding patterns of PcG proteins and their associated H3K27me3 histone mark correlate with repression, whereas chromatin marks such as acetylation and H3K4me3, mediated by the Trithorax Group (TrxG), correlate with the active state. Significant progress has been made toward identifying the multiple factors contributing to these distinct patterns (1–5). However, the biochemical basis for transitions in chromatin state is only partially understood. Here, we bring together disparate but potentially salient information from both fly and mammalian models in an effort to go beyond classical models for answers to fundamental questions.
THE PcG AND TrxG ENCODE OPPOSING CHROMATIN-BASED ACTIVITIES
Mutant developmental phenotypes in Drosophila revealed the original members of the PcG and their critical repression of the Hox genes to pattern fly embryos (6, 7). Likewise, the TrxG was discovered to oppose PcG silencing, allowing proper expression of Hox genes in appropriate regions of the embryo (8). PcG and TrxG proteins are now known to be highly conserved and to function largely through the modification and modulation of chromatin (5). Alteration of cell type–specific chromatin organization, via both loss-of-function and gain-of-function mutations in members of the PcG and TrxG families, disrupts differentiation and has been implicated in a wide range of cancers (9, 10).
The PcG proteins assemble mainly into two types of biochemically defined Polycomb Repressive Complexes: PRC1 and PRC2 (Figure 1). Each family of complexes has many orthologous or alternative subunits. PRC1 is an E3 ubiquitin ligase that mono-ubiquitylates histone H2A (H2AK118ub1 in flies and H2AK119ub1 in mammals) (11–13). PRC2 is a methyltransferase that mono-, di-, and trimethylates histone H3 at lysine 27 (H3K27me1, me2, or me3) (14–17). H3K27me3 is coincident with stable binding profiles of PcG, and its requirement for silencing has been demonstrated in Drosophila tissues in which all histone H3 genes remaining after mitotic recombination were mutant at position 27 (18). H2Aub was not required for stable Hox gene silencing in the same clonal assay. However, H2Aub is required for efficient H3K27 trimethylation by PRC2 early in Drosophila embryogenesis (19) and is essential for PRC2 targeting in mouse embryonic stem cells (mESCs) (20, 21).
Figure 1.
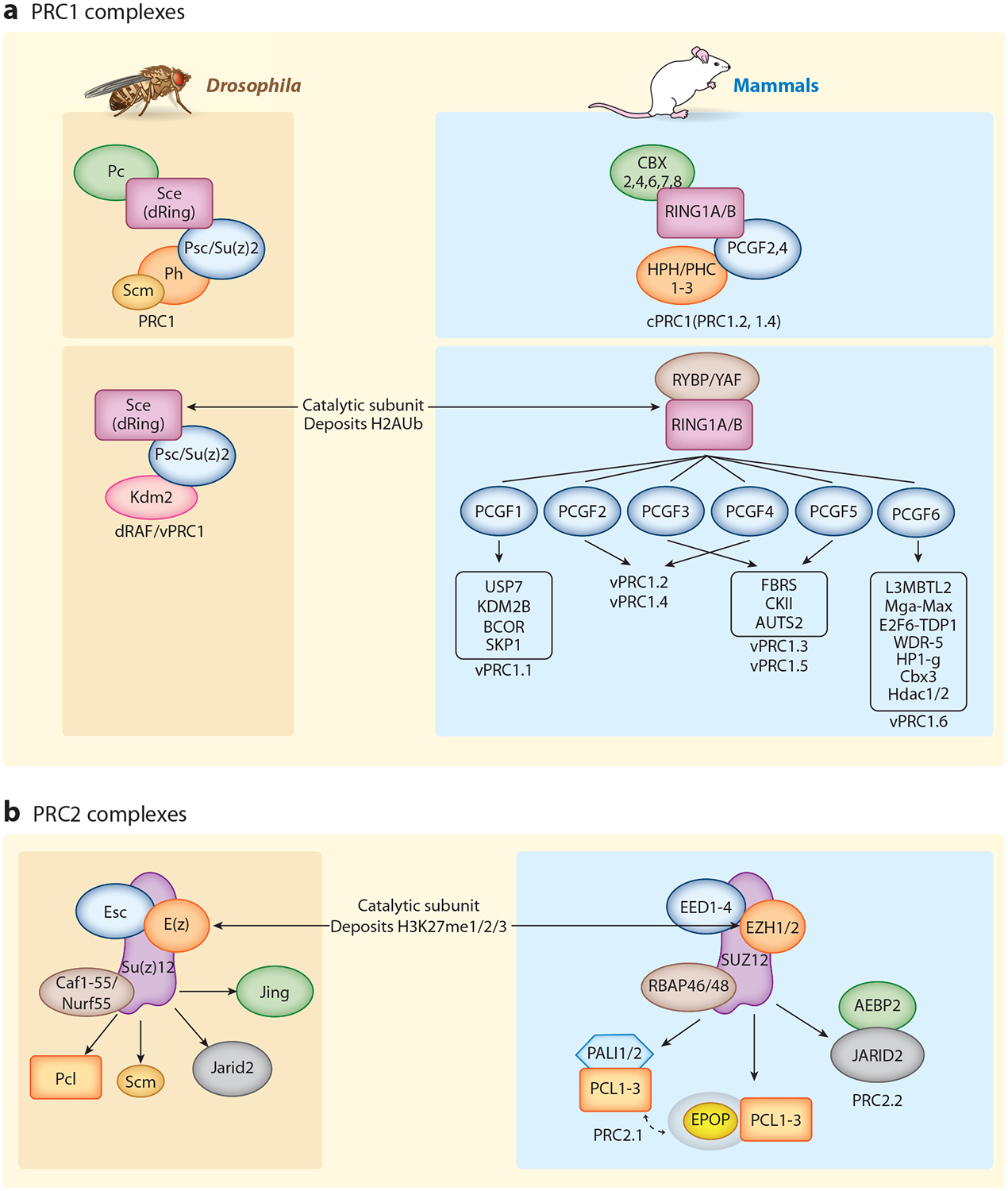
PcG complexes in Drosophila and mammals. PcG proteins are classified into two major complexes: (a) PRC1 and (b) PRC2. Homologous core complex subunits are color coded between Drosophila (left, brown) and mammals (right, blue), and their common catalytic subunits are indicated [dRing or RING1A/B in all PRC1 complexes and E(z) or EZH1/2 in PRC2]. The core complexes are diversified by interactions with accessory proteins, especially in mammals. Accessory subunits can be mutually exclusive as in mammalian PRC2.1 and PRC2.2. Similarly, mammalian PRC1 is divided into cPRC1 (canonical PRC1) and vPRC1 or ncPRC1 (variant or noncanonical PRC1), as initially defined by Gao et al. (38). Analogous in-depth studies of Drosophila vPRC1/ncPRC1 have not been reported. Protein–protein contacts presented here are not meant to be accurate. In many cases, they are not known in detail, although substantial progress has occurred recently (55–60, 81). Likewise, depicted complexes are meant to represent a general view, but the existence of additional configurations or cell type– and tissue type–divergent versions is also likely. Abbreviations: PcG, Polycomb Group; PRC1 and PRC2, Polycomb Repressive Complexes.
The TrxG comprises a more biochemically diverse set of proteins with the common feature of promoting the active state. Enzymatic activities include methylation of histone H3 at lysine 4 at promoters (H3K4me2 and me3) and enhancers (H3K4me1) by MLL family members (22). In Drosophila, the TrxG protein Ash1 is a methyltransferase that counteracts PcG silencing (23, 24). Additional TrxG members assemble into large SWI/SNF or BRG ATP-dependent remodeling complexes that can mobilize nucleosomes and promote chromatin accessibility (10). TrxG factors that have been mapped typically enrich on accessible regions such as genes and regulatory elements (22).
Genome-wide, PRC2 binds stably to silenced regions and is coincident with H3K27me3. Functionally, PRC2 is also responsible for H3K27me1 found on active genes and abundant intergenic H3K27me2 but is typically not seen at those locations in chromatin immunoprecipitation (ChIP) profiles (25–27). The importance of intergenic H3K27 modification may be to protect the genome from aberrant activation. The prevalence of these modifications provides clear evidence that PRC2 is capable of scanning the genome for its targets.
Interestingly, genome-wide profiles in both flies and mammals detect PRC1 enrichment in stably silenced regions, but it is also detectable on active genes (28–35). After initially ignoring this paradoxical observation, the field is starting to consider how this may fit into the overall PcG/TrxG dynamic (36, 37).
HIGHLY MODULAR PcG COMPLEXES WITH MUTUALLY REINFORCING INTERACTIONS
Much of the recent activity in the PcG field has focused on the subunit interactions and structures of PRC1 and PRC2. The core complexes are conserved between flies and mammals (Figure 1), whereas numerous orthologs and additional vertebrate-specific subunits presumably add regulatory versatility (38–47). One common theme is the identification of feed-forward, self-reinforcing interactions. For example, variant or noncanonical PRC1 (vPRC1 or ncPRC1) recognizes its own H2Aub mark through the RYBP subunit (48), and core PRC2 is not fully activated until encountering its own H3K27me3 mark in a feed-forward interaction likely deployed during spreading (49–51). Furthermore, PRC1 and PRC2 reinforce each other, as a specific configuration of PRC2, containing the JARID2 subunit, recognizes the PRC1-dependent H2Aub mark (52, 53), and the chromodomain of canonical PRC1 recognizes the PRC2-dependent H3K27me3 mark (14, 54). Many recent articles and reviews have featured informative new structural studies (55–60). The number of alternative PRC complexes suggests many avenues for feedback and feed-forward regulation during development.
DISCOVERY OF POLYCOMB RESPONSE ELEMENTS IN KEY DEVELOPMENTAL LOCI
Early studies in Drosophila identified DNA segments in the Hox cluster and other developmental genes that can render transgenes PcG responsive (61, 62). These segments are termed Polycomb response elements (PREs). Within broad H3K27me3 domains, candidate fly PREs can be recognized as strong peaks of PRC1 and PRC2 enrichment. PREs consist of binding sites for a number of different DNA binding proteins (63). Recruitment of PcG complexes requires a combination of these DNA binding proteins, and no single factor, even when its binding site is multimerized, is sufficient for PRE function (64).
The best-understood DNA binding factor found at PREs is Pho, a zinc-finger protein related to the mammalian transcription factor (TF) YY1 (65). Mutations in pho display a homeotic phenotype, and its protein product forms the PhoRC complex with Sfmbt (66). Biochemical, genetic, and structural studies support a model in which PhoRC directly recruits PRC1 (67). In agreement with this, PRC1 can be recruited to some PREs in the absence of PRC2 (68). The protein Scm, which interacts with PhoRC, PRC1, and PRC2 may serve as a link between all three complexes (69) (Figure 2).
Figure 2.
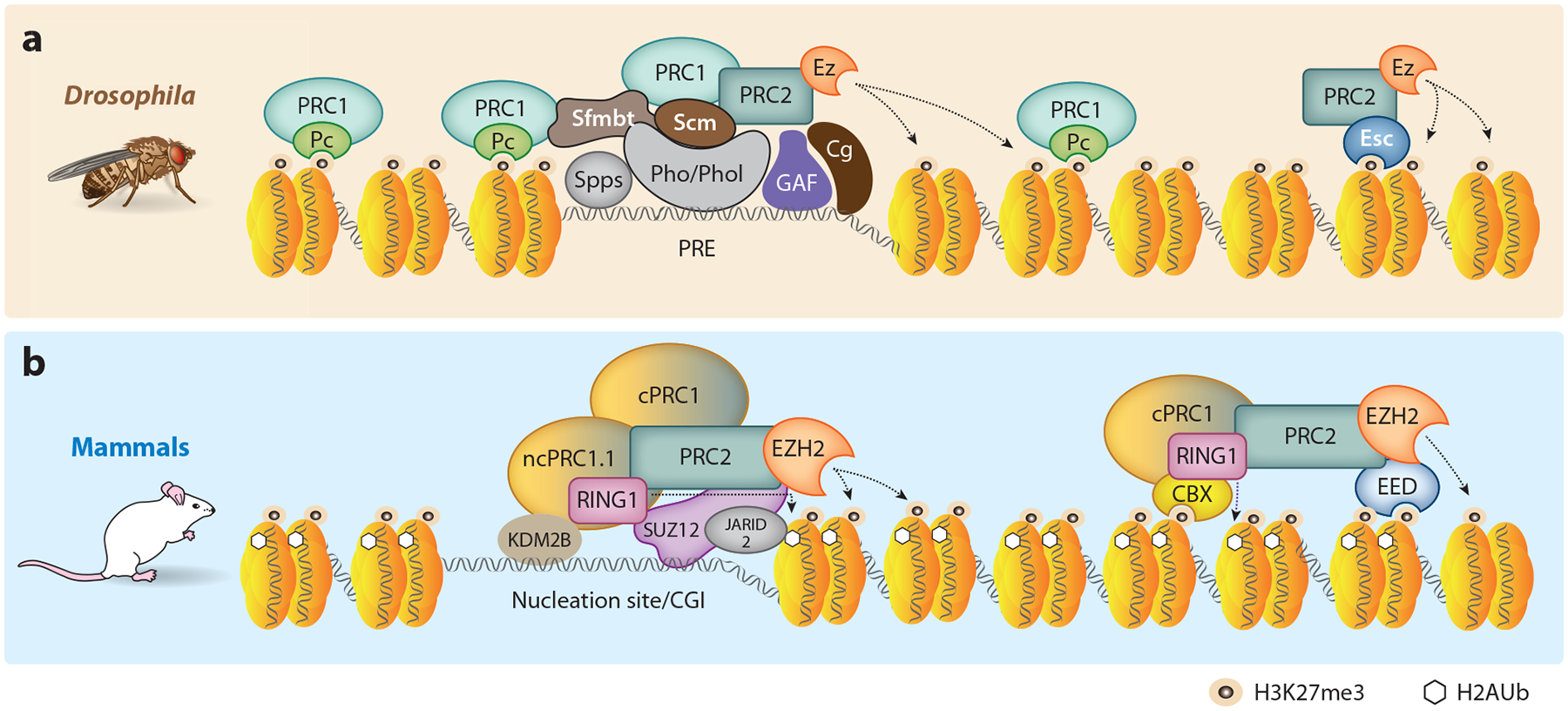
Assembly of PcG complexes at target loci. (a) Drosophila PREs bind many different DNA binding proteins including Pho/Phol, Spps, GAF, and Cg (63), and a combination of these PRE binding proteins is typically required for recruitment of PRC1 and PRC2. Genome-wide ChIP studies have shown that, although Pc (a component of PRC1) can spread beyond the recruitment sites via binding to H3K27me3, the highest concentration is near PREs. The genomic binding pattern of H2AK118ub is less well defined in flies (26). (b) In mammals, PcG complexes are enriched at unmethylated CGIs. KDM2B of vPRC1.1 and SUZ12, JARID2, and MTF2 of PRC2 all have affinity for GC-rich DNA sequences that may help drive assembly at nucleation sites. vPRC1 ubiquitinylates H2AK119 in mammals, which facilitates recruitment of PRC2 via JARID2. Further, EED of PRC2 and CBX of cPRC1 bind H3K27me3 to drive self-propagation or spreading of the repressive mark. Abbreviations: CGI, CpG island; ChIP, chromatin immunoprecipitation; cPRC1, canonical PRC1; Pc, Polycomb; PcG, Polycomb Group; PRC1 and PRC2, Polycomb Repressive Complexes; PRE, Polycomb response element; vPRC1, variant PRC1.
On the basis of the successful studies in flies, the search for PREs in mammals began in the HOX loci. However, unlike in Drosophila, easily recognized peaks of PcG complexes were not found; instead, the proteins were bound broadly throughout the large H3K27me3 domains. A few PRE-like fragments were identified, but they did not reveal a common principle (70–73). Mammals do not appear to have the PhoRC protein complex. Nevertheless, genetic deletion experiments revealed that Hox clusters in mice must have redundant fragments that can act independently to recruit PcG proteins and facilitate H3K27me3 spreading over the whole cluster (73). In this way, mammalian and Drosophila Hox loci are likely to be similar.
STABLE SILENCING AND SPREADING OF PcG AND H3K27me3
HOX genes and other key developmental loci in mammals and Drosophila are known as canonical PcG targets. They are strongly detectable genetically, as their derepression when PcG function is mutant leads to striking developmental defects (6, 7). Furthermore, they are strong targets of PcG enzymatic function, as the loci are bound by H3K27me3 that can extend over tens to hundreds of kilobases (Figure 3). The discovery of PREs as discrete targeting elements within silenced chromatin extending across large regions strongly suggests a spreading mechanism. Furthermore, these regions typically are flanked by boundary/insulator elements that stop the apparent spreading of the H3K27me3 mark (74–76). Actively transcribed genes can also stop the H3K27me3 spreading (77). For example, H3K36me3, a mark deposited during the process of transcription, inhibits PRC2 activity (78). Having discreet limits or boundaries could help facilitate the formation of the 3D structure of the Polycomb domain and contribute to its stability.
Figure 3.
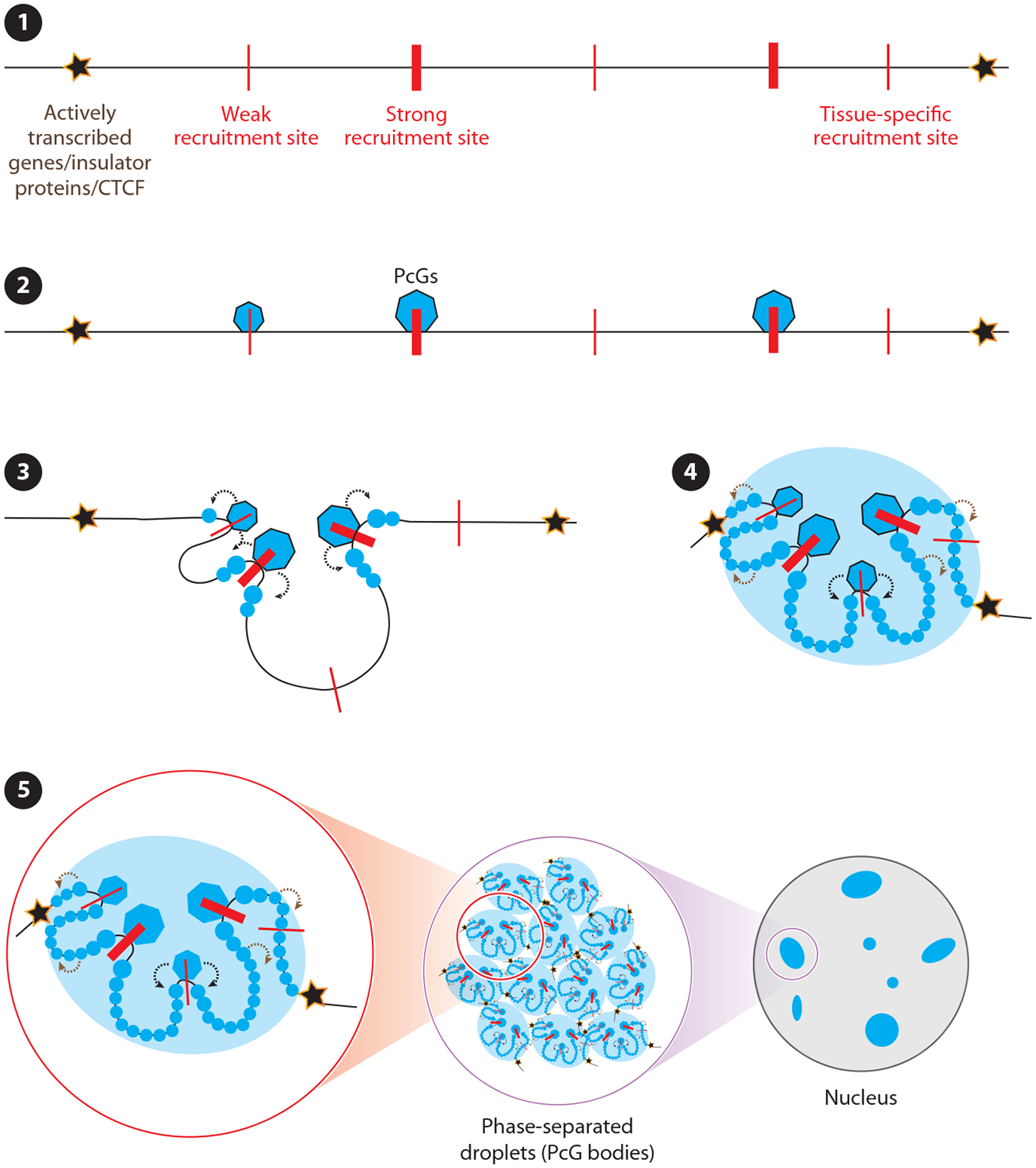
Model for Polycomb Group (PcG) domain establishment. (❶) PcG domain boundaries are defined in Drosophila by either actively transcribed genes or insulator proteins and in mammals by CTCF proteins, and these boundaries may form similarly in flies and in mammals (3). (❷) In both organisms, PcG proteins appear to first engage with recruitment sites (red lines) within PcG target domains. (❸) The recruitment sites can be either strong or weak and either cell type–, tissue type–, or developmental stage–specific. After the initial recruitment, two parallel events happen: (❹) PcG complexes modify flanking histone tails (modified histones are shown with blue spheres) and interactions between the PcG proteins bound to the recruitment sites drive changes in 3D structure of the domain. The initial histone modifications and changes in 3D architecture of the domain drive further recruitment of PcG complexes and cause modification of the rest of the histones to establish the PcG domain. (➎) Finally, the PcG domains form phase-separated droplets either individually or by fusing with each other. These phase-separated droplets are also known as PcG bodies.
How and when might spreading occur? In Drosophila, experiments in early development have not yielded a consensus. One group reported no detectable H3K27me3 during the rapid early embryonic cell divisions (79). Another group found that H3K27me3 marks can be inherited through the germ line, although they were at least partially erased during the rapid early cycles of embryogenesis (80). In either case, the best evidence for spreading comes during cell cycle 14, as H3K27me3 gradually accumulates over the large chromatin domains, bidirectionally, until the entire domain is covered (79).
Two seminal studies in mESCs were able to document inducible spreading after CRISPR-mediated knockout of PRC2 components and apparent removal of all detectable H3K27me3 (51, 81). Interestingly, both groups showed that normal H3K27me3 patterns were restored after reintroduction of the deleted PRC2 subunit. Oksuz et al. (51) examined a time course of H3K27me3 restoration and identified nucleation sites that initiated and spread H3K27me3 locally and to distant intrachromosomal sites. The first nucleation sites to appear were very stable, whereas sites that appeared later were more labile to PRC2 loss, perhaps analogous to strong and weak PREs present in Drosophila genes (51, 82). Sequence analysis of the nucleation sites showed enrichment for CpG- and GA-rich motifs, with the majority located within 5 kb of a transcription start site. Nucleation was dependent on the presence of either JARID2 or MTF2, two PRC2 accessory proteins previously implicated in affinity for CpG-rich DNA sequences (51, 83, 84). Because PRC2 was able to reassemble at its original sites in the apparent absence of residual H3K27me3, it would be very interesting to test those locations for retention of PRC1, H2Aub, and other potential chromatin marks that, in addition to DNA sequence composition, might mediate PRC2 attraction back to its appropriate cell type–specific locations.
How does spreading occur at a biochemical level? PRC2 enzymatic activity is stimulated by binding to the product of its own catalysis (H3K27me3) through an aromatic cage present in its Eed/Esc subunit, suggesting a feed-forward mechanism that could progress along the chromatin (50, 85). Consistent with this hypothesis, a mutant Eed subunit lacking the ability to bind H3K27me3 also failed to support inducible spreading (51). Further, H3K27me3 facilitates spreading of the Pc/CBX components of PRC1, which contain chromodomains that bind to H3K27me3 (54). Like Drosophila PREs, which are known to interact with each other, it is also proposed that recruitment sites in mammals interact with each other and facilitate the spreading of H3K27me3 (51) (Figure 3). A model has emerged that interactions between PREs stabilize Polycomb domains—this is supported by recent data in Drosophila (86). Sufficient enrichment of PRC1 components, including subunits such as Ph and Scm that have the potential to oligomerize, may promote compaction of the domains visualized as large Polycomb bodies within the nucleus (87). Recently, an intrinsically disordered domain in the CBX2 protein, also linked to the ability to compact chromatin, has been shown to cause phase separation in vitro (88).
Spreading of Polycomb proteins is also a notable feature of mammalian X chromosome inactivation. Through interactions with the noncoding RNA Xist, PcG proteins facilitate X chromosome inactivation through a spreading mechanism involving histone deacetylation, vPRC1, H2Aub, PRC2, and H3K27me3 (89). Finally, nucleation and spreading are not unique to the PcG but appear to be important general mechanisms in chromatin-based regulation. This is evident from studies of HP1-dependent heterochromatin (90) and from all three models of X chromosome dosage compensation (fruit flies, nematodes, and mice) (91).
PcG BINDING MUST BE DYNAMIC, AS EACH CELL TYPE DISPLAYS A DIFFERENT PATTERN
Stable silencing through PcG spreading and reinforcement is well established, especially for the early decisions that coordinate the body plan. However, the majority of repression requires reversibility, to allow development to transition through distinct transcriptional programs (92, 93). For all of those regulated loci, how does PcG targeting change with cell type (1, 94)?
To date, we are aware of two general classes of models for how cell type–specific patterns of PcG targeting occur (95). The instructive model posits that combinatorial protein–protein (or protein–noncoding RNA) interactions allow TFs to attract the appropriate members of the PcG or TrxG for regulation in each situation and at each gene. Supportive evidence for the role of sequence-specific DNA binding factors comes mainly from studies in Drosophila (63), whereas reports of the involvement of noncoding RNAs come from studies in mammalian cells and are still controversial (1). In either case, to have distinct yet specific multivalent interactions at each gene in each cell type is a very complicated scenario in biochemical terms.
The responsive model instead relies on a default affinity of PcG-associated proteins for unmethylated CpG islands upstream of mammalian genes, coupled with incompatibility of transcriptional activity with PcG function (95). In this model, PcG and TrxG competitively sample upstream regulatory sequences, with PcG prevailing at nonexpressed genes but inhibited locally at each expressed gene. This model alleviates the need for the complicated locus-specific interactions required of the instructive model and is supported by the apparent sufficiency of unmethylated DNA of high CpG content to attract PRC2 and H3K27me3 to ectopic, nonexpressed locations (96–99). Interestingly, default targeting was also an early model for PcG repression in flies that could explain how PcG maintained silencing of diverse loci that were initially repressed by many different TFs (100, 101). Although Drosophila genes do not typically have upstream CpG islands, PcG binding is enriched near the 5′ transcription start sites in flies, similar to mammals (102). Further, in Drosophila, at least for the canonical Polycomb target genes such as the Hox loci, early TFs set the ON or OFF state of the gene. Transcriptional activity then determines whether Polycomb or Trithorax prevail to maintain the transcription state.
Although flies and mammals may have diverged considerably in their mechanisms of PcG targeting at the 5′ ends of genes, the conservation of the protein complexes, enzymatic functions, and unexpected localization of PRC1 on active genes (discussed below) suggests there could still be unifying concepts to be discovered.
BIVALENCY MARKS A SWITCHABLE STATE
As stated earlier, the chromatin marks found on active versus silent genes are generally mutually exclusive. The canonical active mark, H3K4me3, is catalyzed by the MLL family of TrxG proteins at the promoters of expressed genes, whereas the signature PRC2 mark, H3K27me3, is found instead on silent genes. Many years after recognition of this universality, developmental genes in mammalian pluripotent cells were found, unexpectedly, to carry both active and silent marks (103, 104). The bivalent marks can be detected on the same histone octamer but on opposing histone H3 tails (105, 106). Bivalent genes typically are not expressed until the coexistence of opposing marks resolves into either the active or silent state upon differentiation (Figure 4). These observations support a model in which bivalency represents a key poised state during pluripotency that can resolve independently at each gene (103, 104). Although conceptually pleasing, testing the functional significance of bivalency has remained challenging (107, 108). For example, determining whether bivalency exists in model organisms such as flies has been difficult because a pluripotent cell culture system is lacking. However, an increasingly strong case for conservation is now emerging (109–112).
Figure 4.
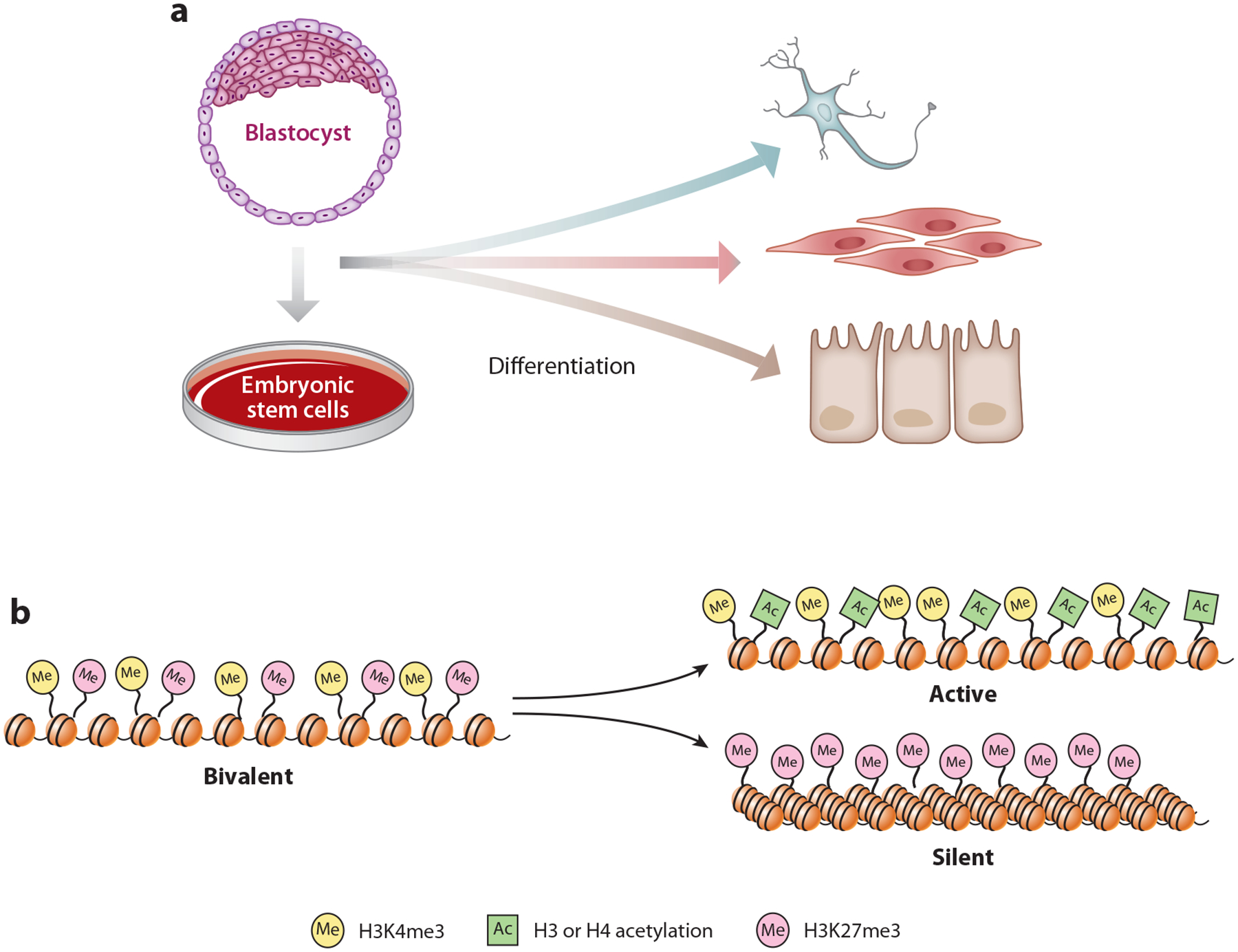
Bivalent chromatin marks and their resolution upon differentiation. (a) Mammalian embryonic stem cells are derived from the inner cell mass of blastocysts. Embryonic stem cells not only are capable of self-renewal but also are pluripotent, meaning they can give rise to many cell types in the body. (b) Many developmental genes in pluripotent cells are marked with bivalent chromatin in which both active H3K4me3 and silent H3K27me3 modifications coexist. This bivalency typically is resolved into either active or silent states during differentiation.
Interestingly, PRC1 and PRC2 are colocalized on bivalent genes in embryonic stem cells, but upon differentiation those patterns can diverge considerably. As mentioned earlier, in both mammals and in flies PRC1 is bound to large numbers of active genes (28–35). This result was strongly foreshadowed by a classic early study. In one of the first comparisons of PcG ChIP at a gene in its active versus silent state, it became clear that PhoRC, PRC1, and PRC2 proteins still occupied the Ubx locus even when expressed, contrary to expectations (113). Instead of a loss of PcG binding, the clearest difference was an increase in the Ash1 TrxG protein at the active promoter, suggesting that the balance between PcG and TrxG opposing functions might be critical.
Consistent with the possibility that PRC1 and PRC2 have both shared and unique functions, mutants in PRC1 and PRC2 can have overlapping but distinct mutant phenotypes in both flies and mammals (32, 114–116). Considering this divergence within the bivalency framework, PRC1 could plausibly retain a regulatory role after bivalent genes are resolved into the active state.
A MASTER SWITCH MODEL FOR PcG TARGETING AND REVERSIBILITY
Here, we combine ideas derived from experiments in flies and mammals to propose a new version of the responsive model for PcG targeting. Central to this model is a relationship between bivalency, discovered in mammalian embryonic stem cells (103, 104), and protein–protein interactions between PcG and TrxG seen in fly embryos (109, 117). If these relationships/interactions competitively mark the 5′ ends of all developmental genes, they could provide the critical ability to independently maintain or reverse the transcriptional state at each locus (Figure 5). The discovery that PRC1 complexes copurify and cross-link efficiently to select coactivator proteins in fly embryos was unexpected (109, 117). Interestingly, these coactivators, orthologs of mammalian BRD4 and the MOZ/MORF histone acetyltransferase, contain protein domains that bind acetyl groups and the TrxG H3K4me3 mark and are implicated in TrxG function through genetic analyses in Drosophila (118), zebrafish (119), and mice (120). Notably, PRC1 and PRC2 both recognize the H3K27me3 silent mark at bivalent genes. If PcG and TrxG opposing activities interact with bivalent genes and their chromatin marks in dynamic equilibrium, throughout development they may not need to be attracted de novo to their targets, which is the aspect of PcG targeting that has presented the most significant conceptual difficulty in classical models (1).
Figure 5.
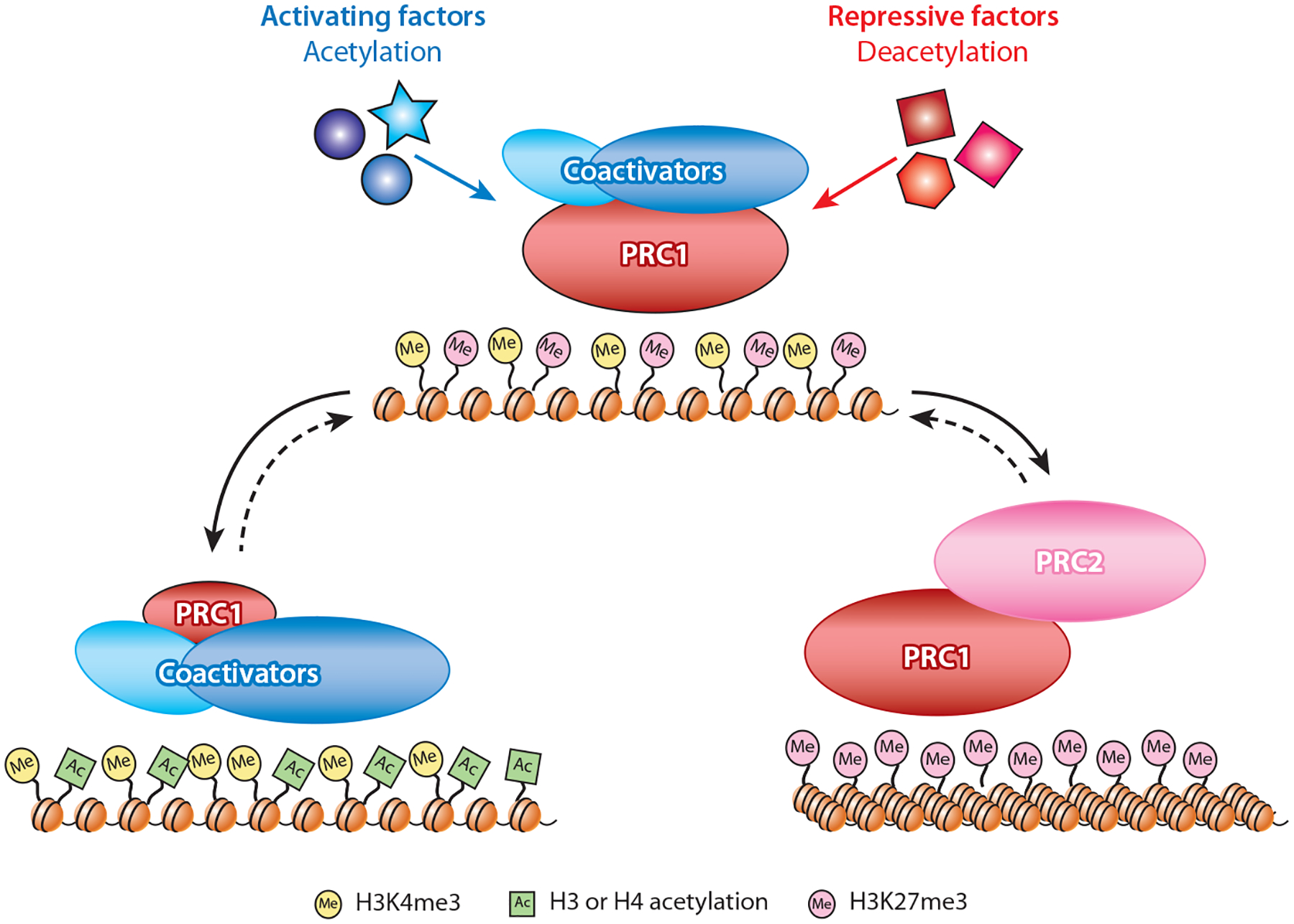
The bivalent master switch model. During embryonic development in Drosophila, PRC1 and specific coactivator proteins are proposed to form bivalent protein complexes on transcriptionally poised genes. The coactivator module includes a histone acetyltransferase that can both catalyze and recognize acetylation marks, whereas a separate subunit can bind the H3K4me3 chromatin mark at active or bivalent promoters. In contrast, PRC1/PRC2 enrichment is reinforced by the H3K27me3 silencing mark and unacetylated nucleosomes. The choice between a transcriptionally active or silent state may be triggered by combinations of specific transcription factors that alter the acetylation state of the local chromatin environment, favoring increased association of coactivators or PRC2, respectively. If some level of co-occupancy is maintained, the resulting transcriptional states may be reversible (dotted lines) by changes in critical thresholds of competing transcription factors during subsequent differentiation. Although this model rests largely on protein interactions observed in Drosophila, it is compatible with the responsive model for PcG targeting in mammalian cells (95). In an extension of that model, mammalian PcG and TrxG proteins may compete for interaction at CpG islands and be influenced by local acetylation, without necessarily interacting physically as observed in Drosophila embryos. Abbreviations: PcG, Polycomb Group; PRC1 and PRC2, Polycomb Repressive Complexes; TrxG, Trithorax Group.
Known PcG behavior appears to be compatible with this model. The overlap of cis-acting elements responsive to PcG or TrxG, or their colocalization, has been observed repeatedly in Drosophila (113, 121–124). Furthermore, detection of PcG and TrxG proteins at the Ubx locus in both the expressed and the repressed states in Drosophila larval imaginal discs suggests that lack of full resolution may underlie reversibility (109, 113). Most exciting are the multiple observations of abundant bivalency in mammalian germ cells, which are consistent with the idea that genes may not need to be marked de novo in embryogenesis but may instead be marked throughout the life of the organism (125–129). If so, the PcG targeting question becomes greatly simplified.
COULD PcG–TrxG INTERCHANGE PLAY THE PIVOTAL ROLE IN REVERSIBILITY?
If the retention of a bivalent PcG–TrxG memory underlies the ability for genes to switch during transcriptional programming, how might transitions occur on a biochemical level? One intriguing possibility is that transitions occur in response to enzymatic activities typically attracted by TFs, with acetylation as a strong candidate (109, 121). TFs attract a set of coactivators, typically histone acetyltransferases, that together with histone deacetylases influence the local balance of acetylation/deacetylation activities (Figure 5). In this model, posttranslational modifications would influence the locally bound PcG–TrxG balance, defining the correct activity state independently for each gene while retaining reversibility when local TF combinations and levels change. For example, in fly embryos, increased acetylation would favor enhanced binding of the bromodomain-containing dMoz/Morf and dBrd4 (109) and repel the PcG (78, 130, 131). Experiments showing that PRC2 targeting is more dependent on PRC1 than vice versa are also compatible with the idea of competition that can either be resolved toward stabilizing PRC2 on chromatin (20, 32, 68, 132) or acquiring more TrxG functions (23, 133–137).
The ability to regulate genes while only partially resolving bivalent complexes would be critical for the reversibility seen with changes in the repertoire and binding of cell type–specific TFs (Figure 5). This could explain how memory can persist after the temporary loss of a PcG factor (51, 81, 138) and how new bivalent genes can arise during development (139, 140). Our focus here has been on promoters and genes, as these seem to be where regulation is first apparent, but this same logic is likely to hold for enhancers (22, 141). PREs in flies may fit in this category, with dual enhancer and silencer activities (123); they could also serve as tethering sites to stabilize PRC2 spreading via protein–protein interactions, consistent with more classical models (67, 82).
Key elements in the master switch model are (a) the central importance of PRC1 (30, 109); (b) the likely importance of acetylation, linking regulation of the PcG–TrxG master switch to TFs (109, 121, 130, 131, 142, 143); and (c) the simple idea that regulatory elements may always be marked by competing activities (141, 144). Like any chemical equation, these series of interactions would be governed by critical thresholds of competing factors, equilibria, and thus reversibility (145). Finally, the reliance on a prelocalized PcG–TrxG master switch might explain how widespread binding of TFs can result in precise readouts at developmental genes.
If a reversible PcG–TrxG interaction is prevalent throughout development, why was it not identified previously? One likely reason is that PRC1 occupancy on active genes is typically lower than on silent regions; thus, methods needed to be highly reproducible before this paradoxical result could be recognized (2). Likewise, skewed PcG–TrxG balances may lead to occupancy that is below the threshold for detectability with current technologies, and the key protein–protein interactions may require cross-linking within the context of chromatin for robust recovery (109). It is also not clear how many coactivators and PRC1 subunits might be involved and when critical transitions might occur, especially in mammals. With the evolution of CpG islands, PcG–TrxG competition at 5′ ends of genes in mammals may rely on subunits with affinity for GC-rich sequences (95–99) and not require the observable protein interactions seen in flies. Importantly, we lack comprehensive methods to identify complex compositions, protein interactions, and posttranslational modifications that differ at individual loci. And it is already clear that substantial redundancy governs regulation, resulting in considerable difficulty in testing any of the current models (47).
Although the master switch idea is still only a hypothesis, could it also shed light on how the PcG actually interferes with gene expression? Stably silenced regions are more compact when probed for chromatin accessibility or by imaging (146–149). Yet, cell type–specific transcriptional programming at the majority of loci does not appear to depend on compaction (150). Consistent with the master switch model, is it possible that PRC1 and PRC2 function primarily to counteract acetylation? Acetylation and deacetylation have been discounted as stable epigenetic states because of their dynamic turnover, but this characteristic would not preclude a model in which PcG functions by keeping acetylation consistently below a threshold required for productive transcription. If so, prolonged PcG enrichment might eventually lead to complete deacetylation, 3D compaction, and irreversible silencing, as seen at epigenetically silenced Hox clusters. Those canonical PcG targets also have multiple recruiting elements that facilitate spreading of H3K27me3, thus favoring stable maintenance of the repressive mark.
ACKNOWLEDGMENTS
A major goal of this review is to stimulate consideration of ideas that synergize across the fly and mammalian models. In this process, we thank the readers for tolerating a significant amount of speculation. We also apologize in advance for unintentional omissions in referencing this very deep and talented field. PcG research in the Kuroda laboratory is supported by National Institutes of Health (NIH) grant R35GM126944. S.D. and J.A.K. are supported by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, NIH.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Laugesen A, Hojfeldt JW, Helin K. 2019. Molecular mechanisms directing PRC2 recruitment and H3K27 methylation. Mol. Cell 74:8–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vidal M. 2019. Polycomb assemblies multitask to regulate transcription. Epigenomes 3:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu JR, Lee CH, Oksuz O, Stafford JM, Reinberg D. 2019. PRC2 is high maintenance. Genes Dev. 33:1–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holoch D, Margueron R. 2017. Mechanisms regulating PRC2 recruitment and enzymatic activity. Trends Biochem. Sci 42:531–42 [DOI] [PubMed] [Google Scholar]
- 5.Schuettengruber B, Bourbon HM, Di Croce L, Cavalli G. 2017. Genome regulation by Polycomb and Trithorax: 70 years and counting. Cell 171:34–57 [DOI] [PubMed] [Google Scholar]
- 6.Lewis EB. 1978. A gene complex controlling segmentation in Drosophila. Nature 276:565–70 [DOI] [PubMed] [Google Scholar]
- 7.Struhl G. 1981. A gene product required for correct initiation of segmental determination in Drosophila. Nature 293:36–41 [DOI] [PubMed] [Google Scholar]
- 8.Ingham PW. 1983. Differential expression of bithorax complex genes in the absence of the extra sex combs and trithorax genes. Nature 306:591–93 [DOI] [PubMed] [Google Scholar]
- 9.Pasini D, Di Croce L. 2016. Emerging roles for Polycomb proteins in cancer. Curr. Opin. Genet. Dev 36:50–58 [DOI] [PubMed] [Google Scholar]
- 10.Poynter ST, Kadoch C. 2016. Polycomb and trithorax opposition in development and disease. WIREs Dev. Biol 5:659–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, et al. 2004. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev. Cell 7:663–76 [DOI] [PubMed] [Google Scholar]
- 12.Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, et al. 2004. Role of histone H2A ubiquitination in Polycomb silencing. Nature 431:873–78 [DOI] [PubMed] [Google Scholar]
- 13.Lagarou A, Mohd-Sarip A, Moshkin YM, Chalkley GE, Bezstarosti K, et al. 2008. dKDM2 couples histone H2A ubiquitylation to histone H3 demethylation during Polycomb group silencing. Genes Dev 22:2799–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, et al. 2002. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298:1039–43 [DOI] [PubMed] [Google Scholar]
- 15.Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. 2002. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111:185–96 [DOI] [PubMed] [Google Scholar]
- 16.Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, et al. 2002. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111:197–208 [DOI] [PubMed] [Google Scholar]
- 17.Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. 2002. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev 16:2893–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pengelly AR, Copur O, Jackle H, Herzig A, Muller J. 2013. A histone mutant reproduces the phenotype caused by loss of histone-modifying factor Polycomb. Science 339:698–99 [DOI] [PubMed] [Google Scholar]
- 19.Pengelly AR, Kalb R, Finkl K, Muller J. 2015. Transcriptional repression by PRC1 in the absence of H2A monoubiquitylation. Genes Dev 29:1487–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blackledge NP, Fursova NA, Kelley JR, Huseyin MK, Feldmann A, Klose RJ. 2019. PRC1 catalytic activity is central to Polycomb system function. bioRxiv. 10.1101/667667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tamburri S, Lavarone E, Fernandez-Perez D, Zanotti M, Manganaro D, et al. 2019. Histone H2AK119 mono-ubiquitination is essential for Polycomb-mediated transcriptional repression. bioRxiv 690461. 10.1101/690461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Piunti A, Shilatifard A. 2016. Epigenetic balance of gene expression by Polycomb and COMPASS families. Science 352:aad9780. [DOI] [PubMed] [Google Scholar]
- 23.Schmähling S, Meiler A, Lee Y, Mohammed A, Finkl K, et al. 2018. Regulation and function of H3K36 di-methylation by the trithorax-group protein complex AMC. Development 145:dev163808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang C, Yang F, Zhang Z, Zhang J, Cai G, et al. 2017. Mrg15 stimulates Ash1 H3K36 methyltransferase activity and facilitates Ash1 Trithorax group protein function in Drosophila. Nat. Commun 8:1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang L, Joshi P, Miller EL, Higgins L, Slattery M, Simon JA. 2018. A role for monomethylation of histone H3-K27 in gene activity in Drosophila. Genetics 208:1023–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee HG, Kahn TG, Simcox A, Schwartz YB, Pirrotta V. 2015. Genome-wide activities of Polycomb complexes control pervasive transcription. Genome Res 25:1170–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferrari KJ, Scelfo A, Jammula S, Cuomo A, Barozzi I, et al. 2014. Polycomb-dependent H3K27me1 and H3K27me2 regulate active transcription and enhancer fidelity. Mol. Cell 53:49–62 [DOI] [PubMed] [Google Scholar]
- 28.Schaaf CA, Misulovin Z, Gause M, Koenig A, Gohara DW, et al. 2013. Cohesin and polycomb proteins functionally interact to control transcription at silenced and active genes. PLOS Genet 9:e1003560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao Z, Lee P, Stafford JM, von Schimmelmann M, Schaefer A, Reinberg D. 2014. An AUTS2-Polycomb complex activates gene expression in the CNS. Nature 516:349–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Creppe C, Palau A, Malinverni R, Valero V, Buschbeck M. 2014. A Cbx8-containing Polycomb complex facilitates the transition to gene activation during ES cell differentiation. PLOS Genet 10:e1004851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kloet SL, Makowski MM, Baymaz HI, van Voorthuijsen L, Karemaker ID, et al. 2016. The dynamic interactome and genomic targets of Polycomb complexes during stem-cell differentiation. Nat. Struct. Mol. Biol 23:682–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loubiere V, Delest A, Thomas A, Bonev B, Schuettengruber B, et al. 2016. Coordinate redeployment of PRC1 proteins suppresses tumor formation during Drosophila development. Nat. Genet 48:1436–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pemberton H, Anderton E, Patel H, Brookes S, Chandler H, et al. 2014. Genome-wide co-localization of Polycomb orthologs and their effects on gene expression in human fibroblasts. Genome Biol 15:R23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van den Boom V, Maat H, Geugien M, Rodriguez Lopez A, Sotoca AM, et al. 2016. Non-canonical PRC1.1 targets active genes independent of H3K27me3 and is essential for leukemogenesis. Cell Rep 14:332–46 [DOI] [PubMed] [Google Scholar]
- 35.Brown JL, Sun MA, Kassis JA. 2018. Global changes of H3K27me3 domains and Polycomb group protein distribution in the absence of recruiters Spps or Pho. PNAS 115:E1839–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pherson M, Misulovin Z, Gause M, Mihindukulasuriya K, Swain A, Dorsett D. 2017. Polycomb repressive complex 1 modifies transcription of active genes. Sci. Adv 3:e1700944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cohen I, Zhao D, Bar C, Valdes VJ, Dauber-Decker KL, et al. 2018. PRC1 fine-tunes gene repression and activation to safeguard skin development and stem cell specification. Cell Stem Cell 22:726–39.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao Z, Zhang J, Bonasio R, Strino F, Sawai A, et al. 2012. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol. Cell 45:344–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.De Cegli R, Iacobacci S, Flore G, Gambardella G, Mao L, et al. 2013. Reverse engineering a mouse embryonic stem cell-specific transcriptional network reveals a new modulator of neuronal differentiation. Nucleic Acids Res 41:711–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smits AH, Jansen PW, Poser I, Hyman AA, Vermeulen M. 2013. Stoichiometry of chromatin-associated protein complexes revealed by label-free quantitative mass spectrometry-based proteomics. Nucleic Acids Res 41:e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alekseyenko AA, Gorchakov AA, Kharchenko PV, Kuroda MI. 2014. Reciprocal interactions of human C10orf12 and C17orf96 with PRC2 revealed by BioTAP-XL cross-linking and affinity purification. PNAS 111:2488–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liefke R, Shi Y. 2015. The PRC2-associated factor C17orf96 is a novel CpG island regulator in mouse ES cells. Cell Discov 1:15008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beringer M, Pisano P, Di Carlo V, Blanco E, Chammas P, et al. 2016. EPOP functionally links Elongin and Polycomb in pluripotent stem cells. Mol. Cell 64:645–58 [DOI] [PubMed] [Google Scholar]
- 44.Liefke R, Karwacki-Neisius V, Shi Y. 2017. EPOP interacts with Elongin BC and USP7 to modulate the chromatin landscape. Mol. Cell 65:202. [DOI] [PubMed] [Google Scholar]
- 45.Conway E, Jerman E, Healy E, Ito S, Holoch D, et al. 2018. A family of vertebrate-specific Polycombs encoded by the LCOR/LCORL genes balance PRC2 subtype activities. Mol. Cell 70:408–21.e8 [DOI] [PubMed] [Google Scholar]
- 46.Lee CH, Holder M, Grau D, Saldana-Meyer R, Yu JR, et al. 2018. Distinct stimulatory mechanisms regulate the catalytic activity of polycomb repressive complex 2. Mol. Cell 70:435–48.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fursova NA, Blackledge NP, Nakayama M, Ito S, Koseki Y, et al. 2019. Synergy between variant PRC1 complexes defines Polycomb-mediated gene repression. Mol. Cell 74:P1020–36.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arrigoni R, Alam SL, Wamstad JA, Bardwell VJ, Sundquist WI, Schreiber-Agus N. 2006. The Polycomb-associated protein Rybp is a ubiquitin binding protein. FEBS Lett 580:6233–41 [DOI] [PubMed] [Google Scholar]
- 49.Hansen KH, Bracken AP, Pasini D, Dietrich N, Gehani SS, et al. 2008. A model for transmission of the H3K27me3 epigenetic mark. Nat. Cell Biol 10:1291–300 [DOI] [PubMed] [Google Scholar]
- 50.Margueron R, Justin N, Ohno K, Sharpe ML, Son J, et al. 2009. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 461:762–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oksuz O, Narendra V, Lee CH, Descostes N, LeRoy G, et al. 2018. Capturing the onset of PRC2-mediated repressive domain formation. Mol. Cell 70:1149–62.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cooper S, Grijzenhout A, Underwood E, Ancelin K, Zhang T, et al. 2016. Jarid2 binds mono-ubiquitylated H2A lysine 119 to mediate crosstalk between Polycomb complexes PRC1 and PRC2. Nat. Commun 7:13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kalb R, Latwiel S, Baymaz HI, Jansen PW, Muller CW, et al. 2014. Histone H2A monoubiquitination promotes histone H3 methylation in Polycomb repression. Nat. Struct. Mol. Biol 21:569–71 [DOI] [PubMed] [Google Scholar]
- 54.Fischle W, Wang Y, Jacobs SA, Kim Y, Allis CD, Khorasanizadeh S. 2003. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev 17:1870–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kasinath V, Faini M, Poepsel S, Reif D, Feng XA, et al. 2018. Structures of human PRC2 with its cofactors AEBP2 and JARID2. Science 359:940–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen S, Jiao L, Shubbar M, Yang X, Liu X. 2018. Unique structural platforms of Suz12 dictate distinct classes of PRC2 for chromatin binding. Mol. Cell 69:840–52.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chittock EC, Latwiel S, Miller TC, Muller CW. 2017. Molecular architecture of polycomb repressive complexes. Biochem. Soc. Trans 45:193–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vann KR, Kutateladze TG. 2018. Architecture of PRC2 holo complexes. Trends Biochem. Sci 43:487–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kasinath V, Poepsel S, Nogales E. 2019. Recent structural insights into Polycomb repressive complex 2 regulation and substrate binding. Biochemistry 58:346–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McGinty RK, Henrici RC, Tan S. 2014. Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome. Nature 514:591–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kassis JA. 2002. Pairing-sensitive silencing, polycomb group response elements, and transposon homing in Drosophila. Adv. Genet 46:421–38 [DOI] [PubMed] [Google Scholar]
- 62.Ringrose L, Paro R. 2004. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu. Rev. Genet 38:413–43 [DOI] [PubMed] [Google Scholar]
- 63.Kassis JA, Brown JL. 2013. Polycomb group response elements in Drosophila and vertebrates. Adv. Genet 81:83–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Americo J, Whiteley M, Brown JL, Fujioka M, Jaynes JB, Kassis JA. 2002. A complex array of DNA-binding proteins required for pairing-sensitive silencing by a polycomb group response element from the Drosophila engrailed gene. Genetics 160:1561–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brown JL, Mucci D, Whiteley M, Dirksen ML, Kassis JA. 1998. The Drosophila Polycomb group gene pleiohomeotic encodes a DNA binding protein with homology to the transcription factor YY1. Mol. Cell 1:1057–64 [DOI] [PubMed] [Google Scholar]
- 66.Klymenko T, Papp B, Fischle W, Kocher T, Schelder M, et al. 2006. A Polycomb group protein complex with sequence-specific DNA-binding and selective methyl-lysine-binding activities. Genes Dev 20:1110–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Frey F, Sheahan T, Finkl K, Stoehr G, Mann M, et al. 2016. Molecular basis of PRC1 targeting to Polycomb response elements by PhoRC. Genes Dev 30:1116–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kahn TG, Dorafshan E, Schultheis D, Zare A, Stenberg P, et al. 2016. Interdependence of PRC1 and PRC2 for recruitment to Polycomb Response Elements. Nucleic Acids Res 44:10132–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kang H, McElroy KA, Jung YL, Alekseyenko AA, Zee BM, et al. 2015. Sex comb on midleg (Scm) is a functional link between PcG-repressive complexes in Drosophila. Genes Dev 29:1136–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sing A, Pannell D, Karaiskakis A, Sturgeon K, Djabali M, et al. 2009. A vertebrate Polycomb response element governs segmentation of the posterior hindbrain. Cell 138:885–97 [DOI] [PubMed] [Google Scholar]
- 71.Woo CJ, Kharchenko PV, Daheron L, Park PJ, Kingston RE. 2010. A region of the human HOXD cluster that confers Polycomb-group responsiveness. Cell 140:99–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Woo CJ, Kharchenko PV, Daheron L, Park PJ, Kingston RE. 2013. Variable requirements for DNA-binding proteins at Polycomb-dependent repressive regions in human HOX clusters. Mol. Cell. Biol 33:3274–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schorderet P, Lonfat N, Darbellay F, Tschopp P, Gitto S, et al. 2013. A genetic approach to the recruitment of PRC2 at the HoxD locus. PLOS Genet 9:e1003951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fujioka M, Sun G, Jaynes JB. 2013. The Drosophila eve insulator Homie promotes eve expression and protects the adjacent gene from repression by polycomb spreading. PLOS Genet 9:e1003883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Narendra V, Rocha PP, An D, Raviram R, Skok JA, et al. 2015. CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science 347:1017–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hnisz D, Weintraub AS, Day DS, Valton AL, Bak RO, et al. 2016. Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science 351:1454–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.De S, Cheng Y, Sun MA, Gehred ND, Kassis JA. 2019. Structure and function of an ectopic Polycomb chromatin domain. Sci. Adv 5:eaau9739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schmitges FW, Prusty AB, Faty M, Stutzer A, Lingaraju GM, et al. 2011. Histone methylation by PRC2 is inhibited by active chromatin marks. Mol. Cell 42:330–41 [DOI] [PubMed] [Google Scholar]
- 79.Li XY, Harrison MM, Villalta JE, Kaplan T, Eisen MB. 2014. Establishment of regions of genomic activity during the Drosophila maternal to zygotic transition. eLife 3:e03737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zenk F, Loeser E, Schiavo R, Kilpert F, Bogdanovic O, Iovino N. 2017. Germ line–inherited H3K27me3 restricts enhancer function during maternal-to-zygotic transition. Science 357:212–16 [DOI] [PubMed] [Google Scholar]
- 81.Hojfeldt JW, Laugesen A, Willumsen BM, Damhofer H, Hedehus L, et al. 2018. Accurate H3K27 methylation can be established de novo by SUZ12-directed PRC2. Nat. Struct. Mol. Biol 25:225–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.De S, Mitra A, Cheng Y, Pfeifer K, Kassis JA. 2016. Formation of a Polycomb-domain in the absence of strong Polycomb response elements. PLOS Genet 12:e1006200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Peng JC, Valouev A, Swigut T, Zhang J, Zhao Y, et al. 2009. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell 139:1290–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li H, Liefke R, Jiang J, Kurland JV, Tian W, et al. 2017. Polycomb-like proteins link the PRC2 complex to CpG islands. Nature 549:287–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Poepsel S, Kasinath V, Nogales E. 2018. Cryo-EM structures of PRC2 simultaneously engaged with two functionally distinct nucleosomes. Nat. Struct. Mol. Biol 25:154–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ogiyama Y, Schuettengruber B, Papadopoulos GL, Chang JM, Cavalli G. 2018. Polycomb-dependent chromatin looping contributes to gene silencing during Drosophila development. Mol. Cell 71:73–88.e5 [DOI] [PubMed] [Google Scholar]
- 87.Robinson AK, Leal BZ, Chadwell LV, Wang R, Ilangovan U, et al. 2012. The growth-suppressive function of the Polycomb Group protein polyhomeotic is mediated by polymerization of its Sterile Alpha Motif (SAM) domain. J. Biol. Chem 287:8702–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Plys AJ, Davis CP, Kim J, Rizki G, Keenen MM, et al. 2019. Phase separation of Polycomb-repressive complex 1 is governed by a charged disordered region of CBX2. Genes Dev 33:799–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zylicz JJ, Bousard A, Zumer K, Dossin F, Mohammad E, et al. 2019. The implication of early chromatin changes in X chromosome inactivation. Cell 176:182–97.e23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Eissenberg JC, Elgin SC. 2014. HP1a: a structural chromosomal protein regulating transcription. Trends Genet 30:103–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ferrari F, Alekseyenko AA, Park PJ, Kuroda MI. 2014. Transcriptional control of a whole chromosome: emerging models for dosage compensation. Nat. Struct. Mol. Biol 21:118–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kondo T, Ito S, Koseki H. 2016. Polycomb in transcriptional phase transition of developmental genes. Trends Biochem. Sci 41:9–19 [DOI] [PubMed] [Google Scholar]
- 93.Marasca F, Bodega B, Orlando V. 2018. How Polycomb-mediated cell memory deals with a changing environment: variations in PcG complexes and proteins assortment convey plasticity to epigenetic regulation as a response to environment. Bioessays 40:e1700137. [DOI] [PubMed] [Google Scholar]
- 94.Blackledge NP, Rose NR, Klose RJ. 2015. Targeting Polycomb systems to regulate gene expression: modifications to a complex story. Nat. Rev. Mol. Cell Biol 16:643–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Klose RJ, Cooper S, Farcas AM, Blackledge NP, Brockdorff N. 2013. Chromatin sampling—an emerging perspective on targeting Polycomb repressor proteins. PLOS Genet 9:e1003717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lynch MD, Smith AJ, De Gobbi M, Flenley M, Hughes JR, et al. 2012. An interspecies analysis reveals a key role for unmethylated CpG dinucleotides in vertebrate Polycomb complex recruitment. EMBO J 31:317–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mendenhall EM, Koche RP, Truong T, Zhou VW, Issac B, et al. 2010. GC-rich sequence elements recruit PRC2 in mammalian ES cells. PLOS Genet 6:e1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wachter E, Quante T, Merusi C, Arczewska A, Stewart F, et al. 2014. Synthetic CpG islands reveal DNA sequence determinants of chromatin structure. eLife 3:e03397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jermann P, Hoerner L, Burger L, Schubeler D. 2014. Short sequences can efficiently recruit histone H3 lysine 27 trimethylation in the absence of enhancer activity and DNA methylation. PNAS 111:E3415–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pirrotta V 1997. PcG complexes and chromatin silencing. Curr. Opin. Genet. Dev 7:249–58 [DOI] [PubMed] [Google Scholar]
- 101.Paro R 1995. Propagating memory of transcriptional states. Trends Genet 11:295–97 [DOI] [PubMed] [Google Scholar]
- 102.Kharchenko PV, Alekseyenko AA, Schwartz YB, Minoda A, Riddle NC, et al. 2011. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature 471:480–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, et al. 2006. Chromatin signatures of pluripotent cell lines. Nat. Cell Biol 8:532–38 [DOI] [PubMed] [Google Scholar]
- 104.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, et al. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125:315–26 [DOI] [PubMed] [Google Scholar]
- 105.Shema E, Jones D, Shoresh N, Donohue L, Ram O, Bernstein BE. 2016. Single-molecule decoding of combinatorially modified nucleosomes. Science 352:717–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Voigt P, LeRoy G, Drury WJ III, Zee BM, Son J, et al. 2012. Asymmetrically modified nucleosomes. Cell 151:181–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Vastenhouw NL, Schier AF. 2012. Bivalent histone modifications in early embryogenesis. Curr. Opin. Cell Biol 24:374–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Voigt P, Tee WW, Reinberg D. 2013. A double take on bivalent promoters. Genes Dev 27:1318–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kang H, Jung YL, McElroy KA, Zee BM, Wallace HA, et al. 2017. Bivalent complexes of PRC1 with orthologs of BRD4 and MOZ/MORF target developmental genes in Drosophila. Genes Dev 31:1988–2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Akmammedov A, Geigges M, Paro R. 2019. Bivalency in Drosophila embryos is associated with strong inducibility of Polycomb target genes. Fly 13:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Schertel C, Albarca M, Rockel-Bauer C, Kelley NW, Bischof J, et al. 2015. A large-scale, in vivo transcription factor screen defines bivalent chromatin as a key property of regulatory factors mediating Drosophila wing development. Genome Res 25:514–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rickels R, Hu D, Collings CK, Woodfin AR, Piunti A, et al. 2016. An evolutionary conserved epigenetic mark of Polycomb response elements implemented by Trx/MLL/COMPASS. Mol. Cell 63:318–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Papp B, Muller J. 2006. Histone trimethylation and the maintenance of transcriptional ON and OFF states by trxG and PcG proteins. Genes Dev 20:2041–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kassis JA, Kennison JA, Tamkun JW. 2017. Polycomb and Trithorax group genes in Drosophila. Genetics 206:1699–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cohen I, Zhao D, Menon G, Nakayama M, Koseki H, et al. 2019. PRC1 preserves epidermal tissue integrity independently of PRC2. Genes Dev 33:55–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wang D, Yi R. 2019. Is it time to take R(epressive) out of PRC1? Genes Dev 33:4–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Strubbe G, Popp C, Schmidt A, Pauli A, Ringrose L, et al. 2011. Polycomb purification by in vivo biotinylation tagging reveals cohesin and Trithorax group proteins as interaction partners. PNAS 108:5572–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Digan ME, Haynes SR, Mozer BA, Dawid IB, Forquignon F, Gans M. 1986. Genetic and molecular analysis of fs(1)h, a maternal effect homeotic gene in Drosophila. Dev. Biol 114:161–69 [DOI] [PubMed] [Google Scholar]
- 119.Laue K, Daujat S, Crump JG, Plaster N, Roehl HH, et al. 2008. The multidomain protein Brpf1 binds histones and is required for Hox gene expression and segmental identity. Development 135:1935–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sheikh BN, Downer NL, Phipson B, Vanyai HK, Kueh AJ, et al. 2015. MOZ and BMI1 play opposing roles during Hox gene activation in ES cells and in body segment identity specification in vivo. PNAS 112:5437–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Poux S, Horard B, Sigrist CJ, Pirrotta V. 2002. The Drosophila Trithorax protein is a coactivator required to prevent re-establishment of Polycomb silencing. Development 129:2483–93 [DOI] [PubMed] [Google Scholar]
- 122.Kockmann T, Gerstung M, Schlumpf T, Xhinzhou Z, Hess D, et al. 2013. The BET protein FSH functionally interacts with ASH1 to orchestrate global gene activity in Drosophila. Genome Biol 14:R18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Erceg J, Pakozdi T, Marco-Ferreres R, Ghavi-Helm Y, Girardot C, et al. 2017. Dual functionality of cis-regulatory elements as developmental enhancers and Polycomb response elements. Genes Dev 31:590–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chinwalla V, Jane EP, Harte PJ. 1995. The Drosophila trithorax protein binds to specific chromosomal sites and is co-localized with Polycomb at many sites. EMBO J 14:2056–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, Cairns BR. 2009. Distinctive chromatin in human sperm packages genes for embryo development. Nature 460:473–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lesch BJ, Dokshin GA, Young RA, McCarrey JR, Page DC. 2013. A set of genes critical to development is epigenetically poised in mouse germ cells from fetal stages through completion of meiosis. PNAS 110:16061–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sachs M, Onodera C, Blaschke K, Ebata KT, Song JS, Ramalho-Santos M. 2013. Bivalent chromatin marks developmental regulatory genes in the mouse embryonic germline in vivo. Cell Rep 3:1777–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hanna CW, Taudt A, Huang J, Gahurova L, Kranz A, et al. 2018. MLL2 conveys transcription-independent H3K4 trimethylation in oocytes. Nat. Struct. Mol. Biol 25:73–82 [DOI] [PubMed] [Google Scholar]
- 129.Wu SF, Zhang H, Cairns BR. 2011. Genes for embryo development are packaged in blocks of multivalent chromatin in zebrafish sperm. Genome Res 21:578–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Weaver TM, Liu J, Connelly KE, Coble C, Varzavand K, et al. 2019. The EZH2 SANT1 domain is a histone reader providing sensitivity to the modification state of the H4 tail. Sci. Rep 9:987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Tie F, Banerjee R, Stratton CA, Prasad-Sinha J, Stepanik V, et al. 2009. CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development 136:3131–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Blackledge NP, Farcas AM, Kondo T, King HW, McGouran JF, et al. 2014. Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and Polycomb domain formation. Cell 157:1445–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Dorighi KM, Tamkun JW. 2013. The trithorax group proteins Kismet and ASH1 promote H3K36 dimethylation to counteract Polycomb group repression in Drosophila. Development 140:4182–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Denissov S, Hofemeister H, Marks H, Kranz A, Ciotta G, et al. 2014. Mll2 is required for H3K4 trimethylation on bivalent promoters in embryonic stem cells, whereas Mll1 is redundant. Development 141:526–37 [DOI] [PubMed] [Google Scholar]
- 135.Dorafshan E, Kahn TG, Glotov A, Savitsky M, Walther M, et al. 2019. Ash1 counteracts Polycomb repression independent of histone H3 lysine 36 methylation. EMBO Rep 20:e46762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Bracken AP, Brien GL, Verrijzer CP. 2019. Dangerous liaisons: interplay between SWI/SNF, NuRD, and Polycomb in chromatin regulation and cancer. Genes Dev 33:1–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Brand M, Nakka K, Zhu J, Dilworth FJ. 2019. Polycomb/Trithorax antagonism: cellular memory in stem cell fate and function. Cell Stem Cell 24:518–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Beuchle D, Struhl G, Muller J. 2001. Polycomb group proteins and heritable silencing of Drosophila Hox genes. Development 128:993–1004 [DOI] [PubMed] [Google Scholar]
- 139.Jadhav U, Nalapareddy K, Saxena M, O’Neill NK, Pinello L, et al. 2016. Acquired tissue-specific promoter bivalency is a basis for PRC2 necessity in adult cells. Cell 165:1389–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Weiner A, Lara-Astiaso D, Krupalnik V, Gafni O, David E, et al. 2016. Co-ChIP enables genome-wide mapping of histone mark co-occurrence at single-molecule resolution. Nat. Biotechnol 34:953–61 [DOI] [PubMed] [Google Scholar]
- 141.Jadhav U, Cavazza A, Banerjee KK, Xie H, O’Neill NK, et al. 2019. Extensive recovery of embryonic enhancer and gene memory stored in hypomethylated enhancer DNA. Mol. Cell 74:542–54.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Pasini D, Malatesta M, Jung HR, Walfridsson J, Willer A, et al. 2010. Characterization of an antagonistic switch between histone H3 lysine 27 methylation and acetylation in the transcriptional regulation of Polycomb group target genes. Nucleic Acids Res 38:4958–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kehle J, Beuchle D, Treuheit S, Christen B, Kennison JA, et al. 1998. dMi-2, a Hunchback-interacting protein that functions in Polycomb repression. Science 282:1897–900 [DOI] [PubMed] [Google Scholar]
- 144.Kim HS, Tan Y, Ma W, Merkurjev D, Destici E, et al. 2018. Pluripotency factors functionally premark cell-type-restricted enhancers in ES cells. Nature 556:510–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sadasivam DA, Huang DH. 2018. Feedback regulation by antagonistic epigenetic factors potentially maintains developmental homeostasis in Drosophila. J. Cell Sci 131:jcs210179. [DOI] [PubMed] [Google Scholar]
- 146.Zink D, Paro R. 1995. Drosophila Polycomb-group regulated chromatin inhibits the accessibility of a trans-activator to its target DNA. EMBO J 14:5660–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Fitzgerald DP, Bender W. 2001. Polycomb group repression reduces DNA accessibility. Mol. Cell. Biol 21:6585–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Boettiger AN, Bintu B, Moffitt JR, Wang S, Beliveau BJ, et al. 2016. Super-resolution imaging reveals distinct chromatin folding for different epigenetic states. Nature 529:418–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Mateo LJ, Murphy SE, Hafner A, Cinquini IS, Walker CA, Boettiger AN. 2019. Visualizing DNA folding and RNA in embryos at single-cell resolution. Nature 568:49–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.King HW, Fursova NA, Blackledge NP, Klose RJ. 2018. Polycomb repressive complex 1 shapes the nucleosome landscape but not accessibility at target genes. Genome Res 28:1494–507 [DOI] [PMC free article] [PubMed] [Google Scholar]


