Abstract
Endocrine therapy is a systemic therapy and has become the main treatment strategy for patients with estrogen receptor (ER)-positive breast cancer. However, tamoxifen resistance has become an insurmountable clinical challenge, and the underlying mechanisms are still poorly understood. In this study, we explored the roles of CXC chemokine receptor type 4 (CXCR4) in tamoxifen-treated breast cancer and tamoxifen resistance. Based on the Gene Expression Omnibus (GEO) database, high expression of CXCR4 was found to be associated with worse overall survival (hazard ratio [HR] = 4.646, p < 0.001) and cancer-specific survival (HR = 4.480, p < 0.001) in tamoxifen-treated breast cancer. CXCR4 was also positively correlated with the level of AKT phosphorylation and the resistance to tamoxifen in breast cancer. AMD3100 is a CXCR4 antagonist and was found to decrease phosphorylated (p)-AKT levels of tamoxifen-resistant cells. The reversal effect of AMD3100 on tamoxifen resistance was also confirmed in vitro and in vivo. Taken together, our study demonstrated that CXCR4 could be a potential prognostic biomarker for tamoxifen-treated breast cancer, and the combination of AMD3100 with tamoxifen could be a more efficacious therapeutic strategy for the treatment of tamoxifen resistance.
Keywords: breast cancer, tamoxifen resistance, CXCR4, AMD3100, AKT phosphorylation
Graphical Abstract
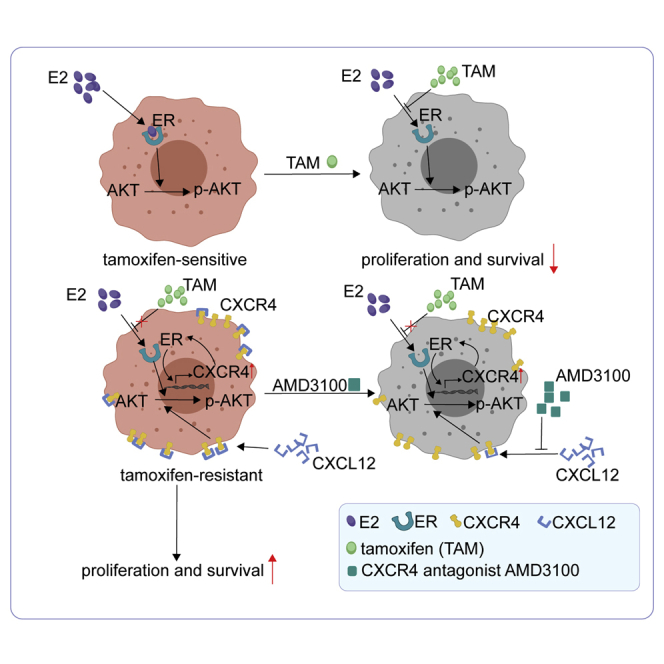
CXCR4 is a good prognostic indicator of tamoxifen-treated breast cancer and promotes the resistance to tamoxifen. CXCR4 antagonist AMD3100 can reverse the resistance to tamoxifen by inhibiting the hyperphosphorylation of AKT. Huang and colleagues provide the feasibility of the combination of tamoxifen and AMD3100 in tamoxifen-resistant breast cancer.
Introduction
Breast cancer is the fifth leading cause of death in women and the most frequently diagnosed malignancy in women worldwide.1 Approximately 70% of breast cancer tumors express estrogen receptor (ER).2 Endocrine therapy is a systemic therapy and has become the main treatment strategy for premenopausal and postmenopausal positive patients.3,4 Tamoxifen is the most widely studied, used, and representative drug blocking ER, which has been used for advanced disease, neoadjuvant and adjuvant therapy, and chemotherapy prevention in high-risk women.5 Despite advances in treatment, endocrine resistance is still present in a subset of patients with recurrent or metastatic disease progression after primary hormone receptor-positive breast cancer diagnosis.6 Therefore, it is important to seek predictive biomarkers for tamoxifen-treated patients and take a deeper insight into the underlying molecular mechanisms of tamoxifen resistance.
CXC chemokine receptor type 4 (CXCR4) is a seven-transmembrane G protein-coupled receptor and promotes tumor growth, angiogenesis, and metastasis in various types of cancers.7, 8, 9, 10 CXCR4 has been reported to be involved in a variety of intracellular signaling pathways, and overexpressed CXCR4 can activate Wnt/β-catenin, Notch, JAK/STAT (Janus kinase-signal transducer and activator of transcription pathway), mitogen-activated protein kinases (MAPKs), and phosphatidylinositol 3-kinase (PI3K)/AKT pathways.11 CXCR4 was reported to be associated with the prognosis of breast cancer patients,12 but it remains unknown whether CXCR4 has prognostic significance in tamoxifen-treated breast cancer or how its expression level changes in tamoxifen-resistant cells. Therefore, it is meaningful to explore the relationship between CXCR4 and tamoxifen resistance in breast cancer from the perspective of clinical significance and mechanism verification. AMD3100 is a specific antagonist of CXCR4, which binds to CXCR4, inhibits the interaction between CXCR4 and CXCL12, and has no cross-reaction with other chemokine receptors.13, 14, 15 Previous studies reported that AMD3100 could inhibit the proliferation of breast cancer cells and increase the sensitivity to radiotherapy in breast cancer in vivo,16,17 but whether AMD3100 could be used to reverse the resistance to tamoxifen has never been reported.
In this study, we aimed to identify a potential predictive biomarker of tamoxifen resistance and explore the role of CXCR4 in tamoxifen resistance. Using multiple gene expression analyses, we identified that high expression of CXCR4 was associated with poor prognosis in tamoxifen-treated breast cancer, and CXCR4 was upregulated in tamoxifen-resistant breast cancer cells. Besides, we also demonstrated that AMD3100, a specific antagonist of CXCR4, could reverse the resistance to tamoxifen via inhibiting AKT phosphorylation.
Results
CXCR4 Is Upregulated in Tamoxifen-Resistant Breast Cancer Cells
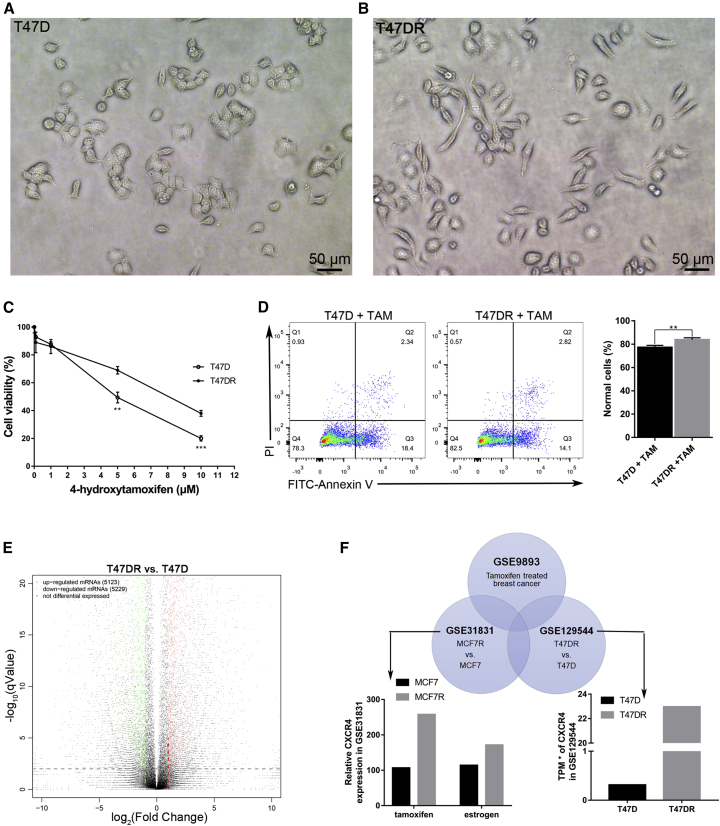
We cultured T47D cells with 1 μM tamoxifen for more than 6 months to establish the tamoxifen-resistant subline T47DR as our previous study has described.18 Observation of morphological changes showed that T47DR cells were fusiform, which was different from T47D (Figures 1A and 1B). In order to verify whether T47DR cells are resistant to tamoxifen, we compared the proliferation and apoptosis of T47D and T47DR cells treated with tamoxifen by Cell Count Kit 8 (CCK8) assays and Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (P)I assays. The results of the CCK8 assay showed that the cell viability of T47DR cells was higher than that of T47D cells when the concentrations of tamoxifen were 5 and 10 μM (Figure 1C). The half-maximal inhibitory concentrations (IC50) of tamoxifen were 4.47 μmol/L in T47D cells and 7.80 μmol/L in T47DR cells. The results of the Annexin V-FITC/PI experiment showed that apoptosis of T47DR cells was reduced compared with T47D cells when treated with tamoxifen (Figure 1D). In order to compare the gene expression profile of T47D and T47DR cells, we conducted the RNA sequencing (GEO: GSE129544). A total of 5,123 upregulated and 5,229 downregulated mRNAs were identified as being differentially expressed between T47D and T47DR (Figure 1E). To obtain an accurate and reliable result, we also conducted bioinformatics analysis based on another two Gene Expression Omnibus (GEO) datasets (GEO: GSE9893 and GSE31831). The intersection of three GEO datasets contained several genes, including CXCR4, and the results showed that CXCR4 was upregulated in tamoxifen-resistant breast cancer cells MCF7R and T47DR (Figure 1F).
Figure 1.
CXCR4 Is Upregulated in Tamoxifen-Resistant Breast Cancer Cells
(A and B) Morphological differences between (A) T47D and (B) T47DR cells. (C) CCK8 assay showed that the cell proliferation ability of T47DR was higher than that in T47D when they were treated with tamoxifen. (D) Annexin V-FITC/PI assay showed the difference of apoptosis between T47D and T47DR cells when they were treated with 5 μM tamoxifen. (E) Differentially expressed genes in T47D and T47DR cells. (F) The intersection of the three datasets (GEO: GSE9893, GSE31831, and GSE129544). CXCR4 was upregulated in tamoxifen-resistant breast cancer cells MCF7R and T47DR. Differences between groups in CCK8 assay were analyzed by Student’s t test. Error bars represent means ± SD of triplicate. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. TAM, tamoxifen.
High CXCR4 Expression Correlated with Poor Prognosis in Tamoxifen-Treated Breast Cancer
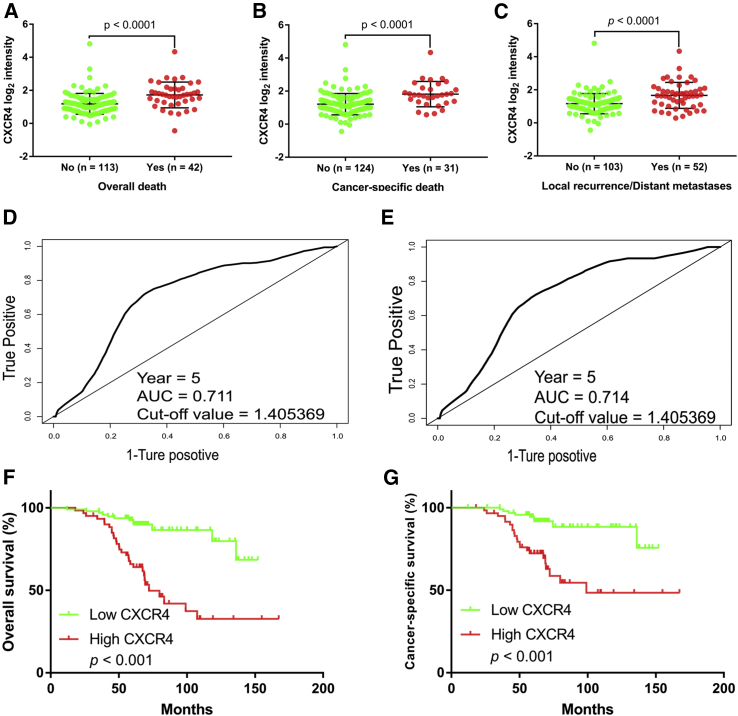
Based on dataset GEO: GSE9893, we analyzed the association between CXCR4 expression and the prognosis of 155 tamoxifen-treated breast cancer patients. We compared the CXCR4 expression in different subgroups: whether local recurrence or distant metastasis, overall death, and breast cancer-specific death occurred. The subgroups with worse outcomes had higher CXCR4 expression, respectively (Figures 2A–2C). Then, we divided the patients into two groups according to the CXCR4 expression level for further analysis. To identify the optimal cutoff value for subdivision, we conducted time-dependent receiver operating characteristic (ROC) curve analyses by using the “survivalROC” package in R to distinguish 5-year survivors from deceased patients. In the overall survival analysis, the cutoff value for CXCR4 (area under the curve [AUC] = 0.711) was 1.405369 (Figure 2D). In cancer-specific survival analysis, the cutoff value for CXCR4 (AUC = 0.714) was 1.405369 (Figure 2E). Finally, we chose 1.405369 as the optimal cutoff value, and patients were divided into CXCR4 low and CXCR4 high groups. In the cohort of 155 tamoxifen-treated patients, patients with high CXCR4 run a higher risk of N category (p = 0.004), local recurrence or distant metastasis (p < 0.001), overall death (p < 0.001), and breast cancer-specific death (p < 0.001) (Table 1). Survival analyses showed that the CXCR4 high group was significantly associated with worse overall survival (p < 0.001; Figure 2F) and cancer-specific survival (p < 0.001; Figure 2G), respectively. The hazard ratios (HRs) for overall mortality and cancer-specific mortality were listed in Table 2 to investigate the clinical significance. Unadjusted HRs of CXCR4 for overall mortality and cancer-specific mortality were 4.646 (p < 0.001) and 4.480 (p < 0.001), respectively. Multivariate Cox analyses adjusted for age at diagnosis, T category, N category, Scarff-Bloom-Richardson (SBR) grade, and adjuvant therapy confirmed the independent prognostic significance of CXCR4 for both overall mortality (HR = 3.027, p = 0.002) and cancer-specific mortality (HR = 2.498, p = 0.031).
Figure 2.
High Expression of CXCR4 Was Correlated with Poor Outcomes in GEO: GSE9893 Cohort
(A–C) CXCR4 was significantly higher in the overall death group (A), cancer-specific death group (B), and local recurrence or distant metastases group (C) compared with other groups, respectively. (D and E) ROC curves for 5-year overall survival (D) and cancer-specific survival (E) according to CXCR4 gene expression. Cutoff values and AUC values are described in the figures. (F and G) Overexpression of CXCR4 was correlated with worse overall survival (F) and cancer-specific survival (G).
Table 1.
Clinical Characteristics of 155 Patients according to CXCR4 High or Low Expression
| Characteristics | Levels | CXCR4 |
||
|---|---|---|---|---|
| Low (n = 95) | High (n = 60) | p Value | ||
| Age at diagnosis (years), mean ± SD | 66.96 ± 10.35 | 67.72 ± 10.08 | 0.651 | |
| T category, n | 0.888 | |||
| T1 | 50 | 32 | ||
| T2 | 41 | 26 | ||
| T3/T4 | 1 | 0 | ||
| NA | 3 | 2 | ||
| N category, n | 0.004 | |||
| pN0 | 57 | 21 | ||
| pN1 | 26 | 21 | ||
| pN2/pN3 | 8 | 16 | ||
| NA | 4 | 2 | ||
| SBR grade, n | 0.195 | |||
| 1 | 15 | 6 | ||
| 2 | 61 | 33 | ||
| 3 | 16 | 17 | ||
| NA | 3 | 4 | ||
| Adjuvant therapy, n | 0.067 | |||
| TAM | 18 | 16 | ||
| X-ray + TAM | 70 | 44 | ||
| X-ray + TAM + LHRH | 7 | 0 | ||
| Local recurrence/distant metastases, n | <0.001 | |||
| no | 76 | 27 | ||
| yes | 19 | 33 | ||
| Overall death, n | 12 | 30 | <0.001 | |
| Cancer-specific death, n | 9 | 22 | <0.001 | |
LHRH, luteinizing hormone-releasing hormone; NA, not available; SBR, Scarff-Bloom-Richardson; TAM, tamoxifen.
Table 2.
Risk Factors for Overall Mortality and Cancer-Specific Mortality
| Variables | Overall Mortality |
Cancer-Specific Mortality |
||||||
|---|---|---|---|---|---|---|---|---|
| Univariate Cox Regression |
Multivariate Cox Regression |
Univariate Cox Regression |
Multivariate Cox Regression |
|||||
| HR (95% CI) | p Value | HR (95% CI) | p Value | HR (95% CI) | p Value | HR (95% CI) | p Value | |
| Age at diagnosis (years) | 1.008 (0.979–1.038) | 0.599 | 0.988 (0.954–1.023) | 0.480 | 0.998 (0.965–1.033) | 0.919 | 0.973 (0.934–1.014) | 0.194 |
| T Category | ||||||||
| T1 | ref | ref | ref | ref | ||||
| T2 | 1.310 (0.691–2.482) | 0.408 | 0.881 (0.443–1.749) | 0.716 | 1.198 (0.571–2.514) | 0.633 | 0.737 (0.333–1.633) | 0.452 |
| T3/T4 | ND | ND | ND | ND | ||||
| NA | 2.774 (0.824–9.336) | 0.099 | 0.716 (0.142–3.622) | 0.686 | 2.511 (0.574–10.579) | 0.221 | 0.942 (0.154–5.771) | 0.949 |
| N Category | ||||||||
| pN0 | ref | Ref | ref | ref | ||||
| pN1 | 2.240 (1.240–8.466) | 0.016 | 3.114 (1.172–8.278) | 0.023 | 5.437 (1.516–19.492) | 0.009 | 5.690 (1.555–20.816) | 0.009 |
| pN2/pN3 | 11.152 (4.389–28.338) | <0.001 | 8.386 (3.171–22.182) | <0.001 | 18.021 (5.171–62.798) | <0.001 | 15.374 (4.186–56.457) | <0.001 |
| NA | 16.557 (4.630–59.199) | <0.001 | 28.684 (5.552–148.200) | <0.001 | 16.326 (2.712–98.292) | 0.002 | 31.264 (3.561–274.503) | 0.002 |
| Adjuvant Therapy | ||||||||
| TAM | ref | Ref | ref | ref | ||||
| X-ray + TAM | 0.772 (0.386–1.544) | 0.465 | 0.504 (0.229–1.107) | 0.088 | 0.582 (0.272–1.247) | 0.164 | 0.320 (0.134–0.763) | 0.010 |
| X-ray + TAM + LHRH | ND | ND | ND | ND | ||||
| CXCR4 | ||||||||
| Low | ref | ref | ref | ref | ||||
| High | 4.646 (2.374–9.093) | <0.001 | 3.027 (1.480–6.191) | 0.002 | 4.480 (2.059–9.748) | <0.001 | 2.498 (1.089–7.730) | 0.031 |
CI, confidence interval; HR, hazard ratio; NA, not available; ND, not done; ref, reference.
Confirmation of Overexpression of CXCR4 in Tamoxifen-Resistant Breast Cancer
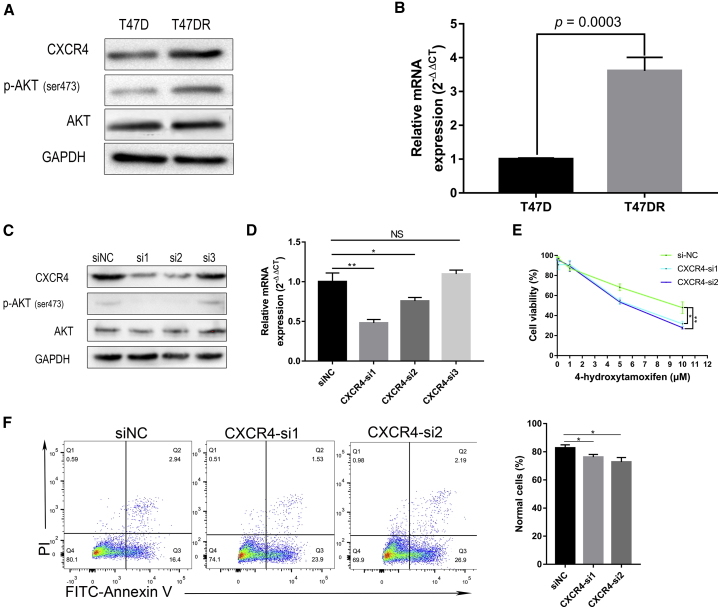
In order to verify the results of sequencing and bioinformatics analysis, we detected the protein and mRNA levels of CXCR4 in T47D and T47DR cell lines, and found that CXCR4 was significantly overexpressed in T47DR cells (Figures 3A and 3B). Also, the phosphorylated forms of AKT (ser473) were increased in T47DR cells. Taken together, our results strongly suggested that CXCR4 was upregulated in tamoxifen-resistant T47DR cells and may play an important role in tamoxifen resistance.
Figure 3.
CXCR4 Is Required for Sustaining Tamoxifen Resistance in Breast Cancer Cells
(A and B) Western blot (A) and quantitative real-time PCR (B) confirmed CXCR4 overexpression in tamoxifen-resistant breast cancer T47DR cells. (C and D) CXCR4 was significantly downregulated in T47DR cells after transfection with CXCR4-si1 and CXCR4-si2 detected by western blot (C) and quantitative real-time PCR (D). (E) CCK8 assay showed that CXCR4 knockdown reversed the resistance of T47DR cells to tamoxifen. (F) Annexin V-FITC/PI assay showed that CXCR4 knockdown increased the apoptosis of T47DR cells treated with tamoxifen. Differences between groups were analyzed by Student’s t test or two-way ANOVA test. Error bars represent means ± SD of triplicate. ∗p < 0.05, ∗∗p < 0.01. NS, not significant.
Knockdown of CXCR4 Expression in T47DR Cells Reversed Tamoxifen Resistance
Given the above results that CXCR4 was upregulated in tamoxifen-resistant cells, we then explored the potential involvement of CXCR4 in tamoxifen resistance of T47DR cells. To this end, we knocked down the expressing levels of CXCR4 in T47DR cells with exogenous introduction of CXCR4 siRNAs (CXCR4-si1, CXCR4-si2, and CXCR4-si3). Western blot and quantitative real-time PCR assays were conducted to detect the knockdown efficiency, and the results showed that CXCR4-si1 and CXCR4-si2 led to a significant reduction of CXCR4 expression in T47DR cells (Figures 3C and 3D). There was no significant difference in total AKT, but phosphorylated (p)-AKT was significantly lower, along with CXCR4 knockdown, compared with the control group. Then T47DR cells transfected with CXCR4-si1 and CXCR4-si2 were selected to further investigate the effects of CXCR4 knockdown on the resistance to tamoxifen. The results of CCK8 indicated that CXCR4 knockdown reduced the resistance of T47DR cells to tamoxifen (Figure 3E). Besides, Annexin V-FITC/PI assay confirmed that knockdown of CXCR4 reversed the resistance to tamoxifen in T47DR cells (Figure 3F). These results indicated that CXCR4 knockdown led to T47DR cells re-sensitization to tamoxifen, and CXCR4 is required for sustaining tamoxifen resistance in breast cancer cells.
CXCR4 Antagonist AMD3100 Reverses the Resistance to Tamoxifen via Inhibiting AKT Phosphorylation
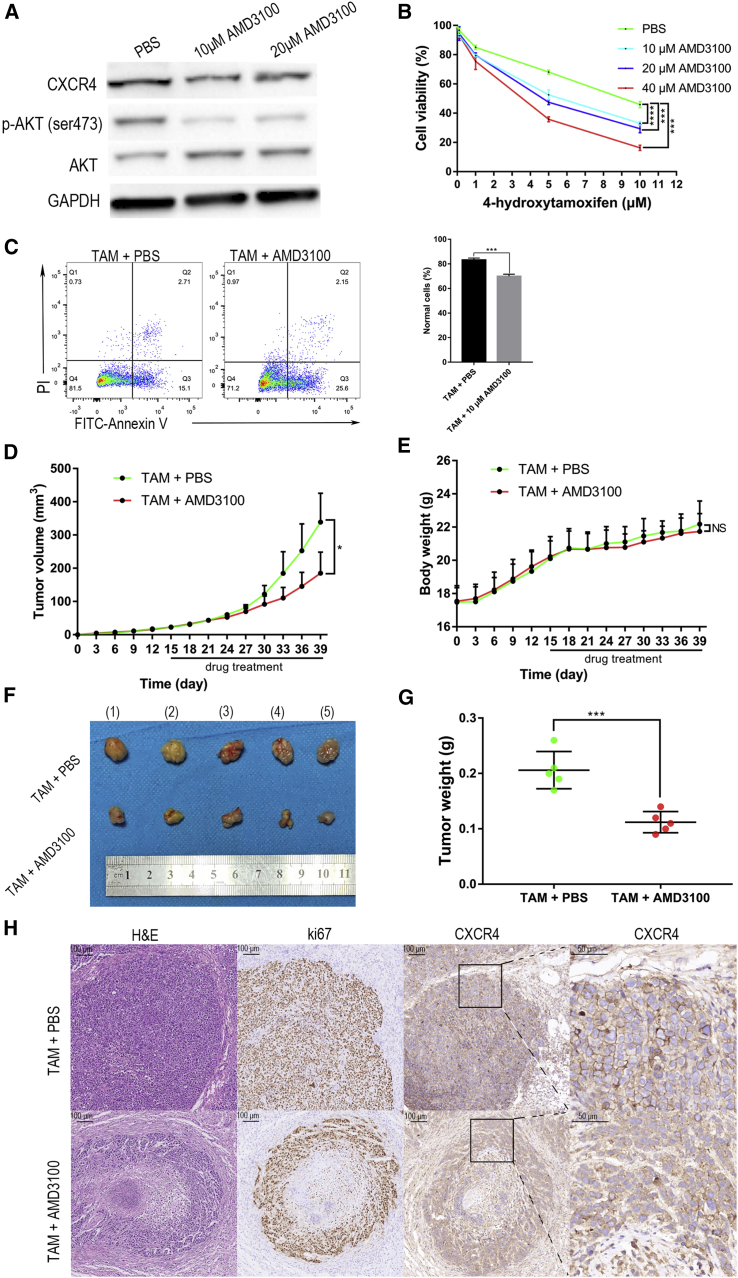
Because we have observed that knockdown of CXCR4 reversed tamoxifen resistance in T47DR cells, we used CXCR4 antagonist AMD3100 against T47DR cells overexpressing CXCR4. As shown in Figure 4A, the phosphorylation levels of AKT were significantly downregulated when T47DR cells were treated with AMD3100. The results of CCK8 assay indicated that AMD3100 reversed the resistance of T47DR cells to tamoxifen (Figure 4B). Furthermore, Annexin V-FITC/PI assay confirmed that AMD3100 reduced the resistance to tamoxifen in T47DR cells (Figure 4C). By means of synergistic or additive inhibitory effects, AMD3100 was capable of enhancing the activity of tamoxifen on tamoxifen-resistant breast cancer cells in vitro via inhibiting AKT phosphorylation. To evaluate whether AMD3100 can reverse the resistance to tamoxifen in vivo, we injected the T47DR cells into the dorsal flank of female nude mice to construct a murine breast cancer xenograft model. Fifteen days after injecting, we divided the tumor-bearing mice into two groups: tamoxifen plus PBS-treated group and tamoxifen plus AMD3100-treated group. Each group was composed of five mice, and all of the mice were treated for 24 days. Tumor growth curves showed that tumor progression was significantly slowed in mice treated with tamoxifen plus AMD3100 compared with another group (Figure 4D), but there was no significant difference in body weight (Figure 4E). The isolated tumors were weighed, and the tamoxifen plus AMD3100-treated group had lighter tumors compared with the control group (Figures 4F and 4G). Besides, we used hematoxylin and eosin (H&E), Ki67, and CXCR4 staining to investigate the pathological and proliferation status of these tumors. Compared with the control group, AMD3100 inhibited the function of CXCR4, increased the necrosis of tumor cells, and decreased the expression of Ki67 (Figure 4H). In brief, the animal experiments suggested that the combination of tamoxifen and AMD3100 could effectively reverse tamoxifen resistance, but without significant side effects in vivo.
Figure 4.
AMD3100 Overcomes Tamoxifen Resistance via Inhibiting AKT Phosphorylation
(A) p-AKT was significantly downregulated in T47DR cells after T47DR cells were treated with AMD3100 for 24 h. (B) CCK8 assay showed that AMD3100 reversed the resistance of T47DR cells to tamoxifen. (C) Annexin V-FITC/PI assay showed that AMD3100 increased the apoptosis of T47DR cells treated with tamoxifen. (D–G) Compared with the other groups, the TAM + AMD3100-treated mice had significantly smaller tumors, but there was no significant difference in body weight. (H) Compared with the control group, AMD3100 inhibited CXCR4, increased the necrosis of tumor cells, and decreased the expression of Ki67. Differences between groups were analyzed by Student’s t test or two-way ANOVA test. Error bars represent means ± SD of triplicate. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Discussion
Tamoxifen, as an effective endocrine therapy, significantly improves the prognosis of patients with ER-positive breast cancer.19 However, the resistance to tamoxifen makes it difficult for some breast cancer patients to get a good prognosis. Studies reported that several components were overexpressed or activated in tamoxifen-resistant breast cancer.20,21 The AKT/mammalian target of rapamycin (mTOR) signaling pathway is one of the alternative oncogenic signaling pathways that are often activated in endocrine-resistant breast cancer. Two different endocrine therapy strategies are being adopted to tamoxifen resistance: enhancement of endocrine therapy and the combination of endocrine therapy and other inhibitors of components that mediate endocrine resistance.3 A phase II clinical trial showed that mTOR inhibitor everolimus significantly improved the efficacy of letrozole in neoadjuvant therapy for ER-positive breast cancer.22 However, there are always many twists and turns in the process of new drug development. Many clinical trials of inhibitors against endocrine therapy of breast cancer, such as AZD8931, ganitumab, and selumetinib, have failed.23, 24, 25 Therefore, the exploration of mechanisms of resistance, the search for new therapeutic targets, and the development of new drugs are still of great significance for the treatment of tamoxifen-resistant breast cancer.
CXCR4 encodes a CXC chemokine receptor specific for stromal cell-derived factor-1. It is well known for its earlier role as a coreceptor for HIV entry.26 In addition to CXCL12, the only known chemokine that binds CXCR4, extracellular ubiquitin also acts as an immune regulator through CXCR4-mediated signal transduction.27,28 When CXCL12 combines with CXCR4, various downstream signaling pathways are activated, resulting in various reactions, such as intracellular calcium content increase, gene transcription, chemotaxis, cell survival, and proliferation.29 Some recent studies have reported the relationship between CXCR4 and the estrogen-dependent tumor progression. ER could promote the expression of CXCR4 when ER combined with estrogen, and the combination of CXCR4 and CXCL12 could also promote the expression of ER.30,31 These findings suggest that the interaction between CXCR4 and ER may be a key mechanism of tumor progression in ER-positive breast cancer. In addition, tamoxifen has also been found to inhibit the expression of CXCR4, which may be one of the mechanisms by which tamoxifen could inhibit the proliferation of ER-positive breast cancer cells.32 Although some studies have found that CXCR4 can promote the progress of ER-positive breast cancer, these studies cannot explain the relationship between CXCR4 and estrogen-independent tumor progression. Rhodes et al.32 reported that CXCR4 could mediate estrogen-independent tumor progression of breast cancer, and suggested that CXCR4 might be associated with the resistance to endocrine therapy in breast cancer.
Furthermore, AMD3100 was shown to specifically inhibit CXCR4-mediated events.15 A previous study33 reported that CXCR4 promotes tamoxifen resistance via aryl hydrocarbon receptor (AhR) signaling, but the prognostic significance and the expression differences of CXCR4 were not found. We further explored the role of CXCR4 in tamoxifen resistance and found the effect of CXCR4 on the AKT signaling pathway, as well as the inhibitory effect of AMD3100 on AKT phosphorylation and tamoxifen resistance from another perspective. The results of this study showed that CXCR4 exhibited differential expression levels in three datasets from the GEO database. Based on the clinical information of GEO: GSE9893, high expression of CXCR4 was found to be associated with poor prognosis in tamoxifen-treated breast cancer and a good prognostic indicator of these patients. Cell experiments showed that CXCR4 promoted tamoxifen resistance through the AKT pathway and might be a potential predictive biomarker of tamoxifen resistance. Besides, CXCR4 antagonist AMD3100 reversed the resistance to tamoxifen in breast cancer via inhibiting AKT phosphorylation. As mentioned above, the combination of endocrine therapy and other inhibitors of components that mediate endocrine resistance is one of the suggested strategies for endocrine therapy-resistant breast cancer. The combination of tamoxifen and AMD3100 could increase tumor-suppressing effects and reduce the potential side effects caused by drug dosage. We observed no significant side effects of the combination, particularly in the body weight of nude mice. However, the more detailed mechanisms by which CXCR4 promotes tamoxifen resistance and AMD3100 reverses tamoxifen resistance have not been studied deeply, and we aim to further explore the downstream molecules that are regulated by CXCR4 and its antagonist AMD3100 in tamoxifen-resistant breast cancer in future studies.
In conclusion, our results will help us to further understand the mechanisms of how CXCR4 mediated the biological process of tamoxifen resistance in breast cancer. We demonstrated that AMD3100 is capable of improving the efficacy of tamoxifen treatment by inhibiting the hyperphosphorylation of AKT in tamoxifen-resistant breast cancer. Our findings provide the feasibility for using the combination of tamoxifen and AMD3100 as a more efficacious therapeutic strategy in the treatment of tamoxifen-resistant breast cancer.
Materials and Methods
Reagents and Establishment of Tamoxifen-Resistant Subline
ER-positive breast cancer T47D cells were purchased from the American Type Culture Collection (ATCC, USA), and 4-hydroxytamoxifen was purchased from Sigma-Aldrich (H7904; USA). To establish the tamoxifen-resistant subline T47DR, we treated T47D cells with 1 μM 4-hydroxytamoxifen continuously and cultured them in RPMI-1640 medium (GIBCO, USA) supplemented with 10% fetal bovine serum (GIBCO, USA) and 1% penicillin/streptomycin (Beyotime, China) in a humidified condition containing 5% CO2 at 37°C for more than 6 months. After 6 months, the resistance of T47DR cells to tamoxifen was verified by CCK8 assay. AMD3100 was purchased from Selleck (Shanghai, China) and dissolved in phosphate-buffered saline (PBS).
High-Throughput Sequencing and Bioinformatics Analysis
High-throughput sequencing was performed using T47D and T47DR cell lines. A total of 5106 cells of each cell line were prepared and subjected to Sangon Biotech (Shanghai, China). The paired-end sequencing on an Illumina HiSeq platform was performed following the manufacturer’s recommended protocol. Transcripts per million (TPMs) of each gene were calculated for relative quantitative analysis. |Fold change| >2 and q <0.05 between two cell lines were selected to represent statistical significance. Volcano plots were performed by the R package “ggplot2.” The raw sequence data have been submitted to the GEO database (accession code GEO: GSE129544). Another two datasets (GEO: GSE9893 and GSE31831) related to tamoxifen treatment or resistance were obtained from GEO. Standardized gene expression levels in 155 samples of tamoxifen-treated breast cancer patients were analyzed based on GEO: GSE9893. Differential gene expression analysis between tamoxifen-sensitive cells (MCF7) and tamoxifen-resistant cells (MCF7R) was based on GEO: GSE31831 at the same time.
CCK8 Assay
To evaluate the response of different cell lines to tamoxifen treatment, we also performed a CCK8 assay according to the manufacturer’s protocol. Different cells were seeded into 96-well plates at a density of 5,000 cells/well with different concentrations of tamoxifen or AMD3100 and then cultured for 48 h. Subsequently, RPMI-1640 containing 10% CCK8 solution (Dojindo, Japan) was supplemented into each well and incubated in a 37°C incubator for 1–3 h. The spectrometric absorbance of each well at 450 nm was measured on a microplate reader (Thermo Fisher, USA) to evaluate the cytotoxicity of tamoxifen to cell lines.
Apoptosis Assay
Annexin V-FITC Apoptosis Kit (556547; BD, USA) was used to determine the apoptotic cells according to the manufacturer’s instruction. Cells in the logarithmic growth phase were collected and seeded into six-well plates in triplicates and then treated with 5 μM tamoxifen for 24 h. In the antagonist function experiment, 10 μM AMD3100 or PBS was used in combination with tamoxifen. Finally, we used a flow cytometer (Beckman Coulter, Fullerton, CA, USA) to identify apoptotic cells.
Western Blot
The total protein of cells was isolated with protein extraction reagent radioimmunoprecipitation assay (RIPA) buffer (Beyotime, China) and quantified by the BCA Protein Assay Kit (Beyotime, China). After SDS-PAGE electrophoresis and polyvinylidene fluoride (PVDF) membrane transfer, the target protein was detected with primary antibodies against CXCR4 (Proteintech, China), AKT (Cell Signaling Technology, USA), p-AKT (Cell Signaling Technology, USA), and GAPDH (Cell Signaling Technology, USA). The protein signals were determined with the ChemiDoc XRS+ System (Bio-Rad, USA) using the ECL detection kit (Beyotime, China).
RNA Extraction and Quantitative Real-Time PCR
Total RNA was extracted from cultured cells using RNAiso Plus reagent (TaKaRa, Japan) according to the manufacturer’s protocol. mRNAs were reverse transcribed to cDNAs with a PrimeScript RT Master Mix Kit (TaKaRa, Japan). Quantitative real-time PCR was performed in triplicate using synthesized primers (Tsingke, China) with an SYBR Premix Ex TaqII Kit (TaKaRa, Japan) to detect the mRNA levels. The expression levels of the target genes were quantitated utilizing the method of 2−ΔΔCT normalized according to the mean levels of GAPDH.
Primers for CXCR4 and GAPDH were as follows: CXCR4 forward: 5′-ACTACACCGAGGAAATGGGCT-3′; CXCR4 reverse: 5′-CCCACAATGCCAGTTAAGAAGA-3′; GAPDH forward: 5′-GGAGCGAGATCCCTCCAAAAT-3′; GAPDH reverse: 5′-GGCTGTTGTCATACTTCTCATGG-3′.
Cell Transfection
Short interference RNAs for CXCR4 and corresponding siRNA negative controls were purchased from RiboBo (China). Transient transfection was performed using Lipofectamine 3000 (Thermo Fisher, USA) according to the manufacturer’s protocol. The transfection efficiency was evaluated using western blot and quantitative real-time PCR. The siRNA target sequences for CXCR4 were as follows: CXCR4-si1: 5′-GCCTCAAGATCCTCTCCAA-3′; CXCR4-si2: 5′-GGAACCCTGTTTCCGTGAA-3′; CXCR4-si3: 5′-GAAGCATGACGGACAAGTA-3′.
Tumor Growth Model in Nude Mice
Athymic female BALB/c nude mice (4–6 weeks old) were obtained from Beijing Vital River Laboratory Animal Technology and maintained under pathogen-free conditions. All of the procedures of animal experiments were approved by the Institutional Animal Care and Use Committee of Tongji Medical College, Huazhong University of Science and Technology (approval number: S2124), and carried out following NIH Guidelines for the Care and Use of Laboratory Animals. A total of 5106 cells were suspended in 0.1 mL of a 1:1 mixture of Matrigel (356234; BD, USA) and PBS, and then injected into the flanks of mice. The health status of nude mice was recorded every 3 days, and the tumor size was measured. From the 15th day after the inoculation of tumor cells, each nude mouse was given 10 mg/kg tamoxifen by oral gavage every day. All of the mice were divided into two groups randomly and administered intraperitoneally every 2 days the following: (1) 5 mg/kg AMD3100 dissolved in 100 μL PBS, or (2) 100 μL PBS alone. At 39 days after injection, mice were killed, and the tumor was removed from the back and weighed.
Immunohistochemistry (IHC)
The primary antibodies against CXCR4 (Proteintech, China) and anti-Ki67 (Novocastra, UK) were used for IHC analyses. IHC was performed as previously described.34 In addition, the slide was stained with H&E.
Statistical Analysis
SPSS 23.0, R 3.5.3, and GraphPad Prism 7.0 were used for statistical analysis. The experiment data are presented as the mean ± standard deviation (SD). Differences between groups in CCK8 assay were analyzed by the Student’s t test or two-way ANOVA test. Survival rates of patients were estimated using the Kaplan-Meier method, and differences between groups were assessed using the log rank test. Cox proportional hazard regression model was used to analyze the prognostic significance of CXCR4. A p value less than 0.05 was considered statistically significant.
Data Availability
The high-throughput sequencing data (GEO: GSE129544, GSE9893, and GSE31831) used to support the findings of this study are included in the article or GEO database.
Author Contributions
S.R. and T.H. designed the experiments. J.Z. and K.L. conducted the experiments and contributed to data analysis and the revision of the manuscript. J.Z., Q.Z., and L.L. contributed to bioinformatics analysis. J.Z., W.Y., and Z.X. conducted the animal assays. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
Our research was supported by National Natural Science Foundation of China (grant 81672611). We thank all those who contributed to this research. We thank the Chinese government and citizens for their quick response to the COVID-19 outbreak and generous support to the healthcare system in Wuhan, China. With their help, the situation in Wuhan has improved quickly, and we had enough time and energy to revise this article.
Contributor Information
Shengnan Ruan, Email: ruanshengnan@yahoo.com.
Tao Huang, Email: huangtaowh@163.com.
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Clark G.M., Osborne C.K., McGuire W.L. Correlations between estrogen receptor, progesterone receptor, and patient characteristics in human breast cancer. J. Clin. Oncol. 1984;2:1102–1109. doi: 10.1200/JCO.1984.2.10.1102. [DOI] [PubMed] [Google Scholar]
- 3.Tryfonidis K., Zardavas D., Katzenellenbogen B.S., Piccart M. Endocrine treatment in breast cancer: Cure, resistance and beyond. Cancer Treat. Rev. 2016;50:68–81. doi: 10.1016/j.ctrv.2016.08.008. [DOI] [PubMed] [Google Scholar]
- 4.Harbeck N., Gnant M. Breast cancer. Lancet. 2017;389:1134–1150. doi: 10.1016/S0140-6736(16)31891-8. [DOI] [PubMed] [Google Scholar]
- 5.Orlando L., Schiavone P., Fedele P., Calvani N., Nacci A., Rizzo P., Marino A., D’Amico M., Sponziello F., Mazzoni E. Molecularly targeted endocrine therapies for breast cancer. Cancer Treat. Rev. 2010;36(Suppl 3):S67–S71. doi: 10.1016/S0305-7372(10)70023-2. [DOI] [PubMed] [Google Scholar]
- 6.Jacot W., Dalenc F., Lopez-Crapez E., Chaltiel L., Durigova A., Gros N., Lozano N., Lacaze J.L., Pouderoux S., Gladieff L. PIK3CA mutations early persistence in cell-free tumor DNA as a negative prognostic factor in metastatic breast cancer patients treated with hormonal therapy. Breast Cancer Res. Treat. 2019;177:659–667. doi: 10.1007/s10549-019-05349-y. [DOI] [PubMed] [Google Scholar]
- 7.Chatterjee S., Behnam Azad B., Nimmagadda S. The intricate role of CXCR4 in cancer. Adv. Cancer Res. 2014;124:31–82. doi: 10.1016/B978-0-12-411638-2.00002-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Darash-Yahana M., Pikarsky E., Abramovitch R., Zeira E., Pal B., Karplus R., Beider K., Avniel S., Kasem S., Galun E., Peled A. Role of high expression levels of CXCR4 in tumor growth, vascularization, and metastasis. FASEB J. 2004;18:1240–1242. doi: 10.1096/fj.03-0935fje. [DOI] [PubMed] [Google Scholar]
- 9.Furusato B., Mohamed A., Uhlén M., Rhim J.S. CXCR4 and cancer. Pathol. Int. 2010;60:497–505. doi: 10.1111/j.1440-1827.2010.02548.x. [DOI] [PubMed] [Google Scholar]
- 10.Vandercappellen J., Van Damme J., Struyf S. The role of CXC chemokines and their receptors in cancer. Cancer Lett. 2008;267:226–244. doi: 10.1016/j.canlet.2008.04.050. [DOI] [PubMed] [Google Scholar]
- 11.Xu C., Zhao H., Chen H., Yao Q. CXCR4 in breast cancer: oncogenic role and therapeutic targeting. Drug Des. Devel. Ther. 2015;9:4953–4964. doi: 10.2147/DDDT.S84932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Z., Ni C., Chen W., Wu P., Wang Z., Yin J., Huang J., Qiu F. Expression of CXCR4 and breast cancer prognosis: a systematic review and meta-analysis. BMC Cancer. 2014;14:49. doi: 10.1186/1471-2407-14-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Clercq E. The bicyclam AMD3100 story. Nat. Rev. Drug Discov. 2003;2:581–587. doi: 10.1038/nrd1134. [DOI] [PubMed] [Google Scholar]
- 14.Fricker S.P., Anastassov V., Cox J., Darkes M.C., Grujic O., Idzan S.R., Labrecque J., Lau G., Mosi R.M., Nelson K.L. Characterization of the molecular pharmacology of AMD3100: a specific antagonist of the G-protein coupled chemokine receptor, CXCR4. Biochem. Pharmacol. 2006;72:588–596. doi: 10.1016/j.bcp.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 15.Hatse S., Princen K., Bridger G., De Clercq E., Schols D. Chemokine receptor inhibition by AMD3100 is strictly confined to CXCR4. FEBS Lett. 2002;527:255–262. doi: 10.1016/s0014-5793(02)03143-5. [DOI] [PubMed] [Google Scholar]
- 16.Zhou K.X., Xie L.H., Peng X., Guo Q.M., Wu Q.Y., Wang W.H., Zhang G.L., Wu J.F., Zhang G.J., Du C.W. CXCR4 antagonist AMD3100 enhances the response of MDA-MB-231 triple-negative breast cancer cells to ionizing radiation. Cancer Lett. 2018;418:196–203. doi: 10.1016/j.canlet.2018.01.009. [DOI] [PubMed] [Google Scholar]
- 17.Lefort S., Thuleau A., Kieffer Y., Sirven P., Bieche I., Marangoni E., Vincent-Salomon A., Mechta-Grigoriou F. CXCR4 inhibitors could benefit to HER2 but not to triple-negative breast cancer patients. Oncogene. 2017;36:1211–1222. doi: 10.1038/onc.2016.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou J., Li W., Ming J., Yang W., Lu L., Zhang Q., Ruan S., Huang T. High expression of TRAF4 predicts poor prognosis in tamoxifen-treated breast cancer and promotes tamoxifen resistance. Anticancer Drugs. 2020;31:558–566. doi: 10.1097/CAD.0000000000000943. [DOI] [PubMed] [Google Scholar]
- 19.Early Breast Cancer Trialists’ Collaborative Group (EBCTCG) Aromatase inhibitors versus tamoxifen in early breast cancer: patient-level meta-analysis of the randomised trials. Lancet. 2015;386:1341–1352. doi: 10.1016/S0140-6736(15)61074-1. [DOI] [PubMed] [Google Scholar]
- 20.Fu X., Creighton C.J., Biswal N.C., Kumar V., Shea M., Herrera S., Contreras A., Gutierrez C., Wang T., Nanda S. Overcoming endocrine resistance due to reduced PTEN levels in estrogen receptor-positive breast cancer by co-targeting mammalian target of rapamycin, protein kinase B, or mitogen-activated protein kinase kinase. Breast Cancer Res. 2014;16:430. doi: 10.1186/s13058-014-0430-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baselga J., Campone M., Piccart M., Burris H.A., 3rd, Rugo H.S., Sahmoud T., Noguchi S., Gnant M., Pritchard K.I., Lebrun F. Everolimus in postmenopausal hormone-receptor-positive advanced breast cancer. N. Engl. J. Med. 2012;366:520–529. doi: 10.1056/NEJMoa1109653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baselga J., Semiglazov V., van Dam P., Manikhas A., Bellet M., Mayordomo J., Campone M., Kubista E., Greil R., Bianchi G. Phase II randomized study of neoadjuvant everolimus plus letrozole compared with placebo plus letrozole in patients with estrogen receptor-positive breast cancer. J. Clin. Oncol. 2009;27:2630–2637. doi: 10.1200/JCO.2008.18.8391. [DOI] [PubMed] [Google Scholar]
- 23.Johnston S., Basik M., Hegg R., Lausoontornsiri W., Grzeda L., Clemons M., Dreosti L., Mann H., Stuart M., Cristofanilli M. Inhibition of EGFR, HER2, and HER3 signaling with AZD8931 in combination with anastrozole as an anticancer approach: Phase II randomized study in women with endocrine-therapy-naïve advanced breast cancer. Breast Cancer Res. Treat. 2016;160:91–99. doi: 10.1007/s10549-016-3979-5. [DOI] [PubMed] [Google Scholar]
- 24.Robertson J.F., Ferrero J.M., Bourgeois H., Kennecke H., de Boer R.H., Jacot W., McGreivy J., Suzuki S., Zhu M., McCaffery I. Ganitumab with either exemestane or fulvestrant for postmenopausal women with advanced, hormone-receptor-positive breast cancer: a randomised, controlled, double-blind, phase 2 trial. Lancet Oncol. 2013;14:228–235. doi: 10.1016/S1470-2045(13)70026-3. [DOI] [PubMed] [Google Scholar]
- 25.Zaman K., Winterhalder R., Mamot C., Hasler-Strub U., Rochlitz C., Mueller A., Berset C., Wiliders H., Perey L., Biaggi Rudolf C. Fulvestrant with or without selumetinib, a MEK 1/2 inhibitor, in breast cancer progressing after aromatase inhibitor therapy: a multicentre randomised placebo-controlled double-blind phase II trial, SAKK 21/08. Eur. J. Cancer. 2015;51:1212–1220. doi: 10.1016/j.ejca.2015.03.016. [DOI] [PubMed] [Google Scholar]
- 26.Feng Y., Broder C.C., Kennedy P.E., Berger E.A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 27.Tripathi A., Saini V., Marchese A., Volkman B.F., Tang W.J., Majetschak M. Modulation of the CXC chemokine receptor 4 agonist activity of ubiquitin through C-terminal protein modification. Biochemistry. 2013;52:4184–4192. doi: 10.1021/bi400254f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saini V., Marchese A., Majetschak M. CXC chemokine receptor 4 is a cell surface receptor for extracellular ubiquitin. J. Biol. Chem. 2010;285:15566–15576. doi: 10.1074/jbc.M110.103408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ganju R.K., Brubaker S.A., Meyer J., Dutt P., Yang Y., Qin S., Newman W., Groopman J.E. The alpha-chemokine, stromal cell-derived factor-1alpha, binds to the transmembrane G-protein-coupled CXCR-4 receptor and activates multiple signal transduction pathways. J. Biol. Chem. 1998;273:23169–23175. doi: 10.1074/jbc.273.36.23169. [DOI] [PubMed] [Google Scholar]
- 30.Boudot A., Kerdivel G., Habauzit D., Eeckhoute J., Le Dily F., Flouriot G., Samson M., Pakdel F. Differential estrogen-regulation of CXCL12 chemokine receptors, CXCR4 and CXCR7, contributes to the growth effect of estrogens in breast cancer cells. PLoS ONE. 2011;6:e20898. doi: 10.1371/journal.pone.0020898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sauvé K., Lepage J., Sanchez M., Heveker N., Tremblay A. Positive feedback activation of estrogen receptors by the CXCL12-CXCR4 pathway. Cancer Res. 2009;69:5793–5800. doi: 10.1158/0008-5472.CAN-08-4924. [DOI] [PubMed] [Google Scholar]
- 32.Rhodes L.V., Short S.P., Neel N.F., Salvo V.A., Zhu Y., Elliott S., Wei Y., Yu D., Sun M., Muir S.E. Cytokine receptor CXCR4 mediates estrogen-independent tumorigenesis, metastasis, and resistance to endocrine therapy in human breast cancer. Cancer Res. 2011;71:603–613. doi: 10.1158/0008-5472.CAN-10-3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dubrovska A., Hartung A., Bouchez L.C., Walker J.R., Reddy V.A., Cho C.Y., Schultz P.G. CXCR4 activation maintains a stem cell population in tamoxifen-resistant breast cancer cells through AhR signalling. Br. J. Cancer. 2012;107:43–52. doi: 10.1038/bjc.2012.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ming J., Ruan S., Wang M., Ye D., Fan N., Meng Q., Tian B., Huang T. A novel chemical, STF-083010, reverses tamoxifen-related drug resistance in breast cancer by inhibiting IRE1/XBP1. Oncotarget. 2015;6:40692–40703. doi: 10.18632/oncotarget.5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The high-throughput sequencing data (GEO: GSE129544, GSE9893, and GSE31831) used to support the findings of this study are included in the article or GEO database.