Abstract
Coronavirus disease 2019 (COVID-19) is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and has resulted in hundreds of thousands of deaths worldwide. While the majority of people with COVID-19 won't require hospitalization, those who do may experience severe life-threatening complications, including acute respiratory distress syndrome. SARS-CoV-2 infects human cells by binding to the cellular surface protein angiotensin-converting enzyme 2 (ACE2); in addition, the cellular transmembrane serine protease 2 (TMPRSS2) is needed for priming of the spike (S) protein of the virus. Virus entry may also depend on the activity of the endosomal/lysosomal cysteine proteases cathepsin B, L (CTSB, CTSL) although their activity is likely dispensable. Given that the uncertainty of how COVID-19 kills, hampers doctors' ability to choose treatments the need for a deep understanding of COVID-19 biology is urgent. Herein, we performed an expression profiling meta-analysis of ACE2, TMPRSS2 and CTSB/L genes (and proteins) in public repository databases and found that all are widely expressed in human tissues; also, the ACE2 and TMPRSS2 genes tend to be co-regulated. The ACE2 and TMPRSS genes expression is (among others) suppressed by TNF, and is induced by pro-inflammatory conditions including obesity, Barrett's esophagus, stomach infection by helicobacter pylori, diabetes, autoimmune diseases and oxidized LDL; by exercise, as well as by growth factors, viruses' infections, cigarette smoke, interferons and androgens. Regarding currently investigated therapies interferon-beta induced ACE2 gene expression in bronchial epithelial cells, while chloroquine tends to upregulate CTSB/L genes. Finally, we analyzed KEGG pathways modulated by ACE2, TMPRSS2 and CTSB/L and probed DrugBank for drugs that target modules of the affected pathways. Our data indicate possible novel high-risk groups for COVID-19; provide a rich resource for future investigations of its pathogenesis and highlight the therapeutic challenges we face.
Keywords: ACE2, COVID-19, CTSB/L, Gene expression, SARS-CoV-2, TMPRSS2
Abbreviations: ACE, angiotensin-converting enzyme; ACE2, angiotensin-converting enzyme 2; COVID-19, coronavirus disease 2019; CTSB/L, cathepsins B/L; GEO, Gene Expression Omnibus; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; TMPRSS2, transmembrane protease, serine 2
Graphical abstract
Highlights
-
•
ACE2 and TMPRSS2 genes tend to be co-regulated and are suppressed by TNF.
-
•
Pro-inflammatory conditions, interferons and androgens induce ACE2, TMPRSS2 genes.
-
•
Cathepsins CTSB/L are widely expressed in human tissues.
-
•
Various druggable pathways can be targeted to mitigate SARS-CoV-2 infectivity.
1. Introduction
Coronavirus disease 2019 (COVID-19) is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and since the initiation of the pandemic it has caused globally hundreds of thousands of deaths [1]. While the majority of people with COVID-19 won't require hospitalization (being asymptomatic or with mild symptoms) for those who need the clinical features may evolve to acute respiratory distress syndrome (ARDS) and cardiac injury. Major risk factors for developing severe symptoms is age, sex (with a prevalence in males), as well as obesity, diabetes, hypertension and respiratory or cardiovascular diseases [2,3]. As no COVID-19 specific drugs are available [4] a better understanding of the underlying COVID-19 pathobiology is required.
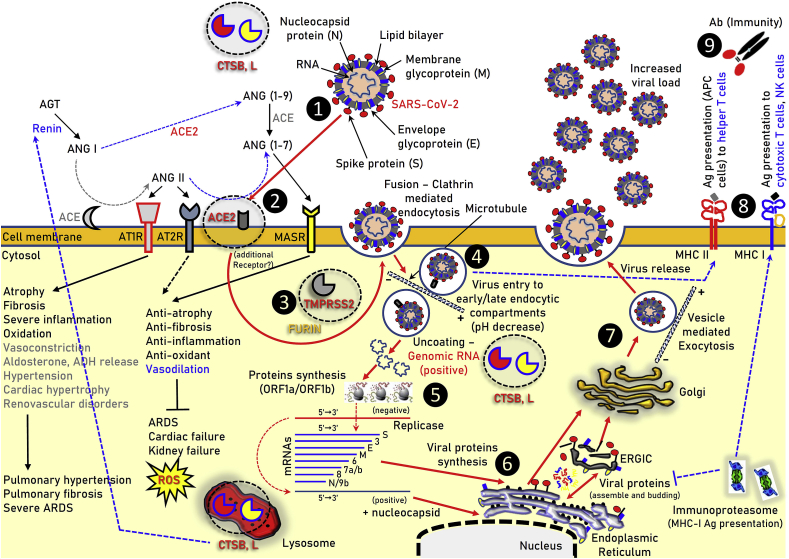
At the molecular level SARS-CoV-2 entry into target cells is facilitated by the spike (S) protein of the coronavirus. SARS-CoV-2 infection starts with binding to zinc metallopeptidase angiotensin-converting enzyme 2 (ACE2) which enables viral attachment to the surface of target cells; an additional step refers to S protein priming by the transmembrane protease serine 2 (TMPRSS2) [5,6] (Fig. 1). Reportedly, furin protease is also likely involved in the SARS-CoV-2 infection process [7]. These processes are similar to those used for SARS-CoV transmissibility [8]. Notably, as for SARS-CoV where the endosomal cysteine proteases cathepsins B/L (CTSB, CTSL) are also needed for viral cell entry [8,9], ammonium chloride treatment strongly inhibited SARS-CoV-2 cell entry into TMPRSS2- 293T cells suggesting that SARS-CoV-2 can likely use either TMPRSS2 or CTSB/L for cell infection [5]. TMPRSS2 expression in lung tissues may be a determinant of viral tropism and pathogenicity at the initial site of SARS-CoV-2 infection [10], and, as for SARS-CoV [11], it may reduce viral control by the humoral immune response. Notably, in common with SARS-CoV, SARS-CoV-2 infection is enhanced by TMPRSS2 [10].
Fig. 1.
Illustration of the main components of the ACE/ANGII/AT1R and ACE2/ANG(1–7)/MASR axes along with the other molecular components reported to be involved in SARS-CoV-2 infection (e.g. TMPRSS2 or FURIN) in human cells. The pathway of SARS-CoV-2 endocytosis and replication in cells is also indicated. ❶ SARS-CoV-2 (extracellular or in circulation); ❷ binding to ACE2; ❸ TMPRSS2 priming; ❹ clathrin-mediated endocytosis (entry to early and acidic late -microtubule bound-endosomes); ❺, ❻ uncoating and viral proteins synthesis in free and endoplasmic reticulum (ER) attached ribosomes; ❼ vesicle-mediated exocytosis; ❽ MHC class II and MHC class I antigen (Ag) presentation by endocytic compartments and proteasome respectively; ❾ immune cells attraction and development of immunity or elimination of infected cells. The hemagglutinin-esterase dimmer component of SARS-CoV-2 is not shown. → induction, ┤inhibition.
ACE2 is a critical component of the renin-angiotensin system (RAS) which plays a key role in maintaining blood pressure homeostasis, as well as fluid and salt balance [12]. The components of the system mainly include renin, angiotensinogen (AGT), angiotensin II (ANGII), angiotensin converting enzyme (ACE), angiotensin II type 1 receptor (AT1R) and angiotensin II type 2 receptor (AT2R) (Fig. 1). ANGII is generated by ANGI from ACE and is a key effector peptide of the system causing vasoconstriction. RAS overactivation has been implicated (among others) in the induction and progression of hypertension, atherosclerosis, diabetes, heart failure, renovascular disorders, pulmonary hypertension and fibrosis, pneumonia and sepsis [13,14]. ACE2, a highly homologous to ACE metalloprotease, functions as a negative regulator of the AGT system as it reduces ANGII levels by producing the sorter ANG(1-7) and ANG(1-9) peptides, which can then activate the anti-inflammatory and vasodilation-promoting receptor MAS (MASR) (Fig. 1) [14]. Recombinant ACE2 protects mice from severe acute lung failure and loss of ACE2 expression is likely involved in increased vascular permeability, one of the hallmarks in ARDS pathogenesis [15]. ACE2 also links amino acid malnutrition to intestinal inflammation, as it is a key regulator of dietary amino acid homeostasis, innate immunity and gut microbial ecology [16]. Overall, the balance between the ACE/ANGII/AT1R and the counter-regulatory ACE2/ANG(1–7)/MASR axes is critical in the physiological regulation of neural, cardiovascular, blood pressure and kidney functions [17].
Although ACE2 expression seems to correlate with susceptibility to SARS-CoV infection in cell assays and it is downregulated by SARS-CoV [18], the relationship between ACE2 expression levels and the susceptibly to SARS-CoV-2 infection remains elusive; also, it is not clear whether SARS-CoV-2 infection interferes with ACE2 expression. Nonetheless, it is logical to assume that high ACE2 expression levels and/or the simultaneous co-expression of the ACE2, TMPRSS2 and CTSB/L proteins in SARS-CoV-2 targeted tissues/cells may lead to higher risk for SARS-CoV-2 infection. To get more insights on this issue we mapped ACE2, TMPRSS2, CTSB and CTSL genes and corresponding proteins expression in human tissues and pathology, as well in response to SARS-CoV-2 used therapeutic treatments by analyzing expression datasets from GEO, ArrayExpress and Protein Atlas databases. Our findings provide a rational explanation for most of the clinical features of COVID-19.
2. Materials and methods
2.1. Search strategy and selection of studies
In this gene expression profiling meta-analysis study, the ACE2 [ACE2 (ENTREZ ID 59272, ENSEMBL ENSG00000130234)] and TMPRSS2 [TMPRSS2 (ENTREZ ID 7113, ENSEMBL ENSG00000184012)] genes related microarray datasets were retrieved from NCBI and ArrayExpress databases. The selection of datasets from NCBI or ArrayExpress was based on the terms Homo sapiens [Organism], ACE2 [Gene symbol] or TMPRSS2 [Gene symbol] respectively, along with additional data filtering for up/down genes in the differential expression search filter in GEO Profiles of NCBI. The NCBI and ArrayExpress databases were also searched for currently investigated therapeutic approaches against COVID-19 [e.g. interferon-beta (IFN-β), remdesivir, lopinavir, ritonavir, chloroquine/hydroxychloroquine and a number of immunosuppressants] and the terms ACE2 [Gene symbol] or TMPRSS2 [Gene symbol]. The records collected after database search, include 63 ACE2, 150 TMPRSS2, 36 IFN-β, 34 chloroquine and 13 hydroxychloroquine entries.
2.2. Criteria for included studies
Following the removal of duplicate entries from the NCBI and Array Express databases the unique datasets (n = 250) were collected. Additional datasets were excluded by eliminating the non-GEO entries (n = 69) and those (n = 26) with insufficient data. Specifically, datasets were excluded if these were non-GEO entries or if a very low number of samples per state/condition was provided, thus, the requirements of GEO2R tool were not met. Gene expression analysis of the 155 selected datasets was performed using the NCBI provided GEO2R tool, aiming to identify differential expression of the genes of interest (i.e. ACE2, TMPRSS2, CTSB and CTSL) across different experimental conditions; statistically significant cases (p-value <0.05) were chosen for visualization (see, PRISMA flowchart; Fig. S1) [19].
Additional information about the expression levels of ACE2, TMPRSS2, CTSB and CTSL genes and proteins across human tissues and recorded pathologies was gathered from The Human Protein Atlas database. Finally, the terms ACE2, TMPRSS2, CTSB and CTSL were searched in KEGG pathways and the found pathways/molecules were used to screen DrugBank for drugs (at various phases of development) that modulate identified KEGG pathways.
3. Results
Differential regulation of the SARS-CoV-2 spike protein receptor ACE2 and of the priming protease TMPRSS2 genes in GEO and ArrayExpress databases.
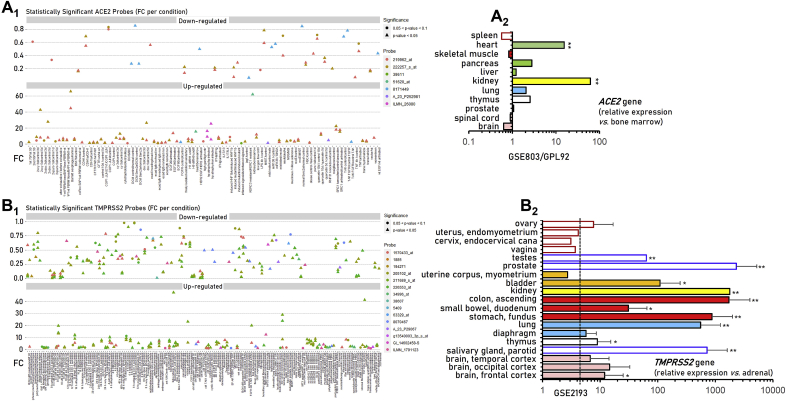
Given that SARS-CoV-2 infection rate in human cells likely depends on the expression levels of its receptor ACE2 and the priming protease TMPRSS2, we initially performed an expression profiling meta-analysis at the GEO and ArrayExpress microarrays databases. We found that the ACE2 gene is significantly less expressed in venous vs. arterial endothelial cells; in biopsies of patients with nephrosclerosis (a hypoxia-related glomerulopathy), as well as in various biological settings following treatment with lipopolysaccharides, tumor necrosis factor (TNF) and interleukins −4 (IL-4) and −13 (IL-13) (Fig. 2A1, Table S1). TMPRSS2 gene was (among others) suppressed by TNF, hepatitis antigen HBsAg and influenza H1N1 virus; hypoxia, methotrexate, serum response factor and estradiol, the primary endothelial receptor LOX-1 for oxidized LDL and oxidized LDL, ARC (a nucleoside analog with anti-cancer activity), sangivamycin (a nucleoside analogue and potent inhibitor of protein kinase C), the BET inhibitor I-BET762, as well as following TGFβ-activated kinase 1 (TAK1) knock down, mycophenolic acid or bicalutamide (an antiandrogen) treatment and prostate castration (Fig. 2B1, Table S2). The TMPRSS2 gene was also found to be significantly downregulated in many tumors or tumor cells.
Fig. 2.
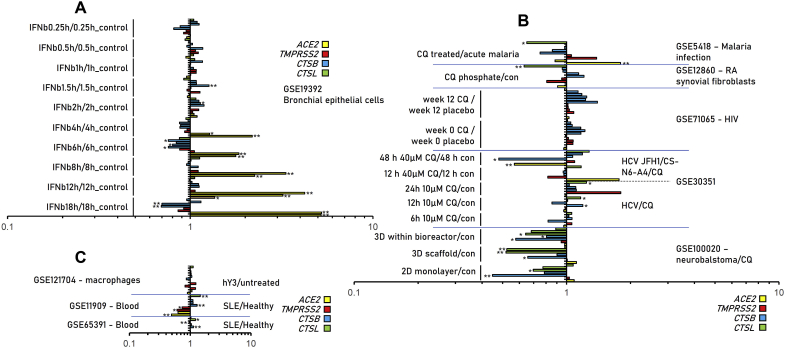
Patterns of differentially expressed ACE2 (A1) and TMPRSS2 (B1) genes, along with expression levels in human tissues [ACE2, (A2); TMPRSS2, (B2)] in GEO and ArrayExpress deposited experiments. Dashed line in (B2) represents the mean TMPRSS2 gene expression in female tissues. See also Tables S1 and S2. Bars, ± SD. *, P < 0.05, **, P < 0.01.
On the other hand, ACE2 gene expression is (among others; see Fig. 2A1, Table S1) induced by obesity, stomach infection by helicobacter pylori and interferons IFN-α and IFN-γ; also, it is upregulated following treatment with retinoic acid, the p160 steroid coactivator protein SRC-1, EGF/serum or FGFR stimulation, knock down of the RNF31 atypical E3 ligase and by hepatocyte nuclear factor 1-beta. The expression of the TMPRSS2 gene is (among others; see Fig. 2B1, Table S2) elevated by androgens, androgen receptor (AR) activation/overexpression or by AR agonists; by cigarette smoke extract; in rectal suction specimens of cystic fibrosis patients, in nasal lavage samples obtained from asthmatic children during an acute picornavirus-induced exacerbation, following rhinovirus infection, as well as in juvenile rheumatoid arthritis, diabetes and Barrett's esophagus. Notably, the ACE2 and TMPRSS2 genes showed a trend for similar regulation in GEO, ArrayExpress experiments (Fig. S2; Tables S1 and S2).
Our search in GEO and ArrayExpress databases showed that the ACE2 gene is highly expressed in thymus, lung, kidney, pancreas and heart (Fig. 2A2, Table S1); while, the TMPRSS2 gene is overexpressed in bladder, kidney, as well as in the tissues of the gastrointestinal and respiratory tract (Fig. 2B2, Table S2). Thus, the ACE2 and TMPRSS2 genes are widely expressed in human tissues where they are induced in pro-inflammatory conditions and by androgens.
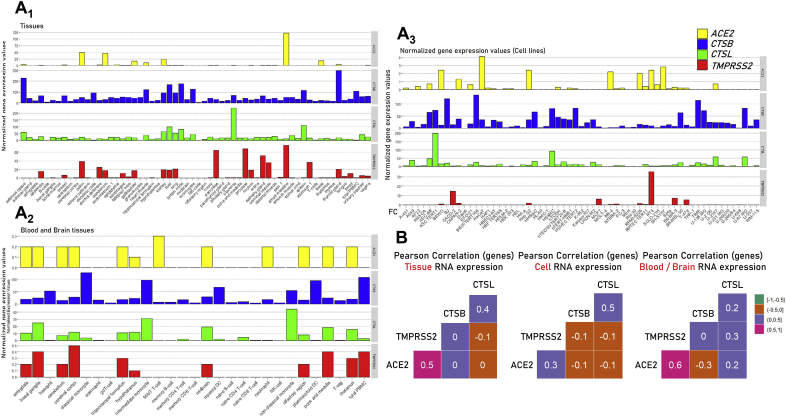
Comparative analysis of ACE2, TMPRSS2, CTSB and CTSL genes and corresponding proteins expression patterns as depicted in Protein Atlas.
We then collected data for ACE2, TMPRSS2, CTSB and CTSL genes and proteins expression data from The Protein Atlas database. The CTSB and CTSL genes are in general more homogeneously expressed and in higher levels, as compared to ACE2 and TMPRSS2, across different tissues (Fig. 3A, Table S3). For ACE2 and TMPRSS2 genes very low expression levels were found in brain and blood tissues, as well as in most cell lines studied; on the contrary, high ACE2 and TMPRSS2 genes co-expression was found in kidney, gallbladder, colon duodenum, small intestine, salivary and thyroid glands. Also, the ACE2 gene is highly expressed in heart and in testis.
Fig. 3.
Comparative ACE2, TMPRSS2, CTSB and CTSL genes (mRNA) expression and correlative co-expression analyses (B) at the shown tissues or cell types/lines (Protein Atlas data). See also, Table S3.
Similarly, according to Protein Atlas database the ACE2 protein is expressed in adrenal gland and testis and in high levels at the gastrointestinal tract; while TMPRSS2 in highly expressed in kidney and in the gastrointestinal tract (Fig. S3; Table S4). A notable observation in these analyses was again a tendency for a positive correlation of the ACE2 and TMPRSS2 genes expression patterns (Fig. 3B); thus, an intervention that modulates either ACE2 or TMPRSS2 expression would likely also affect the non-targeted gene.
Regulation of the ACE2, TMPRSS2, CTSB and CTSL genes following treatment with IFN1-β, chloroquine or hydroxychloroquine and screening of KEGG pathways and DrugBank for the identification of possible anti COVID-19 treatments.
Given that a number of combinational therapeutic schemes [4] are currently being prioritized for COVID-19 treatment, we also performed a more focused search in GEO and ArrayExpress databases for these drugs and the ACE2, TMPRSS, CTSB/L genes. Our findings are shown in Fig. 4 (see also Tables S5–S7) and refer to experiments containing information for IFN-β, chloroquine and hydroxychloroquine. Interestingly, IFN-β significantly induced ACE2 gene expression in bronchial epithelial cells (Fig. 4A; Table S5). On the other hand, chloroquine administration in HIV-infected patients tended (non-significant) to induce CTBS gene expression, likely due to a negative feedback loop aiming to restore physiological cathepsins activity (Fig. 4B, Table S6). Finally, hydroxychloroquine suppressed ACE2 and TMPRSS2 genes expression and induced CTSB and CTSL genes expression in PBMC cells of systemic lupus erythematosus patients (Fig. 4C, Table S7).
Fig. 4.
Differential regulation of ACE2, TMPRSS2, CTSB and CTSL genes following treatment in shown biological settings with IFN1-β (A), chloroquine (B) or hydroxychloroquine (C). Data from GEO and ArrayExpress. See also Tables S5–S7. Bars, ± SD. *, P < 0.05, **, P < 0.01.
We then analyzed KEGG pathways modulated by ACE2, TMPRSS2 and CTSB/L and found ACE2 to be involved in renin-angiotensin system and protein digestion; TMPRSS2 in transcriptional mis-regulation in cancer, prostate cancer and influenza A; and CTSB/L, in renin secretion, antigen processing/presentation, autophagy, lysosome, phagosome, apoptosis, NOD-like receptor signaling pathway, proteoglycans in cancer, rheumatoid arthritis, fluid shear stress and atherosclerosis (Table S8). Modules of these pathways were used to probe DrugBank in order to identify drugs (experimental, in trials or approved) that target components of these pathways; we also searched for clathrin-mediated endocytosis-, actin- and tubulin-polymerization inhibitors (Tables S9–S26; see also, Fig. 1). A number of potentially useable drugs include anti-androgens or AR inhibitors, vasodilators, TNF blockers or interleukins’ activity modulators (Tables S9–S26).
4. Discussion
Our findings indicate that the major cellular components (i.e. ACE2, TMPRSS2 and CTSB/L) that enable SARS-CoV-2 infection in human cells are widely expressed in human tissues; being enriched in kidney, heart as well as in the tissues of the gastrointestinal and respiratory tract, with minimal expression (for ACE2 and TMPRSS2 genes) levels in brain and blood cells. These data are in accordance to recent similar approaches focused solely on the ACE2 gene [20,21]. Regarding lung (a primary site of infection) it was recently found that both the ACE2 and TMPRSS2 genes are primarily expressed in bronchial transient secretory cells [22]; it was also found that SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells [23]. The enrichment of all SARS-CoV-2 infection-related cellular components in the gastrointestinal tract, explain diarrhoea and SARS-CoV-2 sustained isolation from stool as a major symptom of COVID-19 [2,24], and suggest that the intestine may represent a major entry site for the virus due to consumption of contaminated food. Considering that ACE2 expression correlates with SARS-CoV (the close relative of SARS-CoV-2) infection rate in cells [15,18], it is likely that even in patients with no underlying morbidities most major human organs are potentially vulnerable to COVID-19.
In addition of tissues/organs enrichment to ACE2, TMPRSS2 and CTSB/L genes expression, we also found that the ACE2 (likely as a counterbalancing anti-inflammatory response) and TMPRSS2 genes are induced in various pro-inflammatory conditions including obesity, Barrett's esophagus, stomach infection by helicobacter pylori, diabetes, autoimmune diseases and oxidized LDL. Several of these morbidities have been already recognized as risk factors for severe disease and death [2] while others indicate novel possible risk factors for severe COVID-19. Notably, in the particular biological settings assayed, these genes were induced by exhaustive and likely moderate exercise, as well as by cigarette smoke, growth factors, viruses' infections and interferons (see below); we also found a strong positive regulation by androgens largely explaining the increased sensitivity of men to COVID-19 [2].
In high-risk COVID-19 conditions like hypertension, respiratory disease and cardiovascular disease the ACE/ANGII/AT1R axis is overactive and likely ACE2 expression (and thus SARS-CoV-2 available binding sites) increases when patients are treated with drugs that suppress ACE/ANGII/AT1R (e.g. AT1R blockers) [25]. So, when a hypertensive patient is infected by SARS-CoV-2 it is in high risk simply because the ACE/ANGII/AT1R is overactive (but partly suppressed by AT1R blockers) and ACE2 is overexpressed. The infection by SARS-CoV-2 likely eliminates the ACE2/ANG(1–7)/MASR regulatory axis (Fig. 1) leading to sudden exaggeration of the ACE/ANGII/AT1R activity, causing thus ARDS and at the systemic level (among others) kidney and cardiac failure (Fig. 5). Indeed, SARS-CoV infection in mice downregulates ACE2 protein (but not ACE) contributing to severe lung injury [12]. Also, loss of ACE2 expression and locally increased ANGII production triggers leakage of pulmonary blood vessels through AT1R stimulation in ARDS [15]. Interestingly, in a model of nanoparticles-mediated lung injury due to direct binding to ACE2 (that led to suppression of ACE2 activity and expression levels) the administration of losartan (an AT1R antagonist) can ameliorate nanoparticles-induced lung injury [26].
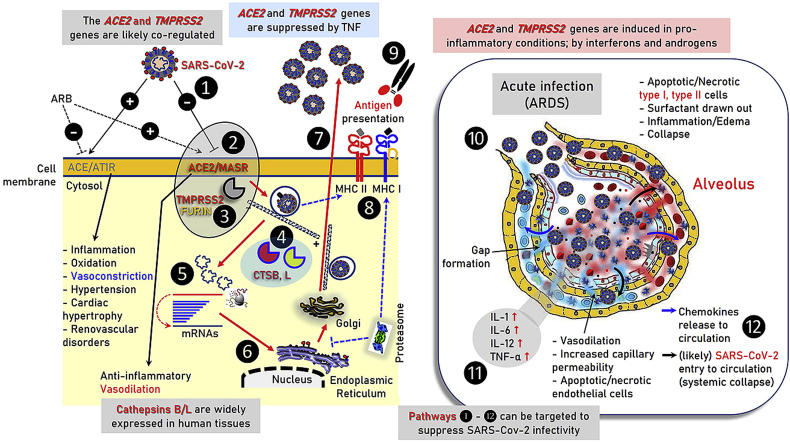
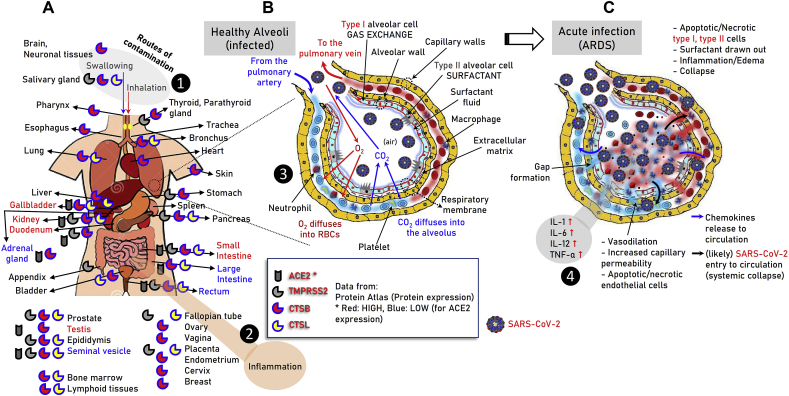
Fig. 5.
Graphical illustration of ACE2, TMPRSS2, CTSB and CTSL proteins expression in various somatic tissues as per the Protein Atlas database (A) (see also, Fig. S3, Table S4). The distinct, i.e. early (B) and acute (C) phases of SARS-CoV-2 infection in lung alveoli are also shown. The major cell types, local tissue alterations and inflammatory cytokines that relate to the disease in the lung are indicated. ❶ Sites of SARS-CoV-2 entry in the body; ❷ likely inflammatory responses at the gastrointestinal tract; ❸ early phases of virus entry to the lung – binding to the alveolar epithelial cells; ❹ acute phase of ARDS, cytokine storm and likely virus entry to the circulation – systemic collapse.
Since ACE2, TMPRSS2 and CTSB/L genes are widely expressed in human tissues (e.g. heart and kidney) leakage of pulmonary blood vessels during ARDS (Fig. 5) would likely allow virus spreading in the circulation enabling direct infection of these organs, decreased ACE2/ANG(1–7)/MASR activity and systemic failure. Although there is likely no detectable viremia (i.e., infectious virus in plasma) during asymptomatic infection or prior to clinical disease [27], SARS-CoV-2 RNA has been detected in blood donations from patients with mild symptoms [28] and detectable SARS-CoV-2 viral RNA in blood is a strong indicator for further clinical severity [24]. Overall, and since aging (another risk factor for COVID-19) correlates with increased ACE1/ACE2 ratio in a rat model [29], high risk COVID-19 patients (or even not hypertensive patients as a prophylactic measure) should likely receive additional RAS inhibitors to decrease systemic damage risk.
Given the delicate balance between the ACE/ANGII/AT1R and ACE2/ANG(1–7)/MASR regulatory axes along with the aforementioned morbidities being induced due to disruption of this balance in favor of ACE/ANGII/AT1R activation, the development of effective COVID-19 therapeutics seems particularly puzzling. Specifically, COVID-19 is a two-phase disease, namely a. infection and spreading (life cycle) of the virus (Fig. 1; ❶ - ❾) in the respiratory and, likely, gastrointestinal tracts, and b. ARDS induction which can then lead to systemic failure (Fig. 5; ❶ - ❹). Thus, as it is seemingly both the virus and the uncontrolled response of a patient's immune system that promote lethality; effective treatments therefore, should probe both the life cycle and the side-effects induced by SARS-CoV-2. Current, under investigation or in clinical trials, therapeutics for COVID-19 include azithromycin (bacterial protein synthesis inhibitor), chloroquine/hydroxy-chloroquine (lysosomotropic agents inhibiting the activity of endosomal/lysosomal compartments), lopinavir/lavipiravir/remdesivir (antiviral drugs) and Τocilizumab (a humanized monoclonal antibody against the IL-6 receptor) [4]. For alternatives and/or additional combinatorial approaches, we searched KEGG pathways for the ACE2, TMPRSS2, CTSB and CTSL terms (Figs. S4-S6) and DrugBank for agents (at various phases of development) (Tables S9–S23) that target modules of the found pathways; additional information for clathrin-mediated endocytosis, as well as for actin or tubulin polymerization inhibitors in shown in Tables S24–S26.
For targeting the virus life cycle the magic bullet will be the development of an effective vaccine; here, cell lines that express ACE2 (see Fig. 3A2, Table S3) and facilitate viral replication may be most efficient in large-scale vaccine production. In another approach, soluble ACE2 (e.g. rhACE2; APN01, GSK2586881) can be likely used to neutralize the virus by competitive binding and also rescue cellular ACE2 activity.
Towards blocking SARS-CoV-2 binding to ACE2 or reducing available binding sites by ACE2 downregulation possible approaches may involve, a. antibodies or small molecules that target ACE2, e.g. SSAA09E2 which blocks early interactions of SARS-CoV with ACE2 [30], or b. TNF, androgen or AR inhibitors (see Tables S9–S26) as we found that these interventions can strongly suppress ACE2 and TMPRSS2 genes expression. Yet, given the anticipated toxic effects of systemic loss-of ACE2 activity whatever intervention should be transient and with constant monitoring of the clinical output. An alternative (and likely safer as compared to ACE2 inhibition) approach would be the usage of TMPRSS2 specific inhibitors. TMRPSS2 is druggable and camostat mesylate partially blocked SARS-CoV-2 entry into Caco-2 and Vero-TMPRSS2 cells [5]. Additional benefits of TMRPSS2 inhibition would be the reduced viral tropism at the initial site of SARS-CoV-2 infection and enhanced humoral immune responses (see above). Next steps to be targeted at the SARS-CoV-2 life cycle include membrane fusion and clathrin-mediated endocytosis. Here, the clathrin-mediated endocytosis inhibitor ikarugamycin [31], the dynamin inhibitor dynasore (or its analogs) [32] or the actin depolymerizing drug latrunculin B [33] can be tested in preclinical models. Also, SSAA09E3 prevents fusion of the viral membrane with the host cellular membrane in a SARS-CoV infection model [30]. Intracellularly, novel SARS-CoV-2 specific antiviral drugs will target the SARS-CoV-2 main protease due to its essential role in processing the polyproteins that are translated from the viral RNA [34]; alternatively, existing antiviral can be tested, as according to molecular docking studies they bind tightly to RNA dependent RNA polymerase [35].
Other intracellular components of the virus life cycle that can be targeted, include tubulin and/or CTSB/L (Fig. 1). For the former, colchicine (a tubulin polymerization inhibitor [36]) is a promising (at least in patients with ARDS [37]) drug, while for the latter, chloroquine or hydroxy-chloroquine are already used against COVID-19 [38]. Yet, in spite of promising clinical data obtained for chloroquine/hydroxy-chloroquine, which could also relate to reduced renin production (Fig. S4), these lysosomotropic agents inhibit the activity of endosomal/lysosomal compartments non-specifically. Thus, existing CTSB/L specific inhibitors, some of which are of endogenous origin and function as regulators of cathepsin B activity in the cell, such as the cystatins [39], can be also considered. Notably, cathepsins B/L activity increases significantly with age [40] and has been associated with arterial stiffening and atherosclerotic vascular disease [41]; these observations further support the notion that age and cardiac dysfunction are risk factors for COVID-19. At the dark side of inhibiting the acidic cellular endosomal compartments or CTSB/L is their functional involvement in MHC class II antigen (Ag) presentation (Fig. 1) [42]. Thus, the usage of chloroquine/hydroxychloroquine; CTSB/L specific inhibitors or even the excessive contamination of the endosomal compartments by the virus itself could suppress MHC class II antigen (Ag) presentation. This functional output along with the presence of O-linked glycans at the surface of the virus [43] could result in immunoevasion. In this case, clearance of the virus may proceed via proteasome-mediated MHC-I related Ag presentation (Fig. 1) [44] and the activity of specific immune cells (e.g. NK cells). Proteasome is downregulated during aging [45] and is less responsive to IFN-γ [46], which would then result in reduced Ag presentation in MHC I molecules and consequently reduced immune responses (Fig. 1) in the elderly; in this context drugs and/or small molecules that activate proteasome could provide useful additive therapeutics against COVID-19. Our previous high-throughput screenings for the identification of natural products modulating proteostatic pathways has revealed promising candidates that exert anti-oxidant activity and a parallel combinatorial action as CTSB/L inhibitors and proteasome activators (unpublished data).
In relation to phase 2 of the disease i.e. ARDS induction and the cytokine storm which can then lead to systemic failure [47] along with the downregulation of ACE2 due to SARS-CoV-2 infection we propose that the use of the ANG(1-7) peptide (or non-peptide analogs) could bypass the loss of ACE2 and reactivate the anti-inflammatory/vasodilatory MASR signaling pathway (Fig. 1). In support, ANG(1-7) protects endothelial cells from high glucose-induced injury and inflammation [48], improves the action of insulin [49] and has shown protective action in heart failure [50] and stroke [51]. Due to the likely COVID-19 infection-mediated overactivation of the pro-oxidative ACE/ANGII/AT1R axis which can then promote endothelial dysfunction due to dysbalanced nitric oxide and reactive oxygen species ratios in the vessel wall [52], the use of drugs that activate anti-oxidant cellular defenses or act as radicals scavengers could be an additional prophylactic intervention. Finally, regarding COVID-19 induced cytokine storm, i.e. the uncontrolled systemic inflammatory response resulting from the release of large amounts of pro-inflammatory cytokines [2,47], the idea of inhibiting key pro-inflammatory cytokines (e.g. the IL-6 receptor by tocilizumab) makes sense [47]; obviously however, an excessive inhibition of the immune system by corticosteroids should be avoided [53]. Our findings that interferons, and especially IFN-β and IFN-γ, induce the ACE2 gene (Figs. 2A and 4Α; Tables S1 and S5) indicate that their therapeutic activation could upregulate the bindings sites and hence infection rates of the virus in targeted tissues. Thus, the regulation of the ACE2, TMPRSS2 and CTSB/L genes from immune system key effector molecules should be studied in details.
In conclusion, our data indicate possible novel high-risk groups for COVID-19; they also provide a rich resource for a. future investigations of COVID-19 pathogenesis and b. possible combinatorial therapeutic approaches.
Author contributions
IPT -supervised the study, interpreted the data and wrote the paper; EG, GB and KV collected and analyzed data. All authors edited the manuscript.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgments
IPT acknowledges funding from the Hellenic GSRT projects BIOIMAGING-GR (MIS 5002755) and PlantUP-GR (MIS 5002803).
Footnotes
Supplementary Information includes Figures and Tables with the GEO, ArrayExpress and human Protein Atlas data; KEGG pathways, DrugBank data and data for clathrin-mediated endocytosis, actin and tubulin polymerization inhibitors.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2020.101615.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplementary Information includes Figures and Tables with the GEO, ArrayExpress and human Protein Atlas data; KEGG pathways, DrugBank data and data for clathrin-mediated endocytosis, actin and tubulin polymerization inhibitors.
References
- 1.Shereen M.A., Khan S., Kazmi A., Bashir N., Siddique R. COVID-19 infection: origin, transmission, and characteristics of human coronaviruses. J. Adv. Res. 2020;24:91–98. doi: 10.1016/j.jare.2020.03.005.eCollection.2020.Jul. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. Feb 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y., Zhao Y., Li Y., Wang X., Peng Z. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in wuhan, China. J. Am. Med. Assoc. 2020 Feb 7 doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mahase E. Covid-19: what treatments are being investigated? BMJ. 2020 Mar26;368 doi: 10.1136/bmj.m1252. m1252. [DOI] [PubMed] [Google Scholar]
- 5.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. Mar 4. pii: S0092-8674(20)30229-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020 Mar 30 doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 7.Hoffmann M., Kleine-Weber H., Pöhlmann S.A. Multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol. Cell. 2020 doi: 10.1016/j.molcel.2020.04.022. S1097-2765(20)30264-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matsuyama S., Nagata N., Shirato K., Kawase M., Takeda M., Taguchi F. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J. Virol. 2010;84(24):12658–12664. doi: 10.1128/JVI.01542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang I.C., Bosch B.J., Li F., Li W., Lee K.H., Ghiran S., Vasi, Dermody T.S., Harrison S.C., Dormitzer P.R., Farzan M., Rottier P.J., Choe H. SARS coronavirus, but not human coronavirus NL63, utilizes cathepsin L to infect ACE2-expressing cells. J. Biol. Chem. 2006 Feb 10;281(6):3198–3203. doi: 10.1074/jbc.M508381200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsuyama S., Nao N., Shirato K., Kawase M., Saito S., Takayama I., Nagata N., Sekizuka T., Katoh H., Kato F., Sakata M., Tahara M., Kutsuna S., Ohmagari N., Kuroda M., Suzuki T., Kageyama T., Takeda M. Enhanced isolation of SARS-CoV-2 byTMPRSS2-expressing cells. Proc. Natl. Acad. Sci. U. S. A. 2020 Mar31;117(13):7001–7003. doi: 10.1073/pnas.2002589117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glowacka I., Bertram S., Müller M.A., Allen P., Soilleux E., Pfefferle S., Steffen I., Tsegaye T.S., He Y., Gnirss K., Niemeyer D., Schneider H., Drosten C., Pöhlmann S. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J. Virol. 2011 May;85(9):4122–4134. doi: 10.1128/JVI.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuba K., Imai Y., Penninger J.M. Angiotensin-converting enzyme 2 in lung diseases. Curr. Opin. Pharmacol. 2006 Jun;6(3):271–276. doi: 10.1016/j.coph.2006.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bindom S.M., Lazartigues E. The sweeter side of ACE2: physiological evidence for a role in diabetes. Mol. Cell. Endocrinol. 2009 Apr 29;302(2):193–202. doi: 10.1016/j.mce.2008.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Imai Y., Kuba K., Ohto-Nakanishi T., Penninger J.M. Angiotensin-converting enzyme 2 (ACE2) in disease pathogenesis. Circ. J. 2010 Mar;74(3):405–410. doi: 10.1253/circj.cj-10-0045. [DOI] [PubMed] [Google Scholar]
- 15.Imai Y., Kuba K., Rao S., Huan Y., Guo F., Guan B., Yang P., Sarao R., Wada T., Leong-Poi H., Crackower M.A., Fukamizu A., Hui C.C., Hein L., Uhlig S., Slutsky A.S., Jiang C., Penninger J.M. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature. 2005 Jul 7;436(7047):112–116. doi: 10.1038/nature03712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hashimoto T., Perlot T., Rehman A., Trichereau J., Ishiguro H., Paolino M., Sigl V., Hanada T., Hanada R., Lipinski S., Wild B., Camargo S.M., Singer D., Richter A., Kuba K., Fukamizu A., Schreiber S., Clevers H., Verrey F., Rosenstiel P., Penninger J.M. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature. 2012 Jul 25;487(7408):477–481. doi: 10.1038/nature11228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li X.C., Zhang J., Zhuo J.L. The vasoprotective axes of the renin-angiotensin system: physiological relevance and therapeutic implications in cardiovascular, hypertensive and kidney diseases. Pharmacol. Res. 2017 Nov;125(Pt A):21–38. doi: 10.1016/j.phrs.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuba K., Imai Y., Rao S., Gao H., Guo F., Guan B., Huan Y., Yang P., Zhang Y., Deng W., Bao L., Zhang B., Liu G., Wang Z., Chappell M., Liu Y., Zheng D., Leibbrandt A., Wada T., Slutsky A.S., Liu D., Qin C., Jiang C., Penninger J.M. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat. Med. 2005 Aug;11(8):875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moher D., Liberati A., Tetzlaff J., Altman D.G., PRISMA Group Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6(7) doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao Y., Zhao Z., Wang Y., Zhou Y., Ma Y., Zuo W. bioRxiv; April 2020. Single-cell RNA Expression Profiling of ACE2, the Putative Receptor of Wuhan 2019-nCov. 2020.01.26.919985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zou X., Chen K., Zou J., Han P., Hao J., Han Z. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front. Med. 2020 Mar 12 doi: 10.1007/s11684-020-0754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lukassen S., Chua R.L., Trefzer T., Kahn N.C., Schneider M.A., Muley T., Winter H., Meister M., Veith C., Boots A.W., Hennig B.P., Kreuter M., Conrad C., Eils R. SARS-CoV-2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. EMBO J. 2020 Apr 14 doi: 10.15252/embj.2020105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sungnak W., Huang N., Bécavin C., Berg M., Queen R., Litvinukova M., Carlos Talavera-López C., Maatz H., Reichart D., Sampaziotis F., Worlock K.B., Yoshida M., Barnes J.L. HCA Lung Biological. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020 May;26(5):681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen W., Lan Y., Yuan X., Deng X., Li Y., Cai X., Li L., He R., Tan Y., Deng X., Gao M., Tang G., Zhao L., Wang J., Fan Q., Wen C., Tong Y., Tang Y., Hu F., Li F., Tang X. Detectable 2019-nCoV viral RNA in blood is a strong indicator for the further clinical severity. Emerg. Microb. Infect. 2020 Feb 26;9(1):469–473. doi: 10.1080/22221751.2020.1732837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marin G.H. Facts and reflections on COVID-19 and anti-hypertensives drugs. Drug Discov. Ther. 2020 Mar 26 doi: 10.5582/ddt.2020.01017. [DOI] [PubMed] [Google Scholar]
- 26.Sun Y., Guo F., Zou Z., Li C., Hong X., Zhao Y., Wang C., Wang H., Liu H., Yang P., Han Z., Liu K., Kuba K., Song B., Gao J., Mo Z., Li D., Li B., Li Q., Zhong N., Wang C., Penninger J.M., Jiang C. Cationic nanoparticles directly bind angiotensin-converting enzyme 2 and induce acute lung injury in mice. Part. Fibre Toxicol. 2015 Mar 7;12:4. doi: 10.1186/s12989-015-0080-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brünink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020 Apr 1 doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 28.Chang L., Zhao L., Gong H., Wang L., Wang L. Severe acute respiratory syndrome coronavirus 2 RNA detected in blood donations. Emerg. Infect. Dis. 2020;(7):26. doi: 10.3201/eid2607.200839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pasanen L., Launonen H., Siltari A., Korpela R., Vapaatalo H., Salmenkari H., Forsgard R.A. Age-related changes in the local intestinal renin-angiotensin system in normotensive and spontaneously hypertensive rats. J. Physiol. Pharmacol. 2019 Apr;70(2) doi: 10.26402/jpp.2019.2.03. [DOI] [PubMed] [Google Scholar]
- 30.Adedeji A.O., Severson W., Jonsson C., Singh K., Weiss S.R., Sarafianos S.G. Novel inhibitors of severe acute respiratory syndrome coronavirus entry that act by three distinct mechanisms. J. Virol. 2013 Jul;87(14):8017–8028. doi: 10.1128/JVI.00998-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elkin S.R., Oswald N.W., Reed D.K., Mettlen M., MacMillan J.B., Schmid S.L. Ikarugamycin: a natural product inhibitor of clathrin-mediated endocytosis. Traffic. 2016 Oct;17(10):1139–1149. doi: 10.1111/tra.12425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCluskey A., Daniel J.A., Hadzic G., Chau N., Clayton E.L., Mariana A., Whiting A., Gorgani N.N., Lloyd J., Quan A., Moshkanbaryans L., Krishnan S., Perera S., Chircop M., von Kleist L., McGeachie A.B., Howes M.T., Parton R.G., Campbell M., Sakoff J.A., Wang X., Sun J.Y., Robertson M.J., Deane F.M., Nguyen T.H., Meunier F.A., Cousin M.A., Robinson P.J. Building a better dynasore: the dyngo compounds potently inhibit dynamin and endocytosis. Traffic. 2013 Dec;14(12):1272–1289. doi: 10.1111/tra.12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weir D.L., Laing E.D., Smith I.L., Wang L.F., Broder C.C. Host cell virus entry mediated by Australian bat lyssavirus G envelope glycoprotein occurs through a clathrin-mediated endocytic pathway that requires actin and Rab5. Virol. J. 2014 Feb 27;11:40. doi: 10.1186/1743-422X-11-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020 Mar 20 doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elfiky A.A. Ribavirin, Remdesivir, Sofosbuvir, Galidesivir, and Tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): a molecular docking study. Life Sci. 2020 Mar 25 doi: 10.1016/j.lfs.2020.117592. 117592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaur R., Kaur G., Gill R.K., Soni R., Bariwal J. Recent developments in tubulin polymerization inhibitors: an overview. Eur. J. Med. Chem. 2014 Nov 24;87:89–124. doi: 10.1016/j.ejmech.2014.09.051. [DOI] [PubMed] [Google Scholar]
- 37.Deftereos S.G. The Greek study in the Effects of Colchicine in COvid-19 complications prevention (GRECCO-19 study): rationale and study design. Hellenic J. Cardiol. 2020 Apr 3:S1109–9666–0. doi: 10.1016/j.hjc.2020.03.002. pii: S1109-9666(20)30061-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Devaux C.A., Rolain J.M., Colson P., Raoult D. New insights on the antiviral effects of chloroquine against coronavirus: what to expect for COVID-19? Int. J. Antimicrob. Agents. 2020 Mar 12:105938. doi: 10.1016/j.ijantimicag.2020.105938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frlan R., Gobec S. Inhibitors of cathepsin B. Curr. Med. Chem. 2006;13(19):2309–2327. doi: 10.2174/092986706777935122. [DOI] [PubMed] [Google Scholar]
- 40.Wyczałkowska-Tomasik A., Pączek L. Cathepsin B and L activity in the serum during the human aging process: cathepsin B and L in aging. Arch. Gerontol. Geriatr. 2012 Nov-Dec;55(3):735–738. doi: 10.1016/j.archger.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 41.Mareti A., Kritsioti C., Georgiopoulos G., Vlachogiannis N.I., Delialis D., Sachse M., Sopova K., Koutsoukis A., Kontogiannis C., Patras R., Tual-Chalot S., Koureas A., Gatsiou A., Stellos K., Stamatelopoulos K. Cathepsin B expression is associated with arterial stiffening and atherosclerotic vascular disease. Eur. J. Prev. Cardiol. 2019 Dec 4 doi: 10.1177/2047487319893042. 2047487319893042. 2047487319893042. [DOI] [PubMed] [Google Scholar]
- 42.Keller C.W., Loi M., Ligeon L.A., Gannagé M., Lünemann J.D., Münz C. Endocytosis regulation by autophagy proteins in MHC restricted antigen presentation. Curr. Opin. Immunol. 2018 Jun;52:68–73. doi: 10.1016/j.coi.2018.04.014. [DOI] [PubMed] [Google Scholar]
- 43.Andersen K.G.1, 2, Rambaut A.3, Wi4 Lipkin, Holmes E.C.5, Garry R.F.6, 7 The proximal origin of SARS-CoV-2. Nat. Med. 2020 Apr;26(4):450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zanker D., Chen W. Standard and immunoproteasomes show similar peptide degradation specificities. Eur. J. Immunol. 2014 Dec;44(12):3500–3503. doi: 10.1002/eji.201445272. [DOI] [PubMed] [Google Scholar]
- 45.Tsakiri E.N., Trougakos I.P. The amazing ubiquitin-proteasome system: structural components and implication in aging. Int. Rev. Cell Mol. Biol. 2015;314:171–237. doi: 10.1016/bs.ircmb.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 46.Stratford F.L., Chondrogianni N., Trougakos I.P., Gonos E.S., Rivett A.J. Proteasome response to interferon-gamma is altered in senescent human fibroblasts. FEBS Lett. 2006 Jul 10;580(16):3989–3994. doi: 10.1016/j.febslet.2006.06.029. [DOI] [PubMed] [Google Scholar]
- 47.Moore J.B., Hune C.H. Cytokine release syndrome in severe COVID-19. Science. 2020 doi: 10.1126/science.abb8925. [DOI] [PubMed] [Google Scholar]
- 48.Zhang K., Meng X., Li D., Yang J., Kong J., Hao P., Guo T., Zhang M., Zhang Y., Zhang C. Angiotensin (1-7) attenuates the progression of streptozotocin-induced diabetic renal injury better than angiotensin receptor blockade. Kidney Int. 2015 Feb;87(2):359–369. doi: 10.1038/ki.2014.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Passos-Silva D.G., Verano-Braga T., Santos R.A. Angiotensin-(1-7): beyond the cardio-renal actions. Clin. Sci. (Lond.) 2013 Apr;124(7):443–456. doi: 10.1042/CS20120461. [DOI] [PubMed] [Google Scholar]
- 50.Patel V.B., Zhong J.C., Grant M.B., Oudit G.Y. Role of the ACE2/angiotensin 1-7 Axis of the renin-angiotensin system in heart failure. Circ. Res. 2016;118(8):1313–1326. doi: 10.1161/CIRCRESAHA.116.307708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bennion D.M., Haltigan E., Regenhardt R.W., Steckelings U.M., Sumners C. Neuroprotective mechanisms of the ACE2-angiotensin-(1-7)-Mas axis in stroke. Curr. Hypertens. Rep. 2015 Feb;17(2):3. doi: 10.1007/s11906-014-0512-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alenina N., Xu P., Rentzsch B., Patkin E.L., Bader M. Genetically altered animal models for Mas and angiotensin-(1-7) Exp. Physiol. 2008 May;93(5):528–537. doi: 10.1113/expphysiol.2007.040345. [DOI] [PubMed] [Google Scholar]
- 53.Ritchie A.I., Singanayagam A. Immunosuppression for hyperinflammation in COVID-19: a double-edged sword? Lancet. 2020;395(10230):1111. doi: 10.1016/S0140-6736(20)30691-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.