Abstract
Purpose
To investigate how induced tumor heterogeneity influences immune responses to radiotherapy (RT) with different proportions of mixed immune responsive and unresponsive tumor cells in a triple negative breast cancer model. It is hypothesized that studying the immune environment of mixed tumors and responses to RT could nominate immune active therapies to enhance immune responses after RT.
Experimental Design
Evaluate efficacy and immune responses generated by RT in tumors with different proportions of immunologically responsive and unresponsive tumor cells. Then study the cellular responses and transcriptomic differences between the tumors to nominate immunotherapy combinations with RT and evaluate the combination.
Results
The addition of the responsive cells to unresponsive tumors led to a greater than expected therapeutic response to RT with both innate and adaptive immune components. There was a distinct change in myeloid cells, greater inflammatory macrophage activity, and enhanced antigen presentation with responsive cells after RT. Since differences in matrix components, cell adhesion biology, and innate immune signaling correlated with myeloid cell response and phenotype, we hypothesized that RT combined with CD40 agonist antibody would sensitize unresponsive tumors. The combination therapy resulted improved innate and adaptive immune response. Importantly, CD40 treatment increased tumor response to RT and protected against metastatic spread in a metastatic model.
Conclusion
These data combined with transcriptomics from human patients supports RT and myeloid cell targeting for immunologically cold tumors and presents opportunities to investigate the complex overlapping biologic mechanisms that limit immunotherapy and to implement RT with different immunotherapy combinations.
Introduction
Immune checkpoint therapies are transforming treatment paradigms in many cancers with positive clinical trials not only in metastatic disease but also in the curative intent setting. For example, the Pacific trial demonstrated a significant survival benefit for adjuvant PD-L1 blockade after chemoradiation in locally advanced lung cancer (1). Although focal radiation could provide dramatic results for metastatic disease when coupled with immunotherapy, there is limited evidence in human patients this approach will impact therapeutic outcomes beyond a small proportion of patients with immunogenic tumors (2–5). However, with more studies evaluating immunotherapy in the definitive setting, the combination of radiation, chemotherapy, and immunotherapy is underway (1, 6). Therefore, understanding responses to combination therapy is critical to optimize local and systemic control of disease in the non-metastatic setting. Identification of factors that enhance immune responses after radiation and/or chemotherapy may reveal combination therapies that license immune responses of immunologically cold tumors. Since radiation therapy (RT) is utilized in over half of all solid tumors for definitive treatment, RT and optimal immunotherapy combinations may be impactful. To this end, characterization of the responses of the complex tumor microenvironment (TME) is essential to drive advancements in the treatment of immunologically unresponsive tumors using RT.
RT can induce adaptive immune responses and result in enhanced efficacy of immune checkpoint blockade; unfortunately, many tumors are not immunologically responsive to RT at baseline (3, 7–9). Dendritic cell activation is key for effective adaptive immune responses through presenting tumor antigens, detecting pattern recognition molecules that activate innate pathways and sensing cytosolic DNA (10–12). However, understanding how RT induces productive immune responses through this critical step is often bypassed in preclinical studies that use tumor cells that express model antigens or innate ligands that trigger dominant adaptive immune responses (12–15). Furthermore, since RT can induce tolerogenic cell death instead of immunogenic cell death, neoantigens may not be dominant and dendritic cell activation may not subsequently occur (16). Thus, a better understanding of different competing signals in the microenvironment is critical.
In this study, we modeled intrinsic intratumoral immune heterogeneity by mixing clonal tumor cells that induce different immune microenvironments prior to tumor implantation. With this approach we determined how different mixtures of tumor cells can induce an immune licensing phenotype over a dominant suppressive phenotype. Analysis of the microenvironmental factors that induced a licensing effect indicated that cell intrinsic factors such as induced antigen presentation and infiltrating myeloid cell phenotype were associated with immune licensing. These findings led us to hypothesize that the CD40 agonist (anti-CD40) antibody in combination with RT can transform immunologically unresponsive tumors. Interestingly, we found that CD40 activation was robust and was associated with dendritic cell proliferation and a increased T-cell infiltration. In immunologically unresponsive tumors our data suggests that an immune licensing agent such as anti-CD40 could overcome the limited responses of checkpoint therapy alone when combined with RT and efforts are underway to validate this approach prospectively in patients.
Materials and Methods
Cell lines
Py8119 and Py117 clonal cell lines derived from transgenic MMTV-PyMT mice congenic in the C57Bl/6 background were used between passage numbers 15–25. Cells were confirmed to be mycoplasma free and cultured in Hams F12K media, 5% fetal bovine serum, MITO (1:1,000 dilution, Corning), 50 μg ml−1 gentamicin and 2.5 μg ml−1 amphotericin B. Cells were derived as described previously and were not selected in any specific way beyond clonal dilution (17, 18). 4T1 cells expressing GFP and Luciferase were attained by the Contag Lab (Stanford) and grown in RPMI 10% fetal bovine serum.
Orthotopic tumors
Orthotopic implantation of 1×106 PyMT cells in the mammary fat pad #4 in 50% PBS and 50% Matrigel without phenol red and low growth factor (Corning). For mixed ratio experiments a total of 1×106 cells were injected with ratios of cells mixed into Matrigel or as described in the text and figures prior to implantation. A total of 2×105 4T1 cells were used for orthotopic implantation in PBS. Luciferase imaging of 4T1 tumors was performed using an IVIS-200 10 min after IP injection of 3.3 mg of D-luciferin (Biosynth Chemistry & Biology, Itasca, IL)
Tumor treatments
Irradiation of tumors 7–10 days after implantation at the identified dose was performed using a 225 kVp cabinet X-ray irradiator with a 0.5 mm Cu filter (IC-250, Kimtron Inc., CT). Mice were anesthetized using 80 mg kg−1 ketamine hydrochloride and 5 mg kg−1 xylazine intraperitoneally and the tumors were shielded with a 3.2 mm lead shield with a 15 × 20 mm aperture. Anti-CD40 agonist Ab (FGK4.5, Bio-X-Cell) was administered 100 μg IP the starting the day of RT and repeated for 4–5 doses every 3 days. PD-1 blocking Ab (RMP1–14, Bio-X-Cell) was administered 200 μg IP starting the day of treatment and repeated for 4–5 doses every four days. Isotype control IgG2a Ab (2A3, Bio-X-Cell) was administered 100 μg IP starting the day of treatment and repeated for 4–5 doses every 3 days. CSF-1 blocking antibody (5A1, Bio-X-Cell) was administered 100 μg IP starting the day of treatment and repeated for 4–5 doses every four days.
Flow Cytometry
Tumors were harvested after treatment and dissociated by chopping tissue finely and suspended in 1:1 F12K media and DMEM. Tumors were digested with Liberase TL at 20 μg ml−1 (Roche) and DNAse at 400 μg ml−1 (Roche) for 40 min at 37°C then filtered through a 40 μm mesh after diluting in media with 5% FBS. Tumor digestions for Figure 1 were performed using collagenase type I at 200 U ml−1 (Worthington) and Dispase at 0.5 U ml−1 (Stem Cell Technologies). Cells were resuspended in RBC lysis buffer for 10 min at room temperature then counted and diluted in PBS. Cells were then treated with Zombie NIR (Biolegend), CD16/32 Ab (Biolegend) for FC blocking and cell surface antibody panel for 20 min on ice. Cells were washed 2x then fixed and permeabilized using eBiosciences Foxp3/ Transcription factor Staining Buffer set then labeled with intracellular antibodies including Arg1, iNOS, Ki67, FoxP3, for 20 min.
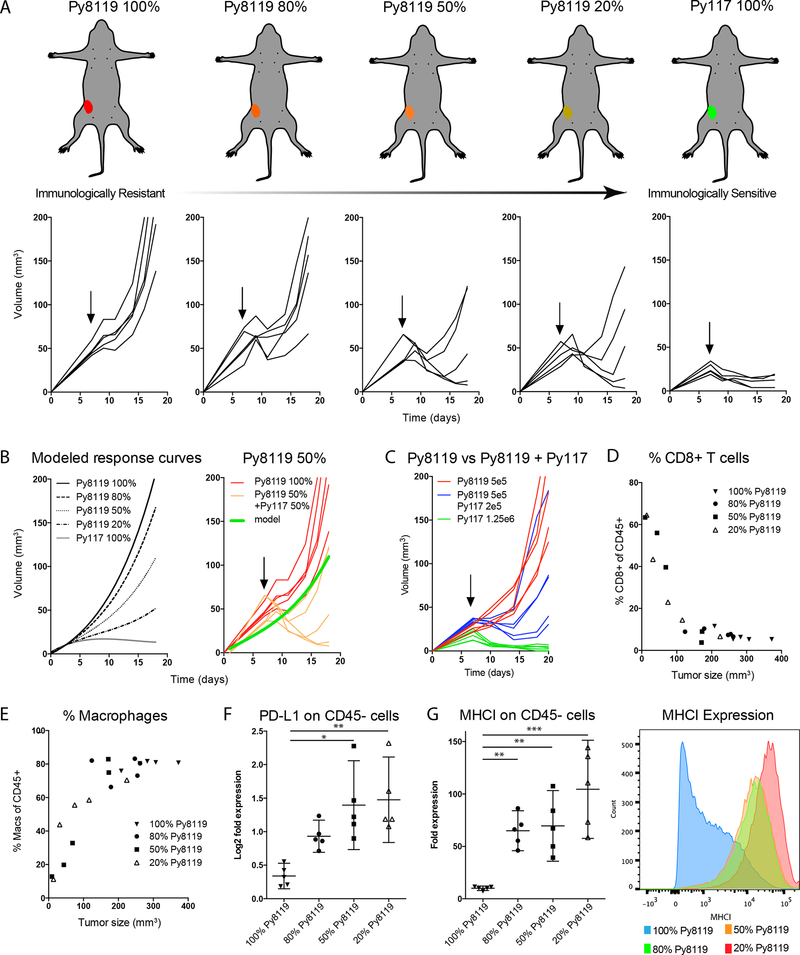
Figure 1:
Increasing the proportion of py117 cell that are sensitive to radiation and checkpoint therapy results in increased radiation sensitivity (18). A) Increasing the proportion of Py117 tumor cells out of a total of 1×106 increased RT sensitivity to 12 Gy (arrow). Images of mice depict immunosuppressive Py8119 tumors (red) down to the immune sensitive Py117 tumors (green) and the accompanying growth curves after RT are show below. B) Response curves were modeled using single cell line growth curves revealing that tumors with 50% or greater Py117 cells responded better to RT than predicted. C) Tumors with 5×105 Py8119 cells with an additional 2×105 Py117 cells responded better than tumors with 5×105 Py8119 cells alone, confirming the sensitizing or licensing effect of Py117 cells. D-E) Tumors with the greatest response and smaller size 10 days after 12 Gy exhibited an increased proportion of CD8+ T cells and decreased proportion of CD45+CD11b+F4/80+ macrophages. F) Relative PD-L1 expression on tumor cells increased with increasing proportions of Py117 cells, suggesting an immune response form increased interferon. G) The presence of Py117 cells in tumors resulted in greater than expected MHCI antigen presentation on tumor cells compared to tumors with only Py8119 tumor cells alone. * p<0.05, **<0.01, and *** p<0.0001 on analysis of variance with pairwise comparisons.
Flow cytometry was performed on a four laser LSRII (BD Biosciences) or a LSRFortessa X-20 (BD Biosciences). In all analyses at least 100,000 events of 2–4 million cells that were stained were recorded to the determine the relative abundance of respective cells per total live cells or CD45+ cells. Analysis was performed using FlowJo v10 Software (Tree Star). Rat/Hamster compensation beads (Life Technologies) and cells stained with live dead stain were used for compensations. The following Ab clones targeting mouse epitopes were used for analysis. Biolegend: CD45 (1:160, 103133, 30-F11), CD3 (1:80, 100237, 17A2), CD4 (1:400, 100406/100432, GK1.5), CD8 (1:160, 100708, 53–6.7), CD69 (1:100, 104509, H1.2F3), CD11b (1:400, 101243, M1/70), CD11c (1:200, 117310, N418), F4/80 (1:60, 123110/123133, BM8), MHCII IA I-e (1:160, 107606, M5/114.15.2), GR-1 (1:160, 108408, RG6–8C5) and PD-L1 (1:160, 124310, 10 F.9G2). BD Biosciences: MHCI H2Kb (1:160, 553570, AF6–88.5), Arg-1 (3 μL per sample, IC5868A, and Ki-67 (4 μL per sample, 561126, B56). eBiosciences: FoxP3 (1:100, 61–5773, FJK165), TCR-β (1:200, 45−5961, Η57−597), and NOS2 (1:500, 25–5920, CXNFT). Gating strategy for immune cells, myeloid cells in Py117 and Py8119, and T cells in Py117 tumors are shown in Supplemental Figure 1A–C. Gating strategy for leukocytes in the draining lymph nodes are show in Supplemental Figure 2A–C.
Luminex multiplex cytokine assay
Py117 and Py8119 tumors implanted into C57Bl/6 mice and one group was irrradiated with 12 Gy or a second group was not irradiated when tumors were 75–125 mm^3. Tumors were harvested 10 days after radiation and tumor tissue was homogenized in RIPA lysis buffer with protease inhibitor 100 mg per 500 μL of buffer. Supernatants were collected after centrifugation and frozen at −80 °C. On day of assay samples were thawed on ice and diluted to 1 μg/μL after measurement by BCA assay.
Affymetrix mouse 38-plex kits were used according to manufacturer’s recommendations and processed by the Stanford Human Immune Monitoring Core. Samples were further diluted 3-fold to a final volume of 60 μL and incubated with antibody linked beads in a 96-well plate the incubated for 1 hour at room temperature followed by overnight incubation at 4 °C. Samples were further process as previously described (18).
Microarray Analysis
For tissue culture microarray Py117 cells and Py8119 cells were plated in 6 cm disses with 3.33 × 105 cells, media was changed after 24 hours and cells were lysed on ice in RIPA buffer at 48 hours. For homogenates, tumors were harvested from mice on the day RT would typically occur for the Py117, Py8119, and the Py5050 mix tumors then tissue was homogenized 100 mg per 500 μL centrifuged, and RNA was extracted from the supernatants. Samples were measured by the Bioanalyzer QC to verify the quality, underwent in vitro transcription, fragmentation, labeling, hybridization, and scanning performed by the Stanford Protein and Nucleic Acid Facility. The Affymetrix mouse gene 2.0 ST whole transcriptome array was used for all samples. All microarray data is uploaded to GEO accession ID GSE118181.
The array data was analyzed using R\Bioconductor package oligo and limma version 1.42.0 and 3.36.1 respectively, RMA for normalization (19–21). All differential gene expression was reported at corrected p value equal or less than 0.01 and fold change equal or greater than 2. Annotation and collapse dataset to gene symbols were determined using annotation package specifically for Mouse gene 2.0 ST array mogene20sttranscriptcluster.db in R. Hierarchical clustering and heat map visualization was generated using ComplexHeatmap version 1.17.1 in R (22). Cell intrinsic genes and genes induced in microenvironment were determined as following. First, we compared 117 and 8119 cells in culture and list of genes that were differentially expressed named as DE genes at cell level, and all other genes named as non-DE at cell level. Then, we compared tissues from Py117, Py5050 and Py8119 and differential gene expression was calculated as we mentioned above. After that, we filtered these genes by DE genes at cell level and shared genes were named cell intrinsic genes. We then filtered by non-DE at cell level and shared genes was named genes induced in microenvironment. Gene enriched in cells and tumors was analyzed by Gene set enrichment analysis software GSEA Desktop v3.0 (23) and enriched terms were selected from terms at FDR p ≤0.25. Principal components analysis was calculated in R and visualized using pca3d package version 3.4.4. Gene ontology networks were generated and visualized using the ClueGO v2.5.1-CluePedia v.1.5.1 Cytoscape (v3.3.6) plugin (24, 25).
Publicly available data analysis
Transcriptomic data from PD-1 responsive and non-responsive melanoma from Hugo et al, 2016 was obtained from accession number GEO: GSE78220. Data was re-analyzed using STAR and DEseq2 pipeline and GRCh38 as a reference genome (26). Gene enriched was obtained by using GSEA Desktop v3.0 as we described above. TCGA transcriptomic data of Breast (BRCA), Skin (SCKM) and pancreatic (PAAD) cancer patients along with clinical data were obtained using TCGAbiolinks R package (version 2.9.5) for harmonized data (27). Before calculating survival curves data was cleaned from any censored incident before first death. Samples from BRCA male patients were removed. Neuroendocrine carcinoma patients were removed from PAAD data. Multivariate Cox regressions was performed with the coxph function from the R survival library, Kaplan-Meier plot, a logrank p-value were plotted using survfit and ggsurvplot R packages. Correlation between CD40, CD40LG and MAP3K14 expression level and infiltration of immune cells and tumor purity were calculated and plotted using TIMER (28, 29). Oncolnc data for CD40LG was obtained from TCGA BRCA, SKCM, and PAAD from oncolnc.org (30). Chen et al. normalized Nanostring values were obtained from supplementary tables (31). Riaz et al. normalized RNAseq data was obtained from GEO: GSE91061 and significance was measured by a two-sided T test (32).
Statistics
Flow cytometry and Luminex multiplex cytokine arrays were quantified using an analysis of variance model and post hoc comparison with a Tukey adjustment using GraphPad6 (GarphPad Software Inc, La Jolla, CA). Tumor growth curves were analyzed in a repeated measure mixed model allowing for errors to be correlated within mice using R (R Core Team, Vienna, Austria)
Study Approval
All animal studies were approved by the Stanford IACUC protocol number 9984.
Results
Mixed tumors with responsive and unresponsive clones license a greater than expected response to RT
We previously described differences in the epithelial to mesenchymal transition (EMT) phenotype and AXL receptor tyrosine kinase expression between the more immunogenic Py117 and immune resistant Py8119 PyMT tumors derived from the MMTV-PyMT mouse model (18). Using this model, we investigated how mixed tumor clones influence immune infiltrates in the microenvironment and studied the impact of immunologic heterogeneity on therapeutic responses. We hypothesized that the Py117 cells would cause a sensitizing or licensing effect over the suppressive effect of Py8119 cells. Orthotopic tumors with varying ratios of immunologically resistant Py8119 and sensitive Py117 tumor clones were implanted as depicted in Figure 1A. Seven days after tumor implantation, tumors were treated with 12 Gy and exhibited tumor growth delay by RT that increased proportionally to the number of Py117 cells. The expected tumor growth curves were modeled using responses derived from single clone growth curves. Interestingly, the response of tumors with 50% Py117 cells were greater than predicted from modeling and the addition of Py117 cells to tumors with the same number of Py8119 lead to greater response compared to Py8119 alone, supporting a dominant sensitizing effect (Figure 1B and 1C). In concordance with our past study, when mixed tumors were implanted into athymic nude mice, tumor growth increased and radiation response diminished in the mixed tumors, indicating the importance of the immune response (Supplemental Figure 3A) (18).
To evaluate the immunologic response of these tumors in more detail, tissues were dissociated and analyzed by flow cytometry. The proportion of CD45+ leukocytes was unchanged across the 20–100% Py8119 containing tumors (Supplemental Figure 3B). However, tumors with the greatest RT response displayed significantly increased numbers of CD8+ T-cells and reduced CD45+CD11b+F480+ macrophages (Figure 1D, E). No differences in CD4+ T cells or immature myeloid cells (CD45+CD11b+Gr1hi) were observed in the mixed tumors (Supplemental Figure 3C, D).
In mixed tumor populations, we found increased expression of PD-L1 that tracked with increased proportion of Py117 cells (Figure 1F). These findings suggest that specific subclones within a heterogeneous tumor can support a more robust immunogenic microenvironment leading to induced PD-L1 expression. A similar observation was made with MHCI, where mixed tumor populations had higher MHCI expression compared to tumors with 100% Py8119 cells. Moreover, as low as 20% of Py117 cells in the mixture was enough to induce a homogeneous increase in MHCI (Figure 1G, Right), which can lead to enhanced antigen presentation in the microenvironment.
Clonal tumors have different tumor infiltrating myeloid cell, cytokine, and chemokine profiles
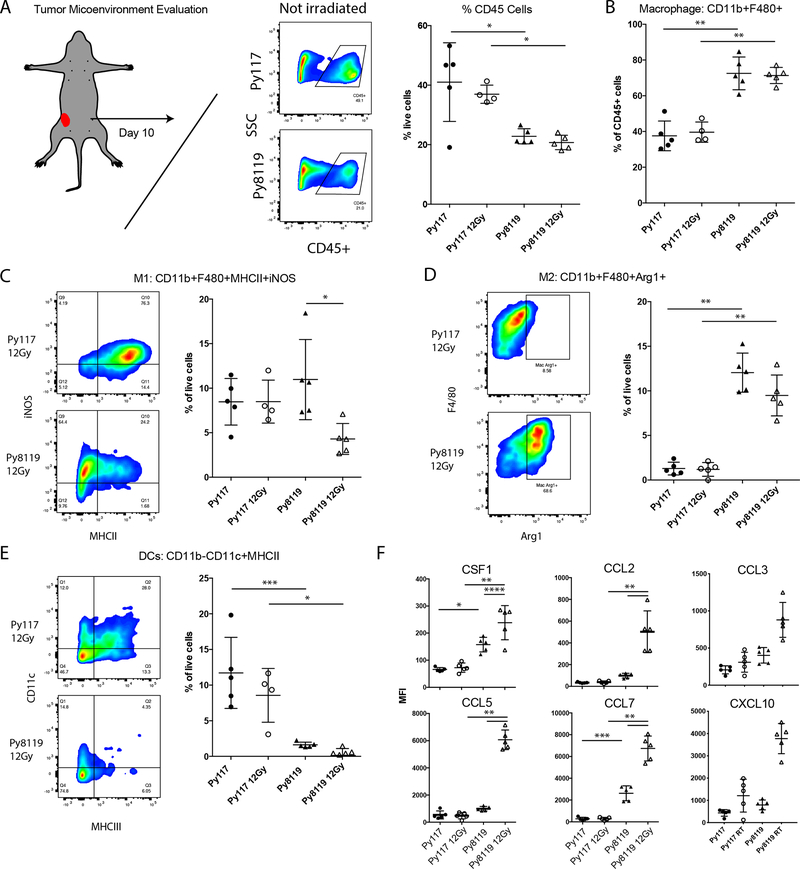
We hypothesized that the greater than expected reduction in tumor volume after RT in mixed tumors is due to an initial innate response triggered by Py117 tumor cells followed by an adaptive T cell response. Although NK cell depletion did not result in a reduced response in Py117 tumors (18), we did observe differences in CD11b myeloid populations, as highlighted in Figure 1E. Therefore, we assessed the different M1-like macrophage subtypes (M1: CD45+CD11b+F4/80+MHCII+iNOS+), M2-like macrophages (M2: CD45+CD11b+F4/80+Arg1+), and dendritic cells (DCs: CD45+CD11b-CD11c+MHCII+). Ten days after treatment with 12 Gy RT, tumors were harvested, dissociated, and analyzed by flow cytometry. Regardless of RT, the proportion of infiltrating CD45+ leukocytes were greater in the Py117 tumors, and the number of macrophages as a proportion of CD45+ cells was higher in Py8119 tumors (Figure 2A–B). Within the Py8119 tumors, we observed similar proportions of M1-like macrophage cells expressing MHCII and iNOS that decreases after RT (Figure 2C). In contrast, we observed significantly more Arg1+ M2-like macrophages in the Py8119 tumors regardless of RT (Figure 2D). Thus, Py117 tumors possess a greater innate inflammatory M1-like response than a protumor M2-like response. In addition to the proinflammatory M1 environment, increased numbers of DCs were found in the Py117 tumors (Figure 2E).
Figure 2:
Evaluation of the cellular tumor microenvironment reveals greater suppressive myeloid cells in Py8119 tumors. A) Greater CD45+ infiltrates in Py117 than Py8119 tumors after 1×106 cells were inoculated then harvested 10 days after 12 Gy RT treatment would have been given; B) the relative proportion of macrophages was greater in Py8119 tumors. C) The total percentage of inflammatory M1-like CD11b+F4/80+MHCII+iNOS+ macrophages remained the same but decreased in Py8119 tumors 10 days after RT. D) The number of M2-like CD11b+F4/80+Arg1+ macrophages were much higher in the Py8119 tumors regardless of RT. E) Alternatively, the tumor microenvironment showed greater numbers of dendritic cells CD11b-CD11c+MHCII+ in the sensitive Py117 tumors and was nearly absent in Py8119 tumors. F) The macrophage-promoting growth factor CSF1 and chemokines CCL2, CCL5, CCL7, CCL3, and CXCL10 were elevated in Py8119 tumors and were significantly induced after RT, as measured by the Luminex multiplex assay. * <0.01, ** <0.0001, *** <0.001, and **** <0.05 on analysis of variance with pairwise comparisons.
Since macrophage infiltration supports an immunosuppressive microenvironment that can impede adaptive immune responses by various mechanisms, we sought to determine how cytokine and chemokine profiles correlated with these differences. Py8119 tumors had significantly higher CSF1, CCL2, CCL5, and CCL7 levels that were further elevated after RT compared to immune responsive Py117 tumors (Figure 2F). We also observed that CCL3 another substrate for CCR5 and CXCL10 that is often elevated in cancer. In contrast, Py117 tumors express reduced levels of these factors, have greater antigen presentation, increased dendritic cell infiltration, and generate immune responses that can be enhanced with RT and immunotherapy. The cytokine and chemokine profile differences between these tumors is similar to the profile of individual cultured tumor cells, suggesting that most of the factors present in vivo are in part generated endogenously by the tumor cells (18).
Mixed tumors generate greater adaptive immune responses compared to resistant Py8119 tumors
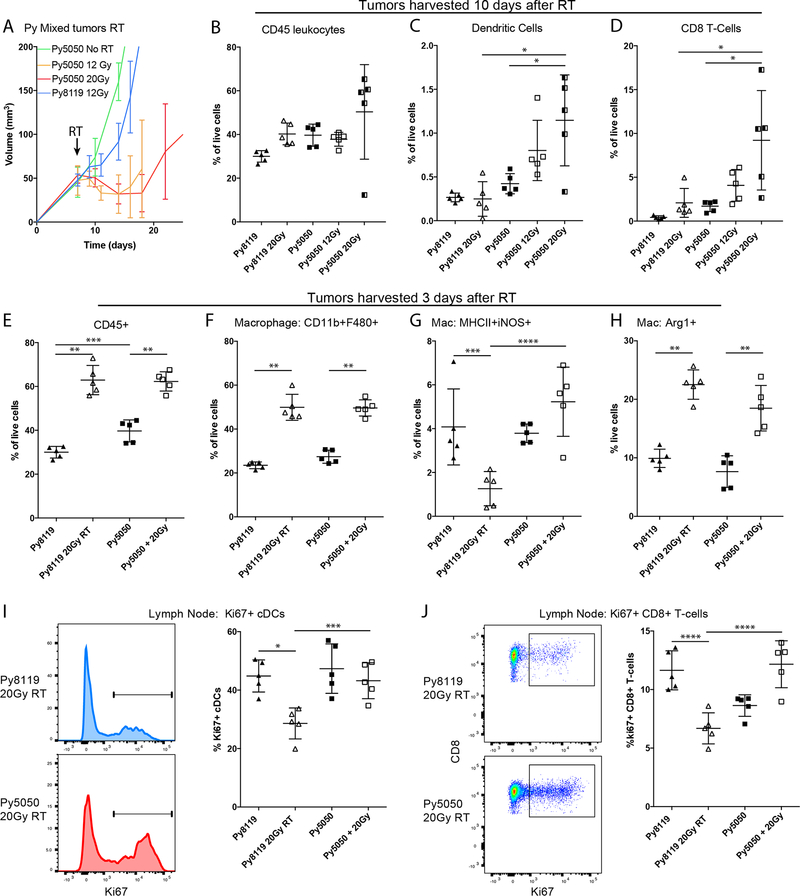
A range of single fraction radiation doses of 8 Gy up to 30 Gy have been observed to induce adaptive immune responses which depend on immunogenicity, and the immune microenvironment amongst other factors (33–35). Tumors containing 50% Py8119 cells and 50% Py117 cells (Py5050) were treated with either 12 Gy or 20 Gy in a single fraction which resulted in no significant difference in tumor growth between the two RT doses (Figure 3A). However, there was a trend toward greater infiltrating CD45, DCs, and CD8 T-cells in the mixed Py5050 mixed tumors ten days after 20 Gy RT compared to 12 Gy. Since all cell infiltrates in the Py5050 mixed tumors were greater than in Py8119 tumors alone after 20 Gy, we focused on 20 Gy for the subsequent experiments (Figure 3B–D). In comparing different tumor populations, the abundance of myeloid cells was identified as a key difference ten days after RT, we next evaluated the response three days after RT. We choose three days since previous studies indicate that mice can develop robust responses within three days after effective immunotherapy that are greatly dampened eight to ten days after treatment despite change in immune cell such as T cell influx after an adaptive response (36). We found that levels of CD45+ cells and macrophages were significantly elevated in Py8119 and Py5050 tumors three days after treatment (Figure 3E–F). However, M1-like cells were significantly reduced in Py8119 tumors but not in Py5050 tumors despite an increase in M2-like macrophages in both tumors (Figure 3G–H). No differences in T-cells or DCs were observed in either tumor type three days after treatment (Supplemental Figure 4A, B). We then assessed adaptive responses in the lymph node three days after RT since there were greater numbers of infiltrating DCs and CD8+ T-cells in mixed tumors ten days after RT. We observed increased proportion of Ki67+ DCs and CD8+ T-cells in Py5050 lymph nodes after RT compared to Py8119 tumors. While the DCs were unchanged in the nodes, the relative proportions of Ki67+ CD8+ T-cells were increased, suggesting a greater adaptive immune response by Py5050 tumors (Figure 3I, J).
Figure 3:
Mixed tumors show enhanced radiation sensitivity consistent with licensing a differential immediate innate response and an adaptive T cell response. A) Tumor growth curves of 50% Py8119/Py117 (Py5050) tumors revealed similar responses with either 12 or 20Gy or RT after 1×106 total cells injected for each tumor. B) Ten days after RT increased CD45+ infiltrating leucocytes with or without RT in the Py5050 tumors were observed compared to Py8119. C-D) The proportion of absolute dendritic cells and CD8+ T cells in the tumor at 10 days was higher in Py5050 tumors without RT and increased with increasing 12 and 20Gy RT. E-F) To evaluate the immediate immune response, tumors were harvested 3 days after RT, revealing an influx of CD45+ leucocytes and macrophages in both tumors after 20Gy RT. G) Despite the increase in macrophages, M1-like cells were maintained in Py5050 tumors and were decreased in the Py8119 tumors. H) Alternatively, activated M2-like macrophages were induced in both tumors 3 days after RT. I) Evaluation of the draining lymph nodes 3 days after RT revealed an increased proportion of Ki67+ dividing dendritic cells and J) an increased proportion of CD8+ T cells compared to a decreased proportion after RT, supporting the finding of increased CD8+ T cells infiltrating tumors 10 days after RT. *<0.01, **<0.0001,***<0.05, or ****<0.001 by analysis of variance with pairwise comparison.
Cell intrinsic genes influence the immune microenvironment
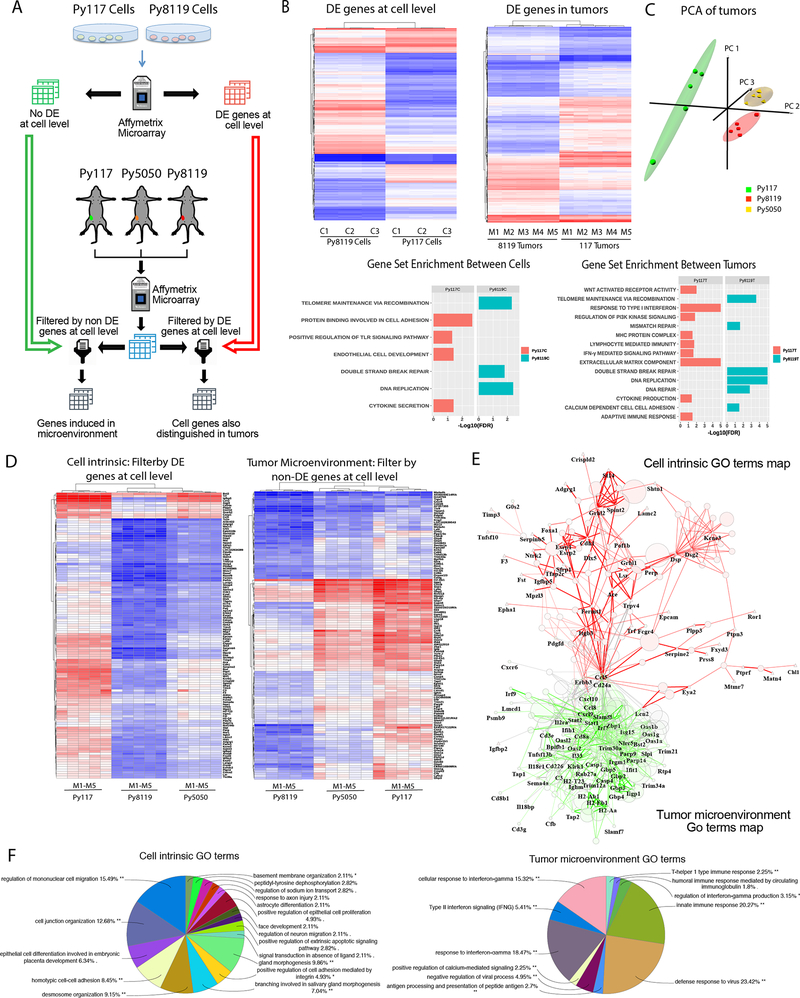
To better characterize the tumor microenvironment of mixed tumors we evaluated changes in gene expression of cells and tumors individually (Figure 4A). Whole transcriptome microarrays for both cultured Py117 and Py8119 tumor cells and Py117 and Py8119 tumors revealed 1079 and 1760 differentially expressed (DE) genes at adjusted p-value < 0.001. Gene set enrichment analysis (GSEA) showed that Py8119 cells are enriched for cell proliferation and DNA repair, whereas Py117 cells are enriched for cell adhesion, endothelial cell development, and innate immune signaling. GSEA of the tumors further confirmed the tumor cell findings in Py8119 tumors and revealed enrichment of extracellular matrix components, antigen presentation and adaptive immune responses in Py117 tumors, which are all consistent with our biologic findings (Figure 4B). A full list of gene set enrichment analysis could be found in Supplementary Table 1–4.
Figure 4:
Microarray analysis of cells and tumors revealed factors of immunologic sensitivity and resistance. A) Microarray experiment evaluating Py117 and Py8119 cells in culture as well as Py117, Py8119, and Py5050 mixed tumors. Differentially expressed (DE) genes from cells P<0.001 and > 2-fold difference were used to generate DE gene list and non-DE gene list. These lists were then used to evaluate DE genes from microarray of the three tumors to determine DE genes induced in the microenvironment or cell genes also DE in tumors. B) Hierarchical cluster analysis and gene set enrichment analysis (GSEA) performed on Py117 and Py8119 cells and tumors. C) Principal component analysis for Py117, Py8119, and Py5050 tumors. D) Hierarchical clustering of cell intrinsic DE genes at cell level and tumor microenvironment representing non-DE genes at the cell level evaluated for differential expression between Py117 and Py5050 from Py8119 tumors. E) GO term network maps of gene lists from Figure 4 D to reveal GO terms (circles) associated with cell intrinsic gene names and their associations. Cell intrinsic genes are identified by triangles, tumor microenvironment induced genes are identified by squares and the red network represents connection of cell intrinsic associated GO terms where the green represents tumor microenvironment associated GO terms. F) Shows a pie chart the most commonly represented GO terms from the cell intrinsic gene list and tumor microenvironment gene list.
Principal component analysis (PCA) was performed from microarrays of Py8119, Py117, and Py5050 tumors to determine if Py5050 global gene expression was more similar to Py8119 or Py117 tumors. PCA revealed that Py5050 tumors are more similar to Py8119 tumors (Figure 4C). To understand why Py5050 tumors are more immunologically sensitive than Py8119 tumors and their transcriptomics cluster more closely, we filtered all tumor sets by DE and non-DE genes at the cellular level to identify immune sensitivity enhancers inherited from cell intrinsic gene or by tumor microenvironment interactions, as depicted in Figure 4A. This analysis revealed that by using DE cell intrinsic genes, Py5050 tumors clustered with Py8119 tumors with few overlapping genes induced in Py5050 and Py117. However, by using the tumor microenvironment induced genes or non-DE genes at the cellular level, or cells not differentially expressed between the cells in culture, Py5050 tumors clustered with the Py117 tumors and we found a greater number of overlapping genes between the Py5050 and Py117 tumors (Figure 4D). We conclude that most of the immunological sensitivity of Py5050 tumors result from tumor microenvironment induced genes controlled by intrinsic characteristics of the Py117 cells.
A gene ontology network map was generated using the ClueGo-CluePedia with significant GO terms (circles), cell intrinsic genes (triangle-DE genes in cells and tumors) and tumor microenvironment genes (hexagon- non-DE genes at the cellular level induced in the tumor microenvironment). The green network represents the major cell intrinsic gene significant GO terms represented in the microenvironment cluster, where the red network represents the induced tumor microenvironment gene significant GO terms map (Figure 4E). The significant go terms are shown in the pie charts to depict the relevant cellular processes associated with the intrinsic genes and then induced microenvironment genes. These data demonstrate cell migration, epithelial differentiation, cell adhesion, and junction organization are key intrinsic differences between tumor populations, and then the TME differences are associated with innate and adaptive immune responses consistent with our findings described above (Figure 4F).
Targeting myeloid cells with anti-CD40 sensitizes immune resistant tumors to radiation
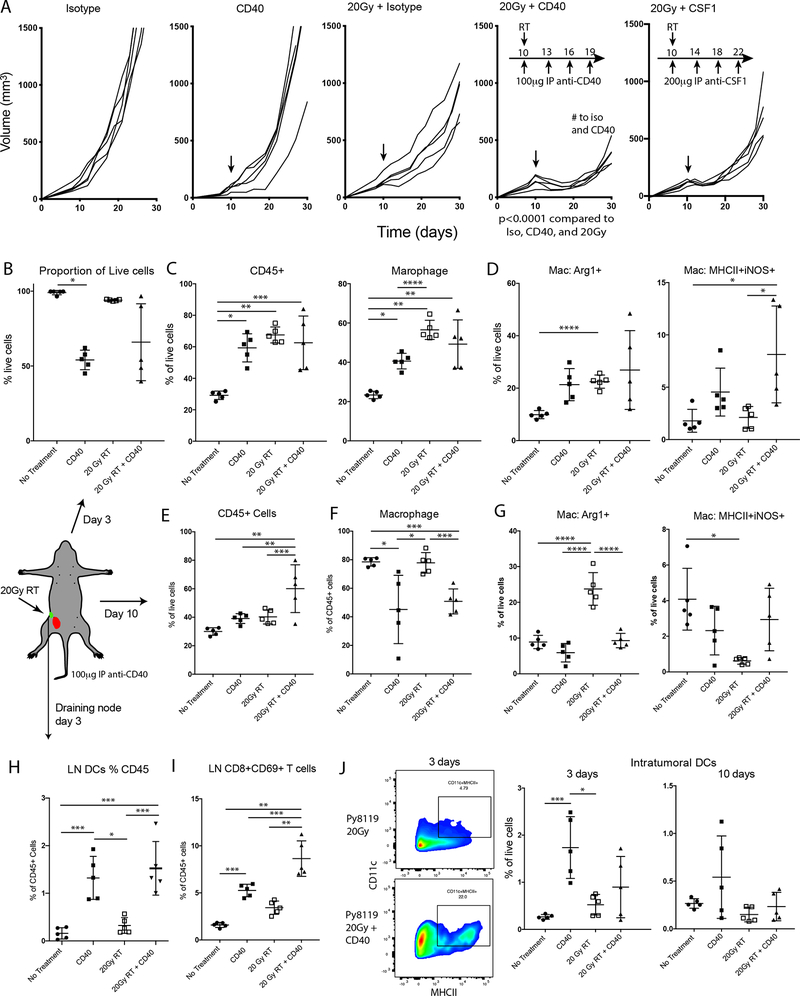
Multiple features in the Py117 and the Py5050 tumors contribute to the observed adaptive immune response. Py117 cells at baseline seem to have an adaptive immune response that is downstream of cell intrinsic innate immune pathways and antigen presentation as suggested by our microarray data (Figure 4). Based upon the differences in the immune microenvironment including increased M2-like macrophages, fewer M1-like macrophages, lower antigen presentation, fewer dendritic cells, and minimal innate and adaptive transcripts in the Py8119 tumors, we hypothesized that targeting myeloid and antigen presenting cells using a CD40 agonist antibody (anti-CD40) would generate an improved immune response. Anti-CD40 could address all the immunosuppressive factors observed from the Py8119 tumors by enhancing antigen presentation, activating M1 versus M2-like macrophages and mature dendritic cells in the tumor and draining nodes (37). We tested this hypothesis by treating Py8119 tumors with isotype control antibody, anti-CD40, 20 Gy RT, or a combination of these treatments. The combination of RT and anti-CD40 therapy revealed a more robust response than either treatment alone (Figure 5A). We also examined whether targeting myeloid cells through a different mechanism by blocking CSF-1 would also result in an increased response. Targeting CSF-1 to decrease macrophage numbers also led to enhanced radiation response. However, the treatment was less effective than anti-CD40, so we focused on the anti-CD40 agonist (Figure 5A).
Figure 5:
The addition of the CD40 agonist antibody (anti-CD40) in Py8119 tumors induced greater responses and early signs of an adaptive immune response. A) Tumor growth curves after injection of 1×106 cells treated with isotype antibody, anti-CD40, 20 Gy + isotype, 20Gy + anti-CD40, or 20Gy + anti-CSF1. B) Three days after treatment the anti-CD40 resulted in broad cell death either as monotherapy or in combination with RT. C) Three days after initiating treatment there was increase in CD45+ infiltrates and macrophages. D) All treatments increased the proportion of Arg1+ M2-like macrophages, and anti-CD40 treatments increased the proportion of M1-like MHCII+iNOS+ inflammatory macrophages. E-G) Ten days after treatment there were increased immune cells but decrease relative macrophage in the combination arm. However, macrophage expressing MHCII and iNOS was increased after combination therapy. H) Evaluation of the draining lymph nodes revealed that anti-CD40 and combination of 20Gy RT increased in the number of dendritic cells. I) Similarly, a significant increase in CD45+SSClowTCRβ+CD8+CD69+ T cells in the lymph node suggests that a greater adaptive immune response developed. J) A greater proportion of dendritic cell infiltrates were found in the anti-CD40 treated tumors 3 days after treatment, though the increase was not as significant as with RT. #<0.0001 by repeated measures. *<0.01, **<0.0001, ***<0.001, or ****<0.05 by analysis of variance with pairwise comparison.
Tumors were harvested three days after initiating each treatment alone or the combination of anti-CD40 + RT to investigate the impact on innate immune cells. Anti-CD40 and anti-CD40 + RT resulted in a greater immediate and cytotoxic response to that which was previously reported for anti-CD40 therapy (Figure 5B) (38). All treatments induced a significant influx of CD45+ leucocytes and macrophages (Figure 5C). The macrophage change consisted of an increase in Arg1+ cells in all treatment groups, while the M1-like MHCII+iNOS+ phenotype specifically increased in the anti-CD40 containing groups (Figure 5D). Tumors were then harvested ten days after anti-CD40 and RT treatment and infiltrating leukocytes remained elevated in all treatment groups as a proportion of live cells, being 40% in anti-CD40 alone and RT alone groups, compared to 60% after anti-CD40 + RT (Figure 5E). The relative proportion of macrophages remained high after treatment but significantly decreased in the anti-CD40 and combination treatment groups (Figure 5F). The group treated with RT alone had the most unfavorable macrophage profile, showing the highest M2-like and lowest M1-like phenotypes. Even ten days after starting treatment, the groups receiving anti-CD40 maintained a relatively low proportion Arg1+ M2-like and higher proportion of MHCII+iNOS+ M1-like macrophages (Figure 5G).
In addition to the favorable macrophage response, we next evaluated the lymph node 3 days after therapy initiation and observed a greater proportion of DCs in the draining lymph node only in the anti-CD40 and RT combination (Figure 5H). At this early time point, anti-CD40 treatment resulted in an increased proportion of CD8+ T-cells that were expressing CD69, a marker for lymphoid activation, and was significantly greater in combination with RT (Figure 5I). When we evaluated the DCs within the microenvironment, we found a greater number of DCs after CD40 administration alone at 3 days, and a similar level at 10 days (Figure 5J). This result suggests that in these tumors the anti-CD40 agonist is effective in enhancing DCs in the TME, but that the combination is required for more efficient lymph node migratory capacity and activation of T cells.
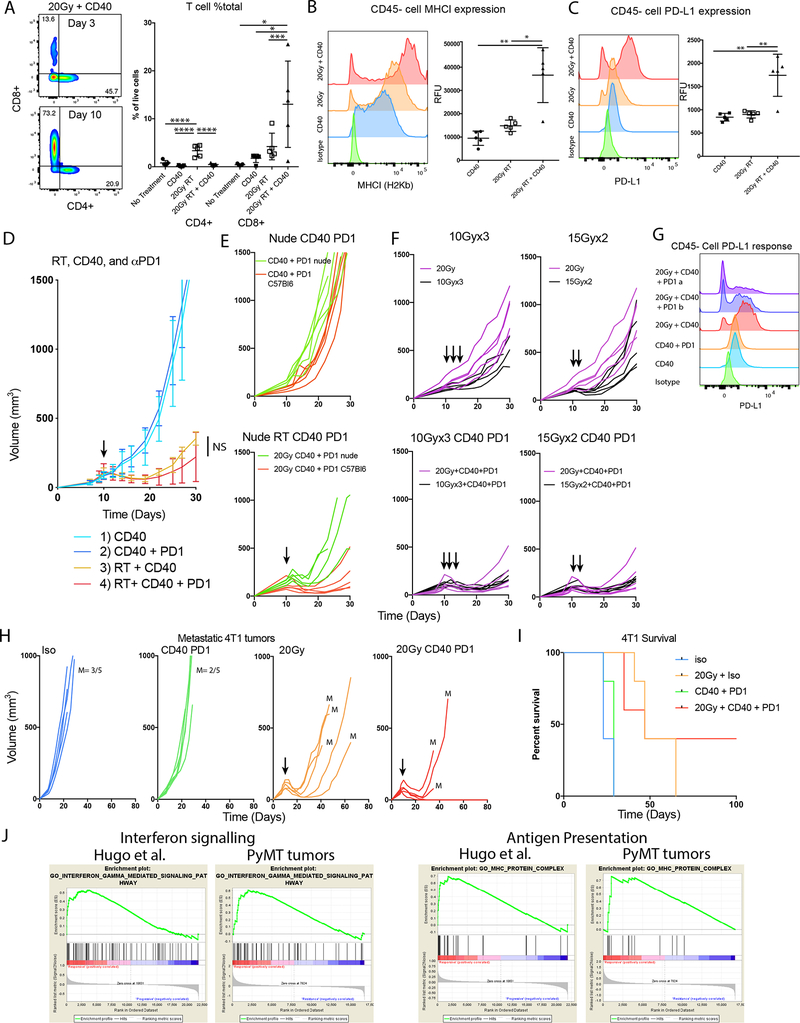
RT and anti-CD40 combination have innate and immune responses that could improve survival
Our data suggest that in resistant Py8119 tumors, anti-CD40 in combination with RT induces a rapid innate and early adaptive immune response three days after initiating treatment. Tumors harvested ten days after anti-CD40 and RT treatment were evaluated for evidence of an adaptive immune response in the lymph node after three days of treatment. There was no significant increase in infiltrating T-cells detected after three days, but at day ten, a 30-fold increase in infiltrating CD8+ T-cells was detected with the combination treatment compared to control treated mice. The number of CD4+ T-cells only increased after RT treatment alone (Figure 6A). Comparing CD40, RT, and the combination we observed greater MHCI levels on CD45 negative cells after combination treatment (Figure 6B). In addition, CD45- cells exhibited increased expression of PD-L1 after combination therapy, suggesting that interferon induction of PD-L1 was induced after the antitumor immune response (Figure 6C).
Figure 6:
The combination immunotherapy enhanced responses with RT in immune resistant tumors. A) CD8 T cell infiltration was greater at 10 days post treatment and significantly high in the combination compared to anti-CD40, or 20 Gy RT. B-C) MHCI and PD-L1 expression on tumor cells 10 days after RT. D) The addition of PD1 immunotherapy to anti-CD40 and RT. E) The combination of CD40 and PD1 without and with RT in immunodeficient mice reveals innate and adaptive components. F) Combination therapy with different fractionations did not show significant improvement over 20 Gy in a single fraction. G) Flow cytometry histograms revealed that 20Gy + anti-CD40 resulted in the greatest induction of PD-L1 on tumor cells, but the addition of PD-1 Ab increased a population of cells that did not express PD-L1. H) The 4T1 breast cancer model was treated with a combination of anti-CD40 + anti-PD-1 + 20 Gy, resulting in greater response to the combination. Though 20 Gy slowed tumor growth, most mice developed distant metastases (lung metastases detected= M). I) Long term survival without detectable metastases for more than 100 days was observed in 40% of the mice with primary tumors that had received the combination treatment. J) GSEA of PyMT Py117 vs Py8119 tumors and Hugo et al melanoma PD-1 responders and non-responders have similar pattern of enrichment for interferon signaling and antigen presentation. *<0.01, **<0.001, ***<0.05, ****<0.0001 by analysis of variance with pairwise comparison.
Given the induction of PD-L1 and improved T-cell infiltration we aimed to evaluate the addition of a PD-1 blocking antibody but observed a nonsignificant improvement over RT + anti-CD40 in the therapeutic response in mice (Figure 6D). However, there was a greater response compared to combination therapy with and without radiation in nude mice supporting an adaptive component to the therapeutic response (Figure 6E). Because fractionated RT at lower doses per fraction has been suggested to be more effective for generating adaptive responses, mice were treated with 10 Gy x 3 and 15 Gy x 2 with anti-CD40 and PD-1 Ab to determine if cures could be obtained. This combination resulted in no obvious additive or synergistic effects when adding anti-CD40 and PD-1 in these tumors though the overall response was robust (Figure 6F). Despite evidence of an adaptive immune response with triple combination therapy, complete regression was still not observed. Interestingly, PD-L1 expression was increased by anti-CD40 + RT treatment, but the addition of PD-1 Ab resulted in an increased population of PD-L1- CD45- cells that may support outgrowth of PD-L1 negative cells (Figure 6G).
Complete responses were unable to be obtained in Py8119 tumors, so we next tested the triple combination in 4T1 mammary carcinoma tumors that are highly metastatic, more sensitive to radiation, and more immunogenic. Although these tumors were resistant to combination immunotherapy with anti-CD40 and PD-1 Ab inhibition, complete responses were observed when immunotherapy was combined with a single dose of 20 Gy RT. Mice in all treatment groups had to be euthanized due to the development of lung metastases. However, in the combination therapy group, 40% of 4T1 tumor-bearing animals exhibited a complete response and no evidence of metastases occurred (Figure 6H, I).
CD40 may be a target in immunologically unresponsive human malignancies
In this work we describe that a RT and anti-CD40 combination therapy could generate robust adaptive immune responses in immunologically unresponsive tumors even if complete eradication was not accomplished. We next sought to determine if the transcriptomic phenotypes found in murine tumors are representative of human tumors that are immunologically unresponsive and if CD40 could be a good target in this setting. In the PyMT tumors, GSEA revealed enrichment in interferon signaling and antigen presentation suggesting a greater adaptive immune response in the Py117 tumors. We found a similar pattern of enrichment differentiating melanoma patients that were PD-1 responders and non-responders after analyzing RNAseq data from Hugo et al. using the STAR-DESeq2 pipeline (Figure 6J)(26). Since anti-CD40 can enhance adaptive immune responses and tumor infiltrating leukocytes we sought to evaluate if CD40LG that is primarily presented on activated helper T-cells, CD40, and downstream MAP3K14 kinase expression correlated with immune cell infiltration into the TME (39). We plotted RNA expression of these three genes from breast cancer and melanoma TCGA datasets against relative immune cell infiltration as determined by the TIMER deconvolution algorithm showing a positive correlation with CD8, CD4 and dendritic cell populations in both tumor types (Supplemental Figure 5A, B) (29).
Given that key CD40 pathway genes correlate with immune cell infiltration in melanoma and breast cancer, we next investigated whether the CD40 pathway was enriched in breast cancer patients with longer survival. We compared survival of patients from the TCGA that died within three years of diagnosis (66 patients) vs patients that lived at least three years (429 patients) and found significant enrichment of the CD40 pathway in the latter group confirming favorable association with outcome (Supplemental Figure 6A). Since CD40LG is expressed on helper T-cells and is the only known ligand for CD40 we hypothesized its expression in the TME could predict survival. We observed a correlation with CD40LG with outcome in the OncoLnc pipeline using a multivariate cox regression model (breast p=0.015, melanoma p=0.010, and pancreatic p=0.42). Of the differentially expressed genes CD40LG was high in the rank for breast and melanoma but not pancreatic adenocarcinoma (Supplemental Figure 6B) (30). We also investigated how expression levels of CD40LG related to survival in breast, melanoma, and pancreatic cancer in the TCGA datasets (Supplemental Figure 6C). Representative Kaplan-Meier curves with cut offs at the median CD40LG expression levels are shown for each cancer type and the number of patients at risk with log rank p-value over a broad range of cut offs show that high expression level is positively correlated with improved survival except in pancreatic cancer (Supplemental Figure 6D). Having found that CD40 pathway expression correlates with myeloid cell infiltration and CD40LG correlates with survival, we hypothesized that a high CD40LG/CD40 ratio correlates with immunotherapy response and that high ligand would favor an overall antigen presentation phenotype leading to improved adaptive immunity. We found that before therapy the CD40LG/CD40 ratio was not significantly higher in melanoma patients who ultimately responded to CTLA-4 therapy or PD-1 therapy, but CTLA-4 unresponsive patients had a lower ratio before than after and a trend for higher ratio remained in patients that responded to PD-1 therapy (31). A significant difference in the ratio was observed for on-treatment biopsies and the difference in the ratio was even greater in patients progressing on PD-1 therapy (Supplemental Figure 4E). This observation is consistent comparing pretreatment and on-treatment melanoma patients treated with PD-1 therapy from Riaz et al. as there was a higher CD40LG/CD40 ratio from on-treatment biopsies, not observed from pre-treatment biopsies, in patients that obtained a CR, PR, or SD compared to progressive disease (Supplemental Figure 6F) (32). Though these data are only hypothesis generating, immune resistance and the role of CD40 in antigen presenting cells could play an important role in enhancing immune responses in tumors not responsive to checkpoint immunotherapy.
Discussion
Ionizing radiation can function as an immune stimulator because it can release tumor antigens, HMGB1, calreticulin, and activate the cGAS/STING pathway (40). However, evidence that RT triggers local and systemic immune responses in patients either alone or combined with immunotherapy is minimal except for dramatic responses in a limited number of patients (3, 5, 41–43). In this work, using a model of induced heterogeneity by mixing ratios of immunologically cold tumor cells with responsive cells, we show tumor derived factors impact the immune response to RT and that anti-CD40, which targets myeloid cells, antigen presentation, and innate immune responses is sufficient to overcome limitations of Py8119 cold tumors. The Py8119 tumors have a resistant EMT phenotype, elevated expression of immunosuppressive Axl tyrosine kinase, heavy M2 macrophage infiltration, low levels of dendritic cells, and few infiltrating T-cells (18). The immune stimulating activity of Py117 cells altered the TME, recapitulating many factors targeted by anti-CD40, thereby converting the unresponsive Py8119 tumors to an immune responsive tumor. The combination of CD40 agonist and RT resulted in a 25-fold induction of Ki67 expressing dendritic cells in the draining lymph node three days after treatment, as compared to either no response for RT or 11-fold induction after CD40 alone. This further supports that a constellation of immunosuppressive and immune activating factors can tip the balance toward a better antitumor immune response. Thus, targeting the myeloid compartment will depend greatly upon the specific TME of a given tumor.
There is increasing support for combining RT and anti-CD40 to treat immune unresponsive tumors. In our poorly immunogenic tumors, RT did not provide immune stimulating activity, but targeting the myeloid compartment increased tumor response. This data is consistent with the results from the recently reported Tonic trial that aimed to detect enhanced immunotherapy responses in unirradiated tumors in triple negative breast cancer which showed very little evidence of focal radiation improving systemic responses to nivolumab (44). We show that the immune stimulating effect is robust in the draining lymph node days after treatment despite only modest shifts in the balance of the macrophage phenotype in the TME. Previous studies suggest that total body irradiation with anti-CD40 can lead to enhanced infiltration of adoptively transferred T-cells into pancreatic tumors (45), that single fraction RT with intratumorally anti-CD40 could lead to T-cell responses in unirradiated tumors (46), and that anti-CD40 in combination with RT and immune checkpoint therapy complement each other in resistant pancreatic cancer models (47). Our data complements prior work and proposes a clinical approach to combine anti-CD40 with RT for locally advanced disease to enhance direct antitumor responses that may provide systemic protection if sufficient immunogenicity is generated.
We hypothesized that combining RT with immunotherapy has the greatest potential to treat local disease at high risk for developing metastases, especially when undetectable micrometastatic disease likely exists at the time of treatment. To test this concept, we used the mouse 4T1 syngeneic breast cancer tumor model, which is highly metastatic. A significant portion of mice with primary 4T1 tumors treated with RT, anti-CD40, and PD-1 Ab never developed metastatic disease compared to control mice. This approach could have promise in breast cancer as high dose stereotactic radiation is currently proving to be safe in partial breast irradiation and a single fraction dose escalated neoadjuvant trial have completed and are being planned (NCT03366844, NCT0116220, NCT02685332 and NCT03872505) (48). Testing combination therapies this setting would provide an opportunity to evaluate the efficacy of radiation and immunotherapy combinations to improve progression free survival with a potential impact on local and distant disease. Two additional approaches for combination therapy are to use RT to consolidate minimal residual metastatic sites after initial systemic therapy in metastatic patients or as adjuvant treatment for locally advanced definitive therapy, as reported in the Pacific trial using Durvalumab, a PD-L1 Ab after chemoradiation in stage III lung cancer (1, 49, 50).
Supplementary Material
Translational Relevance.
The majority of tumor are resistant to checkpoint immunotherapy and some key reasons stem from tumor cell intrinsic signaling to set up an unresponsive immunologic microenvironment. Such factors likely limit the generation of antitumor innate and adaptive immune responses after radiation therapy (RT). With a many immunotherapy approaches in development we evaluated an experimental model of induced heterogeneity to assess cellular and transcriptomic factors that influence immune responses to RT. We found that RT responsive cells can induce greater than expected responses to unresponsive tumors through factors that influenced myeloid cell recruitment and phenotype, interferon responses including antigen presentation, and innate immune signaling. This suggested, as we observed that anti-CD40 RT combination can overcome limitations in the unresponsive tumor. This approach and the findings support the role of RT to support antitumor immune responses but effects of RT on an individual microenvironment will be important to better identify ideal combinations.
Acknowledgments
We thank the Stanford Shared FACS Facility, the Stanford Human Immune Monitoring Core, the Stanford Protein and Nucleic Acid Facility, and the UT Southwestern Bioinformatics Core Facility and the fellowship program, Min Kim, and Venkat Malladi for use of their facilities and services. We thank Sharon Clarke and Damiana Chiavolini for assistance with editing the manuscript.
Funding: This work was supported by the ASTRO Resident Seed Grant# RA2014-3 (T.A.A); CPRIT first-time tenure track recruitment award #RR170051 (T.A.A/E.E.), Ruth L. Kirschstein National Research Service Award PA-14-015 Grant# T32 CA 121940 (T.A.A/M.R.); Katherine McCormick Advanced Postdoctoral Fellowship and NIH grant# K99CA201304 (M.R.); NIH Grants CA-67166 and CA-197713, the Silicon Valley Foundation, the Sydney Frank Foundation and the Kimmelman Fund (A.J.G.); NIH grant K08CA88035 (L.G.E.); and the Blue Dot Fund (A.K.)
Footnotes
Data and materials availability: All relevant data is available from the authors. All microarray data is uploaded to GEO accession ID GSE118181. PyMT cells are available through ATCC or MTA with UC San Diego.
References
- 1.Antonia SJ, Villegas A, Daniel D, Vicente D, Murakami S, Hui R, et al. Durvalumab after Chemoradiotherapy in Stage III Non-Small-Cell Lung Cancer. The New England journal of medicine. 2017;377(20):1919–29. [DOI] [PubMed] [Google Scholar]
- 2.Postow MA, Callahan MK, Barker CA, Yamada Y, Yuan J, Kitano S, et al. Immunologic correlates of the abscopal effect in a patient with melanoma. The New England journal of medicine. 2012;366(10):925–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Twyman-Saint Victor C, Rech AJ, Maity A, Rengan R, Pauken KE, Stelekati E, et al. Radiation and dual checkpoint blockade activate non-redundant immune mechanisms in cancer. Nature. 2015;520(7547):373–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brooks ED, and Chang JY. Time to abandon single-site irradiation for inducing abscopal effects. Nature reviews Clinical oncology. 2018. [DOI] [PubMed] [Google Scholar]
- 5.Formenti SC, Rudqvist N-P, Golden E, Cooper B, Wennerberg E, Lhuillier C, et al. Radiotherapy induces responses of lung cancer to CTLA-4 blockade. Nature Medicine. 2018;24(12):1845–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharma P, and Allison JP. Immune checkpoint targeting in cancer therapy: toward combination strategies with curative potential. Cell. 2015;161(2):205–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Demaria S, Coleman CN, and Formenti SC. Radiotherapy: Changing the Game in Immunotherapy. Trends in cancer. 2016;2(6):286–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spranger S, and Gajewski TF. Tumor-intrinsic oncogene pathways mediating immune avoidance. Oncoimmunology. 2016;5(3):e1086862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aguilera TA, and Giaccia AJ. Molecular Pathways: Oncologic Pathways and Their Role in T-cell Exclusion and Immune Evasion- A New Role for the AXL Receptor Tyrosine Kinase. Clin Cancer Res. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woo SR, Fuertes MB, Corrales L, Spranger S, Furdyna MJ, Leung MY, et al. STING-dependent cytosolic DNA sensing mediates innate immune recognition of immunogenic tumors. Immunity. 2014;41(5):830–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deng L, Liang H, Xu M, Yang X, Burnette B, Arina A, et al. STING-Dependent Cytosolic DNA Sensing Promotes Radiation-Induced Type I Interferon-Dependent Antitumor Immunity in Immunogenic Tumors. Immunity. 2014;41(5):843–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Apetoh L, Ghiringhelli F, Tesniere A, Obeid M, Ortiz C, Criollo A, et al. Toll-like receptor 4-dependent contribution of the immune system to anticancer chemotherapy and radiotherapy. Nat Med. 2007;13(9):1050–9. [DOI] [PubMed] [Google Scholar]
- 13.Lugade AA, Moran JP, Gerber SA, Rose RC, Frelinger JG, and Lord EM. Local radiation therapy of B16 melanoma tumors increases the generation of tumor antigen-specific effector cells that traffic to the tumor. J Immunol. 2005;174(12):7516–23. [DOI] [PubMed] [Google Scholar]
- 14.Lee YJ, Auh SL, Wang YG, Burnette B, Wang Y, Meng YR, et al. Therapeutic effects of ablative radiation on local tumor require CD8(+) T cells: changing strategies for cancer treatment. Blood. 2009;114(3):589–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zitvogel L, Apetoh L, Ghiringhelli F, Andre F, Tesniere A, and Kroemer G. The anticancer immune response: indispensable for therapeutic success? The Journal of clinical investigation. 2008;118(6):1991–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zitvogel L, Kepp O, and Kroemer G. Decoding cell death signals in inflammation and immunity. Cell. 2010;140(6):798–804. [DOI] [PubMed] [Google Scholar]
- 17.Bao L, Cardiff RD, Steinbach P, Messer KS, and Ellies LG. Multipotent luminal mammary cancer stem cells model tumor heterogeneity. Breast Cancer Res. 2015;17(1):137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aguilera TA, Rafat M, Castellini L, Shehade H, Kariolis MS, Hui AB, et al. Reprogramming the immunological microenvironment through radiation and targeting Axl. Nat Commun. 2016;7:13898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carvalho BS, and Irizarry RA. A framework for oligonucleotide microarray preprocessing. Bioinformatics. 2010;26(19):2363–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–64. [DOI] [PubMed] [Google Scholar]
- 22.Gu Z, Eils R, and Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32(18):2847–9. [DOI] [PubMed] [Google Scholar]
- 23.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(43):15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, et al. ClueGO: a Cytoscape plugin to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25(8):1091–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hugo W, Zaretsky JM, Sun L, Song C, Moreno BH, Hu-Lieskovan S, et al. Genomic and Transcriptomic Features of Response to Anti-PD-1 Therapy in Metastatic Melanoma. Cell. 2016;165(1):35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Colaprico A, Silva TC, Olsen C, Garofano L, Cava C, Garolini D, et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016;44(8):e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li B, Severson E, Pignon JC, Zhao H, Li T, Novak J, et al. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol. 2016;17(1):174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer research. 2017;77(21):e108–e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anaya J OncoLnc: linking TCGA survival data to mRNAs, miRNAs, and lncRNAs. Peerj Comput Sci. 2016. [Google Scholar]
- 31.Chen PL, Roh W, Reuben A, Cooper ZA, Spencer CN, Prieto PA, et al. Analysis of Immune Signatures in Longitudinal Tumor Samples Yields Insight into Biomarkers of Response and Mechanisms of Resistance to Immune Checkpoint Blockade. Cancer Discov. 2016;6(8):827–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Riaz N, Havel JJ, Makarov V, Desrichard A, Urba WJ, Sims JS, et al. Tumor and Microenvironment Evolution during Immunotherapy with Nivolumab. Cell. 2017;171(4):934–49 e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dewan MZ, Galloway AE, Kawashima N, Dewyngaert JK, Babb JS, Formenti SC, et al. Fractionated but not single-dose radiotherapy induces an immune-mediated abscopal effect when combined with anti-CTLA-4 antibody. Clin Cancer Res. 2009;15(17):5379–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Filatenkov A, Baker J, Mueller AM, Kenkel J, Ahn GO, Dutt S, et al. Ablative Tumor Radiation Can Change the Tumor Immune Cell Microenvironment to Induce Durable Complete Remissions. Clin Cancer Res. 2015;21(16):3727–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vanpouille-Box C, Alard A, Aryankalayil MJ, Sarfraz Y, Diamond JM, Schneider RJ, et al. DNA exonuclease Trex1 regulates radiotherapy-induced tumour immunogenicity. Nat Commun. 2017;8:15618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spitzer MH, Carmi Y, Reticker-Flynn NE, Kwek SS, Madhireddy D, Martins MM, et al. Systemic Immunity Is Required for Effective Cancer Immunotherapy. Cell. 2017;168(3):487–502 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vonderheide RH, and Glennie MJ. Agonistic CD40 antibodies and cancer therapy. Clin Cancer Res. 2013;19(5):1035–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beatty GL, Chiorean EG, Fishman MP, Saboury B, Teitelbaum UR, Sun W, et al. CD40 agonists alter tumor stroma and show efficacy against pancreatic carcinoma in mice and humans. Science. 2011;331(6024):1612–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Elgueta R, Benson MJ, de Vries VC, Wasiuk A, Guo Y, and Noelle RJ. Molecular mechanism and function of CD40/CD40L engagement in the immune system. Immunol Rev. 2009;229(1):152–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Demaria S, Golden EB, and Formenti SC. Role of Local Radiation Therapy in Cancer Immunotherapy. JAMA oncology. 2015;1(9):1325–32. [DOI] [PubMed] [Google Scholar]
- 41.Hiniker SM, Reddy SA, Maecker HT, Subrahmanyam PB, Rosenberg-Hasson Y, Swetter SM, et al. A Prospective Clinical Trial Combining Radiation Therapy With Systemic Immunotherapy in Metastatic Melanoma. International journal of radiation oncology, biology, physics. 2016;96(3):578–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tang C, Welsh JW, de Groot P, Massarelli E, Chang JY, Hess KR, et al. Ipilimumab with stereotactic ablative radiation therapy: Phase I results and immunologic correlates from peripheral T-cells. Clin Cancer Res. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kwon ED, Drake CG, Scher HI, Fizazi K, Bossi A, van den Eertwegh AJ, et al. Ipilimumab versus placebo after radiotherapy in patients with metastatic castration-resistant prostate cancer that had progressed after docetaxel chemotherapy (CA184–043): a multicentre, randomised, double-blind, phase 3 trial. The Lancet Oncology. 2014;15(7):700–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Voorwerk L, Slagter M, Horlings HM, Sikorska K, van de Vijver KK, de Maaker M, et al. Immune induction strategies in metastatic triple-negative breast cancer to enhance the sensitivity to PD-1 blockade: the TONIC trial. Nat Med. 2019;25(6):920–8. [DOI] [PubMed] [Google Scholar]
- 45.Ward-Kavanagh LK, Kokolus KM, Cooper TK, Lukacher AE, and Schell TD. Combined sublethal irradiation and agonist anti-CD40 enhance donor T cell accumulation and control of autochthonous murine pancreatic tumors. Cancer Immunol Immunother. 2018;67(4):639–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yasmin-Karim S, Bruck PT, Moreau M, Kunjachan S, Chen GZ, Kumar R, et al. Radiation and Local Anti-CD40 Generate an Effective in situ Vaccine in Preclinical Models of Pancreatic Cancer. Front Immunol. 2018;9:2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rech AJ, Dada H, Kotzin JJ, Henao-Mejia J, Minn AJ, Twyman-Saint Victor C, et al. Radiotherapy and CD40 Activation Separately Augment Immunity to Checkpoint Blockade in Cancer. Cancer research. 2018;78(15):4282–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rahimi A, Thomas K, Spangler A, Rao R, Leitch M, Wooldridge R, et al. Preliminary Results of a Phase 1 Dose-Escalation Trial for Early-Stage Breast Cancer Using 5-Fraction Stereotactic Body Radiation Therapy for Partial-Breast Irradiation. International journal of radiation oncology, biology, physics. 2017;98(1):196–205 e2. [DOI] [PubMed] [Google Scholar]
- 49.Gomez DR, Blumenschein GR Jr., Lee JJ, Hernandez M, Ye R, Camidge DR, et al. Local consolidative therapy versus maintenance therapy or observation for patients with oligometastatic non-small-cell lung cancer without progression after first-line systemic therapy: a multicentre, randomised, controlled, phase 2 study. The Lancet Oncology. 2016;17(12):1672–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iyengar P, Wardak Z, Gerber DE, Tumati V, Ahn C, Hughes RS, et al. Consolidative Radiotherapy for Limited Metastatic Non-Small-Cell Lung Cancer: A Phase 2 Randomized Clinical Trial. JAMA oncology. 2018;4(1):e173501. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.