Abstract
Actinomycetes bacteria produce diverse bioactive molecules that are useful as drug seeds. To improve their yield, researchers often optimize the fermentation medium. However, exactly how the extracellular chemicals present in the medium activate secondary metabolite gene clusters remains unresolved. BR-1, a β-carboline compound, was recently identified as a chemical signal that enhanced reveromycin A production in Streptomyces sp. SN-593. Here we show that BR-1 specifically bound to the transcriptional regulator protein RevU in the reveromycin A biosynthetic gene cluster, and enhanced RevU binding to its promoter. RevU belongs to the LuxR family regulator that is widely found in bacteria. Interestingly, BR-1 and its derivatives also enhanced the production of secondary metabolites in other Streptomyces species. Although LuxR-N-acyl homoserine lactone systems have been characterized in Gram-negative bacteria, we revealed LuxR-β-carboline system in Streptomyces sp. SN-593 for the production of secondary metabolites. This study might aid in understanding hidden chemical communication by β-carbolines.
Subject terms: Chemical ecology, Mechanism of action, Small molecules, Applied microbiology, Soil microbiology
Introduction
Microorganisms utilize a variety of communication systems that enable them to send and receive chemical signals to one another1. Quorum sensing is one cell-cell communication mechanism. Gram-negative bacteria produce hormone-like signals, such as the release of acyl-homoserine lactones, to control cellular responses2,3. Streptomyces, a soil-dwelling gram-positive bacteria, utilize autoregulatory hormones to produce a variety of secondary metabolites (SMs)4. The classical example of such hormone is A-factor in Streptomyces griseus5, and the structurally diverse autoregulators such as 2-alkyl-4-hydroxymethylfuran-3-carboxylic acids in Streptomyces coelicolor6, PI factor in Streptomyces natalensis7, avenolide in Streptomyces avermitilis8, VB-A in Streptomyces virginiae9, IM-2 in Streptomyces sp. FRI-510, SCB1 in Streptomyces coelicolor11, and SRB1 in Streptomyces rochei12 are also known to promote morphological development and regulating secondary metabolism. These have been extensively reviewed elsewhere13–15. On the other hand, chemical signals derived from extra-species/environmental stimuli such as hormaomycin16, goadsporin17, promomycin18, antibiotic-remodeling compounds (ARCs)19, and rare earth elements20 also induce morphogenesis and SM production in Streptomyces species at higher concentration range compared to autoregulators. Co-culturing Streptomyces with mycolic acid-containing bacteria was found to also induce SM production21. Except for some ARCs, which inhibit fatty acid biosynthesis, the mechanisms of how extracellular chemical signals activate SM biosynthesis have not been clarified.
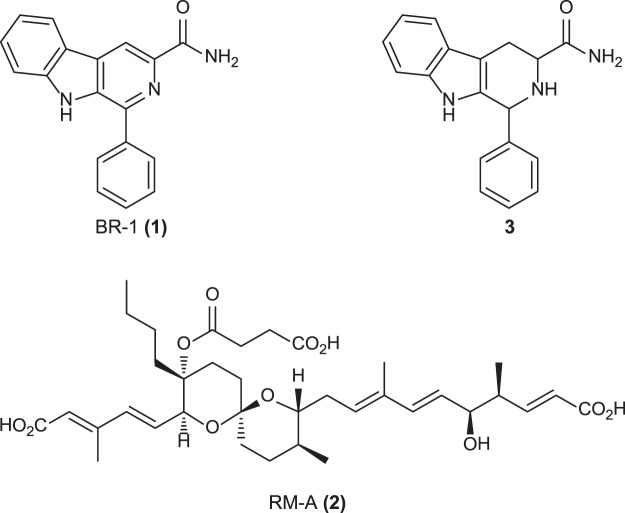
Reveromycin (RM) A was identified from Streptomyces sp. SN-593 as an inhibitor of the mitogenic activity of epidermal growth factor22. It also inhibits bone resorption specifically in osteoclast23. The RM biosynthetic gene cluster consists of 21 genes, including three transcriptional regulators24. Among the regulators, RevU belongs to the LuxR family regulators that harbour a Walker A and B motifs at the N-terminus and a DNA-binding helix-turn-helix domain at the C-terminus25. We found that RM production was triggered by the addition of tomato juice to the culture medium, and uncovered the biosynthetic gene cluster for RM-A production in Streptomyces sp. SN-59324. This led us to speculate that extracellular chemical signals present in nature can enhance SM production. Such chemicals should facilitate the isolation of novel natural products without genetic engineering. However, all attempts to purify chemical signals from tomato did not succeed due to its low-level presence. As an alternative approach, we screened small molecules from the RIKEN Natural Products Depository (NPDepo) consisting of natural products, natural product derivatives, and synthetic chemicals26,27. Then, we successfully identified a β-carboline lead compound that enhanced RM production. Based on the structure-activity relationship study, we succeeded to create BR-1 (1), which induced RM-A (2) production at as little as 0.35 μM concentration in Streptomyces sp. SN-593 (Fig. 1)28. BR-1 enhanced RM production without affecting cell growth and was demonstrated as an external chemical trigger for increasing SM28. In this study, we define BR-1 as biomediator to distinguish them from autoregulators, which are produced intracellularly to regulate functions in the cells that produced them. Based on a chemical biology approach, we identified that the target of β-carboline was a LuxR family transcriptional regulator in the RM gene cluster. Production of autoinducer and subsequent cell responses through LuxR regulators are well characterized in Gram-negative bacteria29,30. Here, we discovered that the hidden chemical signal by β-carboline alkaloids through LuxR family regulator was linked with the production of RMs in Streptomyces sp. SN-593.
Figure 1.
Chemical structures of BR-1 (1), RM-A (2), and tetrahydro form of BR-1 (3).
Results
Enhancement of RM production by BR-1 requires LuxR family regulator RevU
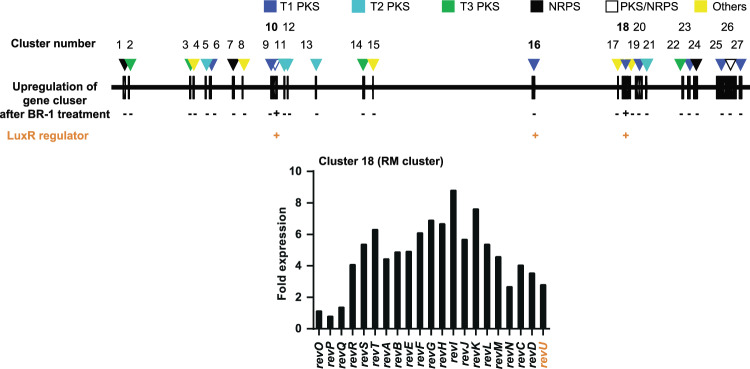
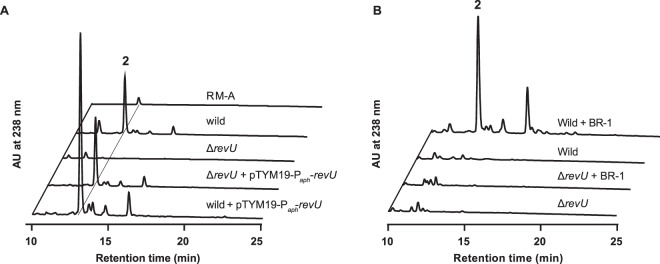
To understand how BR-1 (1) (Fig. 1) signal upregulated the RM-A gene cluster, we first analysed the genome of Streptomyces sp. SN-593 and annotated its gene functions (Table S1). Then we performed RNA-sequence analysis in the presence and absence of BR-1 (Fig. 2). Although we did not observe metabolites, we found that cluster 10, encoding a type I polyketide synthase (PKS), was moderately upregulated after BR-1 treatment (Table S1). mRNA expression of most genes in cluster 18 (RM-A gene cluster) was significantly increased in the presence of BR-1, compared to untreated cells (Fig. 2). Among the 3 regulatory genes, revP, revQ, and revU, the fold-expression values compared to BR-1 untreated cells were 0.8, 1.4, and 2.8 (Table S1). Real-time PCR was also performed to confirm the enhanced expression of the revU gene after BR-1 treatment (Fig. S1A,B). Moreover, revU gene disruption (Fig. S2A,B) resulted in the downregulation of genes involved in RM-A biosynthesis (Fig. S2C,D). We also analysed RM production in the revU gene disruptant. The ΔrevU mutant showed completely abolished RM production, which was recovered by re-introducing the revU gene under the control of the aphII promoter (Fig. 3A). Introducing the revU gene in the wild-type strain led to enhanced production of RMs (Fig. 3A). Next, we examined whether BR-1 treatment could recover the loss of RM production caused by disrupting the revU gene. We found BR-1 treatment did not recover RM production (Fig. 3B), suggesting the importance of RevU in BR-1-mediated RM production. Taken together, these data suggested that RevU might function as a key regulatory molecule linked with the biomediator activity of BR-1.
Figure 2.
Effects of BR-1 on Streptomyces sp. SN-593. Linear map of the 2 producer Streptomyces sp. SN-593 genome with putative 27 gene clusters. Following RNA-sequence analysis, fold-expressions of all gene clusters were analysed (Table S1). Gene-cluster upregulation by BR-1 is indicated by ‘+’ and ‘−’. The presence of LuxR family transcriptional regulator in the cluster is indicated by ‘+’. T1PKS: type I PKS; T2PKS: type II PKS; T3PKS: type III PKS; NRPS: nonribosomal peptide synthetase.
Figure 3.
Involvement of RevU in RM biosynthesis. (A) After culture with RM high producing medium (RM-PM)24, metabolite profiles of wild-type, ΔrevU mutant, and wild-type and ΔrevU mutant transformed with the revU gene controlled by the aphII promoter were analysed. (B) After culture with RM low producing medium (SY-B) in the presence or absence of BR-1, metabolic profiles of wild-type and ΔrevU mutant were analysed.
RevU as the molecular target of BR-1
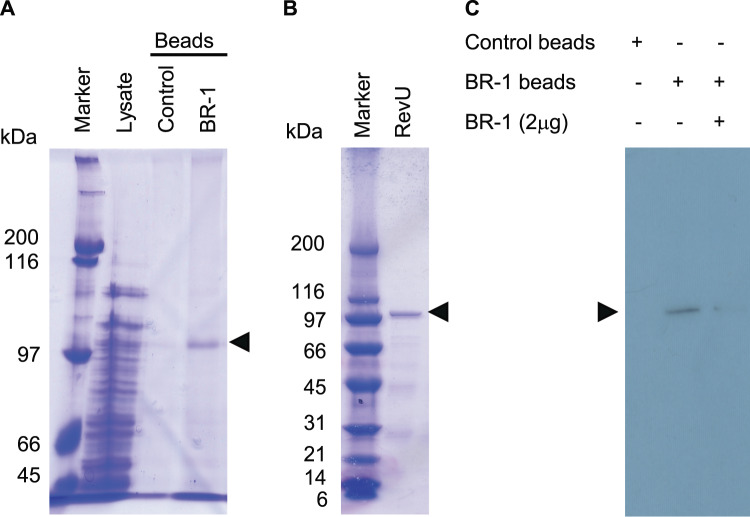
To identify the molecular target of BR-1, we prepared BR-1-conjugated agarose beads using the photoaffinity crosslinking method31,32. The beads were mixed with the cell lysate of Streptomyces sp. SN-593. However, we failed to detect a binding protein because of the low abundance of target protein in the cell lysate. To examine specific interactions between BR-1 and candidate proteins, we expressed RevU protein in Streptomyces lividans strain TK23, and the cell lysate was incubated with the BR-1-conjugated beads. Even in the presence of various proteins in crude extracts, we successfully identified a single protein that bound to the BR-1 beads (Figs. 4A and S3). Matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF/MS) analysis clearly demonstrated that the BR-1-binding protein was RevU (Table S2). Next, we obtained highly purified RevU using the synthetic revU gene optimized for heterologous expression in E. coli (Fig. 4B and Table S3), because RevU expression in S. lividans TK23 was low to obtain a highly purified preparation with high yields. To evaluate the interaction between RevU and BR-1, we tested the competition between BR-1 beads and free BR-1 for RevU binding. We confirmed that His6-tagged RevU bound to BR-1 beads in the absence of free BR-1, but not in the presence of free BR-1 (Fig. 4C), implicating RevU as a molecular target of BR-1. To further characterize the RevU–BR-1 interaction, we conducted surface plasmon resonance (SPR) analysis and observed a quick association and dissociation of BR-1 (Fig. S4A). Interestingly, the equilibrium affinity constant of 1.31 μM calculated for BR-1 was almost identical to its bioactive concentration (Fig. S4B)28. Additionally, we have found that the tetrahydro form of BR-1 (3) (Fig. 1) did not induce the production of RMs28. As expected, the sensorgram for 3 did not reach saturation at a concentration of up to 87 μM, indicative of non-specific binding (Fig. S4C,D).
Figure 4.
Analysis of BR-1-binding proteins. (A) Control and BR-1 beads were mixed with cell lysate (2 mg of protein) from S. lividans TK23 expressing RevU and the bound proteins were analysed by 7.5% SDS-PAGE. Lane 1, protein marker. Lane 2, cell lysate. Lanes 3 and 4, cell lysates incubated with control and BR-1 beads, respectively. (B) SDS-PAGE analysis of the purified His6-tagged RevU. (C) Competition of RevU binding to BR-1 beads using free BR-1. See Fig. S3 for original uncropped images.
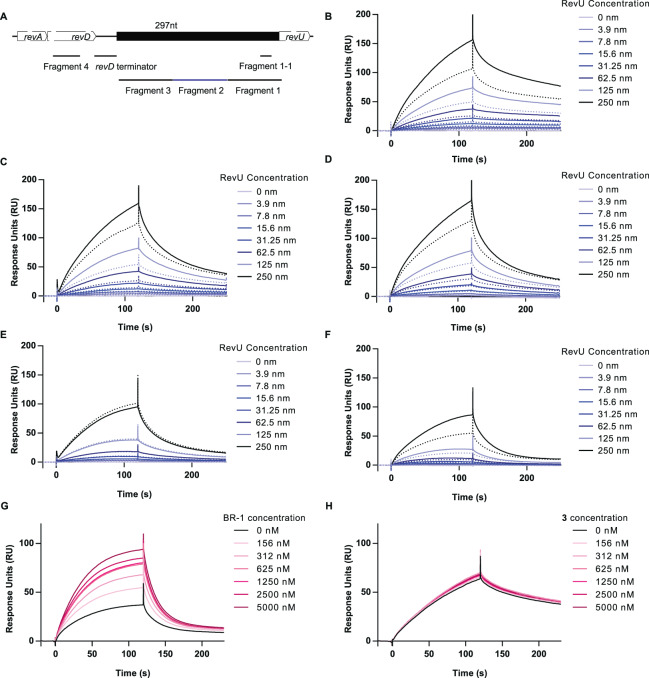
Here, we showed that revU gene expression was enhanced after BR-1 treatment, and BR-1 specifically bound to RevU. Based on all of the observations (Figs. 1–4 and S1–S4), we hypothesized that BR-1 binding to RevU might facilitate binding to the revU gene promoter and activate revU gene expression. To examine whether RevU binding to its promoter region is enhanced by BR-1, we immobilized biotin-labelled, double-stranded DNA fragments on a sensor chip with streptavidin and evaluated RevU-promoter binding activity via SPR. Although RevU bound to all DNA fragments in a concentration-dependent manner, the presence of 1.25 µM BR-1 enhanced the binding activity of RevU to its promoter (Fragment 1–3), but not to another DNA region (Fragment 4) (Figs. 5A–E and S5). Moreover, it is reported that LuxR family regulators positively control secondary metabolism by binding to a conserved 20-bp lux-box CTG-(N10)-CAG in which five of the six bases are essential for LuxR binding33. Upstream of the revU gene, we found a candidate sequence referred to here as fragment 1–1. SPR analysis showed that RevU bound fragment 1-1 in a concentration-dependent manner and the binding was enhanced in the presence of 1.25 µM BR-1 (Fig. 5F). To determine the minimum effective concentration, various concentrations of BR-1 were run over the fragment 1-1-containing chip. We found that 156 nM BR-1 facilitated the DNA binding activity of RevU and the EC50 of BR-1 was 182 nM (Fig. 5G). The EC50 value was about half the concentration required for inducing RM production (0.35 µM)28. We have found that the tetrahydro form of BR-1 (3) did not induce the production of RMs28. Consistently, 3 did not enhance RevU binding to DNA (Fig. 5H). Taken together, these data suggested that activation of the revU gene was triggered by the binding of BR-1 to RevU. Interestingly, we could not find a lux-box sequence in Fragment 2 and 3. An exploration and detail characterization of RevU binding sequence will be our future study.
Figure 5.
SPR analysis of the DNA-binding activity of RevU in the presence of BR-1. (A) Schematic representation of the revU promoter region and DNA fragments used for evaluating the RevU promoter–BR-1 interaction. Fragments 1–3, 99 bp in length, are located immediately upstream of the revU start codon. Fragment 4 (50-bp) located on revD coding sequence (GATTATGCGTCGCATTCGGTGTTTGTGGAGTTGATCGAGGATCGGGTTCT) was used for negative control. The underlined sequences were also found in another part of revA and revD sequence). The revD terminator sequence (CACCCAGCCCTCCCGCGGGAGCCGCCCGGCTCCCGCGGAAGGCGCCCGCG) lies upstream of fragment 3. Fragment 1-1 (22 bp: ACGCCGCAACGACCAACAGAGG) contains the putative lux-box sequence. Blank-subtracted SPR sensorgram showing the binding of biotin-labelled DNA fragment 1 (B), fragment 2 (C), fragment 3 (D), fragment 4 (E), and fragment 1-1 (F) to RevU in the presence (solid lines) or absence (dotted lines) of 1.25 µM BR-1. RevU was injected at various concentrations across the chip surface. Blank-subtracted SPR sensorgram showing the binding of biotinylated DNA fragment 1-1 to RevU (125 nM). Various concentration of BR-1 (G) and 3 (H) were tested. Fold changes in response unit BR-1 (+/−) is shown in Fig. S5.
Moreover, to elucidate whether RevU-BR-1 complex directly enhanced the expression of rev structural genes or not, we searched for lux-box like sequences33 in each operon of rev cluster and found putative sequences in the upstream regions of revA, revB, revJ, revK, revT, and revU genes (Fig. S6A). We evaluated the binding of RevU to these regions in the presence and absence of BR-1using Bio-Layer Interferometry (Fig. S6B–H). We found that the addition of BR-1 resulted in enhanced binding of RevU to three putative lux-boxes for revB, revK, and revU genes. On the other hand, the enhanced RevU binding was not observed for revA, revJ, and revT genes, suggesting that revB, revK, and revU are mainly regulated through RevU.
β-carbolines enhanced SM biosynthesis on Streptomyces sp. RK95-74 and Streptomyces sp. RK10-A626
We speculated that BR-1 and its derivatives might have an ability to induce SM in other Streptomyces. To test this hypothesis, Streptomyces sp. RK95-74 was cultured in the presence or absence of BR-1 or its derivatives. Interestingly, we found enhanced production of neomediomycin B, with a molecular mass of 1193 from biomediator treated Streptomyces sp. RK95-74 (Fig. S7)34. Similar to Streptomyces sp. SN-59328, the optimum β-carboline structures responsible for biomediator activity varied (Fig. S8A–C). We also cultured other Streptomyces spp. in the presence of β-carbolines and analysed the metabolites profiles using LC-MS. Treatment of Streptomyces sp. RK10-A626 with BR-1 resulted in the enhanced production of unidentified SM with m/z ratios ranging from 1032 to 1291 (Fig. S8D).
We also tested the production of stambomycins from Streptomyces ambofaciens in the presence of BR-1 or its derivatives because stambomycins from S. ambofaciens were obtained by expression of a LuxR family regulator (SamR0484) associated with the sleeping gene cluster35. However, the treatment of BR-1 derivatives did not result in the production of stambomycins. The alignment amino acid sequences between SamR0484 and RevU showed 34% identity and 47% similarity with a 7% gap, and phylogenetic analysis indicated that SamR0484 did not clade with RevU (Fig. S9). Because of the structural diversity of LuxR regulators, we expect that β-carbolines chemical signals are functional for a limited number of Streptomyces strains.
Discussion
Understanding of chemical signals and the related mode of action is of profound importance. In this paper, we discovered that the β-carboline compound (BR-1) enhanced the production of RMs from Streptomyces sp. SN-593 through selective activation of the rev gene cluster (Fig. 2). Based on revU gene disruption and RT-qPCR analysis, we found that RevU, LuxR family regulator, regulates structural genes involved in RM-A biosynthesis (Figs. 3 and S2). Moreover, BR-1 enhances RevU binding to its own promoter and leads to self-activation of revU gene expression (Fig. 5). We also demonstrated that RevU-BR-1 complex is involved in the enhanced expression of rev structural genes such as revB and revK, but not for revA, revJ, and revT genes (Fig. S6).
RevU contains an N-terminal, ATP-binding region with Walker A and B motifs and a C-terminal DNA-binding region (helix-turn-helix domain)25. LuxR family regulators found in Gram-negative bacteria respond to acyl-homoserine lactone-type compounds, and ligand binding at their N-terminal regions facilitates their dimerization36. In the case of Streptomyces sp. SN-593, the binding of BR-1 to RevU may also cause structural changes or dimerization to facilitate RevU binding to its promoter and accelerate gene expression. In this paper, we also showed that BR-1 enhanced the production of SMs from Streptomyces sp. RK95-74 and Streptomyces sp. RK10-A626 (Figs. S7 and S8). To elucidate a mode of action of BR-1 in these strains, genome sequence, transcriptome analysis after treatment of BR-1, and gene disruption of transcriptional regulators will be our future study. Moreover, it is of interest to perform extended screening of actinomycetes which show the ability to enhance SM production by treatment of BR-1 and its derivatives because there are many RevU homologs in the database (Fig. S9).
Natural soil environments consist of diverse plants and microbial communities, where the presence of a wide range of chemical communications may trigger a variety of responses, including the production of bioactive molecules. For instance, in response to flavonoids released from plant roots, Rhizobium induces the expression of nod genes encoding enzymes for Nod factors37. Then, it induced the development of nodules in plant roots to promote growth. Plants provide a variety of organic compounds derived from photosynthesis in the rhizosphere38,39, a process known as rhizodeposition. Subsequently, microorganisms may provide benefits for plant growth40. β-carboline alkaloids widely found in plants are also isolated from fungi, marine tunicate, and actinomycetes41–43. However, their physiological roles in nature are not well understood. Interestingly, RMs produced from Streptomyces sp. SN-593 in response to β-carboline treatment have the antifungal activity against plant pathogenic fungi22,44. The metabolites may protect the producing strain itself and/or plants. In this study, our findings might shed light on hidden roles of β-carboline in nature. Searching for natural counterparts of β-carboline alkaloids and their relationship with symbiosis will be the subjects of future studies.
To summarize, we discovered that β-carboline-based chemical signals enhanced the production of SMs in some Streptomyces spp. In the case of Streptomyces sp. SN-593, we demonstrated that the activation of a LuxR family regulator was responsible for the phenomena. In general, LuxR regulators found in Gram-negative bacteria respond to acyl-homoserine lactone. Data generated in this study suggested that previously unknown interactions occur between β-carboline compounds and the LuxR regulator in Streptomyces sp. SN-593, a Gram-positive bacterium. Hidden chemical communication signals similar to β-carboline might act to induce secondary metabolites and our study may open avenues for enhanced production of natural products without genetic engineering.
Methods
Bacterial strains, plasmids, and culture medium
The bacterial strains and plasmids used are shown in Table S3. Luria–Bertani broth (Nacalai Tesque, Japan) was used to culture E. coli. The media were supplemented with appropriate antibiotics (100 µg ml−1 ampicillin, 50 µg ml−1 kanamycin, 50 µg ml−1 streptomycin, 100 µg ml−1 spectinomycin; 30 µg ml−1 chloramphenicol; 50 µg ml−1 ribostamycin; 5 µg ml−1 carumonam). Spores of Streptomyces species were prepared on MS plates and cultured using several media, including synthetic medium45, SK224, MS24, RM high producing medium (RM-PM)24, YMG46, SY46, and RM low producing medium (SY-B) (1% soluble starch and 0.1% yeast extract).
Metabolite profiles of RMs from Streptomyces sp. SN-593
Wild-type Streptomyces sp. SN-593, the ΔrevU mutant, and strains transformed with pTYM-Paph-revU (Table S3) were cultured in SY medium (70 ml) at 28 °C in a cylindrical 500-ml flask at 150 rpm for ~2 days. The pre-culture medium was supplemented with kanamycin (1 µg ml−1) or thiostreptone (25 µg ml−1). One-millilitre cultures were added to RM-PM (70 ml) in 500-ml cylindrical flasks and cultured at 28 °C for 5 days. RM produced in the culture broth was extracted in an equal volume of acetone. After sonication and centrifugation at 5,000 × g for 10 min, the supernatant (8 ml) was concentrated to remove the acetone. The pH of the aqueous extract was adjusted to 4 with acetic acid and extracted twice with an equal volume of ethyl acetate. The dried residue was dissolved in methanol (1.2 ml), and 1 µl was analysed by LC-MS.
Culture conditions and metabolite extraction from Streptomyces species after treatment with β-carboline compounds
Streptomyces sp. SN-593, the ΔrevU mutant, and Streptomyces sp. RK10-A626 were cultured in 70 ml SY medium at 28 °C for 2 days. One ml of each culture was diluted in 100 ml SY-B medium, and 1-ml aliquots were cultured in 2.2-ml wells of a 96-well plate (4titude, reorder # 4ti-0130) for 3 days in the presence or absence of 1 μg ml−1 β-carboline compounds. Streptomyces sp. RK95-74 was cultured for 2 days at 28 °C in 70 ml of YMG medium. Then, 1 ml culture was diluted into 100 ml of YMG medium. The 1-ml aliquots were cultured in separate 2.2-ml wells for 3 days in the presence or absence of β-carboline compounds. Subsequently, the broth was mixed with 0.5 ml acetone, sonicated, and centrifuged at 36,200 × g for 10 min. Metabolites in the supernatant were analysed by LC-MS or UPLC-MS. The treatment of 1 μg ml−1 carboline compounds had no obvious effect on cell growth28.
LC-MS analysis
Analysis of metabolites from Streptomyces sp. SN-593, Streptomyces sp. RK95-74, and Streptomyces sp. RK10-A626 was performed by ESI-MS using a Waters Alliance high-performance liquid chromatography (HPLC) system equipped with a mass spectrometer (Q-Trap; Applied Biosystems)24. The HPLC system consisted of an XTerra®MSC18 (5-μm, 2.1 mm internal diameter × 150 mm length) column maintained at 0.2 ml min−1. Solvent A was 0.05% aqueous formic acid and solvent B was acetonitrile. The sample was injected into the column after pre-equilibration with 30% solvent B; the column was developed with a linear gradient from 30% to 100% solvent B over 20 min and maintained in 100% solvent B for 20 min. Mass spectra were collected in ESI-negative and -positive modes. Analysis of metabolites from Streptomyces sp. RK95-74 was also performed by UPLC/ESI-MS analysis. An API3200 (Applied Biosystems, CA, USA) was connected to a Waters ACQUITY UPLC-H-class instrument with a photodiode-array detector (Waters, MA, USA) and an ACQUITY UPLC® BEH C18 (1.7-µm, 2.1-mm internal diameter × 50 mm length; Waters). The elution conditions were as follows: Streptomyces sp. RK95-74, 0.6 ml min−1; solvent A: water containing 0.05% formic acid; and solvent B: acetonitrile. After sample injection into a column equilibrated with 30% solvent B, the column was developed with a linear gradient of 30–100% solvent B over 2.5 min and maintained at 100% solvent B for 5 min.
Chemical synthesis of β-carboline derivatives
All the β-carboline derivatives used in this manuscript are listed in Figs. 1 and S8A. The synthesis and confirmation of the structure was performed as explained28.
Genome-sequencing analysis
DNA isolation and manipulation were performed as described47. Genomic DNA was isolated from Streptomyces sp. SN-593 and genome-sequence analysis was performed as described24. Briefly, the genomic DNA was cloned into vectors with an average insert size of 2–5 kb and 40 kb, respectively. End sequencing was performed using BigDye terminator 3.1 (Applied Biosystems). Sequencing product were analysed on an automated 3730 × 1 capillary sequencer (Applied Biosystems). Low-quality regions in the assembly were resequenced.
RNA-sequence analysis
Streptomyces sp. SN-593 was cultured in the presence or absence of 3.5 µM BR-1. After 1 day, cultures from 8-well plates were harvested by centrifugation at 5,000 × g for 10 min. Total RNA was isolated using the RNeasy Protect Bacteria Mini Kit (Qiagen). The integrity and purity of the RNA was analysed using an Agilent Bioanalyzer. Five micrograms of total RNA was subjected to rRNA depletion using the RiboZero Meta-Bacteria Kit (Epicentre Biotechnologies, Madison, WI, USA), and Complementary DNA (cDNA) was generated using the NEBNext mRNA Sample Prep Kit (New England Biolabs, Ipswich, MA USA). cDNA was subjected to Illumina library preparation using NEBNext reagents (New England BioLabs, Ipswich, MA USA). The quality, quantity, and size distribution of the Illumina libraries were determined using an Agilent Bioanalyzer 2100. The libraries were submitted for Illumina HiSeq 2000 sequencing following a standard protocol. Paired-end 90- or 100-nucleotide reads were generated, and the data quality was checked using FastQC software (Babraham Institute, Cambridge, UK). The obtained data were analysed using CLC Genomics Workbench Server (Qiagen). Reads per kilobase per million mapped sequence reads (RPKM) values were used to determine gene-expression levels. Fold-expression levels were calculated as the RPKM in the presence of BR-1/the RPKM in the absence of BR-1.
Quantitative PCR (qPCR) analysis
Total RNA was isolated using the RNeasy Protect Bacteria Mini Kit (Qiagen). RNA samples were treated with the RNase-Free DNase Set (Qiagen) and the RNeasy MinElute Cleanup Kit (Qiagen). The concentration and RNA integrity number (average: 8) were checked using an Agilent Bioanalyzer 2100. First-strand cDNA was synthesized using random hexamers and Superscript III reverse transcriptase (Invitrogen), and the cDNA was subsequently diluted with 5 volumes of nuclease-free water. qPCR amplification mixtures contained template cDNA, SsoFast™ EvaGreen® Supermix (BioRad), and forward and reverse primers (Table S4). Reactions were run on a CFX96 Real-Time PCR Analysis System (BioRad). Thermocycling proceeded at 95 °C for 30 s and 40 cycles at 95 °C for 5 s and 61 °C for 10 s.
Gene disruption and complementation of revU
To construct the revU gene-disruption plasmid, a 7,784-bp region encoding the revU gene was amplified from pCC1FOSTM clone (30B04)24 using the primers, 5′-CCCAAGCTTGGATCACCTTGCTGGACCTTGGTG-3′ and 5′-CCCAAGCTTGGAACGCGGTCCTGACGATCGAGC-3′. The underlined sequences indicate HindIII site. After enzyme digestion, the insert was ligated into the HindIII site of the pIM vector46 to construct pIM-revU. To create the disruption cassette, the aphII gene was amplified from the pKD13 plasmid48 using the primers, 5′-GCGGCGATGAAGATCCTCACCGAGTCCCGGCTGATCGAGATTCCGGGGATCCGTCGACC-3′ and 5′-GATCTCGACGCCCCATGCCTCCACCGACATACTGCGCAGTGTAGGCTGGAGCTGCTTC-3′). The underlined text (39-bp) show sequences homologous to the target-DNA region. The revU gene was replaced by the aphII gene using λ-red-mediated recombination to construct the gene-disruption plasmid pIM-ΔrevU. The vector was then transformed into Streptomyces sp. SN-593 by conjugal gene transfer24,49. Gene disruption was confirmed by Southern hybridization. To complement the gene disruption, the revU gene was amplified from the 30B04 fosmid clone using the primers, 5′-CGCGGATCCATGCTGGAGCGCGACCAATGCCTC-3′ and 5′-CCCAAGCTTTCAGGACGCGAGGCTGGCCGGCA-3′. The underlined sequences indicate BamHI and HindIII sites, respectively. After enzyme digestion, the insert was ligated into the BamHI/HindIII sites of pTYM-Paph to construct pTYM-Paph-revU. The plasmid was introduced into the revU gene disruptant and wild-type Streptomyces sp. SN-593. The selection of exconjugants was performed on SY agar plates containing 25 µg ml−1 thiostreptone and 5 µg ml−1 carumonam.
Southern hybridization
Thirty micrograms of genomic DNA was digested with 50 units NcoI. The digested DNA (1 μg) was loaded onto a 0.9% agarose gel, run in Tris-acetate-ethylenediaminetetraacetic acid (EDTA) buffer, and stained with ethidium bromide. The DNA was then transferred to a Hybond N+ membrane (GE Healthcare) and fixed by baking overnight at 80 °C. The DNA probe was amplified from the gene-disruption plasmid (pIM-ΔrevU) by PrimeSTAR®HS DNA polymerase (TaKaRa) at 98 °C for 10 s; 25 cycles of 98 °C for 10 s, 65 °C for 5 s, and 68 °C for 3 min; and a final extension at 68 °C for 6 min, using the oligonucleotide primers RevU-F (5′-ATGCTGGAGCGCGACCAATGC-3′) and ORF1-F (5′-ATGGTCCGCGAACTCGCGTAC-3′). The amplified DNA (4,249 bp) was purified by QIAquick® Gel Extraction Kit (Qiagen) and labelled for Southern hybridisation with AlkPhos Direct Labeling Reagents (GE Healthcare). The hybridized DNA was detected using CDP-StarTM reagent (GE Healthcare). The chemiluminescent signal on the membrane was exposed to X-ray film (Kodak).
Vector construction, transformation, and expression of RevU in S. lividans TK23
The revU gene was amplified from the 30B04 fosmid clone using PrimeSTAR®HS DNA polymerase (TaKaRa) and thermocycling at 98 °C for 10 s and 25 cycles of 98 °C for 10 s, 62 °C for 5 s, and 68 °C for 1.5 min. The amplification primer sequences were: 5′-GGAATTCCATATGCTGGAGCGCGACCAATGCCTC-3′ and 5′-CCGCTCGAGTCAGGACGCGAGGCTGGCCGGCA-3′. The underlined sequences indicate NdeI and XhoI sites, respectively. After enzyme digestion, the insert was ligated into the pET28b(+) vector to construct pET28b(+)-revU. The β lactamase gene in the pWHM3 vector was replaced with the aphII gene using λ Red recombination to form the pWK vector. Then, the tipA promoter (PtipA) was inserted into the EcoRI and BamHI sites to construct pWK-PtipA50. To facilitate protein purification, the revU gene containing a His-tag was amplified from pET28b (+)-revU. The primers used were 5′-CGCGGATCCATGGGCAGCAGCCATCATCAT-3′ and 5′-CCCAAGCTTAGCCGGATCTCAGTGGTGGTG-3′. The underlined sequences indicate the BamHI and HindIII sites. After enzyme digestion, the insert was ligated into pWK-PtipA to generate pWK-PtipA-revU. The vector was introduced into S. lividans TK23 using a polyethylene glycol-mediated protoplast method51. The resulting transformants were grown in 70 ml SK2 medium in a 500-ml cylindrical flask at 28 °C at 150 rpm. RevU expression was induced in the presence of 50 μg ml−1 thiostrepton. Cells were harvested by centrifugation at 5,000 × g for 10 min and sonicated in 5 ml binding buffer (50 mM Tris-HCl [pH 7.5], 500 mM NaCl, 10% glycerol, and 5 mM imidazole) containing a protease inhibitor cocktail (Roche). The cell lysate was centrifuged at 36,200 × g for 10 min at 4 °C, and the supernatant was used for BR-1 bead-binding experiments.
revU gene expression in E. coli and purification
The synthetic revU gene (revUsyn) in the pMA-RQ vector (Life Technologies) was ligated into the NdeI and XhoI sites of pColdI (TaKaRa) to create pColdI-revUsyn, and transformed into E. coli BL21 cells. The cells were cultured overnight in LB medium containing 100 µg ml−1 ampicillin. The preculture was added to 100 ml of LB-ampicillin (100 µg ml−1) medium in a 500-ml cylindrical flask. The cells were cultured at 37 °C until the OD600 reached 0.2. The cells were cooled on ice for 30 min, and isopropyl β-D-1-thiogalactopyranoside was added at to 0.5 mM. The cells were incubated at 18 °C for 24 h and collected by centrifugation at 5,000 × g for 10 min. Purification of the His6-tagged RevU protein was performed as described46. The purity was confirmed by 5–20% SDS-PAGE.
BR-1 chemical bead preparation and protein binding detection
BR-1 and control beads were prepared as described31,32. Briefly, 100 μl of photoaffinity linker-coated agarose beads was suspended in 200 μl of isopropyl alcohol and dried in vacuo. Methanol (control beads) or BR-1 solution (1 mg/150 μl methanol) was added, and the mixture was dried in vacuo. The beads were irradiated at 365 nm using an ultraviolet (UV)-activated crosslinker (4 J/cm2). The irradiated beads were sequentially washed in 50% methanol, methanol, DMSO, and methanol (3 × 400 μl/wash) and suspended in 200 μl PBS (1.8 mM KH2PO4, 10 mM Na2HPO4•12H2O, 137 mM NaCl, and 2.7 mM KCl). To examine protein binding, 25 µl control beads or BR-1-conjugated beads was mixed with cell lysates (2 mg protein) from S. lividans TK23 cells expressing the RevU protein and incubated overnight at 4 °C in 1 ml binding buffer. Then, the beads were washed 5 times with binding buffer containing 0.1% Tween-20. The bound proteins were eluted with sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer. The eluted protein was analysed by 7.5% SDS-PAGE. To identify proteins, MALDI-TOF/MS analysis was performed using an Ultraflex instrument (Bruker Daltonics).
Competition assay
The purified His6-tagged RevU protein (100 ng) was dissolved in 500 µl binding buffer containing 20 μg ml−1 bovine serum albumin. The solution was mixed with 10 µl of control and BR-1 beads in the presence or absence of 2 µg free BR-1. After overnight incubation at 4 °C, the beads were washed 5 times with binding buffer containing 0.1% Tween-20, and the bound proteins were eluted in SDS-PAGE buffer. The eluted proteins were separated by 5–20% SDS-PAGE and transferred to a nitrocellulose membrane. The membrane was treated with a mouse anti-His antibody (GE healthcare) and a horseradish peroxidase-conjugated AffiniPure goat anti-mouse IgG (H + L) (Jackson ImmunoResearch). After treatment with PierceTM ECL Western Blotting Substrate (ThermoFisher), the membrane was exposed to X-ray film (Kodak).
SPR analysis
SPR measurements were performed on a BIAcore T200 instrument (GE Healthcare) maintained at 25 °C. The SPR buffer (50 mM Tris-HCl [pH 7.5], 500 mM NaCl, and 0.05% (v/v) Tween-20) was used to evaluate BR-1 binding to RevU. The purified His6-tagged RevU protein was diluted to 5 μg ml−1 in running buffer and immobilized on a Series S Sensor Chip NTA (GE Healthcare) at 5 μl min−1 for 600 s, with a 600-s stabilization period. β-carboline compounds were diluted in SPR buffer and injected over the sensor chip for 60 s at 30 μl min−1. Dissociation proceeded for 120 s. RevU binding to the RevU promoter was measured in 10 mM HEPES (pH 7.4), 3 mM EDTA, 150 mM NaCl, and 0.05% (v/v) Tween-20. Double-stranded, 5′ biotinylated DNA was synthesized by Hokkaido System Sciences. The labelled promoter DNA was diluted to 30 nM in SPR buffer and immobilized on a Series S Sensor Chip SA (GE Healthcare), with immobilized streptavidin, at 15 μl min−1 for 180 s. RevU, dissolved in 10 mM HEPES (pH 7.4) and 3 mM EDTA, was diluted in SPR buffer. Various RevU concentrations (ranging from 3.9 nM to 250 nM) were injected over the surface for 120 s at 15 μl min−1. Dissociation occurred for 120 s. Regeneration of the SA sensor surface was performed with 2 M NaCl for 30 s at 30 μl min−1. The effect of BR-1 on RevU–RevU promoter binding was studied by injecting 1.25 µM BR-1 pre-mixed with various RevU concentrations. Sensorgrams were analysed using BiaEvaluation software (GE Healthcare), and affinity constants were calculated using sample values obtained by subtracting blank values.
Bio-Layer Interferometry analysis
BLI measurements were performed on a BLItz system (ForteBio), a label-free technology that analyses the interference pattern of white light reflected from two surfaces, maintained at room temperature. The BLItz buffer contained 10 mM HEPES (pH 7.3), 3 mM EDTA (pH 8.0) (both diluted from 1 M concentrate); 150 mM NaCl, and 0.05% (v/v) Tween-20. The labelled promoter DNA was diluted to 200 nM in BLItz buffer and immobilized on a streptavidin biosensor, followed by 120 s each of association and dissociation with 125 nM RevU in the presence and absence of 5 µM BR-1.
Supplementary information
Acknowledgements
We thank A. Toyoda and Y. Sakaki for genome sequencing; M. Muroi and N. Dohmae for proteomic analysis; T. Onuki and S. Matsuoka for help in SPR experiments; T. Nogawa and M. Uramoto for metabolite analysis; Y. Kondo and Ms. K. Honda for preparation of chemical beads; and E. Oowada, H. Takagi, and A. Okano for technical assistance. This work was supported by the JSPS KAKENHI (Grant 24658088), JSBBA Innovative Research Program Award, and in part by the Program for Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry of the Science and Technology Research Promotion Program for Agriculture, Forestry, Fisheries and Food Industry.
Author contributions
S.P. and S.T. designed the experiments, performed gene transformations, vector constructions, and southern hybridizations and wrote the manuscript. S.P. performed heterologous expression, chemical beads experiments, and the SPR study. S.T., N.K. and J.I. performed gene annotations for Streptomyces sp. SN-593. T.S. and T.H. performed chemical synthesis of β-carboline compounds. N.K. and S.P. performed reverse transcriptase-PCR analysis. S.P and H.H. performed the BLI experiments. H.O. and S.T. integrated the overall research project. All authors discussed the results and commented on the manuscript.
Data availability
The data that support the finding of this study are available from the corresponding author upon request. Sequence data have been deposited to DNA data bank (accession number AP018365 RVR, AP018366 pRVR1, and AP018367 pRVR2). The RNA-seq data were deposited in SRA database with accession code SRR6715512 and SRR6715514. The data that support the finding of this study are available from the corresponding authors upon request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Hiroyuki Osada, Email: cb-secretary@ml.riken.jp.
Shunji Takahashi, Email: shunjitaka@riken.jp.
Supplementary information
is available for this paper at 10.1038/s41598-020-66974-y.
References
- 1.Bassler BL, Losick R. Bacterially speaking. Cell. 2006;125:237–246. doi: 10.1016/j.cell.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 2.Bassler BL. Small talk: cell-to-cell communication in bacteria. Cell. 2002;109:421–424. doi: 10.1016/S0092-8674(02)00749-3. [DOI] [PubMed] [Google Scholar]
- 3.Waters CM, Bassler BL. Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell. Dev. Biol. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 4.Daniel-Ivad M, Pimentel-Elardo S, Nodwell JR. Control of specialized metabolism by signaling and transcriptional regulation: opportunities for new platforms for drug discovery? Annu. Rev. Microbiol. 2018;72:25–48. doi: 10.1146/annurev-micro-022618-042458. [DOI] [PubMed] [Google Scholar]
- 5.Horinouchi S, Beppu T. Hormonal control by A-factor of morphological development and secondary metabolism in Streptomyces. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2007;83:277–295. doi: 10.2183/pjab.83.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corre C, Song L, O’Rourke S, Chater KF, Challis GL. 2-Alkyl-4-hydroxymethylfuran-3-carboxylic acids, antibiotic production inducers discovered by Streptomyces coelicolor genome mining. Proc. Natl. Acad. Sci. USA. 2008;105:17510–17515. doi: 10.1073/pnas.0805530105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Recio, E., Colinas, Á., Rumbero, A., Aparicio, J. F. & Martin, J. F. PI factor, a novel type quorum-sensing Inducer elicits pimaricin production in Streptomyces natalensis. J. Biol. Chem.279, 41586–41593 (2004). [DOI] [PubMed]
- 8.Kitani S, et al. Avenolide, a Streptomyces hormone controlling antibiotic production in Streptomyces avermitilis. Proc. Natl. Acad. Sci. USA. 2011;108:16410–16415. doi: 10.1073/pnas.1113908108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamada Y, Sugamura K, Kondo K, Yanagimoto M, Okada H. The structure of inducing factors for virginiamycin production in Streptomyces virginiae. J. Antibiot. 1987;40:496–504. doi: 10.7164/antibiotics.40.496. [DOI] [PubMed] [Google Scholar]
- 10.Sato K, Nihira T, Sakuda S, Yanagimoto M, Yamada Y. Isolation and structure of a new butyrolactone autoregulator from Streptomyces sp. FRI-5. J. Ferment. Bioeng. 1989;68:170–173. doi: 10.1016/0922-338X(89)90131-1. [DOI] [Google Scholar]
- 11.Takano E, et al. Purification and structural determination of SCB1, a γ-Butyrolactone that elicits antibiotic production in Streptomyces coelicolor A3(2) J. Biol. Chem. 2000;275:11010–11016. doi: 10.1074/jbc.275.15.11010. [DOI] [PubMed] [Google Scholar]
- 12.Arakawa K, Tsuda N, Taniguchi A, Kinashi H. The butenolide signaling molecules SRB1 and SRB2 induce lankacidin and lankamycin production in Streptomyces rochei. Chembiochem. 2012;13:1447–1457. doi: 10.1002/cbic.201200149. [DOI] [PubMed] [Google Scholar]
- 13.Takano, E. γ-Butyrolactones: Streptomyces signalling molecules regulating antibiotic production and differentiation. Curr. Opin. Microbiol.9, 287–294 (2006). [DOI] [PubMed]
- 14.Chater, K. F., Biró, S., Lee, K. J., Palmer, T. & Schrempf, H. The complex extracellular biology of Streptomyces. FEMS Microbiol. Rev. 34, 171–198 (2010). [DOI] [PubMed]
- 15.Niu G, Chater KF, Tian Y, Zhang J, Tan H. Specialised metabolites regulating antibiotic biosynthesis in Streptomyces spp. FEMS Microbiol. Rev. 2016;40:554–573. doi: 10.1093/femsre/fuw012. [DOI] [PubMed] [Google Scholar]
- 16.Andres N, Wolf H, Zahner H. Hormaomycin, a new peptide lactone antibiotic effective in inducing cytodifferentiation and antibiotic biosynthesis in some Streptomyces species. Z Naturforsch C. 1990;45:850–855. doi: 10.1515/znc-1990-7-817. [DOI] [Google Scholar]
- 17.Onaka H, Tabata H, Igarashi Y, Sato Y, Furumai T. Goadsporin, a chemical substance which promotes secondary metabolism and morphogenesis in Streptomycetes I. purification and characterization. J. Antibiot. 2001;54:1036–1044. doi: 10.7164/antibiotics.54.1036. [DOI] [PubMed] [Google Scholar]
- 18.Amano S, et al. Promomycin, a polyether promoting antibiotic production in Streptomyces spp. J. Antibiot. 2010;63:486–491. doi: 10.1038/ja.2010.68. [DOI] [PubMed] [Google Scholar]
- 19.Craney A, Ozimok C, Pimentel-Elardo S, Capretta A, Nodwell J. Chemical perturbation of secondary metabolism demonstrates important links to primary metabolism. Chem. Biol. 2012;19:1020–1027. doi: 10.1016/j.chembiol.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 20.Kawai K, Wang G, Okamoto S, Ochi K. The rare earth, scandium, causes antibiotic overproduction in Streptomyces spp. FEMS Microbiol. Lett. 2007;274:311–315. doi: 10.1111/j.1574-6968.2007.00846.x. [DOI] [PubMed] [Google Scholar]
- 21.Onaka H, Mori Y, Igarashi Y, Furumai T. Mycolic acid-containing bacteria induce natural-product biosynthesis in Streptomyces species. Appl. Environ. Microbiol. 2011;77:400–406. doi: 10.1128/AEM.01337-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Osada H, Koshino H, Isono K, Takahashi H, Kawanishi G. Reveromycin A, a new antibiotic which inhibits the mitogenic activity of epidermal growth factor. J. Antibiot. 1991;44:259–261. doi: 10.7164/antibiotics.44.259. [DOI] [PubMed] [Google Scholar]
- 23.Woo J-T, et al. Reveromycin A, an agent for osteoporosis, inhibits bone resorption by inducing apoptosis specifically in osteoclasts. Proc. Natl. Acad. Sci. USA. 2006;103:4729–4734. doi: 10.1073/pnas.0505663103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takahashi S, et al. Reveromycin A biosynthesis uses RevG and RevJ for stereospecific spiroacetal formation. Nat. Chem. Biol. 2011;7:461–468. doi: 10.1038/nchembio.583. [DOI] [PubMed] [Google Scholar]
- 25.De Schrijver A, De Mot R. A subfamily of MalT-related ATP-dependent regulators in the LuxR family. Microbiology. 1999;145:1287–1288. doi: 10.1099/13500872-145-6-1287. [DOI] [PubMed] [Google Scholar]
- 26.Kato N, Takahashi S, Nogawa T, Saito T, Osada H. Construction of a microbial natural product library for chemical biology studies. Curr. Opin. Chem. Biol. 2012;16:101–108. doi: 10.1016/j.cbpa.2012.02.016. [DOI] [PubMed] [Google Scholar]
- 27.Osada H, Nogawa T. Systematic isolation of microbial metabolites for natural products depository (NPDepo) Pure Appl. Chem. 2012;84:1407–1420. doi: 10.1351/PAC-CON-11-08-11. [DOI] [Google Scholar]
- 28.Panthee S, Takahashi S, Hayashi T, Shimizu T, Osada H. β-carboline biomediators induce reveromycin production in Streptomyces sp. SN-593. Sci. Rep. 2019;9:5802. doi: 10.1038/s41598-019-42268-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hawver LA, Jung SA, Ng WL. Specificity and complexity in bacterial quorum-sensing systems. FEMS Microbiol. Rev. 2016;40:738–752. doi: 10.1093/femsre/fuw014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Papenfort K, Bassler BL. Quorum sensing signal-response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 2016;14:576–588. doi: 10.1038/nrmicro.2016.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawatani M, et al. The identification of an osteoclastogenesis inhibitor through the inhibition of glyoxalase I. Proc. Natl. Acad. Sci. USA. 2008;105:11691–11696. doi: 10.1073/pnas.0712239105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanoh N, Honda K, Simizu S, Muroi M, Osada H. Photo-cross-linked small-molecule affinity matrix for facilitating forward and reverse chemical genetics. Angew. Chem. Int. Ed. Engl. 2005;44:3559–3562. doi: 10.1002/anie.200462370. [DOI] [PubMed] [Google Scholar]
- 33.Lechner A, Eustáquio AS, Gulder TAM, Hafner M, Moore BS. Selective overproduction of the proteasome inhibitor salinosporamide A via precursor pathway regulation. Chem. Biol. 2011;18:1527–1536. doi: 10.1016/j.chembiol.2011.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang LH, et al. Characterization of giant modular PKSs provides insight into genetic mechanism for structural diversification of aminopolyol polyketides. Angew. Chem. Int. Ed. Engl. 2017;56:1740–1745. doi: 10.1002/anie.201611371. [DOI] [PubMed] [Google Scholar]
- 35.Laureti L, et al. Identification of a bioactive 51-membered macrolide complex by activation of a silent polyketide synthase in Streptomyces ambofaciens. Proc. Natl. Acad. Sci. USA. 2011;108:6258–6263. doi: 10.1073/pnas.1019077108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Churchill MEA, Chen L. Structural basis of acyl-homoserine lactone-dependent signaling. Chem. Rev. 2010;111:68–85. doi: 10.1021/cr1000817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peters NK, Frost JW, Long SR. A plant flavone, luteolin, induces expression of Rhizobium-meliloti nodulation genes. Science. 1986;233:977–980. doi: 10.1126/science.3738520. [DOI] [PubMed] [Google Scholar]
- 38.Dakora FD, Phillips DA. Root exudates as mediators of mineral acquisition in low-nutrient environments. Plant Soil. 2002;245:35–47. doi: 10.1023/A:1020809400075. [DOI] [Google Scholar]
- 39.Jones DL, Nguyen C, Finlay RD. Carbon flow in the rhizosphere: carbon trading at the soil-root interface. Plant Soil. 2009;321:5–33. doi: 10.1007/s11104-009-9925-0. [DOI] [Google Scholar]
- 40.Bulgarelli D, Schlaeppi K, Spaepen S, van Themaat EVL, Schulze-Lefert P. Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant. Biol. 2013;64:807–838. doi: 10.1146/annurev-arplant-050312-120106. [DOI] [PubMed] [Google Scholar]
- 41.Huang HB, et al. Antimalarial beta-carboline and indolactam alkaloids from marinactinospora thermotolerans, a deep sea isolate. J. Nat. Prod. 2011;74:2122–2127. doi: 10.1021/np200399t. [DOI] [PubMed] [Google Scholar]
- 42.Aroonsri A, Kitani S, Ikeda H, Nihira T. Kitasetaline, a novel β-carboline alkaloid from Kitasatospora setae NBRC 14216 T. J. Biosci. Bioeng. 2012;114:56–58. doi: 10.1016/j.jbiosc.2012.02.027. [DOI] [PubMed] [Google Scholar]
- 43.Chen Q, et al. Discovery of McbB, an enzyme catalyzing the β-carboline skeleton construction in the marinacarboline biosynthetic pathway. Angew. Chem. Int. Ed. Engl. 2013;52:9980–9984. doi: 10.1002/anie.201303449. [DOI] [PubMed] [Google Scholar]
- 44.Lyu, A. et al. Reveromycins A and B from Streptomyces sp 3–10: antifungal activity against plant pathogenic fungi In vitro and in a strawberry food model system. Front. Microbiol. 8 (2017). [DOI] [PMC free article] [PubMed]
- 45.Komatsu M, Uchiyama T, Omura S, Cane DE, Ikeda H. Genome-minimized Streptomyces host for the heterologous expression of secondary metabolism. Proc. Natl. Acad. Sci. USA. 2010;107:2646–2651. doi: 10.1073/pnas.0914833107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takahashi S, et al. Biochemical characterization of a novel indole prenyltransferase from Streptomyces sp. SN-593. J. Bacteriol. 2010;192:2839–2851. doi: 10.1128/JB.01557-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sambrook, J. & Russell, D. W. Molecular Cloning: a Laboratory Manual. 3rd ed, (Cold Spring Harbor Laboratory Press, 2001).
- 48.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Panthee S, et al. Furaquinocins I and J: novel polyketide isoprenoid hybrid compounds from Streptomyces reveromyceticus SN-593. J. Antibiot. 2011;64:509–513. doi: 10.1038/ja.2011.41. [DOI] [PubMed] [Google Scholar]
- 50.Miyazawa T, et al. Identification of middle chain fatty acyl-CoA ligase responsible for the biosynthesis of 2-alkylmalonyl-CoAs for polyketide extender unit. J. Biol. Chem. 2015;290:26994–27011. doi: 10.1074/jbc.M115.677195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kieser, T., Bibb, M. J., Buttner, M. J., Chater, K. F. & Hopwood, D. A. Practical Streptomyces Genetics. (The John Innes Foundation, 2000).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the finding of this study are available from the corresponding author upon request. Sequence data have been deposited to DNA data bank (accession number AP018365 RVR, AP018366 pRVR1, and AP018367 pRVR2). The RNA-seq data were deposited in SRA database with accession code SRR6715512 and SRR6715514. The data that support the finding of this study are available from the corresponding authors upon request.