Abstract
Purpose
The aim of the study is to investigate presence and role of the gene encoding the maternally contributed nucleotide-binding oligomerization domain (NOD)–like receptors with a pyrin domain (PYD)–containing protein 9 (NLRP9) in human and mouse ovaries, respectively, and in preimplantation mouse embryo development by knocking down Nlrp9b.
Methods
Expression levels of NLRP9 mRNA in human follicles were extracted from RNA sequencing data from previous studies. In this study, we performed a qPCR analysis of Nlpr9b mRNA in mouse oocytes and found it present. Intracellular ovarian distribution of NLRP9B protein was accomplished using immunohistochemistry. The distribution of NLRP9B was explored using a reporter gene approach, fusing NLRP9B to green fluorescent protein and microinjection of in vitro–generated mRNA. Nlrp9b mRNA function was knocked down by microinjection of short interference (si) RNA targeting Nlrp9b, into mouse pronuclear zygotes. Knockdown of the Nlrp9b mRNA transcript was confirmed by qPCR.
Result
We found that the human NLRP9 gene and its corresponding protein are highly expressed in human primordial and primary follicles. The NLRP9B protein is localized to the cytoplasm in the blastomeres of a 2-cell embryo in mice. SiRNA-mediated knockdown of Nlrp9b caused rapid elimination of endogenous Nlrp9b mRNA and premature embryo arrest at the 2- to 4-cell stages compared with that of the siRNA-scrambled control group.
Conclusions
These results suggest that mouse Nlrp9b, as a maternal effect gene, could contribute to mouse preimplantation embryo development. It remains to investigate whether NLRP9 have a crucial role in human preimplantation embryo and infertility.
Keywords: Nlrp9b, Human ovary, Maternal effect gene, Preimplantation embryo
Introduction
Several lines of evidence have identified reproductive-related NACHT, LRR and PYD domain–containing protein (NLRP) genes, acknowledged for their roles in innate immunity and apoptosis [1, 2], to exert their functions through maternal effects [3]. Maternal effect genes synthesize mRNAs and proteins, which are deposited in the female egg during maturation and upon fertilization. The products of maternal effect genes support early embryo development, and mutations in maternal effect genes can lead to developmental arrest at the earliest stages of embryo development [4]. Mice and humans have 14 and 20 NLRP-encoding genes [5–7], respectively, with a subset being expressed in oocytes and early preimplantation embryos. Of the 14 human NLRP genes, ten NLRPs (2, 4, 5, 7, 8, 9, 11, 12, 13, 14) were detected in oocytes and/or early embryos [7]. Mutations in NLRP genes are associated with different disorders, including developmental diseases [3]. In human, there is only one NLRP9 gene and its corresponding protein product, NLRP9 (also known as NOD6) [8].
In mice, five Nlrp genes (2, 4, 5, 9, 14) have been detected in reproductive cells [6]. Mice have three isoforms of the Nlrp9 gene [2]: Nlrp9a, Nlrp9b and Nlrp9c. Nlrp9a and Nlrp9c are expressed predominantly in oocytes and early embryos [9]. A Cyp2a (4/5) bgs-null mouse model was generated, in which a 1.2–megabase pair genomic fragment containing nine Cyp genes in mouse chromosome 7 (including, sequentially, Cyp2a5, 2g1, 2b19, 2b23, 2a4, 2b9, 2b13, 2b10 and 2s1) was deleted as well as five non-P450 genes (Vmn1r184, Nalp9c, Nalp4a, Nalp9a and Vmn1r185) that included Nlrp4a (Nalp4a), Nlrp9a (Nalp9a) and Nlrp9c (Nalp9c). The resultant mouse strain was viable and fertile, without any developmental deficits or morphologic abnormalities [10]. Since this large genomic deletion contained other genes as well, including Nlrp9a, Nlrp9c and Nlrp4a, the authors concluded that Nlrp9a, Nlrp9c and Nlrp4a have no effect on embryonic development [10]. Moreover, the Nlrp9b transcript was expressed in both mouse oocytes and embryonic stem cells, while Nlrp9a and Nlrp9c transcripts were only expressed in oocytes [11–14].
The expression of the Nlrp9b transcript and protein is detected in mouse ovaries, while in bovine, the protein expression of NLRP9 is determined in both ovaries and testis [14, 15]. Moreover, localization of NLRP9B protein is detected in mouse oocytes, and interestingly, the NLRP9B protein decreased and correlated with oocyte ageing [16]. The NLRP9B protein was detected throughout early mouse embryogenesis, after ovulation and fertilization, but the protein level gradually decreased during embryonic development [11–14, 17]. The NLRP9B protein was located in the cytoplasm of the blastomeres in mouse 2-cell-stage embryos [13, 14, 17]. However, the pattern of Nlrp9b expression showed that Nlrp9b is not expressed as the zygotic genome activation occurs during embryonic development and, thus, has a maternal origin [11–14, 17].
Recently, the role of Nlrp9b was associated with restrictions in rotavirus infection in adult intestinal epithelial cells [18]. Here, it forms an inflammasome including RNA helicase DHX9 and preferentially binds siRNAs [18]. Interestingly, ancient human endogenous retroviruses are linked to regulatory roles in human pluripotent stem cells [19]. MERLV-associated transcripts are already expressed at the 2-cell stage in mice and are crucial for totipotency [20, 21]. This is important in preimplantation development, during which period extensive reprogramming of the genome occurs and cells pass through totipotent and pluripotent states. At this embryonic stage, the main mechanism responsible for retrotransposon silencing—DNA methylation—is inoperative [22].
In this study, we first extracted the gene expression levels of the NLRP9 gene in human primordial and primary follicles from previously published transcriptome studies [23, 24], to precisely note the expression pattern of the NLRP9 gene during the primordial to primary transition. To address the intracellular localization of the NLRP9 protein, we performed immunohistochemistry (IHC) on human ovarian tissue. The expression level of Nlrp9b mRNA was confirmed in mouse oocytes and early embryos. To functionally address NLRP9B during early embryo development, RNA interference (RNAi) was used to specifically deplete Nlrp9b mRNA from newly formed zygotes. This led to a complete knockdown of the corresponding endogenous Nlrp9b mRNA, as confirmed using real-time qPCR. Knockdown of Nlrp9b mRNA function caused premature embryo developmental arrest at the 2- to 4-cell stages compared with that of the siRNA-scrambled control group. This suggests that mouse Nlrp9b mRNA acts as a maternal effect gene and is required for early embryonic development, in line with observations noted for other reproductive-related Nlrp genes [3].
Materials and methods
Immunohistochemistry in human and mouse ovarian tissues
Normal ovarian cortex tissue was donated from three women undergoing oophorectomy followed by cryopreservation before gonadotoxic treatment of non-gynaecological cancer. The patients were aged 26, 34 and 34 years old, respectively. Written informed consent was obtained from all patients. The study was approved by Danish Scientific Ethical Committee (approval number: KF299017 and J7KF/01/170/99) and the Danish Data Protection Agency. Immunohistochemistry was performed as previously described [24]. In brief, ovarian cortical tissues from patients and ovarian tissues from C57BL/6J were fixed in 4% PFA for 12 h, embedded in paraffin and sectioned at 5 μm (Microtome, Leica Microsystems, Wetzlar, Germany). Sections were deparaffinized in xylene (2 × 5 min) and dehydrated through graded alcohols (99%, 96% and 70%). The slides were subjected to heat-induced antigen retrieval in sodium citrate buffer (pH 6.0) for 15 min in a standard microwave oven (750 W). After cooling at room temperature, the sections were washed with PBS (0.15 M NaCl and 0.1 M phosphate buffer, pH 7.4) (2 × 5 min). Next, permeability was obtained in 0.5% Triton X-100 in PBS for 10 min, followed by blocking of non-specific binding of IgG through incubation with normal donkey serum (Chemicon, Millipore) for 30 min at RT. Sections were incubated with primary NOD6 polyclonal antibody (Thermo Fisher, PA5-21019) at 1:100 dilution in PBS overnight at 4 °C. The antibody has been verified to provide specificity for NOD6 in EL4 cell lysate (Thermo Fisher Scientific). On the next day, the slices were washed with PBS (3 × 10 min) and labelled with Alexa-Fluor-488 (Invitrogen, R37118) at 1:300 dilution for 1 h at room temperature in darkness, to avoid photo bleaching. The sections were rinsed in PBS (2 × 5 min) prior to adding 1:1000 DAPI D9542 (Sigma) and incubation for 3 min. The slides were mounted with a fluorescence mounting medium (Dako) on microscope slides and analyzed on an ImageXpress® Pico automated cell imaging system (Molecular Devices) (3 sections per ovary/5 images per section).
Oocyte and embryo isolation
To isolate fully grown GV antral oocytes, female mice were intraperitoneally injected with 3.5 IU Folligon (PMSG; Folligon, Intervet) and were then sacrificed 48 h later. To obtain ovulated MII oocytes, females were injected with 3.5 IU of pregnant mare’s serum gonadotropin (PMSG; Folligon, Intervet), followed by 3.5 IU human chorionic gonadotropin (hCG; Chorulon, Intervet) 48 h later and were then sacrificed after 15 h. Embryo recovery and isolation F1 (C57BL/6xCBA) females were injected with 5 IU PMSG and 5 IU hCG 48 and 24 h later to induce ovulation, respectively, as described previously [25], and they mated with F1 (C57BL/6xCBA) males.
Zygotes and 2-cell and 4-cell embryos were collected from the oviduct at 26, 46 and 56 h after hCG, respectively. Zygotes for RNA injections were collected from female mice 25 h after hCG. All of the procedures were approved by the Ethics Committee for the Use of Laboratory Animals in Aarhus University (2015−15−0201−00800 to KLH). Zygotes were collected from the oviducts and treated with hyaluronidase (Sigma-Aldrich) (50 μl of 3 mg/ml solution in 250 μl M2 medium) to remove surrounding cumulus cells and were then cultured in M2 media (EmbryoMax® M2 medium with phenol red, Specialty Media, Millipore MR-015P-5F) at 37 °C. The embryos were cultured overnight in drops of potassium simplex optimized medium (KSOM) (EmbryoMax KSOM Powdered Media Kit, Specialty Media, Millipore MR-020P-5F) supplemented with amino acids and 4 mg/ml BSA (Millipore), under embryo-tested paraffin oil in an atmosphere of 5% CO2 in air at 37 °C.
RNA isolation
Total RNA extraction was performed using an RNA isolation kit (KIT0312-l Arcturus® PicoPure® RNA Isolation Kit, Applied Biosystems, Life Technologies, Foster City, CA, USA) according to the manufacturer’s protocol. For each stage, 10–20 oocytes or embryos were pooled in each tube (three pools per developmental stage), and RNA was extracted according to Ovation® PicoSL WTA System V2 (Nugen) protocol, and RNA elution was performed with elution buffer (PicoPure® RNA Isolation Kit) (providing 1.5–10 ng RNAtotal per samples). For all RNA extractions, a DNase digestion step was performed using the RNase-Free DNase Set (Qiagen). RNA was subsequently stored at − 80 °C.
NA synthesis
Ovation® PicoSL WTA System V2 (Nugen) protocol was used to generate cDNA. Quantification of RNA was performed based on determined RNA concentration using NanoDrop (NanoDrop 1000, Saveen Werner, Life Science, Sweden). The detection range is ± 2.0 ng/μl for sample concentrations between 2.0 and 100 ng/μl samples. For each cDNA synthesis reaction, 1.5–10 ng RNA was then used.
qPCR
A qPCR analysis of the Nlrp9b gene expression was conducted based on GV and MII oocytes and 2-cell and 4-cell embryos using the TaqMan® Gene Expression Assay (Applied Biosystems). Reactions were set up for the Nlrp9b gene and for the reference gene H2afz [26] (Mm01312681_g1 [Nlrp9b] and Mm05916395_g1 [H2afz], from Thermo Fisher Scientific). The reactions were run on a LightCycler® 96 (Roche) using LightCycler® 480 Probes Master (Roche) (program 50 °C for 2 s, 95 °C for 10 min, 45 cycles of 95 °C for 15 s followed by 60 °C for 60 s and finally 1 cycle of 40 °C for 30 s). All of the reactions were done in triplicate with 100 ng of quantified cDNA, as template, in a total volume of 10 μl containing 2 μl H2O, 0.5 μl TaqMan Gene Expression Assays (Applied Biosystems), 5 μl Probes Master (Roche) and 2.5 μl template cDNA (40 ng/μl). Triplicate expression values of each gene were set relative to the reference gene via the ΔΔCT method [27]. The Ct values for the reference gene H2afz were within 18–26 cycles in the samples. As a negative control, cDNA from no-template RT-PCR reactions was used.
Cloning of Nlrp9b-eGfp
Nlrp9b inserts were generated with a SuperScript III One-Step RT-PCR System with Platinum Taq High Fidelity (Invitrogen) using RNA extracted from a mouse ovary as the template, and the following primers: F: 5′-5′-NNNGCCGGCATGGCGGGCTCATCTGGCTA-3′ and R: 5′-NNNGCCGGCATTCCTGCTGTTCCATACCA-3′. Using NaeI restriction sites, the PCR-amplified Nlrp9b insert was cloned into the NaeI-digested and dephosphorylated pBS_RN3P-eGFP vector [28], in frame with eGfp. The insertion and orientation were verified by DNA sequencing.
RNA synthesis, microinjections and confocal imaging
The mRNAs encoding NLRP9B-eGFP and GFP were generated and microinjected into zygotes as described [29]. Embryos with siRNA (ON-TARGETplus Mouse Nlrp9b, Smartpool, L-061448-01-0005 and L-066417-01-0005, and ON-TARGETplus Non-targeting Pool D-001810-10-05, with a final concentration of 20 μM, together with rhodamine-conjugated dextran as an injection marker) or mRNA (Nlpr9b-eGFP or eGfp, 200–400 ng/μl) were microinjected in M2 media (EmbryoMax® M2 medium with phenol red, Specialty Media, Millipore MR-015P-5F) covered in oil on a glass depression slide using a FemtoJet microinjection system (Eppendorf). The embryos were cultured in KSOM (EmbryoMax KSOM Powdered Media Kit, Specialty Media, Millipore MR-020P-5F) under paraffin oil at 37.5 °C in air enriched with 5% CO2. The microinjected zygotes were incubated overnight (37 °C, 5% CO2) in KSOM (EmbryoMax KSOM Powdered Media Kit, Specialty Media, Millipore MR-020P-5F). On the next day, 2-cell embryos were incubated for 15 min in a 1:7500 dilution of Hoechst (Sigma) in DPBS (Life Technologies), washed three times in PBS-T and fixed for 15 min in 2% PFA before mounting (Fluorescent Mounting Media, Dako S3023). The embryos were analyzed using a fluorescent microscope (Leica DMI400B) and Leica LAS Software. Confocal images were taken using a LSM800 laser scanning confocal microscope with × 20, × 40 and × 63 C-Apochromat water/oil immersion objectives with NA of 1.2 (Carl Zeiss, Jena, Germany). Confocal images were exported to ImageJ for image processing.
Statistical analysis
All qPCR data were analyzed using one-way ANOVA and Tukey’s HSD was used for post hoc tests. The statistical analysis was performed using Prism 6, version 6.0 (GraphPad Software Inc., CA, USA). The data are represented as mean ± SD. A value of p < 0.05 was defined as statistically significant.
Results
Expression of NLRP9 in human primordial and primary follicles
As a first assessment to determine whether the NLRP9 gene is expressed in human ovarian follicles, data on NLRP9 transcript expression were extracted from global gene expression studies performed in oocytes [24] and granulosa [23] cells from primordial and primary follicles. These studies were based on laser capture microdissection of precisely staged oocytes and granulosa cells from primordial and primary follicles, and RNA sequencing of total RNA provided insights into the molecular levels of specific genes. Extracting the fragments per kilobase of exon model per million reads mapped (FPKM) values for NLRP9 revealed that the NLRP9 transcript is highly abundant in both oocytes and granulosa cells from primordial and primary follicles (Table 1). The FPKM values are a normalized estimation of a gene expression and are calculated from the number of reads mapped to each particular gene sequence, taking the gene length into account [24] [23]. A comprehensive description of this is provided in the original RNA sequencing data from human primordial and primary follicles [24] [23]. The gene expression of the NLRP9 gene obtained from published RNA sequencing data [24] [23] ranged from 4.74 to 8.89 FPKM (mean), which suggests that NLRP9 is highly expressed in these early follicles (Table 1). This clearly indicates that NLRP9 is expressed in oocytes and granulosa cells from primordial and primary follicles.
Table 1.
Expression of human NLRP9 mRNA in oocytes and granulosa cells from primordial and primary follicles [23, 24], as noted. FPKM mean values were calculated based on triplicate expression values of the same transcript using a one-sample t test. The p value indicates the consistency in expression pattern across triplicates. p values < 0.05 are considered significant
| Primordial | Primary | |||||||
|---|---|---|---|---|---|---|---|---|
| Oocytes | Granulosa cells | Oocytes | Granulosa cells | |||||
| FPKM (mean) | p value | FPKM (mean) | p value | FPKM (mean) | p value | FPKM (mean) | p value | |
| NLRP9 | 5.29 | 0.05 | 8.89 | 0.01 | 6.95 | 0.02 | 4.74 | 0.04 |
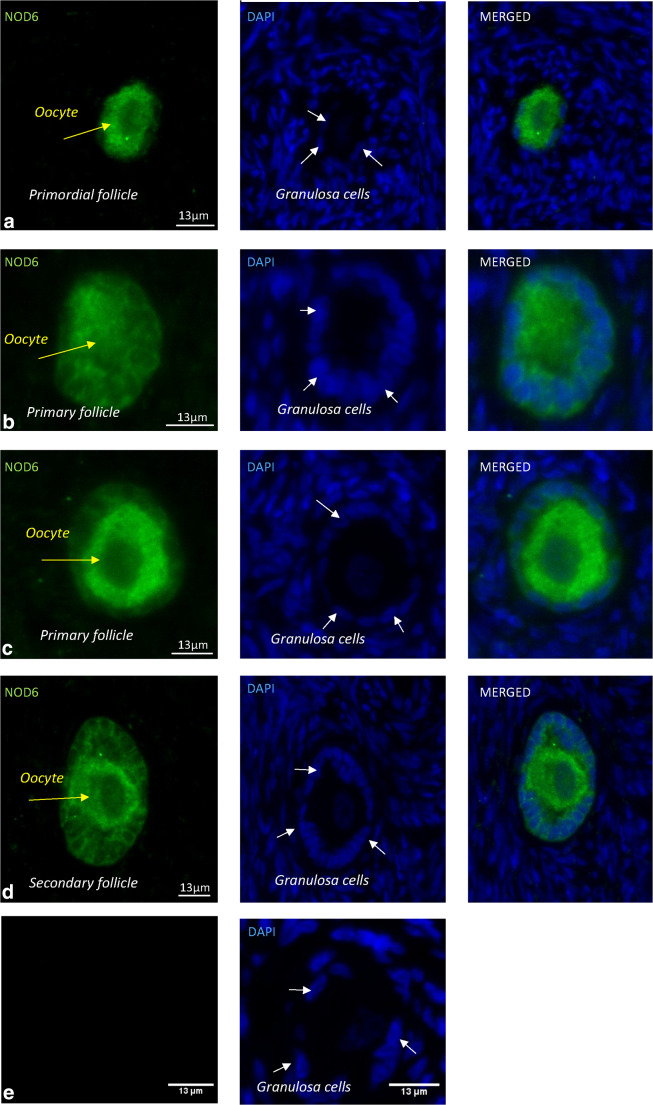
Next, immunohistochemistry using an antibody against NLRP9 (NOD6) was performed on human ovarian tissue to investigate if the NLRP9 protein was likewise present in human primordial and primary follicles. The immunofluorescence revealed strong staining of the NLRP9 protein in human oocytes and granulosa cells from primordial and primary follicles, using the nuclear counterstain DAPI (blue) to identify nuclei (Fig. 1). The stained NLRP9 appeared as cytoplasmic staining in both oocytes and granulosa cells in primordial and both primary follicles with non-detectable and detectable nuclei (Fig. 1), as expected for the NLRP9 receptor (Fig. 1). These results revealed that the staining intensity was high, in agreement with the high FKPM values noted for the NLRP9 gene (Table 1). The empty region in the oocyte is the oocyte nucleus. Immunohistochemistry on human ovarian tissue without the primary antibody was included (Fig. 1) and revealed no specific staining.
Fig. 1.
Intra-ovarian distribution of NLRP9 in human oocytes and granulosa cells from primordial, primary and secondary follicles. Immunohistochemical staining of human ovarian tissue with primary NOD6 antibody and counterstained with DAPI, as indicated. The images show intense staining of NLRP9 protein in human oocytes and granulosa cells from primordial (a) and both primary follicles with non-detectable (b) and detectable nuclei (c). NLRP9 was localized to the cytoplasm of both oocytes (yellow arrows) and granulosa cells (white arrows) in primordial, primary and secondary follicles (d). The DAPI staining identified the cells’ nuclei. Primary follicle in controls without the primary antibody revealed no staining (e). Scale bars, 13 μm
Nlrp9b transcript expressed in mouse GV and MII oocytes as well as zygotes
As the NLRP9 gene and its protein were highly expressed in human ovarian follicle cells, the next step was to evaluate the presence of the mouse Nlrp9b gene in oocytes and early preimplantation embryos. Toward this aim, germinal vesicle (GV) and metaphase II (MII) oocytes as well as early preimplantation embryos (zygotes, 2-cell and 4-cell) were collected. Histone H2afz mRNA was used as the most stable internal reference gene during preimplantation development [29, 30].
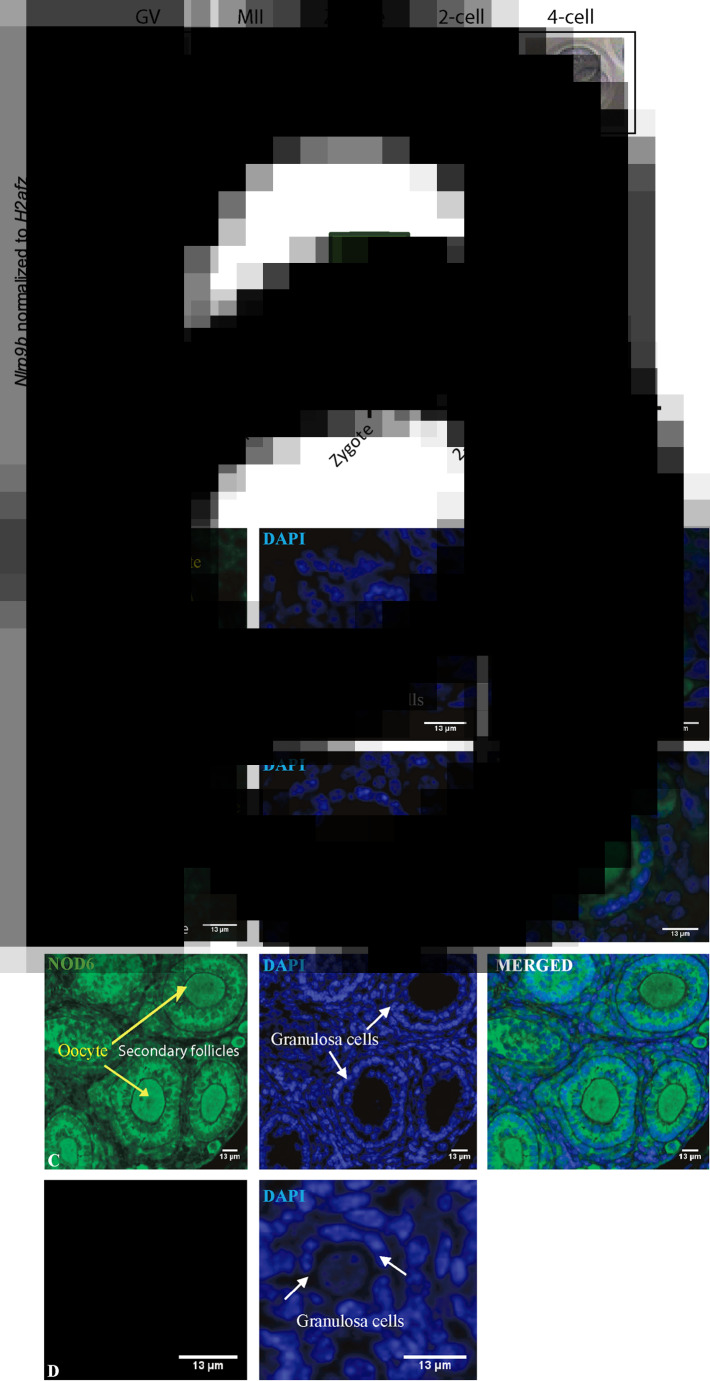
The qPCR analysis revealed that Nlrp9b expression was detectable in GV and MII oocytes (Fig. 2a), as expected. Interestingly, the Nlrp9b transcript was significantly increased a 2-fold from oocyte to the zygote stage, suggesting that transcription of the Nlrp9b is already initiated in the zygote during the first wave of zygotic genome activation. Interestingly, no Nlrp9b mRNA was detected at the 2-cell and 4-cell stages, suggesting that the transcription of Nlrp9b is strictly regulated during the maternal-to-zygotic transition. Then, IHC using an antibody against NLRP9B (NOD6) was performed to investigate if the NLRP9B protein was also present in mouse primordial and primary follicles. This revealed strong staining of the NLRP9B protein in mouse oocytes and granulosa cells from primordial and primary follicles (Fig. 2b). The stained NLRP9B appeared as cytoplasmic staining in both the oocytes and granulosa cells in primordial and primary follicles (Fig. 2b), as also noted in human follicles (Fig. 1), using DAPI for nuclear staining. These results revealed that NlRP9B is already present during early follicle development and that this reserve of maternally loaded NLRP9B might support egg maturation. Immunohistochemistry on mouse ovarian tissue without the primary antibody was included (Fig. 2) and revealed no specific staining.
Fig. 2.
Quantitative RT-PCR analysis of Nlrp9b in mouse oocytes and early preimplantation embryos. Nlrp9b expression and its relative abundance in GV and MII oocytes and preimplantation 1-cell, 2-cell and 4-cell embryos, as indicated (a). The corresponding morphologies of oocytes and embryos are shown. Nlrp9b expression levels were normalized by H2afz, and their relative expressions are displayed. Nlrp9b expression was detected in GV and MII oocytes. There was a significant difference between the Nlrp9b expression in oocytes and zygotes. Moreover, there was no Nlrp9b expression at the 2-cell and 4-cell stages. Data are presented as mean ± standard deviation (SD) (bars) of triplicate measurements including SDs. Immunohistochemical staining of mouse ovarian tissue with primary NOD6 antibody and counterstained with DAPI, as indicated (b). The images show strong staining of the NLRP9B protein in oocytes and granulosa cells from primordial (A) and primary follicles (B). NLRP9B was localized to the cytoplasm of both oocytes (yellow arrows) and granulosa cells (white arrows) in primordial, primary and secondary follicles (c). The DAPI staining identified the cells’ nuclei. Controls without the primary antibody revealed no staining (d). Scale bars, 13 μm
NLRP9B fused to enhanced green fluorescent protein localized to the cytoplasm in mouse 2-cell embryos
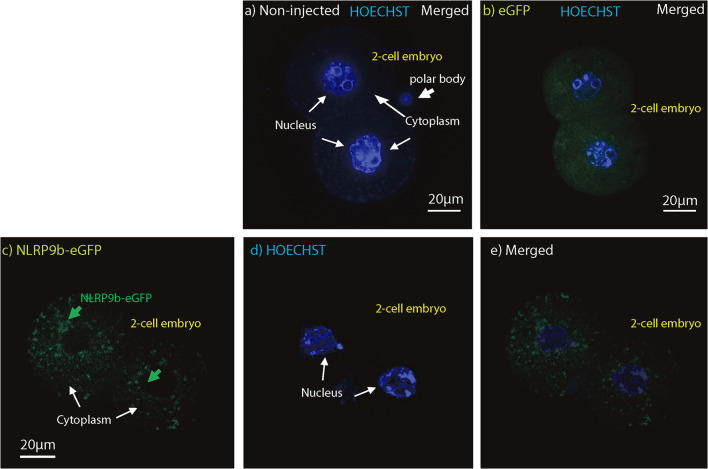
In order to test how the NLRP9B would be distributed intracellularly post-fertilization, NLRP9B was fused to enhanced green fluorescent protein (eGFP), to enable us to follow its dynamics and distribution after the first cell division after fertilization. Non-injected and eGFP protein controls were included and revealed no staining and unspecific widespread distributions, respectively (Fig. 3a, b). In 2-cell-stage embryos, the NLRP9B-eGFP fusion protein is distributed in the cytoplasm (Fig. 3c–e). The cytosolic staining resembles the immunohistochemistry staining observed in the ovarian follicles (Fig. 2); thus, NLRP9B appears to be a cytoplasmic receptor in the early follicles and preimplantation embryos.
Fig. 3.
Intracellular distribution of NLRP9B-eGFP in 2-cell mouse embryos revealing cytoplasm localization. Intracellular immunofluorescent localization and distribution of eGFP in non-injected (a), eGFP control (b) and NLRP9B-eGFP (c–e) in 2-cell mouse embryos, as indicated, using confocal imaging. Non-injected and eGFP protein controls showed no staining and unspecific widespread distributions, respectively. In 2-cell-stage embryos, the NLRP9B-eGFP fusion protein was distributed in dot-like structures within the cytoplasm. Scale bars, 20 μM, as indicated
SiRNA-mediated knockdown of Nlrp9 caused 2-cell developmental arrest
In order to reveal if maternal and zygotic Nlrp9b transcripts are necessary to support early development (alongside the maternally contributed NLRP9B protein), the RNAi approach was used to knock down Nlrp9b transcript.
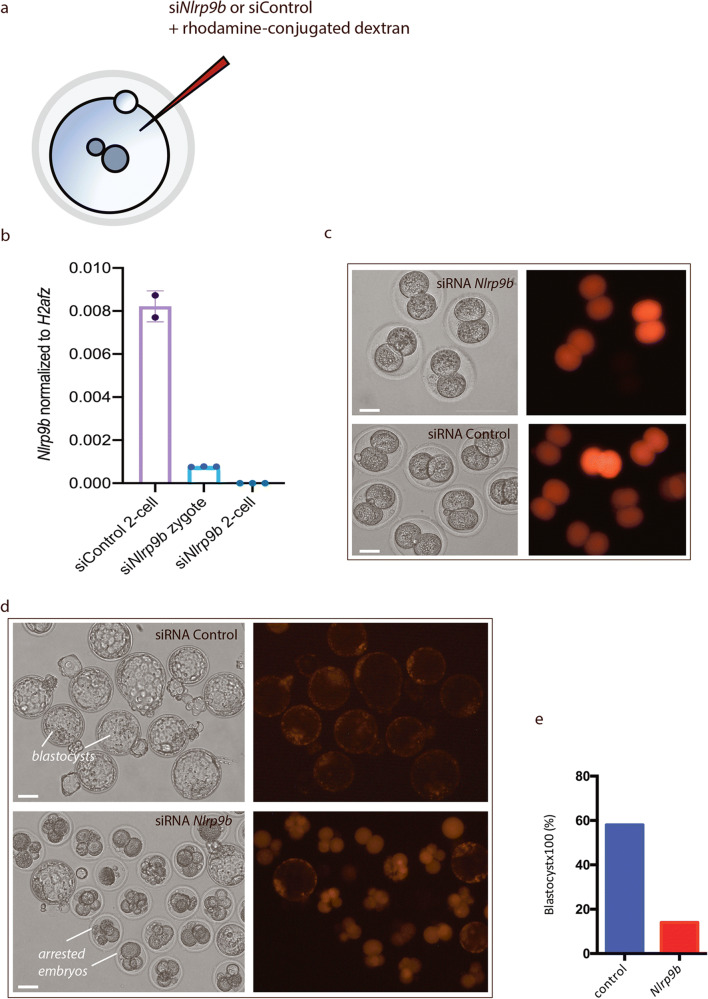
We depleted Nlpr9b in the zygote by injecting siRNAs targeting Nlrp9b, together with rhodamine-conjugated dextran to mark the injected cells by microinjection into mouse pronuclear zygotes (Fig. 4a). The siRNA probes targeting Nlrp9b or scrambled (non-targeting) RNA together with rhodamine-conjugated dextran to identify injected cells that received siRNA were microinjected into zygotes at the pronuclear stage (in three independent experiments).
Fig. 4.
SiRNA-mediated knockdown of Nlrp9b in mouse zygotes causes developmental arrest. Scheme of Nlrp9b knockdown experiment (a). Zygote stage embryos were injected with Nlrp9b siRNA or control siRNA together with the rhodamine-conjugated dextran (injection control). Embryos were cultured in vitro, for qPCR analysis and developmental assessment. qPCR analysis was performed on siRNA control and Nlrp9b siRNA embryos, revealing complete knockdown of the endogenous Nlrp9b at the 2-cell stage (b). Standard deviations are indicated by bars (N = 12 female, n = 20–40 embryos; three independent experiments (dots)). After an overnight in vitro culture, both the siRNA control and Nlrp9b siRNA–injected embryos developed into 2-cell embryos (c). After 3 days of in vitro culture, siRNA-control-injected embryos (RNAi scrambled) developed into blastocysts, in contrast to Nlrp9b siRNA–injected embryos, whose development was arrested at the 2-cell stage (d). The corresponding survival rate for injected embryos shows that while 58% of siRNA control embryos developed to blastocysts (n = 19 [three independent experiments]), Nlrp9b siRNA–injected embryos were severely affected, and only 14% developed into blastocysts (n = 28) (e). Data provided as percentage of developed embryos. Experiments were performed at least three times for each group. Data were reported as mean ± SEM. Scale bars, 25 μM, as indicated
After 6 h and an overnight in vitro incubation, zygotes and 2-cell-stage embryos were collected for qPCR analysis, which revealed that the Nlrp9b transcript was effectively reduced in Nlrp9b siRNA–microinjected embryos (Fig. 4b). In 2-cell-stage embryos, the level of Nlrp9b mRNA was higher (Fig. 4b) compared with that of injected zygotes and 2-cell embryos with Nlrp9b siRNA (Fig. 4b). Already 6 h after Nlrp9b siRNA was injected, the endogenous Nlrp9b mRNA was efficiently knocked down, and by the 2-cell stage, the Nlrp9b mRNA was no longer detectable (Fig. 4b). At this stage, the embryos appeared as healthy 2-cell-stage embryos in both the control siRNA and Nlrp9b siRNA groups (Fig. 4c). The co-injection of rhodamine was included to ensure the selection of embryos receiving siRNA for analysis of both qPCR and their developmental potential. The embryos were subsequently monitored for their ability to develop into blastocysts. In the group of embryos injected with siRNA-scrambled control, the majority of the 2-cell-stage embryos developed into blastocysts (Fig. 4d), whereas in the group that received siRNA targeting Nlrp9b, only a small fraction developed into blastocysts (Fig. 4d). The developmental arrest was noted between the 2- and 8-cell stages, and in line with what has been reported for other Nlrp maternal effect genes in mice (Nlrp2 [17], Nlrp4e [31, 32] or Nlrp14 [6]). The survival rate of 2-cell embryos that developed into blastocysts showed that almost 60% of zygotes that received siRNA-scrambled control formed blastocysts, whereas less than 18% of zygotes microinjected with Nlrp9b siRNA managed to develop to the blastocyst stage (Fig. 4e), likely to be a dose-dependent phenotype, as also observed for Nlrp4 [31].
Discussion
The increasing evidence that NLRPs are emerging as maternal effect genes provides new insights into the functions of these genes during early embryo development. Moreover, it strongly suggests that while some maternal effect genes have already been studied, many new genes are most likely acting as maternal effect genes, and our advances in this area are likely to increase. The NLRPs are an interesting gene family, with roles in both immunology and reproduction. Several of the genes are conserved in human and mice [2]. NLRP9 is one of the NLRP genes duplicated and functionally diversified in mammalian reproductive systems, particularly for reproductive function, such as the specific expansion of Nlrp9 in mice [2].
Surprisingly, recent studies showed that the expression profile of Nlrp9 in mammalian preimplantation development was different from primates [14, 33, 34]. In agreement with mouse, Nlrp9 transcript was lowly expressed in the ovary and testis and highly expressed in oocytes and preimplantation embryos in bovine [15, 28]. Moreover, the expression of the Nlrp9 transcript declined from immature bovine oocytes to 8-cell-stage embryos, which showed there was no reactivation from bovine maternal-to-embryonic genome transition [15, 34]. However, limited expression of the Nlrp9 gene in gonad cells, germ line cells and preimplantation embryonic stages in bovine confirmed the role of this gene in preimplantation development [34]. In addition, the Nlrp9 transcript was, temporally, expressed during early embryogenesis at the same time with genomic transition from oocyte to embryo, in primates which confirmed maternal source of NLRP9 [33].
Human NLRP9 protein appears to be widely expressed throughout the body. In our previous studies, we found the NLRP9 gene highly expressed in human primordial and primary follicles, in agreement with previous data observing NLRP9 in human GV and MII oocytes [7]. In this study, we investigated the intracellular presence and distribution of the human NLRP9 protein using immunohistochemistry on human ovarian tissue, which revealed a high presence of it in primary and secondary follicles. Our observation that both the NLRP9 gene and the NLRP9 protein are loaded into the earliest follicle stages suggests that this gene must be important and most likely required for early development.
Attempts have been made to functionally address the requirement of NLRP9 in early mouse development [10]. The function of maternal Nlrp9b has been partially explored, using electroporation of siRNA probes to target endogenous Nlrp9b in oocytes [14], which included parthenogenetic activation of oocytes after electroporation, and the study concluded that Nlrp9b was not required for early development of parthenogenetically activated 2-cell and 4-cell embryos.
Therefore, to investigate if Nlrp9b is indeed a maternal effect gene, as expected from its high abundance in early reproductive cells, we delivered Nlrp9b siRNA probes by microinjections into newly formed zygotes, to knock down the maternally supplied Nlrp9b mRNA.
We found that the siRNA probe mixture efficiently deleted all endogenous Nlrp9b mRNA, and we observed significantly reduced developmental capacity among the Nlrp9b siRNA–injected embryos, compared with the siRNA-scrambled controls. It should be noted here that we do observe a small expression of Nlrp9b mRNA in early 2-cell stages, as we detected in our siRNA experiments; however, during late 2-cell embryo stages, the Nlrp9b mRNA appears to be completed eliminated, as we detected during our expression assay. The phenotype is similar to that of other reproductive Nlrp genes, which have shown specific and complete elimination of Nlrp5 [5], Nlrp2 [17, 35], Nlrp4e [31, 32], Nlrp4f [36] or Nlrp14 [6], causing early developmental arrest at the 2- to 8-cell stages and indicating these Nlrp genes as maternal effect genes.
In conclusion, we find that the NLRP9 gene and the NLRP9 protein are highly present in human primordial and primary follicles, indicating a requirement for NLRP9. In mice, by using a gene-reporter approach, we found that eGFP-NLRP9B fusion protein is localized to the cytoplasm in 2-cell-stage embryos, in agreement with the intracellular localization noted on human ovarian tissue. Finally, we knocked down endogenous Nlrp9b mRNA in mouse zygotes and observed that this caused a developmental arrest at the 2- to 4-cell stages, indicating that Nlrp9b acts as a maternal effect gene in early mouse development.
Acknowledgements
The authors wish to thank past and current members of the Lykke-Hartmann Laboratory (AU) for their scientific discussions.
Author contributions
KLH and LS conceived the study. AG and AH performed the cloning work. EE provided human tissue. MA performed the IHC. JA and AH performed the qPCR analysis. PS, MA, MSN and KLH performed the confocal analysis. LLS, MA and KLH performed the embryo work and siRNA microinjections. All of the authors analyzed and interpreted the results. KLH wrote the manuscript. All of the authors approved the final manuscript.
Funding information
This work was supported by grants from the Carlsberg Foundation (CF18-0474 to MA), Aarhus University Research Foundation (to LS and KLH) and the Independent Research Fund Denmark – Medical Sciences (grant number 6120-00027B), the Novo Nordisk Foundation (NNF16OC0022480 and NNF170C0026820), Kong Christian Den Tiendes Fond, Th. Maigaards Eft. Fru Lily Benthine Lunds Fond, Toyota Fonden, Augustinus Fonden and Fonden til Lægevidenskabens Fremme (to KLH).
Compliance with ethical standards
Written informed consent was obtained from all patients. The study was approved by Danish Scientific Ethical Committee (approval number: KF299017 and J7KF/01/170/99) and the Danish Data Protection Agency. All of the procedures were approved by the Ethics Committee for the Use of Laboratory Animals in Aarhus University (2015−15−0201−00800 to KLH).
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kim YK, Shin JS, Nahm MH. NOD-like receptors in infection, immunity, and diseases. Yonsei Med J. 2016;57(1):5–14. doi: 10.3349/ymj.2016.57.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tian X, Pascal G, Monget P. Evolution and functional divergence of NLRP genes in mammalian reproductive systems. BMC Evol Biol. 2009;9:202. doi: 10.1186/1471-2148-9-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amoushahi M, Sunde L, Lykke-Hartmann K. The pivotal roles of the NOD-like receptors with a PYD domain, NLRPs, in oocytes and early embryo development. Biol Reprod. 2019. 10.1093/biolre/ioz098. [DOI] [PubMed]
- 4.Kim KH, Lee KA. Maternal effect genes: findings and effects on mouse embryo development. Clin Exp Reprod Med. 2014;41(2):47–61. doi: 10.5653/cerm.2014.41.2.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tong ZB, Gold L, Pfeifer KE, Dorward H, Lee E, Bondy CA, et al. Mater, a maternal effect gene required for early embryonic development in mice. Nat Genet. 2000;26(3):267–268. doi: 10.1038/81547. [DOI] [PubMed] [Google Scholar]
- 6.Hamatani T, Falco G, Carter MG, Akutsu H, Stagg CA, Sharov AA, et al. Age-associated alteration of gene expression patterns in mouse oocytes. Hum Mol Genet. 2004;13(19):2263–2278. doi: 10.1093/hmg/ddh241. [DOI] [PubMed] [Google Scholar]
- 7.Zhang P, Dixon M, Zucchelli M, Hambiliki F, Levkov L, Hovatta O, et al. Expression analysis of the NLRP gene family suggests a role in human preimplantation development. PLoS One. 2008;3(7):e2755. doi: 10.1371/journal.pone.0002755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tschopp J, Martinon F, Burns K. NALPs: a novel protein family involved in inflammation. Nat Rev Mol Cell Biol. 2003;4(2):95–104. doi: 10.1038/nrm1019. [DOI] [PubMed] [Google Scholar]
- 9.Peng H, Zhang W, Xiao T, Zhang Y. Expression patterns of Nlrp9a, Nlrp9b and Nlrp9c during mouse development. Biologia. 2014;69(107):107. doi: 10.2478/s11756-013-0287-y. [DOI] [Google Scholar]
- 10.Wei Y, Li L, Zhou X, Zhang QY, Dunbar A, Liu F, et al. Generation and characterization of a novel Cyp2a(4/5)bgs-null mouse model. Drug Metab Dispos. 2013;41(1):132–140. doi: 10.1124/dmd.112.048736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tong ZB, Gold L, De Pol A, Vanevski K, Dorward H, Sena P, et al. Developmental expression and subcellular localization of mouse MATER, an oocyte-specific protein essential for early development. Endocrinology. 2004;145(3):1427–1434. doi: 10.1210/en.2003-1160. [DOI] [PubMed] [Google Scholar]
- 12.Peng H, Lin X, Li W, Zhang W. Expression and localization of Nlrp4g in mouse preimplantation embryo. Zygote. 2015;23(6):846–851. doi: 10.1017/S0967199414000525. [DOI] [PubMed] [Google Scholar]
- 13.Peng H, Zhang W, Xiao T, Zhang Y. Nlrp4g is an oocyte-specific gene but is not required for oocyte maturation in the mouse. Reprod Fertil Dev. 2014;26(5):758–768. doi: 10.1071/RD12409. [DOI] [PubMed] [Google Scholar]
- 14.Peng H, Lin X, Liu F, Wang C, Zhang W. NLRP9B protein is dispensable for oocyte maturation and early embryonic development in the mouse. J Reprod Dev. 2015;61(6):559–564. doi: 10.1262/jrd.2015-050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dalbies-Tran R, Papillier P, Pennetier S, Uzbekova S, Monget P. Bovine mater-like NALP9 is an oocyte marker gene. Mol Reprod Dev. 2005;71(4):414–421. doi: 10.1002/mrd.20298. [DOI] [PubMed] [Google Scholar]
- 16.Peng H, Zhang W, Xiao T, Zhang Y. Expression patterns of Nlrp9a, Nlrp9b and Nlrp9c during mouse development. Biologia. 2014;69:107–112. doi: 10.2478/s11756-013-0287-y. [DOI] [Google Scholar]
- 17.Peng H, Chang B, Lu C, Su J, Wu Y, Lv P, et al. Nlrp2, a maternal effect gene required for early embryonic development in the mouse. PLoS One. 2012;7(1):e30344. doi: 10.1371/journal.pone.0030344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu S, Ding S, Wang P, Wei Z, Pan W, Palm NW, et al. Nlrp9b inflammasome restricts rotavirus infection in intestinal epithelial cells. Nature. 2017;546(7660):667–670. doi: 10.1038/nature22967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buzdin AA, Prassolov V, Garazha AV. Friends-enemies: endogenous retroviruses are major transcriptional regulators of human DNA. Front Chem. 2017;5:35. doi: 10.3389/fchem.2017.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Macfarlan TS, Gifford WD, Driscoll S, Lettieri K, Rowe HM, Bonanomi D, et al. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature. 2012;487(7405):57–63. doi: 10.1038/nature11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Durruthy-Durruthy J, Sebastiano V, Wossidlo M, Cepeda D, Cui J, Grow EJ, et al. The primate-specific noncoding RNA HPAT5 regulates pluripotency during human preimplantation development and nuclear reprogramming. Nat Genet. 2016;48(1):44–52. doi: 10.1038/ng.3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schoorlemmer J, Perez-Palacios R, Climent M, Guallar D, Muniesa P. Regulation of mouse retroelement MuERV-L/MERVL expression by REX1 and epigenetic control of stem cell potency. Front Oncol. 2014;4:14. doi: 10.3389/fonc.2014.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ernst EH, Franks S, Hardy K, Villesen P, Lykke-Hartmann K. Granulosa cells from human primordial and primary follicles show differential global gene expression profiles. Hum Reprod. 2018;33(4):666–679. doi: 10.1093/humrep/dey011. [DOI] [PubMed] [Google Scholar]
- 24.Ernst EH, Grondahl ML, Grund S, Hardy K, Heuck A, Sunde L, et al. Dormancy and activation of human oocytes from primordial and primary follicles: molecular clues to oocyte regulation. Hum Reprod. 2017;32(8):1684–1700. doi: 10.1093/humrep/dex238. [DOI] [PubMed] [Google Scholar]
- 25.Hogan B, Beddington R, Costantini F, Lacy E. Manipulating the mouse embryo: a laboratory manual. 2. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 26.Mamo S, Gal AB, Bodo S, Dinnyes A. Quantitative evaluation and selection of reference genes in mouse oocytes and embryos cultured in vivo and in vitro. BMC Dev Biol. 2007;7:14. doi: 10.1186/1471-213X-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3(6):1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 28.Lemaire P, Garrett N, Gurdon JB. Expression cloning of Siamois, a Xenopus homeobox gene expressed in dorsal-vegetal cells of blastulae and able to induce a complete secondary axis. Cell. 1995;81(1):85–94. doi: 10.1016/0092-8674(95)90373-9. [DOI] [PubMed] [Google Scholar]
- 29.Albertsen M, Teperek M, Elholm G, Fuchtbauer EM, Lykke-Hartmann K. Localization and differential expression of the Kruppel-associated box zinc finger proteins 1 and 54 in early mouse development. DNA Cell Biol. 2010;29(10):589–601. doi: 10.1089/dna.2010.1040. [DOI] [PubMed] [Google Scholar]
- 30.Jeong YJ, Choi HW, Shin HS, Cui XS, Kim NH, Gerton GL, et al. Optimization of real time RT-PCR methods for the analysis of gene expression in mouse eggs and preimplantation embryos. Mol Reprod Dev. 2005;71(3):284–289. doi: 10.1002/mrd.20269. [DOI] [PubMed] [Google Scholar]
- 31.Chang BH, Liu X, Liu J, Quan FS, Guo ZK, Zhang Y. Developmental expression and possible functional roles of mouse Nlrp4e in preimplantation embryos. In Vitro Cell Dev Biol Anim. 2013;49(7):548–553. doi: 10.1007/s11626-013-9638-9. [DOI] [PubMed] [Google Scholar]
- 32.Pfender S, Kuznetsov V, Pasternak M, Tischer T, Santhanam B, Schuh M. Live imaging RNAi screen reveals genes essential for meiosis in mammalian oocytes. Nature. 2015;524(7564):239–242. doi: 10.1038/nature14568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McDaniel P, Wu X. Identification of oocyte-selective NLRP genes in rhesus macaque monkeys (Macaca mulatta) Mol Reprod Dev. 2009;76(2):151–159. doi: 10.1002/mrd.20937. [DOI] [PubMed] [Google Scholar]
- 34.Ponsuksili S, Brunner RM, Goldammer T, Kuhn C, Walz C, Chomdej S, et al. Bovine NALP5, NALP8, and NALP9 genes: assignment to a QTL region and the expression in adult tissues, oocytes, and preimplantation embryos. Biol Reprod. 2006;74(3):577–584. doi: 10.1095/biolreprod.105.045096. [DOI] [PubMed] [Google Scholar]
- 35.Mahadevan S, Sathappan V, Utama B, Lorenzo I, Kaskar K, Van den Veyver IB. Maternally expressed NLRP2 links the subcortical maternal complex (SCMC) to fertility, embryogenesis and epigenetic reprogramming. Sci Rep. 2017;7:44667. doi: 10.1038/srep44667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qin D, Gao Z, Xiao Y, Zhang X, Ma H, Yu X, et al. The subcortical maternal complex protein Nlrp4f is involved in cytoplasmic lattice formation and organelle distribution. Development. 2019;146(20). 10.1242/dev.183616. [DOI] [PubMed]