Abstract
To assess whether morphokinetic features at the cleavage stage together with specific gene expression in cumulus cells (CCs) may be used to predict whether human embryos are able to achieve the expanded blastocyst stage on day 5. Eighty-one embryos were cultured using the Geri plus® time-lapse system. Twenty-seven embryos progressing to the expanded blastocyst stage (BL group) were compared with thirty-five embryos showing developmental arrest (AR group) and nineteen reaching the stage of early or not fully expanded blastocyst (nBL group). The analyzed morphokinetic variables were pronuclear appearance (tPNa), pronuclear fading (tPNf), and completion of cleavage to two, three, four, and eight cells (t2, t3, t4, and t8). CCs were analyzed by RT-qPCR for bone morphogenetic protein 15 (BMP15), cytochrome c oxidase subunit II (COXII), ATP synthase subunit 6 (MT-ATP6), connexin 43 (Cx43), and heme oxygenase-1 (HO-1). Embryos of BL group showed a significantly faster kinetic. BMP15, COXII, and MT-ATP6 mRNA expression was significantly higher in CCs of BL group embryos, whereas Cx43 and HO-1 mRNA levels were higher in AR group. Kinetic parameters and gene expression were not significantly different between either the BL and nBL groups or the AR and nBL groups. ROC curves showed that the most predictive cut-offs were t2 < 26.25 for morphokinetics and COXII > 0.3 for gene expression. Multivariable logistic regression analysis showed that morphokinetic variables and gene expression were both valuable, independent predictors of embryo development to expanded blastocyst. Our results suggest the possibility of developing integrated prediction models for early embryo selection at the cleavage stage.
Keywords: Time-lapse, Embryo morphology, Cumulus cells, Gene expression, Blastocyst, Human IVF
Introduction
In the last decades, elective single blastocyst transfer progressively became a widespread clinical strategy to reach a fairly good pregnancy rate with very low risk of twin pregnancy [1]. However, extensively applied embryo culture at the blastocyst stage is time- and work-demanding for the in vitro fertilization (IVF) laboratories, and, furthermore, some data raise the suspect that prolonged culture could increase epigenetic alterations in the embryo [2]. An alternative strategy would be transferring in utero a single embryo, at cleavage stage, having the highest chance to reach the blastocyst stage, instead of a single blastocyst. The problem, however, would be how to select, among cleaving embryos, those with the best probability to progress to the blastocyst stage. Some morphological scoring criteria applicable in vitro on day three were proposed, but none of them proved to be fully reliable in predicting embryo further development and competence [3].
In the more recent last years, the morphokinetic evaluation using time-lapse systems (TLS) joined conventional morphological evaluation providing a new tool to identify markers of embryo competence and development potential [4]. Indeed, the use of TLS offers a continuous, dynamic, individualized assessment of embryo growth: information on the timing of specific cleavage events could be used to develop algorithms aimed at improving early embryo selection. The Early Embryo Viability Assessment (Eeva™) software is an example of algorithm that applies embryo growth parameters to identify on day three the embryos with the highest probability to become blastocyst [5–7].
An alternative, non-invasive approach to precociously estimate embryo competence is inspired by the concept that the follicular microenvironment can significantly affect the quality of the oocyte and, as a consequence, of the embryo deriving from each specific oocyte [8]. Within the study of the follicular microenvironment biomarkers, attention was initially given to the follicular fluid (FF) biochemical composition, whereas the study of specific gene expression in the cumulus cells (CCs), potentially affecting embryo development, became progressively more popular in the last years [9, 10].
The quality of CCs can be influenced both by some oocyte-secreted factors and by the intrinsic characteristics of these cells. Indeed, the oocyte produces several growth factors, such as growth differentiation factor 9 (GDF9) and bone morphogenetic protein 15 (BMP15), which are known to regulate growth, differentiation, and function of granulosa and theca cells during follicular development, and plays a fundamental role in oocyte maturation, ovulation, fertilization, and embryonic competence [11–14]. On the other hand, the expression of some structural proteins in CCs is of paramount importance for their nurturing function. For example, some connexins, such as connexin 43 (Cx43), are expressed in human CCs and were claimed to affect oocyte quality [15]; they also represent markers of follicle integrity in both humans and animal models [16]. Moreover, one aspect of CCs still scarcely investigated is the quality of their metabolism, which is likely to be involved in embryo development.
We previously demonstrated that a status of enhanced oxidative stress in CCs, arising when the reactive oxygen species (ROS) production overwhelms the intrinsic antioxidant defense, negatively affects oocyte competence [17, 18]; as the main source of ROS are mitochondria, it is evident that mitochondrial respiration must be strictly regulated and well-coupled to oxidative phosphorylation in order to sustain the energy requirements of CCs and avoid ROS production. Therefore, the analysis of genes involved in this function of CCs could be quite relevant.
In the attempt of finding a multivariable approach taking into account different issues which may be critical in oocyte maturation, we designed the present study aiming at (a) identifying morphokinetic features of cleaving embryos able to predict their timely development to the blastocyst stage, (b) assessing the expression of specific genes in CCs and understanding if they could affect the chance of embryo growth to blastocyst, and (c) checking whether morphokinetic variables and the expression of specific genes in CCs could be independent predictors of embryo development.
Materials and methods
Patients
The study considered 81 embryos obtained from 15 women (mean age: years, range 33–40) with normal body mass index (BMI 18–25), serum day 3 FSH < 12 IU/l, serum AMH > 1.2 ng/ml, and antral follicle count (AFC) > 8, who underwent intracytoplasmic sperm injection (ICSI) at our IVF Unit (Turin, Italy), to treat male-related infertility. Exclusion criteria were the following: presence of polycystic ovary syndrome or endometriosis and history of severe ovarian hyperstimulation syndrome. The patients’ clinical characteristics and the outcome of controlled ovarian stimulation (COS) were recorded, including the total dose of exogenous FSH, the peak estradiol (E2) level, the number of retrieved oocytes, the ovarian sensitivity index (OSI = retrieved oocytes × 1000/total gonadotropin dose) [19], the fertilization rate, and the clinical (ultrasound visualization of the gestational sac) pregnancy rate (CPR) per embryo transfer (ET).
The study was carried out in accordance with the Declaration of Helsinki and was authorized as a prospective observational study by the local Ethical Committee. A signed, written informed consent was obtained from all patients.
Controlled ovarian stimulation, oocyte retrieval, and cumulus cells collection
The gonadotropin-releasing hormone (GnRH)-agonist “long” protocol with recombinant FSH (Gonal-F®, Merck, Germany) at individually tailored daily dose (100–300 IU s.c.) was used to carry out controlled ovarian stimulation (COS). Circulating E2 dosage assessment and transvaginal US examination were performed every second day from stimulation day 7 in order to monitor follicular growth. The dose of FSH was adjusted accordingly. A single s.c. injection of 10,000 IU hCG (Gonasi HP, IBSA, Switzerland) was administered to trigger ovulation, when at least two follicles reached 18 mm mean diameter, with appropriate E2 levels.
US-guided oocyte retrieval (OPU) was performed 35–37 h after hCG trigger under local anesthesia (paracervical block). Follicular fluid was aspirated and immediately observed under a stereomicroscope. Cumulus-oocyte complexes (COCs) were washed in buffered medium (Flushing medium, Cook Ltd., Ireland); 2 h after OPU, oocytes and CCs were separated from each individual COC by gently pipetting in 40-μl HEPES-buffered medium containing 80 IU/ml hyaluronidase (Synvitro Hyadase, Origio Medicult, Denmark) [20]. CCs samples were separated from the corresponding mature oocytes and stored in liquid nitrogen until gene expression analysis.
Preparation of semen samples, ICSI, and fertilization check
Sperm concentration, motility, and morphology were assessed according to the World Health Organization guidelines (WHO laboratory manual 5th ed., 2010). Semen samples were prepared using density gradient centrifugation in order to select normally motile morphologically normal spermatozoa. ICSI was performed with all available mature oocytes within 2 h after CCs removal. Normal fertilization was confirmed when the presence of two pronuclei (2PN) and the extrusion of the second polar body were observed 16–18 h after injection.
Time-lapse embryo culture
Embryos were cultured using the Geri plus® TLS (Genea Biomed, Germany) with integrated embryo monitoring system in microwells (one zygote/microwell). The dish format allowed following each embryo individually even if all embryos shared a common 80-μl drop of medium. Embryos were cultured in pre-equilibrated Cleavage medium (Cook, Ireland) overlaid with mineral oil up to day 3; at this stage, a change of medium was performed and a new one (Blastocyst medium, Cook, Ireland) was added and kept until the blastocyst stage. Bright field images captured by the Geri plus® system were acquired every 5 min from the time of fertilization until the time of embryo transfer (ET), cryopreservation, or discharge.
Embryo morphological evaluation was firstly performed on day 2 using the Integrated Morphology Cleavage Score (IMCS) [21]; embryo selection for ET was performed on day 5 considering both conventional morphology [22] and morphokinetic parameters [4]. All the videos collected by TLS were analyzed, and the following morphokinetic parameters (times) were considered: pronuclear appearance (tPNa); pronuclear fading (tPNf); completion of cleavage to two, three, four, and eight cells (t2, t3, t4, and t8 respectively); and time intervals tPNf-tPNa, t2-tPNf, t3-t2, t4-t3, t4-t2, and t8-t4. Time intervals from t8 (completed cleavage to eight cells) to expanded blastocyst (t cavitation, t blastocyst, t hatching) were annotated, but not considered in our analysis as our aim was to identify early predictive biomarkers of embryo development.
Embryos that progressed to the expanded blastocyst stage on day 5 (score 3 according to [22]) were included in the blastocyst group (BL group; n = 27); embryos that progressed to the early or not fully expanded blastocyst stage on day 5 (score 1 or 2) were included in the not-expanded blastocyst group (nBL group; n = 19), whereas those undergoing developmental arrest, reaching as maximum the morula stage on day 5 and remaining at the same stage until day 6 were included in the arrested group (AR group; n = 35). Blastocyst assessment was performed at the same time interval (116 ± 2 h after injection) for all embryos. Thirteen expanded blastocysts were transferred in utero (one per patient) using the soft catheter Sydney Guardia (Cook, Australia) under transvaginal US guidance, applying the method previously published by our group [23].
RNA extraction, cDNA synthesis, and RT-qPCR
CCs corresponding to each embryo were analyzed by RT-qPCR. Cells-to-CTTM 1-Step PowerSYBR® Green Kit (Ambion; Life Technologies, Italy) was used to extract total RNA following the manufacturer’s instruction. Twenty microliters of RNA were used for retrotranscription using SensiFASTTM cDNA Synthesis Kit (Bioline, Meridian Bioscience, UK) at 42 °C for 15 min and 85 °C for 5 min. cDNAs were stored at – 80 °C until PCR analysis. PCR primers were designed using NCBI/Primer-BLAST and synthesized by Sigma-Merck (UK): bone morphogenetic protein 15 (BMP-15), 5′-GGCTCCTAGGGCATTCACTG-3′, 5′-CCTCGGTTTGGTCTGAGAGG-3′ [24]; connexin 43 (Cx43), 5′-TACCAAACAGCAGCGGAGTT-3′, 5′-TGGGCACCACTCTTTTGCTT-3′ [25]; subunit II of cytochrome C oxidase (COX or respiratory complex IV) (COXII), 5-CGACTACGGCGGACTAATCT-3′, 5′-TCGATTGTCAACGTCAAGGA-3′ [26]; a mitochondrial gene coding for the ATP synthase subunit 6 (MT-ATP6), 5′ CCAATAGCCCTGGCCGTAC-3′, 5′ CGCTTCCAATTAGGTGCATGA-3′ [26]; heme oxygenase 1 (HO-1) [17], 5′-AGGAGGAGATTGAGCGCCAC-3′, 5′-TCTGGTCCTTGGTGTCATGG-3′; beta-2-microglobulin (B2M), 5′-AGCAAGGACTGGTCTTTCTATCTC-3′, 5′-ATGTCTCGATCCCACTTAACTA-3′ [26]; S14 ribosomal protein gene (S14), 5′-AGGTGCAAGGAGCTGGGTAT-3′, 5′-TCCAGGGGTCTTGGTCCTATT-3′ [27]. B2M and S14 were used as endogenous controls as they were previously confirmed to be stable (M value of 0.176 and 0.183, respectively) in human CCs using the CFX Manager Software analysis [27].
SensiFASTTM SYBR® No-ROX Kit (Bioline, Aurogene Srl, Italy) was used to perform RT-qPCR following manufacturer’s protocol: 1 cycle at 95 °C for 2 min, 40 cycles at 95 °C for 5 s, 1 cycle at 60 °C for 30 s and 1 cycle at 95 °C for 1 min. Gene expression analysis was performed by CFX Manager Software (BioRad, CA, USA). Each reaction was performed in triplicate. Melt curve analysis confirmed the specificity of PCRs.
Statistical analysis
Continuous variables are shown as mean ± standard deviation (SD), whereas categorical variables as absolute and relative frequencies. At univariate analysis, the analysis of variance (ANOVA) with Bonferroni correction for multiple comparisons or the non-parametric Kruskal-Wallis rank test, as appropriate, were carried out to compare the BL, the nBL, and the AR groups for all the recorded times and time intervals. The correlation between embryo morphokinetic parameters and CCs gene expression was assessed using the Spearman rank correlation coefficient.
Multivariable analysis models were then fitted to evaluate the independent effect of morphokinetic parameters and gene expression on blastocyst development: blastocyst development at day 5 (arrested embryos – AR group vs. expanded blastocyst – BL group) was set as the dependent variable of logistic regression models, in which different time-lapse parameters and gene expressions in CCs were taken as independent variables.
Significance level was set at α = 0.05. All analyses were performed using STATA 14 (StataCorp. 2015. Stata Statistical Software: Release 14. College Station, TX: StataCorp LP).
GraphPad Prism V7 was used to generate the receiver operating characteristic (ROC) curves in order to assess the performance of different morphokinetic and gene expression variables in identifying embryos with blastocyst development potential. The area under the ROC curve (AUC) was determined to provide a numerical summary of the indicator’s performance. Cut-off points were identified for specificity and sensitivity when the Youden’s Index (sensitivity + specificity − 1) was maximal, considering BL and AR as test and control groups, respectively.
Results
Patients
Table 1 summarizes the clinical characteristics of the patients and the outcome of their IVF cycles. The age and biomarkers were as expected of good prognosis patients; indeed, the retrieved oocytes mean number was 9.3 ± 2.3, and the clinical PR/ET was 61.5%, in line with the predicted outcome after transfer in utero of a single expanded blastocyst in such patients.
Table 1.
Patients’ clinical characteristics and IVF outcome
| Age (years) | 35.8 ± 2.9 |
| BMI (kg/m2) | 23.3 ± 3.2 |
| Basal (day 3) FSH (IU/l) | 7.5 ± 2.8 |
| AMH (ng/ml) | 3.4 ± 2 |
| AFC (n) | 16 ± 8.6 |
| Total exogenous FSH (IU) | 2254.2 ± 734.8 |
| Peak E2 (pg/ml) | 1823.3 ± 486.8 |
| Endometrial thickness (mm) | 11 ± 1.6 |
| Retrieved oocytes (n) | 9.3 ± 2.3 |
| OSI (n) | 4.5 ± 1.7 |
| Maturation rate (%) | 90 ± 14.3 |
| Fertilization rate (%) | 81.7 ± 16.2 |
| Cleavage rate (%) | 97.4 ± 6.3 |
| Blastocyst rate (%) | 61.3 ± 28.4 |
| Implantation rate % (n) | 61.5 (8/13) |
| Clinical PR/ET % (n) | 61.5 (8/13) |
| Live birth rate/ET % (n) | 61.5 (8/13) |
Data are shown as mean ± standard deviation, with the only exception of the last variables which are expressed as a percentage
BMI body mass index, AFC antral follicle count, FSH follicle-stimulating hormone, E2 estradiol, OSI ovarian sensitivity index = number of retrieved oocytes × 1000/total FSH dose
Morphokinetic analysis
Overall, 81 embryos were obtained, and during in vitro culture in the TLS GERI plus®, 27 embryos progressed to the expanded blastocyst stage (BL group), 35 embryos arrested their development on day 5 and remaining at the same stage until day 6 (AR group), whereas 19 embryos reached an intermediate state of development (early blastocyst or not fully expanded blastocyst) on day 5 (nBL group). Interestingly enough, the morphological score IMCS [21] on day 2 was comparable in the three groups (9 ± 4.6 vs. 7.3 ± 1.4 vs. 8.8 ± 4.3 for BL, nBL, and AR groups, respectively; p > 0.05).
Comparing the three groups, we found that times tPNa, tPNf, t2, t3, t4, and t2-tPNf showed significant differences (Table 2): embryos of the BL group showed a significantly quicker early kinetic than embryos in the AR group (p < 0.05) (Fig. 1). We also observed a shorter (although not significantly) t8 time in the BL group.
Table 2.
Morphokinetic parameters recorded by TLS GERI plus®
| BL group (n = 27) |
nBL group (n = 19) |
AR group (n = 35) |
p value* | ∆ (AR group - BL group) time | ∆ (nBL group - BL group) time | ∆ (AR group - nBL group) time | |
|---|---|---|---|---|---|---|---|
| tPNa (h) | 6.6 ± 1.5 | 7.0 ± 1.4 | 7.5 ± 1.5 | 0.0496^ | 0.9 | 0.4 | 0.5 |
| tPNf (h) | 20.9 ± 2.2 | 22.2 ± 2.4 | 24.0 ± 3.5 | 0.0006 | 3.1 | 1.3 | 1.8 |
| t2 (h) | 23.7 ± 1.9 | 24.7 ± 2.4 | 26.5 ± 3.6 | 0.0028 | 2.8 | 1.0 | 1.8 |
| t3 (h) | 33.1 ± 4.0 | 34.9 ± 3.4 | 36.9 ± 6.0 | 0.0031 | 3.8 | 1.8 | 2.0 |
| t4 (h) | 36.0 ± 4.8 | 37.6 ± 3.8 | 41.2 ± 8.7 | 0.0007 | 5.2 | 1.6 | 3.6 |
| t8 (h) | 57.4 ± 7.7 | 62.8 ± 15.2 | 65.0 ± 12.7 | 0.0984 | 7.6 | 5.4 | 2.2 |
| tPNf-tPNa (h) | 14.6 ± 2.4 | 15.3 ± 1.8 | 16.3 ± 3.5 | 0.1485 | 1.7 | 0.7 | 1.0 |
| t2-tPNf (h) | 2.5 ± 0.4 | 2.5 ± 0.4 | 2.8 ± 0.7 | 0.0368 | 0.3 | 0 | 0.3 |
| t3-t2 (h) | 9.4 ± 3.6 | 10.2 ± 3.1 | 10.4 ± 4.6 | 0.1310 | 1.0 | 0.8 | 0.2 |
| t4-t2 (h) | 12.8 ± 3.8 | 12.9 ± 3.4 | 14.2 ± 7.9 | 0.3479 | 1.4 | 0.1 | 1.3 |
| t8-t4 (h) | 21.9 ± 7.6 | 28.3 ± 17.5 | 26.4 ± 10.2 | 0.3557 | 4.5 | 6.4 | -1.9 |
Times and time intervals are shown for embryos reaching the expanded blastocyst stage (BL group), the not-expanded blastocyst group (nBL group) and those whose development arrested (AR group). Values are expressed as mean ± standard deviation. In the ΔTime column, the difference between embryos in groups AR vs. BL, nBL vs. BL and AR vs. nBL were calculated for each parameter. The p-value column shows the significance of the overall comparison among the three groups, performed with analysis of variance (^) or Kruskal-Wallis rank test, as appropriate
Italic p values indicate significant differences
Fig. 1.
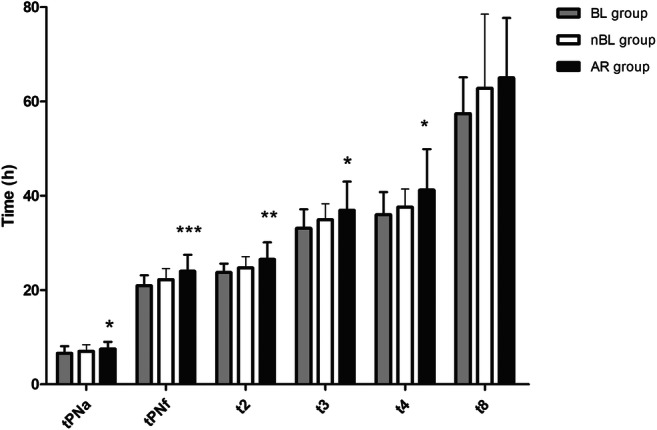
Morphokinetic of embryos reaching the expanded blastocyst stage (BL group), the not-expanded blastocyst stage (nBL group) and of those whose development arrested (AR group). Times legend is the following: pronuclear appearance (tPNa), pronuclear fading (tPNf), completion of cleavage to two, three, four, and eight cells (t2, t3, t4, and t8 respectively). Values are expressed as mean ± standard deviation. *p < 0.05, **p < 0.01, ***p < 0.001 AR group vs. BL group
The interval t2-tPNf, corresponding to the first cleavage, was significantly shorter in the BL group (p = 0.035), and the delay of embryos in AR group vs. those in BL group started as soon as pronuclei appeared (tPNa), progressively increasing until t8 as shown by longer ∆time value (Table 2 and Fig. 1). No significant differences were observed comparing BL vs. nBL group, or nBL vs. AR group (data not shown).
Gene expression in CCs
CCs corresponding to the 81 embryos assessed by TLS GERI plus® were analyzed by RT-qPCR in order to assess the expression of genes BMP15, Cx43, COXII, MT-ATP6, and HO-1. We observed that BMP15, COXII, and MT-ATP6 mRNA expression was significantly lower (p < 0.05 or p < 0.01) in CCs corresponding to embryos of the AR group than in those corresponding to embryos of the BL group (Fig. 2). On the contrary, Cx43 and HO-1 mRNA expression was significantly higher in CCs corresponding to embryos of the AR group (p < 0.001) (Fig. 2). Instead, no significantly different gene expression was observed comparing nBL vs. BL group or nBL vs. AR group (Fig. 2). The observed difference between AR and BL groups was confirmed also by the analysis of gene expression intra-patient variability: within each patient-specific cohort of zygotes, most CC samples of the BL group clustered above the median for BMP15, COXII, and MT-ATP6, and below the median for Cx43 and HO-1 mRNA expression (Fig. 3). The opposite trend was observed for the AR group.
Fig. 2.
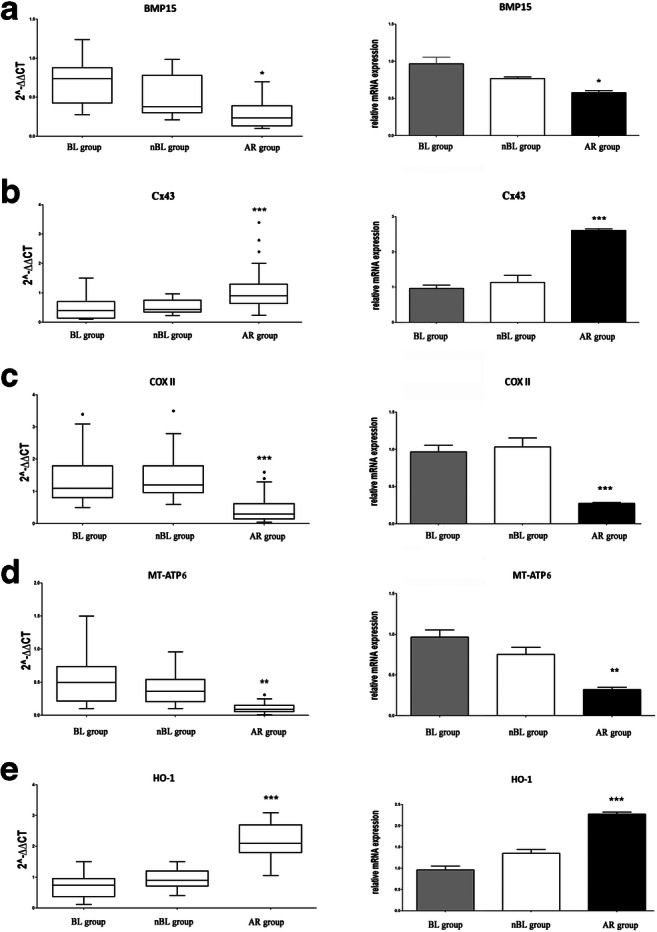
Gene expression of BMP15, Cx43, COXII, MT-ATP6, and HO-1 in the CCs of the three analyzed groups. For each gene, the box-plots of 2−ddCT data (on the left) and the relative mRNA expression (on the right) analyzed by RT-qPCR in individual cumulus cells corresponding to embryos reaching the expanded blastocyst stage (BL group, n = 27), not expanded blastocyst stage (nBL group, n = 19), or those whose development arrested (AR group, n = 35) are shown. Measurements of bone morphogenetic protein 15 (BMP15), cytochrome c oxidase subunit II (COXII), ATP synthase subunit 6 (MT-ATP6), connexin 43 (Cx43), and heme oxygenase-1 (HO-1) were performed in triplicate. *p < 0.05, **p < 0.01, ***p < 0.001 AR group vs. BL group
Fig. 3.
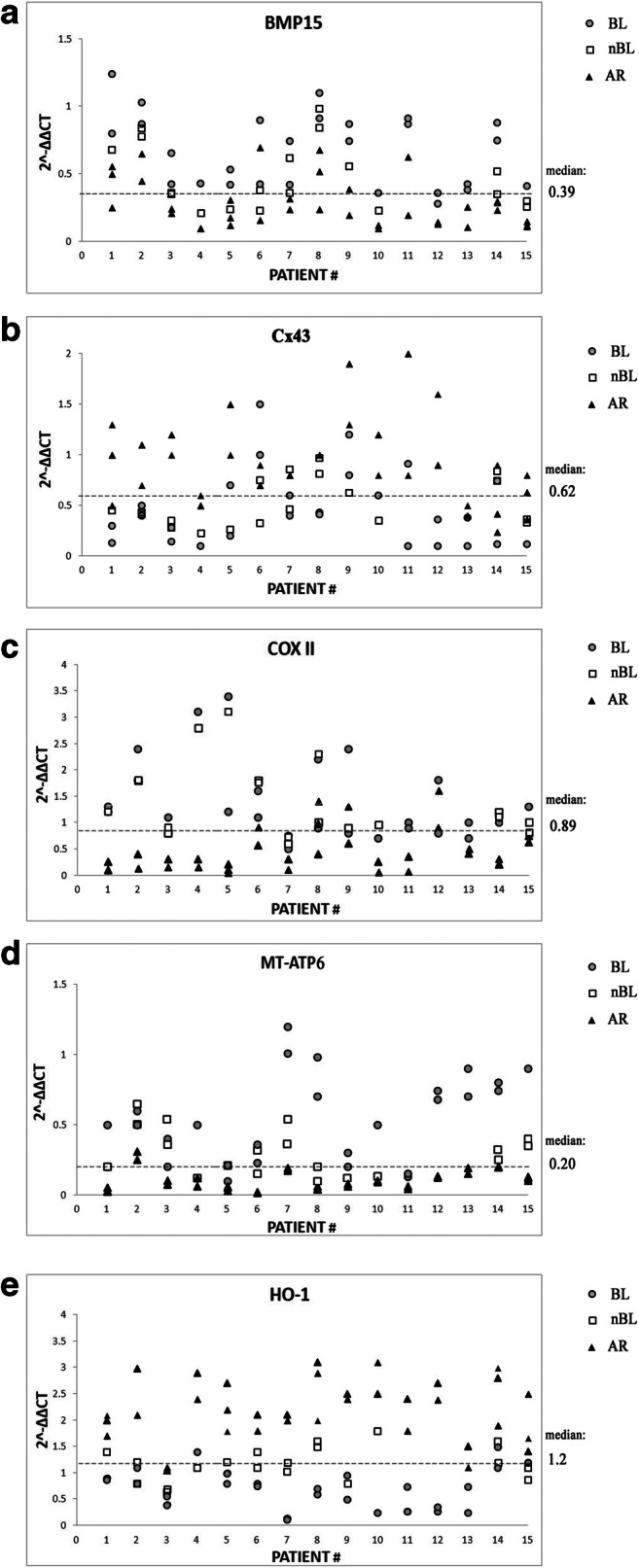
Intra-patient variability analysis for the expression of BMP15, Cx43, COXII, MT-ATP6, and HO-1. For all analyzed genes, the 2−ddCT data of each cumulus cell (CC) sample are grouped by patient and are represented according to embryo development (expanded blastocyst = gray circles, not-expanded blastocyst = white squares, arrested development = black triangles). The median general expression value has been reported for each gene as a dashed black line
Correlation between gene expression in CCs and morphokinetic parameters
Overall, we observed a negative relationship between mRNA levels of the analyzed genes and the evaluated morphokinetic parameters. As shown in Table 3, embryos developing slower had a lower expression of BMP15, COXII, and MT-ATP6 in the corresponding CCs. In fact, we found a statistically significant negative correlation between BMP15 mRNA levels in CCS and the times t3 (rho = − 0.34, p < 0.05), t4 (rho = − 0.39, p < 0.05), t8 (rho = − 0.40, p < 0.05), time intervals tPNf-tPNa (rho = − 0.33, p < 0.05) and t8-t4 (rho = − 0.32, p < 0.05) (Table 3).
Table 3.
Correlation between the expression of specific genes in the CC and embryo morphokinetic parameters
| tPNa | tPNf | t2 | t3 | t4 | t8 | tPNf-tPNa | t2-tPNf | t3-t2 | t4-t2 | t8-t4 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| BMP15 | − 0.0625 | − 0.2921 | − 0.2960 | − 0.3352 | − 0.3885 | − 0.3950 | − 0.3258 | − 0.2946 | − 0.2510 | − 0.1960 | − 0.3160 |
| p value | 0.7014 | 0.0674 | 0.0637 | 0.0345* | 0.0132* | 0.0117* | 0.0402* | 0.0650 | 0.1183 | 0.2255 | 0.0470* |
| Cx43 | 0.0760 | − 0.2700 | − 0.1610 | − 0.1255 | − 0.1067 | 0.0644 | − 0.3578 | 0.4260 | − 0.0359 | 0.0142 | 0.1520 |
| p value | 0.6412 | 0.0920 | 0.3209 | 0.4405 | 0.5121 | 0.6928 | 0.0234 | 0.0061 | 0.8258 | 0.9305 | 0.3490 |
| COXII | 0.0029 | − 0.1443 | − 0.2216 | − 0.3407 | − 0.1485 | − 0.1339 | − 0.2702 | − 0.3664 | − 0.0938 | 0.1354 | − 0.1443 |
| p value | 0.9857 | 0.3745 | 0.1694 | 0.0315* | 0.3604 | 0.4102 | 0.0918 | 0.0200* | 0.5647 | 0.4048 | 0.3743 |
| MT-ATP6 | − 0.2800 | − 0.4024 | − 0.4657 | − 0.5390 | − 0.3334 | − 0.0377 | − 0.3594 | − 0.3995 | − 0.1903 | 0.1465 | 0.1193 |
| p value | 0.0801 | 0.0100 ** | 0.0025** | 0.0003*** | 0.0355 * | 0.8172 | 0.0227 * | 0.0107 * | 0.2396 | 0.3672 | 0.4634 |
| HO-1 | 0.1261 | − 0.0981 | − 0.0029 | − 0.0213 | 0.0013 | 0.0443 | − 0.2178 | 0.1242 | 0.0533 | 0.0156 | 0.0599 |
| p value | 0.4380 | 0.5469 | 0.9860 | 0.8964 | 0.9938 | 0.7860 | 0.1770 | 0.4452 | 0.7441 | 0.9239 | 0.7136 |
Spearman rank coefficients and p-values for correlations of mRNA of bone morphogenetic protein 15 (BMP15), cytochrome c oxidase subunit II (COXII), ATP synthase subunit 6 (MT-ATP6), connexin 43 (Cx43) and heme oxygenase-1 (HO-1) with timelapse parameters (pronuclear appearance (tPNa), pronuclear fading (tPNf), completion of cleavage to two, three, four and eight cells (t2, t3, t4, and t8 respectively), time intervals tPNf-tPNa, t2-tPNf, t3-t2, t4-t3, t4-t2 and t8-t4) are shown: *p < 0.05, **p < 0.01 and ***p < 0.001
Logistic regression models
At multivariable analysis, performed only on the AR and BL groups, the expression of the two CCs genes BMP15 and COXII was independently and significantly associated with a higher probability of development to expanded blastocyst on day 5 (p < 0.05) (Table 4). On the contrary, HO-1 mRNA level in CCs corresponded to a higher risk of embryo developmental arrest, although not statistically significant (not shown).
Table 4.
Multivariable logistic regression analysis
| OR | CI 95% | p | |
|---|---|---|---|
| tPNa | 1.86 | 0.97; 3.6 | 0.063 |
| BMP15 | 0.18 | 0.04; 0.88 | 0.034 |
| tPNf | 1.25 | 0.90; 1.71 | 0.175 |
| BMP15 | 0.22 | 0.05; 1.01 | 0.052 |
| t3 | 1.04 | 0.86; 1.25 | 0.696 |
| BMP15 | 0.19 | 0.04; 0.96 | 0.045 |
| t4 | 1.12 | 0.94; 1.33 | 0.202 |
| BMP15 | 0.17 | 0.03; 0.96 | 0.046 |
| dPN | 1.1 | 0.72; 1.65 | 0.685 |
| COXII | 0.03 | 0.003; 0.30 | 0.003 |
| t2 | 1.3 | 0.82; 2.02 | 0.282 |
| COXII | 0.03 | 0.002; 0.3 | 0.004 |
| t3 | 1.05 | 0.85; 1.30 | 0.632 |
| COXII | 0.03 | 0.003; 0.32 | 0.004 |
| t4 | 1.38 | 0.88; 2.13 | 0.157 |
| COXII | 0.02 | 0.001; 0.32 | 0.005 |
| t8 | 1.40 | 0.99; 1.90 | 0.053 |
| COXII | 0.01 | 0.003; 0.41 | 0.014 |
Independent effect of bone morphogenetic protein 15 (BMP15) and cytochrome c oxidase subunit II (COXII) genes expression in cumulus cells and morphokinetic parameters with respect to the probability of embryo development on day 5 (arrested group vs. blastocyst group). Odds ratio (OR), 95% Confidence Interval (CI 95%) and p-value are shown for each model
Predictive cut-off values of embryo development on day 5
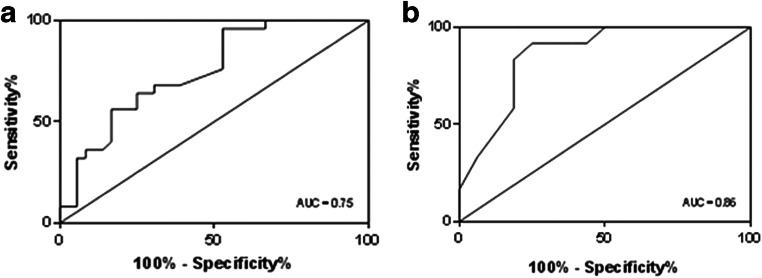
ROC curve analysis was applied to identify optimal cut-off values of both morphokinetic parameters and CCs gene expression predictive of development to expanded blastocyst on day 5. According to maximization of the Youden Index, the parameters showing an optimal cut-off value were t2 for morphokinetic assessment (cut-off at t2 < 26.25 having sensitivity = 96 % and specificity = 47 %, AUC = 0.75; 95 % CI, 0.62–0.87; p = 0.001) (Fig. 4a), and COXII for mRNA expression (cut-off at COXII > 0.3 having sensitivity = 92 %, specificity = 75%, AUC = 0.86; 95 CI, 0.72–0.99; p = 0.001) (Fig. 4b).
Fig. 4.
ROC curves for the predictive potential of TLS time of completion of cleavage to two cells (t2) (a) and cytochrome c oxidase subunit II (COXII) gene expression (b) with respect to the probability of embryo development to expanded blastocyst on day 5. Cut-off values are at t2 < 26.25 (sensitivity = 96% and specificity = 47%, AUC = 0.75; 95% CI, 0.62–0.87; p = 0.001) and at COXII > 0.3 (sensitivity = 92%, specificity = 75%, AUC = 0.86; 95 CI, 0.72–0.99; p = 0.001)
Discussion
The identification of markers able to accurately predict embryo development to the expanded blastocyst stage by studying the early stages of embryo growth (within 72 h from fertilization) is of high practical interest for IVF Labs. In fact, it could allow substituting the selective transfer of a single blastocyst with that of a single day 3 embryo having an optimal potential to become a fully developed, expanded blastocyst on day 5. In practical terms, this would reduce the risk of epigenetic alterations due to a more prolonged embryo permanence in artificial environment [2].
Unfortunately, none of the “classical” morphological scores of the cleaving embryo has proven to be fully reliable in identifying embryos with the best development capability [3]. The rather recent advent of TLS technology provided the possibility to obtain a noticeable bulk of data about the kinetic of human embryo growth in culture, allowing the evaluation of embryo features previously unknown after conventional morphology analysis. Despite this, TLS benefits on embryo selection and IVF outcome, claimed by some authors [28], are disputable according to others [29–33] and, even most important, are partially due to the unperturbed culture conditions rather than to the application of morphokinetic data. Artificial intelligence approach based on deep neural networks has recently been applied to TLS, and would hopefully be useful to develop novel embryo selection algorithms [34]; to date, however, the results of this approach are still unavailable in the clinical practice.
The possibility to use TLS to select the best embryo at the cleavage stage, without waiting day 5, is still hypothetical. Some authors claim that TLS technology may dissect specific events occurring during and soon after fertilization and may identify early putative markers of embryo quality [35]. Recently, our group showed that Eeva™, an algorithm based on the analysis of early embryo growth, is able to improve embryo selection accuracy of standard morphology by effectively predicting which embryo at day 3 will develop into a viable blastocyst on day 5 [36]. Today, however, a panel of specific early morphokinetic features predictive of a favorable embryo development to the blastocyst stage is still lacking. This study was aimed at tackling the issue, providing also a link between embryo morphokinetics and specific molecular markers of gene activation in the corresponding CCs.
Using a last generation TLS, we observed that embryos able to achieve the expanded blastocyst stage on day 5 were characterized by a significantly faster cellular kinetic already during the first 3 days of development; interestingly, these embryos showed a quicker cleavage rate and then proceeded progressively increasing the time gap (∆Time) relative to the embryos who arrested their development; in fact, the gap between embryos reaching the expanded blastocyst stage and those undergoing developmental arrest was maximal on day 3 (ΔTime t8 = 7.6 h). Embryos showing an optimal development on day 5 had a day 2 morphological score comparable to that of the embryos arresting their growth, but very precociously showed shorter and anticipated cleavage times, and kept being faster until day 3, when their DNA started to get its full activation.
The correlation between morphokinetic markers, embryo development, blastocyst formation rates, and ploidy has already reported [37]. Our observations confirm previous studies reporting that a timely blastocyst development on day 5 can be predicted quite early looking at the duration of the first cytokinesis, as well as at the time interval between the end of the first cleavage and the initiation of the second, the time interval between the second and third cleavage [38], the duration of the second [39] and third cleavage divisions [40], and the absence of direct cleavage from one to three cells [41]. Also, Cruz et al., analyzing the morphokinetic data of 834 embryos, found that blastocyst formation rate and blastocyst quality were related to the cleavage kinetic of early embryo [42]. Likewise, Dal Canto et al. showed that embryos that cleave earlier from the 2-cells to 8-cells stage have the highest potential to become blastocyst, expand, and implant [43]. Hashimoto et al. reported that the time intervals between the 3-cells and the 4-cells stage and between the 5-cells and the 8-cells stage were shorter for embryos with high-scored blastocysts compared to low-scored [40]. In addition, Minasi et al., analyzing 928 blastocysts morphokinetic parameters, found that euploid embryos needed shorter intervals to expand to blastocyst and hatch compared to aneuploid embryos [44], while Campbell et al. described that aneuploid embryos showed a delayed initiation of compaction and blastulation, as well as a longer time to reach the full blastocyst stage [45].
Looking at the clinical implications of embryo kinetics, it was reported that the transfer of fresh, slow-growing blastocysts resulted in lower implantation and clinical pregnancy rate compared to the transfer of fully expanded day 5 blastocysts [46, 47], probably due both to a reduced embryo competence and to a loss of embryonic-endometrial synchrony [48]. Moreover, the transfer of a fresh early blastocyst on day 5 was found to obtain success rates comparable to the transfer of a thawed expanded blastocyst cryopreserved on day 6 from the same cohort [49], suggesting that embryo kinetic is more important than the embryo developmental stage itself in determining IVF outcome. In fact, in cases where only early or not fully expanded blastocyst are available on day 5, extending culture until cryopreservation on day 6, and subsequent transfer after thawing may result in successful pregnancy. Taken together, these observations lead us to consider for our analysis the fully expanded blastocyst as the ideal developmental stage that most likely relates to higher embryo implantation. However, the transfer of slower-growing embryos may represent a feasible option in the absence of expanded blastocyst on day 5 or a favorable strategy to increase the cumulative pregnancy rates after frozen-thawed embryo transfer cycles. Using our combined approach, embryos that will develop into early blastocysts cannot be distinguished from those that will arrest, probably due to the limited sample size; however, we observed that embryos achieving the stage of early or not fully expanded blastocyst showed a morphokinetic behavior similar to embryos that reached full expansion on day 5 of development and a quicker, although not significant, kinetics compared to embryos undergoing developmental arrest.
Gene expression in CCs was already proposed as a non-invasive assay useful to predict embryo development and pregnancy [8]. An increased expression of a specific set of genes in CCs was previously related to a higher likelihood of oocyte fertilization, embryo quality, and clinical pregnancy [20, 50–52].
In this study, assessing the gene expression related to oocyte-secreted factors in CCs, we showed that BMP-15 was significantly more expressed, and Cx43 significantly less expressed, in the CCs corresponding to embryos developing to expanded blastocyst on day 5 than in those corresponding to embryos with arrested development. The significant relationship between BMP-15 expression in CCs and optimal embryo development was confirmed by the good correlation between BMP15 and embryo growth speed, as well as by the multivariable logistic regression analysis. Indeed, BMP-15 is an oocyte quality-related gene, playing a fundamental role in normal follicular growth, ovulation, and fertilization: increased BMP-15 levels in FF [53] and in CCs [11] were associated with embryos having good competence for implantation and pregnancy. BMP-15 is also involved in functional changes of GAP junctions and the downregulation of Cx43 expression in granulosa cells [24]. Indeed, previous clinical data showed that CCs reducing Cx43 expression at the time of oocyte collection provided a good embryo competence [54]. In agreement with previous studies, our observations suggest that reduced levels of Cx43 expression in CCs, coupled with high BMP-15 mRNA levels, can be considered a marker of embryo competence.
As some proteins are good bi-directional indicators in the communication between the oocyte and the somatic cells surrounding it, we carried out a preliminary analysis aimed at evaluating the metabolic competence of CCs. Interestingly, previous studies on bovine cumulus-oocyte complexes demonstrated that a metabolic consequence of recombinant human BMP15 addition is the modulation of the oocyte oxidative phosphorylation, acting via CCs [55].
Up to now, no study ever investigated the metabolic consequences of high BMP15 levels in the human cumulus-oocyte complex, notwithstanding that, as member of the transforming growth factor β (TGF-β) superfamily, also BMP15 could exert a metabolic effect similar to TGF-β, which is a key modulator of the respiratory chain [56]. Our approach found interesting novel data related to CCs metabolism. In fact, in the CCs corresponding to embryos with arrested development, we detected a significantly decreased expression of both COXII and MT-ATP6, markers of mitochondrial transcription activity contributing to the modulation of respiratory chain and ATP synthesis. Indeed, COXII mRNA levels were significantly higher in the CCs corresponding to embryos that reached the expanded blastocyst stage and even the logistic regression analysis confirmed a significant, direct correlation between embryo progression to blastocyst and COXII gene expression. As mitochondrial-encoded proteins are required for the formation of active respiratory complexes, this observation unveils the importance of mitochondria in supporting human embryo growth. A downregulation of COXII in CCs, in fact, is associated to reduced ATP levels, decreased mitochondrial membrane potential, and enhanced production of ROS [57]. Likewise, a diminished MT-ATP6 gene expression may result in reduced ATP synthesis rate [58]. The observed decrease of COXII and MT-ATP6 gene expression in CCs in association with embryo developmental arrest suggests that an impaired mitochondrial function in CCs could negatively affect the development potential of embryos since the earliest stages of growth.
Moreover, it was previously reported that the gene expression of HO-1 was significantly higher in lower quality, arrested embryos, consistently with a dysfunctional state of mitochondria [59]. We previously reported that the presence of an oxidative stress status inducing a significantly higher HO-1 expression in CCs may impair oocyte competence [17], suggesting that an excessive ROS production can trigger oocyte damage. The present study further supports the role of ROS, as it shows that a higher expression of HO-1 in CCs is also associated with a higher likelihood of inadequate embryo development.
Notwithstanding the small sample size, the data of gene expression relative to the embryos that achieve the stage of early or not fully expanded blastocyst show a trend similar to what is observed for the expanded blastocyst on day 5 group, suggesting that also these embryos can implant.
A relevant finding of the current study is the evidence that the expression of genes encoding for some oocyte-secreted factors or reflecting mitochondrial status in CCs are significantly related to the morphokinetic parameters of the corresponding embryo. This finding strongly supports the concept that the follicular microenvironment affects not only oocyte maturation, but also the early embryo growth: in fact, embryo development in the first 3 days is almost exclusively due to the expression of oocyte DNA and to metabolic resources of oocyte cytoplasm [58]. Actually, Hammond et al. reported that the expression of CCs specific genes was significantly associated with the achievement of the blastocyst stage on day 5, but differently from our findings, none of these genes was found to significantly correlate with TLS-acquired parameters [39]. On the other side, Scarica et al. observed that a slower embryo growth was associated with developmental arrest and that the assessment of calcium/calmodulin-dependent protein kinase 1D (CAMK1D), prostaglandin-endoperoxide synthase 2 (PTGS2) and hyaluronic acid synthase 2 (HAS2) gene expression in CCs could boost the predictive power of TLS when the two approaches were combined [60].
Despite the limited number of patients considered in this study, gene expression of both BMP15 and COXII resulted strongly associated with a higher probability of blastocyst development on day 5 at multivariable analysis, independently from embryo morphokinetic. This suggests that although early embryo development in the first 3 days relies mostly on resources belonging to the oocyte, the follicular environment where the oocyte develops may partially influence embryo competence to become a blastocyst.
In conclusion, our study shows that embryo morphokinetics, in particular in the first 3 days of in vitro growth, as well as the expression of BMP15 and COXII genes in the corresponding CCs, is able to predict whether an embryo will timely reach the expanded blastocyst stage or will prematurely undergo developmental arrest. ROC curves identified the cut-off values of t2 < 26.25 for kinetics and COXII > 0.3 for gene expression as optimal thresholds to predict a timely blastocyst development; this observation warrants further testing in order to check its potential for embryo selection and implantation prediction in the routine clinical practice. Our data suggest the possibility of developing integrated prediction models and algorithms for early embryo selection at the cleavage stage including both morphokinetic and molecular parameters, even if time-lapse technology is not routinely used in all IVF laboratories for embryo monitoring as support in embryo selection.
Acknowledgments
This work was supported by MIUR in the PhD (for S.C.) and Postdoc Program (for L.B.) of the University of Torino, Italy.
Authors’ contribution
A.R., S.C., and L.B. conceived the study; A.C. and N.D.S. enrolled the patients; S.C., C.M., and C.P. were responsible for the time-lapse data acquisition and analysis; L.B. and C.M. performed real-time polymerase chain reaction experiments; L.C. performed the statistical analysis and S.C. contributed to the statistical analysis; S.C. and L.B. contributed to the validation, the interpretation of the results, the writing and editing of the manuscript; F.S., G.G., C.B., and A.R. contributed to the final interpretation of the data and editing of the manuscript. All authors gave their final approval.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Stefano Canosa and Loredana Bergandi are coauthors
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Glujovsky D, Farquhar C, Quinteiro Retamar AM, Alvarez Sedo CR, Blake D. Cleavage stage versus blastocyst stage embryo transfer in assisted reproductive technology. Cochrane Database Syst Rev. 2016;CD002118. [DOI] [PubMed]
- 2.White CR, Denomme MM, Tekpetey FR, Feyles V, Power SGA, Mann MRW. High frequency of imprinted methylation errors in human preimplantation embryos. Sci Rep. 2015;5:17311. doi: 10.1038/srep17311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gardner DK, Balaban B. Assessment of human embryo development using morphological criteria in an era of time-lapse, algorithms and “OMICS”: is looking good still important? Mol Hum Reprod. 2016;22:704–718. doi: 10.1093/molehr/gaw057. [DOI] [PubMed] [Google Scholar]
- 4.Meseguer M, Herrero J, Tejera A, Hilligsøe KM, Ramsing NB, Remohí J. The use of morphokinetics as a predictor of embryo implantation. Hum Reprod Oxf Engl. 2011;26:2658–2671. doi: 10.1093/humrep/der256. [DOI] [PubMed] [Google Scholar]
- 5.Conaghan J, Chen AA, Willman SP, Ivani K, Chenette PE, Boostanfar R, et al. Improving embryo selection using a computer-automated time-lapse image analysis test plus day 3 morphology: results from a prospective multicenter trial. Fertil Steril. 2013;100:412–419.e5. doi: 10.1016/j.fertnstert.2013.04.021. [DOI] [PubMed] [Google Scholar]
- 6.Diamond MP, Suraj V, Behnke EJ, Yang X, Angle MJ, Lambe-Steinmiller JC, Watterson R, Athayde Wirka K, Chen AA, Shen S. Using the Eeva TestTM adjunctively to traditional day 3 morphology is informative for consistent embryo assessment within a panel of embryologists with diverse experience. J Assist Reprod Genet. 2015;32:61–68. doi: 10.1007/s10815-014-0366-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kieslinger DC, De Gheselle S, Lambalk CB, De Sutter P, Kostelijk EH, Twisk JWR, et al. Embryo selection using time-lapse analysis (Early Embryo Viability Assessment) in conjunction with standard morphology: a prospective two-center pilot study. Hum Reprod Oxf Engl. 2016;31:2450–2457. doi: 10.1093/humrep/dew207. [DOI] [PubMed] [Google Scholar]
- 8.Fragouli E, Lalioti MD, Wells D. The transcriptome of follicular cells: biological insights and clinical implications for the treatment of infertility. Hum Reprod Update. 2014;20:1–11. doi: 10.1093/humupd/dmt044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang Z, Wells D. The human oocyte and cumulus cells relationship: new insights from the cumulus cell transcriptome. Mol Hum Reprod. 2010;16:715–725. doi: 10.1093/molehr/gaq031. [DOI] [PubMed] [Google Scholar]
- 10.Demiray SB, Goker ENT, Tavmergen E, Yilmaz O, Calimlioglu N, Soykam HO, Oktem G, Sezerman U. Differential gene expression analysis of human cumulus cells. Clin Exp Reprod Med. 2019;46:76–86. doi: 10.5653/cerm.2019.46.2.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Y, Li R-Q, Ou S-B, Zhang N-F, Ren L, Wei L-N, Zhang QX, Yang DZ. Increased GDF9 and BMP15 mRNA levels in cumulus granulosa cells correlate with oocyte maturation, fertilization, and embryo quality in humans. Reprod Biol Endocrinol RBE. 2014;12:81. doi: 10.1186/1477-7827-12-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Persani L, Rossetti R, Di Pasquale E, Cacciatore C, Fabre S. The fundamental role of bone morphogenetic protein 15 in ovarian function and its involvement in female fertility disorders. Hum Reprod Update. 2014;20:869–883. doi: 10.1093/humupd/dmu036. [DOI] [PubMed] [Google Scholar]
- 13.Chang H-M, Qiao J, Leung PCK. Oocyte-somatic cell interactions in the human ovary-novel role of bone morphogenetic proteins and growth differentiation factors. Hum Reprod Update. 2016;23:1–18. doi: 10.1093/humupd/dmw039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanfins A, Rodrigues P, Albertini DF. GDF-9 and BMP-15 direct the follicle symphony. J Assist Reprod Genet. 2018;35:1741–1750. doi: 10.1007/s10815-018-1268-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Winterhager E, Kidder GM. Gap junction connexins in female reproductive organs: implications for women’s reproductive health. Hum Reprod Update. 2015;21:340–352. doi: 10.1093/humupd/dmv007. [DOI] [PubMed] [Google Scholar]
- 16.Donfack NJ, Alves KA, Alves BG, Rocha RMP, Bruno JB, Bertolini M, dos Santos RR, Domingues SFS, de Figueiredo JR, Smitz J, Rodrigues APR. Stroma cell-derived factor 1 and connexins (37 and 43) are preserved after vitrification and in vitro culture of goat ovarian cortex. Theriogenology. 2018;116:83–88. doi: 10.1016/j.theriogenology.2018.05.001. [DOI] [PubMed] [Google Scholar]
- 17.Bergandi L, Basso G, Evangelista F, Canosa S, Dalmasso P, Aldieri E, Revelli A, Benedetto C, Ghigo D. Inducible nitric oxide synthase and heme oxygenase 1 are expressed in human cumulus cells and may be used as biomarkers of oocyte competence. Reprod Sci Thousand Oaks Calif. 2014;21:1370–1377. doi: 10.1177/1933719114525268. [DOI] [PubMed] [Google Scholar]
- 18.Revelli A, Canosa S, Bergandi L, Skorokhod OA, Biasoni V, Carosso A, Bertagna A, Maule M, Aldieri E, D’Eufemia MD, Evangelista F, Colacurci N, Benedetto C. Oocyte polarized light microscopy, assay of specific follicular fluid metabolites, and gene expression in cumulus cells as different approaches to predict fertilization efficiency after ICSI. Reprod Biol Endocrinol RBE. 2017;15:47. doi: 10.1186/s12958-017-0265-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huber M, Hadziosmanovic N, Berglund L, Holte J. Using the ovarian sensitivity index to define poor, normal, and high response after controlled ovarian hyperstimulation in the long gonadotropin-releasing hormone-agonist protocol: suggestions for a new principle to solve an old problem. Fertil Steril. 2013;100:1270–1276. doi: 10.1016/j.fertnstert.2013.06.049. [DOI] [PubMed] [Google Scholar]
- 20.Canosa S, Adriaenssens T, Coucke W, Dalmasso P, Revelli A, Benedetto C, Smitz J. Zona pellucida gene mRNA expression in human oocytes is related to oocyte maturity, zona inner layer retardance and fertilization competence. Mol Hum Reprod. 2017;23:292–303. doi: 10.1093/molehr/gax008. [DOI] [PubMed] [Google Scholar]
- 21.Holte J, Berglund L, Milton K, Garello C, Gennarelli G, Revelli A, Bergh T. Construction of an evidence-based integrated morphology cleavage embryo score for implantation potential of embryos scored and transferred on day 2 after oocyte retrieval. Hum Reprod Oxf Engl. 2007;22:548–557. doi: 10.1093/humrep/del403. [DOI] [PubMed] [Google Scholar]
- 22.Alpha Scientists in Reproductive Medicine and ESHRE Special Interest Group of Embryology. The Istanbul consensus workshop on embryo assessment: proceedings of an expert meeting. Hum Reprod Oxf Engl. 2011;26:1270–83. [DOI] [PubMed]
- 23.Revelli A, Rovei V, Dalmasso P, Gennarelli G, Racca C, Evangelista F, Benedetto C. Large randomized trial comparing transabdominal ultrasound-guided embryo transfer with a technique based on uterine length measurement before embryo transfer. Ultrasound Obstet Gynecol. 2016;48(3):289–295. doi: 10.1002/uog.15899. [DOI] [PubMed] [Google Scholar]
- 24.Bayne RAL, Kinnell HL, Coutts SM, He J, Childs AJ, Anderson RA. GDF9 is transiently expressed in oocytes before follicle formation in the human fetal ovary and is regulated by a novel NOBOX transcript. PloS One. 2015;10:e0119819. doi: 10.1371/journal.pone.0119819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang H-M, Cheng J-C, Taylor E, Leung PCK. Oocyte-derived BMP15 but not GDF9 down-regulates connexin43 expression and decreases gap junction intercellular communication activity in immortalized human granulosa cells. Mol Hum Reprod. 2014;20:373–383. doi: 10.1093/molehr/gau001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ricca C, Aillon A, Bergandi L, Alotto D, Castagnoli C, Silvagno F. Vitamin D receptor is necessary for mitochondrial function and cell health. Int J Mol Sci. 2018;19. [DOI] [PMC free article] [PubMed]
- 27.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. [DOI] [PMC free article] [PubMed]
- 28.Pribenszky C, Nilselid A-M, Montag M. Time-lapse culture with morphokinetic embryo selection improves pregnancy and live birth chances and reduces early pregnancy loss: a meta-analysis. Reprod Biomed Online. 2017;35:511–520. doi: 10.1016/j.rbmo.2017.06.022. [DOI] [PubMed] [Google Scholar]
- 29.Armstrong S, Arroll N, Cree LM, Jordan V, Farquhar C. Time-lapse systems for embryo incubation and assessment in assisted reproduction. Cochrane Database Syst Rev. 2015;CD011320. [DOI] [PubMed]
- 30.Armstrong S, Vail A, Mastenbroek S, Jordan V, Farquhar C. Time-lapse in the IVF-lab: how should we assess potential benefit? Hum Reprod Oxf Engl. 2015;30:3–8. doi: 10.1093/humrep/deu250. [DOI] [PubMed] [Google Scholar]
- 31.Chen M, Wei S, Hu J, Yuan J, Liu F. Does time-lapse imaging have favorable results for embryo incubation and selection compared with conventional methods in clinical in vitro fertilization? A meta-analysis and systematic review of randomized controlled trials. PloS One. 2017;12:e0178720. doi: 10.1371/journal.pone.0178720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Polanski LT, Coelho Neto MA, Nastri CO, Navarro PA, Ferriani RA, Raine-Fenning N, Martins WP. Time-lapse embryo imaging for improving reproductive outcomes: systematic review and meta-analysis. Ultrasound Obstet Gynecol Off J Int Soc Ultrasound Obstet Gynecol. 2014;44:394–401. doi: 10.1002/uog.13428. [DOI] [PubMed] [Google Scholar]
- 33.Racowsky C, Kovacs P, Martins WP. A critical appraisal of time-lapse imaging for embryo selection: where are we and where do we need to go? J Assist Reprod Genet. 2015;32:1025–1030. doi: 10.1007/s10815-015-0510-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khosravi P, Kazemi E, Zhan Q, Malmsten JE, Toschi M, Zisimopoulos P, Sigaras A, Lavery S, Cooper LAD, Hickman C, Meseguer M, Rosenwaks Z, Elemento O, Zaninovic N, Hajirasouliha I. Deep learning enables robust assessment and selection of human blastocysts after in vitro fertilization. Npj Digit Med. 2019;2:21. doi: 10.1038/s41746-019-0096-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Coticchio G, Mignini Renzini M, Novara PV, Lain M, De Ponti E, Turchi D, et al. Focused time-lapse analysis reveals novel aspects of human fertilization and suggests new parameters of embryo viability. Hum Reprod Oxf Engl. 2018;33:23–31. doi: 10.1093/humrep/dex344. [DOI] [PubMed] [Google Scholar]
- 36.Revelli A, Canosa S, Carosso A, Filippini C, Paschero C, Gennarelli G, Delle Piane L, Benedetto C. Impact of the addition of Early Embryo Viability Assessment to morphological evaluation on the accuracy of embryo selection on day 3 or day 5: a retrospective analysis. J Ovarian Res. 2019;12:73. doi: 10.1186/s13048-019-0547-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zaninovic N, Irani M, Meseguer M. Assessment of embryo morphology and developmental dynamics by time-lapse microscopy: is there a relation to implantation and ploidy? Fertil Steril. 2017;108:722–729. doi: 10.1016/j.fertnstert.2017.10.002. [DOI] [PubMed] [Google Scholar]
- 38.Wong CC, Loewke KE, Bossert NL, Behr B, De Jonge CJ, Baer TM, et al. Non-invasive imaging of human embryos before embryonic genome activation predicts development to the blastocyst stage. Nat Biotechnol. 2010;28:1115–1121. doi: 10.1038/nbt.1686. [DOI] [PubMed] [Google Scholar]
- 39.Hammond ER, Stewart B, Peek JC, Shelling AN, Cree LM. Assessing embryo quality by combining non-invasive markers: early time-lapse parameters reflect gene expression in associated cumulus cells. Hum Reprod Oxf Engl. 2015;30:1850–1860. doi: 10.1093/humrep/dev121. [DOI] [PubMed] [Google Scholar]
- 40.Hashimoto S, Kato N, Saeki K, Morimoto Y. Selection of high-potential embryos by culture in poly(dimethylsiloxane) microwells and time-lapse imaging. Fertil Steril. 2012;97:332–337. doi: 10.1016/j.fertnstert.2011.11.042. [DOI] [PubMed] [Google Scholar]
- 41.Kirkegaard K, Kesmodel US, Hindkjær JJ, Ingerslev HJ. Time-lapse parameters as predictors of blastocyst development and pregnancy outcome in embryos from good prognosis patients: a prospective cohort study. Hum Reprod. 2013;28:2643–2651. doi: 10.1093/humrep/det300. [DOI] [PubMed] [Google Scholar]
- 42.Cruz M, Garrido N, Herrero J, Pérez-Cano I, Muñoz M, Meseguer M. Timing of cell division in human cleavage-stage embryos is linked with blastocyst formation and quality. Reprod Biomed Online. 2012;25:371–381. doi: 10.1016/j.rbmo.2012.06.017. [DOI] [PubMed] [Google Scholar]
- 43.Dal Canto M, Coticchio G, Mignini Renzini M, De Ponti E, Novara PV, Brambillasca F, et al. Cleavage kinetics analysis of human embryos predicts development to blastocyst and implantation. Reprod Biomed Online. 2012;25:474–480. doi: 10.1016/j.rbmo.2012.07.016. [DOI] [PubMed] [Google Scholar]
- 44.Minasi MG, Colasante A, Riccio T, Ruberti A, Casciani V, Scarselli F, Spinella F, Fiorentino F, Varricchio MT, Greco E. Correlation between aneuploidy, standard morphology evaluation and morphokinetic development in 1730 biopsied blastocysts: a consecutive case series study. Hum Reprod Oxf Engl. 2016;31:2245–2254. doi: 10.1093/humrep/dew183. [DOI] [PubMed] [Google Scholar]
- 45.Campbell A, Fishel S, Bowman N, Duffy S, Sedler M, Hickman CFL. Modelling a risk classification of aneuploidy in human embryos using non-invasive morphokinetics. Reprod Biomed Online. 2013;26:477–485. doi: 10.1016/j.rbmo.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 46.Shapiro BS, Richter KS, Harris DC, Daneshmand ST. A comparison of day 5 and day 6 blastocyst transfers. Fertil Steril. 2001;75:1126–1130. doi: 10.1016/s0015-0282(01)01771-x. [DOI] [PubMed] [Google Scholar]
- 47.Barrenetxea G, López de Larruzea A, Ganzabal T, Jiménez R, Carbonero K, Mandiola M. Blastocyst culture after repeated failure of cleavage-stage embryo transfers: a comparison of day 5 and day 6 transfers. Fertil Steril. 2005;83:49–53. doi: 10.1016/j.fertnstert.2004.06.049. [DOI] [PubMed] [Google Scholar]
- 48.Franasiak JM, Forman EJ, Patounakis G, Hong KH, Werner MD, Upham KM, et al. Investigating the impact of the timing of blastulation on implantation: management of embryo-endometrial synchrony improves outcomes. Hum Reprod Open. 2018;2018:hoy022. doi: 10.1093/hropen/hoy022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tannus S, Cohen Y, Henderson S, Al Ma’mari N, Shavit T, Son W-Y, et al. Fresh transfer of day 5 slow-growing embryos versus deferred transfer of vitrified, fully expanded day 6 blastocysts: which is the optimal approach? Hum Reprod Oxf Engl. 2019;34:44–51. doi: 10.1093/humrep/dey351. [DOI] [PubMed] [Google Scholar]
- 50.Wathlet S, Adriaenssens T, Segers I, Verheyen G, Van de Velde H, Coucke W, et al. Cumulus cell gene expression predicts better cleavage-stage embryo or blastocyst development and pregnancy for ICSI patients. Hum Reprod Oxf Engl. 2011;26:1035–1051. doi: 10.1093/humrep/der036. [DOI] [PubMed] [Google Scholar]
- 51.Wathlet S, Adriaenssens T, Segers I, Verheyen G, Janssens R, Coucke W, et al. New candidate genes to predict pregnancy outcome in single embryo transfer cycles when using cumulus cell gene expression. Fertil Steril. 2012;98:432–439.e1-4. doi: 10.1016/j.fertnstert.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 52.Wathlet S, Adriaenssens T, Segers I, Verheyen G, Van Landuyt L, Coucke W, et al. Pregnancy prediction in single embryo transfer cycles after ICSI using QPCR: validation in oocytes from the same cohort. PloS One. 2013;8:e54226. doi: 10.1371/journal.pone.0054226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu Y-T, Tang L, Cai J, Lu X-E, Xu J, Zhu X-M, Luo Q, Huang HF. High bone morphogenetic protein-15 level in follicular fluid is associated with high quality oocyte and subsequent embryonic development. Hum Reprod Oxf Engl. 2007;22:1526–1531. doi: 10.1093/humrep/dem029. [DOI] [PubMed] [Google Scholar]
- 54.Hasegawa J, Yanaihara A, Iwasaki S, Mitsukawa K, Negishi M, Okai T. Reduction of connexin 43 in human cumulus cells yields good embryo competence during ICSI. J Assist Reprod Genet. 2007;24:463–466. doi: 10.1007/s10815-007-9155-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sutton-McDowall ML, Mottershead DG, Gardner DK, Gilchrist RB, Thompson JG. Metabolic differences in bovine cumulus-oocyte complexes matured in vitro in the presence or absence of follicle-stimulating hormone and bone morphogenetic protein 15. Biol Reprod. 2012;87:87. doi: 10.1095/biolreprod.112.102061. [DOI] [PubMed] [Google Scholar]
- 56.Ricca C, Aillon A, Viano M, Bergandi L, Aldieri E, Silvagno F. Vitamin D inhibits the epithelial-mesenchymal transition by a negative feedback regulation of TGF-β activity. J Steroid Biochem Mol Biol. 2019;187:97–105. doi: 10.1016/j.jsbmb.2018.11.006. [DOI] [PubMed] [Google Scholar]
- 57.Danyu L, Yanran L, Xiuna J, Ying C, Sudan P, Tianen Z, et al. α-Synuclein induced mitochondrial dysfunction via cytochrome c oxidase subunit 2 in SH-SY5Y cells. Exp Cell Res. 2019;378:57–65. doi: 10.1016/j.yexcr.2019.02.006. [DOI] [PubMed] [Google Scholar]
- 58.Ganetzky RD, Stendel C, McCormick EM, Zolkipli-Cunningham Z, Goldstein AC, Klopstock T, et al. MT-ATP6 mitochondrial disease variants: phenotypic and biochemical features analysis in 218 published cases and cohort of 14 new cases. Hum Mutat. 2019;40:499–515. doi: 10.1002/humu.23723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bansal S, Biswas G, Avadhani NG. Mitochondria-targeted heme oxygenase-1 induces oxidative stress and mitochondrial dysfunction in macrophages, kidney fibroblasts and in chronic alcohol hepatotoxicity. Redox Biol. 2014;2:273–283. doi: 10.1016/j.redox.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Scarica C, Cimadomo D, Dovere L, Giancani A, Stoppa M, Capalbo A, Ubaldi FM, Rienzi L, Canipari R. An integrated investigation of oocyte developmental competence: expression of key genes in human cumulus cells, morphokinetics of early divisions, blastulation, and euploidy. J Assist Reprod Genet. 2019;36:875–887. doi: 10.1007/s10815-019-01410-3. [DOI] [PMC free article] [PubMed] [Google Scholar]