Abstract
Raw milk is a frequent vehicle for transmission of thermophilic Campylobacter, leading to reported outbreaks. Milk is a challenging food matrix for pathogen detection, due to its high protein and lipid content. Limited detection of Campylobacter colony-forming unit (CFU) in raw milk might underestimate the pathogen’s infectious potential. We optimized a viability real-time PCR (qPCR) for application with raw milk. The procedure was robust against variations of milk lots and different Campylobacter strains. Various DNA-intercalating dyes were evaluated for their ability to reduce the PCR signal of dead cells. Only propidium monoazide (PMA) and PMAxx qualified for diagnostic use. Different sedimentation properties of viable and dead Campylobacter jejuni and Campylobacter coli strains in 10-fold diluted milk enhanced viable/dead differentiation. The new method enabled to review survival of Campylobacter spp. in raw milk based on viable cells harboring an intact cell membrane. The data were compared to culturability according to ISO10272-2:2017. A difference of up to 4.5 log10 between viable Campylobacter counts and CFU values became apparent. Relevance of viability qPCR values was corroborated by full recovery of CFU under extremely reduced oxygen concentration in the presence of hydrogen. Recovery of CFU was limited, however, upon prolonged exposure in raw milk. The data confirm that Campylobacter survival in raw milk can be largely underestimated when relying on CFU data only. We conclude that raw milk led to oxidative stress-induced growth arrest in thermophilic Campylobacter, which was reversible by reduction of the oxygen partial pressure in a time-limited way.
Keywords: propidium monoazide, viable but non-culturable, resuscitation, food safety, oxidative stress, real-time polymerase chain reaction
Introduction
In the European Union, campylobacteriosis is currently the most frequent bacterial food-borne disease, with 246,571 reported human cases in 2018 (EFSA, 2019). Acute symptoms include watery or bloody diarrhea, abdominal cramps, vomiting, and fever. Associated chronic complications involve reactive arthritis, inflammatory bowel disease, and neurological disorders such as Guillain-Barré syndrome (Keithlin et al., 2014). Rare fatal cases were reported in children less than 5 years of age and in elderly and immunocompromised patients (EFSA, 2019).
Transmission of Campylobacter mainly occurs via contaminated food, by direct contact with colonized animals and also by untreated water (Kaakoush et al., 2015; Mughini-Gras et al., 2016; Rosner et al., 2017). Undercooked poultry meat and an insufficient kitchen hygiene related to raw meat handling are classical causes for the majority of sporadic cases. However, Campylobacter outbreaks are frequently reported upon consumption of raw milk (EFSA, 2018). Due to the repeal of the European milk quotas in 2015, farmers intensified the local sale of raw milk via milk-filling stations. In turn, consumer behavior changed in favor of an increased consumption of raw milk, neglecting prior heating recommendations. In Germany, 26% of all reported food-associated outbreaks with “high evidence” were caused by Campylobacter in 2018 (BVL, 2019). Most of them were caused by consumption of raw milk.
Cultural detection of Campylobacter in raw milk is difficult due to rapid decline of colony-forming unit (CFU) (Barrell, 1981; Doyle and Roman, 1982; Humphrey, 1986). Though considered fastidious in growth, Campylobacter are supposed to resist various environmental stresses. The genus has been suggested to outlast unfavorable environmental conditions in the viable but non-culturable (VBNC) state, putatively able to resuscitate and maintain its infective potential (Rollins and Colwell, 1986; Federighi et al., 1998; Baffone et al., 2006).
Real-time PCR (qPCR) offers a highly sensitive culture-independent quantification method. It can be combined with a DNA intercalating agent, which enters dead cells, crosslinks to DNA, thereby impeding DNA amplification during PCR. This way, viable cells with an intact membrane can be differentiated from dead cells (Nocker et al., 2006; Krüger et al., 2014; Pacholewicz et al., 2019). It allows determining the highest possible residual risk present, while the minimal risk is indicated by CFU.
We established a culture-independent procedure that allows quantification of viable thermophilic Campylobacter in raw milk employing “viability qPCR.” Applying this method, we investigated Campylobacter’s survival in raw milk with intriguing results; a substantial survival of Campylobacter was observed by viability qPCR, while CFU dropped to non-detectable levels. To support our findings based on viability qPCR, recovery of CFU was demonstrated by lowering the partial pressure of oxygen beyond ISO standard recommendations (ISO 10272-2:2017).
Materials and Methods
Strains and Growth Conditions
Campylobacter jejuni DSM 4688, Campylobacter coli DSM 4689, and Campylobacter sputorum DSM 5363 were derived from the DSMZ strain collection (DSMZ, Braunschweig, Germany). C. jejuni BfR-CA-13255, BfR-CA-13290, BfR-CA-14727, BfR-CA-15123, and BfR-CA-15146 had been isolated from raw milk by the Federal State Laboratories in Germany.
Strains stored at −80°C (MAST Group Ltd., Bootle, UK) were cultured on Columbia agar supplemented with 5% sheep blood (ColBA, Oxoid, Thermo Fisher Scientific Inc., Waltham, Massachusetts, USA) at 42°C under microaerobic conditions (5% O2, 10% CO2, and 85% N2) in a microaerobic incubator (Binder GmbH, Tuttlingen, Germany) for 24 h. After subculture for 18 ± 2 h, cells were suspended in Brucella broth (BB, Beckon Dickinsen, Franklin Lakes, New Jersey, USA) at OD600 = 0.2, corresponding to ~9 log10 cell counts per ml (Krüger et al., 2014). The cells were 10-fold diluted to reach 8 log10 cell counts per ml and kept on ice until further use (viable cells). The non-thermophilic C. sputorum was grown at 37°C on ColBA for initially 48 h starting from a −80°C cryoculture. It was subcultured for 24 h and used for the production of a dead cell standard according to Pacholewicz et al. (2019).
Quantification by CFU was performed according to ISO 10272-2:2017 with BB as diluent and plating was performed in duplicate on mCCDA (Oxoid, Thermo Fisher Scientific., Waltham, Massachusetts, USA). The theoretical sensitivity was 1 CFU/ml milk. The standard incubation occurred for 48–72 h at 42°C using an atmosphere of 5% O2, 10% CO2, and 85% N2 (a). Variation of atmospheric conditions was performed using incubators and gas replacement jars (Oxoid™ Anaerobia System, Thermo Fisher Scientific Inc., Waltham, Massachusetts, USA) filled with gas mixtures (Air Liquide, Paris, France). The detection limit for condition b–e. was 10 CFU/ml milk.
Milk Lots and Spiking of the Samples
To assess the effect of milk batch variations, different batches of raw tank milk were either received from the institute’s own farming facility or collected from a regional milk producer. The fresh milk was kept at 4°C until processing, but not longer than 24 h after milking (n = 5). Frozen batches were stored at −20°C for less than 10 days (n = 4) or more than 450 days (n = 2). For all other experiments, one of the long-term frozen milk batches was used in order to improve comparability of the results.
Milk samples were spiked with Campylobacter strains to reach an initial concentration of 6 log10 cells/ml raw milk. The raw milk samples were kept in closed 50-ml tubes and incubated at 5 ± 1°C under normal atmospheric conditions. pH was stable and measured to be 6.7 ± 0.1 throughout the experiment. During a period of 6 days, samples were daily analyzed by CFU determination and viability qPCR analysis in parallel.
Production of Dead and Transiently Inactive Viable Cells
If not otherwise stated, C. jejuni dead cell standards served as sample process control (SPC) during each experiment. Cells were killed by incubating 8 log10 cells in 100 μl PBS supplemented with 5% hydrogen peroxide (Carl Roth GmbH, Karlsruhe, Germany). After 1 h at room temperature, the cell suspension was diluted with 900 μl PBS and the sample was then centrifuged at 16,000 × g for 5 min. The cell pellet was resuspended in 1 ml PBS to obtain 8 log10 inactivated cells per ml, which were kept on ice until further use. In order to verify loss of CFU, 100 μl of each dead cell standard was plated on ColbA.
For the production of transiently inactive cells, the protonophor cyanide-m-chlorophenylhydrazone (CCCP, Sigma-Aldrich, Steinheim, Germany) was applied. An amount of 8 log10 cells in 1 ml PBS was incubated with 100 μM CCCP for 1 h on ice. We stained 6 log10 CCCP-treated cells with intercalating dyes, while keeping 10 μM CCCP during incubation. For CFU determination after recovery from CCCP stress, CCCP-treated cells were centrifuged at 16,000 × g for 5 min, once washed with 1 ml BB and plated as described above.
Differentiation Between Viable and Dead Cells
From each raw milk sample, we took twice 1 ml; one of these two aliquots was processed without intercalating dye (total amount of cells) and the other was used with intercalating dye (only viable cells). Both aliquots were 10-fold diluted in 9 ml of precooled BB and centrifuged at 8,000 × g for 20 min at 4°C using 15 ml centrifugation tubes. The supernatant (BB with the major part of the milk components) was decanted and the remaining supernatant carefully withdrawn with a 1 ml slim-shaft micropipette. The pellet was immediately suspended in 1 ml PBS, transferred to a 1.5-ml tube, and stored on ice until staining with the DNA intercalating dye.
During all experiments, propidium monoazide (PMA, Biotium Inc., Hayward, California, USA) was applied as DNA intercalating agent. When indicated, ethidium monoazide (EMA, Biotium Inc., Hayward, California, USA), PMAxx (Biotium Inc., Hayward, California, USA), or PEMAX (GenIUL, Terrassa, Spain) were used. Intercalating agents were applied at a final concentration of 50 μM. Stock solutions were stored in 20% dimethyl sulfoxide at −20°C. In order to guarantee an efficient reduction of the dead cell signal, control samples either contained dead C. jejuni cells (SPC) or dead C. sputorum cells (ISPC) at 6 log10 cells/ml. The protocol was as previously described (Pacholewicz et al., 2019). In brief, samples were pre-incubated for 10 min at 30°C and 700 rpm in a thermomixer, before addition of the DNA intercalating agent, shortly vortexed, and further incubated for 15 min at 30°C at 700 rpm in the dark. Subsequently, samples were cross-linked for 15 min using the PhAST Blue photo-activation system (GenIUL, Terrassa, Spain) at 100% light intensity. Samples were centrifuged for 5 min at 16,000 × g, the supernatant was discarded, and the cell pellets were stored at −20°C until DNA extraction.
DNA Extraction and qPCR
Genomic DNA was extracted using the GeneJet Genomic DNA Purification kit (Thermo Fisher Scientific Inc., Waltham, Massachusetts, USA) according to the manufacturer’s instructions and extracted DNA was eluted in 100 μl. qPCR was carried out on the same day using 10 μl of extracted DNA per reaction.
qPCR was performed targeting a fragment of the 16S rRNA gene of thermophilic Campylobacter or of C. sputorum (Pacholewicz et al., 2019). In short, the qPCR mastermix contained final concentrations of 1× Platinum Taq buffer, 2.5 mM MgCl2, 0.2 mM of each dNTP, 0.06 × ROX, 500 nM of each Jos-F1 and Jos-R1 primer, 300 nM of each IPC-ntb2-F and IPC-ntb2-R primer (HPLC-grade, Sigma-Aldrich, Steinheim, Germany), 100 nM of each dark-quenched probe (IPC-ntb2-P-TAMRA and Jos-P-FAM, TIB MOLBIOL, Berlin, Germany), 2U Platinum Taq DNA polymerase (Invitrogen, Thermo Fisher Scientific Inc., Waltham, Massachusetts, USA), and 25 copies of the IPC-ntb2 plasmid (Anderson et al., 2011) in a final qPCR reaction volume of 25 μl. qPCR was performed on a 7500 Real-Time PCR system (Applied Biosystems, Thermo Fisher Scientific, Waltham, Massachusetts, USA) using the 7500 Fast Software v2.3. The program started with 3 min at 95°C followed by 45 cycles of 15 s at 95°C, 1 min at 60°C and 30 s at 72°C. For the quantitative detection of C. sputorum the mastermix was as above but contained instead of Jos-F1 and Jos-R1 the primer pair Csput-F and Csput-R. Likewise, the Jos-P probe was exchanged with the C. sputorum-specific probe Csput-P-Cy5 as detailed in Pacholewicz et al. (2019). The sequences of the primers and probes (quenched by the black berry quencher, BBQ) were as follows: Jos-F1 (5′-CCTGCTTAACACAAGTTGAGTAGG-3′), Jos-R1 (5′-TTCCTTAGGTACCGTCAGAATTC-3′), Jos-P (6FAM-TGTCATCCTCCACGCGGCGTTGCTGC-BBQ), IPC-ntb2-F (5′-ACCACAATGCCAGAGTGACAAC-3′), IPC-ntb2-R (5′-TACCTGGTCTCCAGCTTTCAGTT-3′), IPC-ntb2-P (TAMRA-CACGCGCATGAAGTTAGGGGACCA-BBQ), Csput-F (5′-TGGGAAATGTAGCTCTTAATAATATATATC-3′), Csput-R (5′-CCTTACCAACTAGCTGATACAATATAG-3′), and Csput-P (Cy5-CCTCATCCCATAGCGAAAGCTCTT-BBQ).
Each qPCR run contained genomic standards, comprising five decile serial dilutions ranging from 50,000 to 5 genomic copies per reaction in duplicate. The genomic standard was freshly reconstituted from dried stabilized DNA aliquots as described previously (Pacholewicz et al., 2019).
Statistical Analysis
Data were analyzed using GraphPad Prism v. 5.01 software. Statistical analysis was performed using the Mann-Whitney U-test. Graphs display the mean values ± SD, unless stated otherwise.
Results
Establishing Quantification of Viable Campylobacter in Raw Milk by qPCR
We intended to answer the crucial food safety question whether the recovery of thermophilic Campylobacter from raw milk samples by CFU underestimates viable Campylobacter cells. We previously improved a culture-independent viability qPCR optimized for chicken rinses (Pacholewicz et al., 2019). As expected, direct quantification from undiluted raw milk samples was unsuitable because of two reasons – the bacteria could not be centrifuged in a quantitative manner probably due to high protein and lipid contents, and the opaque nature of raw milk rendered staining and proper crosslinking of DNA in dead bacteria inefficient.
Thus, we diluted raw milk samples 10-fold in precooled BB in order to quantitatively harvest Campylobacter upon centrifugation and to reduce the amount of milk compounds putatively deteriorating the staining procedure (Figure 1). After resuspension of the bacterial pellet, various DNA intercalating dyes were tested. A proper method would lead to efficient elimination of the DNA amplification signal of dead cells. Meanwhile, it would most effectively amplify DNA of viable cells, even though these viable cells were transiently inactive in terms of growth. We wondered if the recently commercialized PMAxx and PEMAX agents had improved properties in the viable/dead differentiation protocol. In comparison with the known agents PMA and EMA, we examined their performance in the viability qPCR (Figure 2). For this purpose, C. jejuni DSM 4688 grown on ColBA for 18–20 h was resuspended in BB and used as viable untreated cells. Dead bacteria were produced by inactivation via oxidative stress as previously described (Krüger et al., 2014). Transiently inactivated cells were produced by treatment with 100 μM of the protonophor CCCP, abolishing the proton-motif force of the bacterium. Subsequently, 6 log10 per ml of these differently treated cells were stained with the various dyes and processed via qPCR and CFU determination (Figure 2).
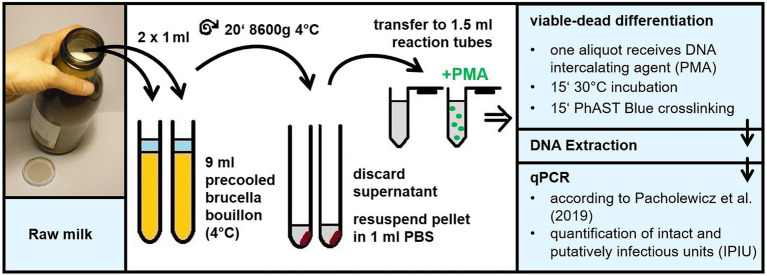
Figure 1.
Schematic overview of the culture-independent method for quantification of viable Campylobacter in raw milk. The method comprises the following steps. Raw milk is diluted 10-fold in precooled Brucella broth to enable quantitative bacterial centrifugation. After careful withdrawal of non-pelleted raw milk residues, the pellet is resuspended in 1 ml PBS for viable/dead differentiation according to Krüger et al. (2014). Subsequently, the DNA intercalating agent is applied to block DNA of dead bacteria from PCR amplification. After DNA extraction qPCR is performed targeting the 16S rRNA sequence in intact and putatively infectious units of thermophilic Campylobacter according to Pacholewicz et al. (2019).
Figure 2.
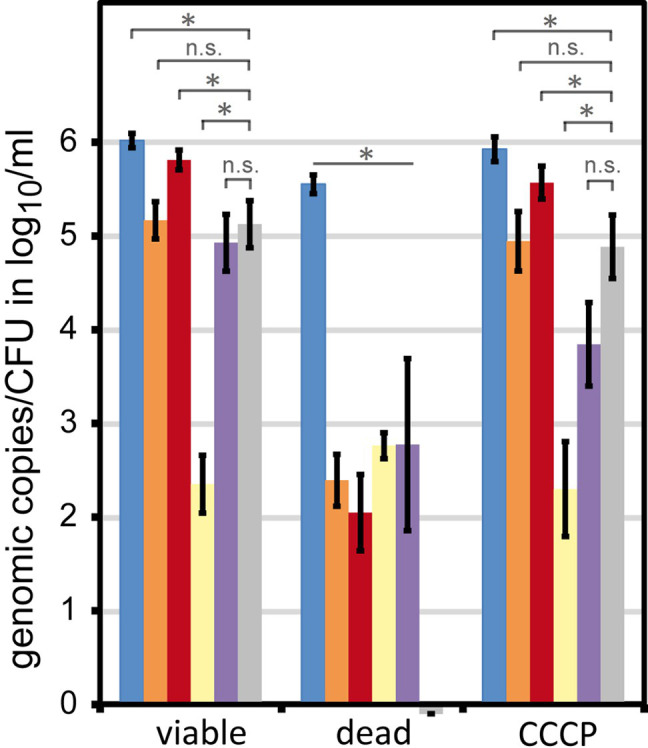
Signal reduction in dead and transiently inactive Campylobacter by different DNA intercalating agents. C. jejuni DSM 4688 were grown on ColbA for 18–20 h (viable bacteria). The bacteria were either killed by H2O2 or transiently inactivated using the protonophore CCCP. Fifty µM of the respective DNA intercalating agent was applied to 6 log10 bacteria per ml PBS. Columns display mean values ± standard deviation of genomic copies (blue, without treatment; orange, propidium monoazide (PMA); red, PMAxx; yellow, EMA; purple, PEMAX) or CFU (gray bars); dataset includes five independent experiments; * p < 0.01; n.s., non-significant (two-tailed Mann-Whitney U-test).
Viable cells showed nearly identical CFU and live qPCR counts (5.17 ± 0.2 log10 live bacteria per ml and 5.13 ± 0.25 log10 CFU per ml qPCR counts in the presence of PMA; Figure 2, orange and gray bar). Around 10-fold more total genomic copies were detected via qPCR (6.02 ± 0.08 log10 per ml without any DNA intercalating dye; Figure 2, blue bar). These results are in agreement with previous data, demonstrating that only around every tenth C. jejuni DSM 4688 precultured under similar conditions displayed CFU capability (Krüger et al., 2014). CCCP-treated cells nearly fully restored their ability to form CFU upon release of CCCP stress (4.89 ± 0.34 log10 CFU/ml; Figure 2, gray bar) and qPCR results with PMA very well-matched CFU determinations (4.94 ± 0.32 log10/ml). In contrast, cells inactivated with 5% H2O2 (dead) were not able to form any CFU (Figure 2, gray bar). Likewise, all tested DNA intercalating agents reduced the detection of dead cells by ≥2.8 log10/ml genomic copies. This confirmed their ability to efficiently reduce the signal of dead cells under the tested conditions.
As observed before (Krüger et al., 2014) and also suggested for other bacteria (Nocker et al., 2006; Codony et al., 2015), EMA was indicative of metabolic activity and was not passively excluded from viable cells. Consistently, EMA largely underestimated transiently inactivated C. jejuni (2.35 ± 0.31 log10/ml and 3.84 ± 0.44 log10/ml genomic copies, respectively) and apparently entered viable bacteria. PMAxx led to slightly higher qPCR results than displayed by CFU, while PEMAX led to lower quantification of transiently inactivated (CCCP-treated) bacteria.
These observations indicate that PMA and PMAxx were passively excluded from intact bacteria. In contrast, PEMAX partially entered de-energized cells, but with less efficiency than EMA. Hence, we confirmed that PMA was the most suitable agent for viable/dead differentiation of C. jejuni cells.
Variation of Milk Lots and Campylobacter Strains
We scrutinized the viable/dead differentiating qPCR with regard to reproducible results upon matrix and strain variations. For this reason, different milk lots (n = 11; Figure 3A) and different Campylobacter strains (n = 7, including six C. jejuni and one C. coli; Figure 3B) were included in the experiments.
Figure 3.
The viability qPCR method is robust against different milk lots and strains variations. Viable or H2O2-inactivated Campylobacter were spiked at 6 log10/ml into PBS or raw milk. Genomic counts were determined by qPCR upon differentiation of viable and dead cells using PMA or PMAxx. (A) Eleven different lots of raw milk (either fresh or after −20°C storage) were tested using the strain C. jejuni BfR-CA-13290; (B) Seven different Campylobacter strains (two reference strains and five field strains) were analyzed using one milk lot. (C) Five experiments were performed with H2O2-inactivated C. sputorum DSM 5363 as potential internal sample process control (Pacholewicz et al., 2019). Absence of rest signal from dead cells was defined as −0.1 log10/ml for enabling calculations. Blue, no treatment; orange, PMA; red, PMAxx. Columns display mean values ± standard deviation. * p < 0.01; n.s., non-significant (two-tailed Mann-Whitney U-test).
Viable cells of C. jejuni BfR-CA-13290 were reliably quantified in both PBS and raw milk using PMA or PMAxx, irrespective of various milk lots (Figure 3A). Likewise, different thermophilic Campylobacter strains could also be consistently quantified with only minor variations (Figure 3B). Interestingly, we observed a partial loss (~2 log10) of dead C. jejuni by centrifugation, which was strain-dependent (Figures 3A,B, dead, blue bars) and most pronounced for strain BfR-CA-13290. In contrast, viable cells were reproducibly retrieved independent of the strain tested. Hence, in combination with PMA or PMAxx treatment, viable/dead differentiation was even improved in raw milk compared to PBS. Dead C. sputorum cells, previously established as internal sample process control (ISPC) (Pacholewicz et al., 2019), were quantitatively retrieved from milk samples (Figure 3C), rendering this ISPC suitable for monitoring DNA loss for the reduction of the dead cell signal in raw milk.
Survival of Campylobacter in Raw Milk at Refrigeration Temperatures
With the new method at hand, we studied the survival of three Campylobacter strains in raw milk stored under aerobic conditions at 5 ± 1°C. Two reference strains (C. jejuni DSM 4688 and C. coli DSM 4689) and the outbreak-associated strain isolated from raw milk (C. jejuni BfR-CA-13290) were each spiked at an initial inoculum of 6 log10/ml. The milk samples were daily analyzed over a period of 6 days by viability qPCR and CFU determination (Figure 4).
Figure 4.
Survival of Campylobacter in raw milk. Three Campylobacter strains were spiked at 6 log10/ml bacterial counts in raw milk and stored aerobically at refrigeration temperatures. Samples were taken over time to quantify the survival by viability qPCR (solid lines) and CFU according to ISO 10272-2:2017 (dotted lines). Blue, C. coli DSM 4689; red, C. jejuni DSM 4688; green, C. jejuni BfR-CA-13290. Absence of CFU or genomic copies was defined as −0.1 log10/ml for enabling calculations. Mean values ± standard deviations are shown from at least five independent experiments.
As expected, CFU significantly decreased over time in a strain-dependent manner. CFU of C. coli DSM 4689 rapidly declined and the strain was uncultivatable within 3 days (Figure 4, dashed blue line). The CFU of C. jejuni BfR-CA-13290 declined most slowly and the strain was undetectable after 6 days in most experiments (Figure 4, dashed green line). CFU loss kinetics of C. jejuni DSM 4688 was intermediate of C. coli and the C. jejuni outbreak strain (Figure 4, dashed red line).
In contrast, the amount of intact and putatively infectious units (IPIU) only marginally decreased by around 1 log10/ml for the two C. jejuni strains and to a level of ~4 log10/ml for the C. coli strains during the first 2 days. Subsequently, IPIU remained on these levels throughout the whole experiment (Figure 4, solid lines). At the end of the experiment, we observed a large discrepancy between CFU and IPIU for all three strains, i.e., absence of CFU and still log 4–4.5 IPIU detected by viability qPCR.
Recovery of CFU at Altered Culture Conditions
We questioned whether the observed prolonged survival in raw milk as assessed by viability qPCR reflected a real existent food safety issue, i.e., that bacteria were indeed viable. For this purpose, we intended to identify parameters critical for Campylobacter growth after exposure to raw milk.
We selected consecutive time points at which several growth conditions were tested in parallel. Campylobacter as a microaerobic organism is susceptible to oxidative stress outside its multiplication site. Thus, we tested whether altered temperature and atmospheric conditions can revert its capability to form CFU in vitro. We tested four alternative conditions (Figure 5B) in parallel to the standard condition of 42°C and an atmosphere of 5% O2, 10% CO2, and rest N2 (a). Condition b only differed by reduction of the temperature to 37°C. Furthermore, we additionally changed the concentrations of O2 (1% for c and d, 6% for e), CO2 (7% for e), and eventually added hydrogen (3.5% for d and e). The incubation time for optimal colony counting varied between the conditions. In the presence of 5–6% oxygen (a, b, and e), growth of Campylobacter led to well visible colonies after 2–3 days. It was confirmed that a prolonged incubation time did not lead to additional CFU. In contrast, at reduced oxygen levels (c and d), colonies grew slower, so that CFU were optimally counted after 4–5 days.
Figure 5.
Recovery of non-culturable Campylobacter from raw milk by reduction of oxygen partial pressure. Campylobacter were spiked at 6 log10/ml bacterial counts into raw milk and stored refrigerated. (A) Colony-forming unit (CFU) was determined under standard conditions (5% O2, 10% CO2, rest N2 at 42°C, condition a.). (B) At selected time points (T1–T4) enumeration was performed under four alternative microaerobic culture conditions. Alternative culture conditions at 37°C and with N2 as background gas: b. 5% O2, 10% CO2; c. 1% O2, 10% CO2; d. 3.5% H2, 1% O2, 10% CO2; e. 3.5% H2, 6% O2, 7% CO2. Mean values of one representative experiment are shown for three strains (blue, Campylobacter coli DSM 4689; red, Campylobacter jejuni DSM 4688; green, C. jejuni BfR-CA-13290). Viability qPCR initially detected 5.0–5.6 log10/ml viable counts and 4.4–4.72 log10/ml after 144 h of exposure in raw milk for the three strains.
Alteration of atmospheric conditions significantly increased CFU in all strains tested in a time point‐ and strain‐dependent manner (Figure 5, conditions c–e). Reduced temperature eventually increased CFU recovery at early time points in C. jejuni strains, but the extent of this increase was less than 1 log10/ml (Figure 5, condition b). The condition, in which hydrogen was present and oxygen was reduced to 1%, resulted in maximal CFU detected for all strains (Figure 5, condition d). In comparison with the original condition (Figure 5, condition a) reduction of oxygen and addition of hydrogen maximally increased CFU by log 4.5 log10/ml in C. jejuni BfR-CA-13290 after 6 days of exposure in milk. Hence, the gap between the initial CFU read-out under standard conditions and the viability qPCR read-out of IPIU was completely bridged (Figure 4). In the other two strains, CFU increased at most by 2.5 (at T2, 66 h) and 3 log10 per ml (T1, 42 h) for C. jejuni DSM 4688 and C. coli DSM 4689, respectively. However, stimulating CFU recovery under reduced oxygen conditions in the presence of hydrogen was dependent on the time point of investigation, since the magnitude of CFU increase diminished with exposure time of Campylobacter in raw milk at 5 ± 1°C.
Our experiments demonstrated, for the first time, that the measured number of IPIU determined by viability qPCR are indeed relevant, since we found distinct conditions under which CFU could completely be restored.
Discussion
Raw milk is an important vehicle for Campylobacter transmission and frequently involved in reported outbreaks. The raw milk matrix is challenging from the food safety perspective, since Campylobacter are difficult to be detected due to rapid decline in CFU. Current alternative methods target detection of Campylobacter after enrichment of raw milk in broth (Razzuoli et al., 2018) or by using in-line milk filters with higher pathogen contamination (FSAI, 2015; Artursson et al., 2018). To our knowledge, no culture-independent direct quantification method is so far available for raw milk. In the present study, we established a viability qPCR method, which allows quantifying IPIU of Campylobacter in raw milk. Furthermore, survival of Campylobacter spp. in raw milk was reassessed by qPCR and CFU. The proposed method was found to be robust against different milk batches and also suitable for different C. jejuni strains, the major species responsible for campylobacteriosis (EFSA, 2019).
While the standardized plate count approach has a quantitative sensitivity of 10 CFU/ml according to ISO 7218:2014, our qPCR-based method is 10-fold less sensitive due to limits of PMA staining and DNA extraction. However, we showed that qPCR can detect 4.5 log10/ml more viable C. jejuni in comparison to CFU enumeration under standard culture conditions (Figure 5). The results indicated an initial 1 log10/ml reduction in viable cells of the outbreak-associated C. jejuni strain within the first 42 h in raw milk. Notably, the remaining population appeared to cease their capacity to form colonies under normal growth conditions. Nevertheless, genomic DNA was protected by an intact cell membrane, which was inaccessible by PMA. In order to prove that qPCR results of viable bacteria are relevant for food safety, we bridged the gap between CFU and viability qPCR.
According to the standard protocols for enhanced enrichment of stressed Campylobacter, growth temperature is decreased for several hours to 37°C (ISO 10272-1:2017) based on observations of improved CFU recovery after cold stress (Humphrey, 1986). However, the effect of temperature reduction from 42°C to 37°C on increased C. jejuni CFU levels after raw milk exposure was negligible (Figure 5B, condition b).
Importantly, despite of decreasing CFU under standard conditions, an extremely low oxygen partial pressure (1% O2 in combination with 3.5% H2) reactivated Campylobacter to quantitatively grow on mCCDA (Figure 5B). These results contradict the traditional interpretation that reduced CFU necessarily reflect reduced viability of the pathogenic bacterium and demonstrate, for the first time, that “viability qPCR” detected IPIU indeed mirror viable cells.
Interestingly, addition of hydrogen (Figure 5, condition c) or lowering oxygen concentration (Figure 5, condition e) both stimulated growth of Campylobacter. The combination of hydrogen and low oxygen concentration had a cumulative effect on CFU recovery (Figure 5, condition d). Since hydrogen functions as electron donor of the respiratory chain (van der Stel et al., 2017), it may also enhance respiratory activity and further consumption of oxygen within the cell. Putatively, these results hint to the presence of a yet unknown oxidative stress response sensor, which regulates replication and/or division in thermophilic Campylobacter. It was shown previously that C. jejuni adapts to various microaerobic atmospheres by significantly changing transcription of specific genes, e.g., involved in detoxification (John et al., 2011). Furthermore, C. jejuni showed prolonged survival under atmospheric conditions in the presence of the oxygen-consuming bacterium Pseudomonas spp. (Hilbert et al., 2010).
Nevertheless, the potential to recover CFU by drastic decrease of oxygen was diminished with time of exposure in milk, suggesting further yet unknown signals/regulators for entry and exit of the non-culturable state of Campylobacter.
Conclusion
The presented work illustrates the underestimation of Campylobacter survival by standard plate count conditions in raw milk and calls for complementary culture-independent quantification of viable cells to improve risk assessment. Incubation under lowered oxygen tension in the presence of hydrogen was demonstrated to improve Campylobacter detection from raw milk in a time-limited way. Consequently, risk assessment of contaminated foods based on CFU might lead to an underestimation of the actual risk to human health. Future studies will decipher further parameters for reactivating Campylobacter culturability upon stress exposure in food matrices and in the environment. They will reveal so far unknown transmission routes as well as persistence niches of the versatile food-borne pathogen.
Data Availability Statement
All datasets generated for this study are included in the article.
Author Contributions
IW performed experiments, analyzed data and co-wrote the manuscript. AG performed experiments. KS designed the study, analyzed data and co-wrote the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Mrs. Lochotzke from the farm facility of BfR for the milk batches and our colleagues of the Federal State Laboratories in Germany for isolation of C. jejuni from raw milk.
Footnotes
Funding. This work was funded by SFB 1322-580 of German Federal Institute for Risk Assessment (BfR). IW was financed by the research project EsRAM (No. 2817701614) of the German Federal Ministry of Food and Agriculture (BMEL). AG received an Erasmus grant during his studies at BfR.
References
- Anderson A., Pietsch K., Zucker R., Mayr A., Müller-Hohe E., Messelhäusser U., et al. (2011). Validation of a duplex real-time PCR for the detection of Salmonella spp. in different food products. Food Anal. Methods 4, 259–267. 10.1007/s12161-010-9142-8 [DOI] [Google Scholar]
- Artursson K., Schelin J., Thisted Lambertz S., Hansson I., Olsson Engvall E. (2018). Foodborne pathogens in unpasteurized milk in Sweden. Int. J. Food Microbiol. 284, 120–127. 10.1016/j.ijfoodmicro.2018.05.015, PMID: [DOI] [PubMed] [Google Scholar]
- Baffone W., Casaroli A., Citterio B., Pierfelici L., Campana R., Vittoria E., et al. (2006). Campylobacter jejuni loss of culturability in aqueous microcosms and ability to resuscitate in a mouse model. Int. J. Food Microbiol. 107, 83–91. 10.1016/j.ijfoodmicro.2005.08.015, PMID: [DOI] [PubMed] [Google Scholar]
- Barrell R. A. (1981). The survival of Campylobacter coli/jejuni in unpasteurised milk. J. Infect. 3, 348–352. 10.1016/S0163-4453(81)91939-3, PMID: [DOI] [PubMed] [Google Scholar]
- BVL (2019). Common national report of BVL and RKI on food-borne infections in Germany in 2018; in German (Gemeinsamer nationaler Bericht des BVL und RKI zu lebensmittelbedingten Krankheitsausbrüchen in Deutschland 2018).
- Codony F., Agusti G., Allue-Guardia A. (2015). Cell membrane integrity and distinguishing between metabolically active and inactive cells as a means of improving viability PCR. Mol. Cell. Probes 29, 190–192. 10.1016/j.mcp.2015.03.003, PMID: [DOI] [PubMed] [Google Scholar]
- Doyle M. P., Roman D. J. (1982). Prevalence and survival of Campylobacter jejuni in unpasteurized milk. Appl. Environ. Microbiol. 44, 1154–1158. 10.1128/AEM.44.5.1154-1158.1982, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFSA (2018). The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2017. EFSA J. 16:e05500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFSA (2019). The European Union One Health 2018 Zoonoses Report. EFSA J. 17:e05926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federighi M., Tholozan J. L., Cappelier J. M., Tissier J. P., Jouve J. L. (1998). Evidence of non-coccoid viable but non-culturable Campylobacter jejuni cells in microcosm water by direct viable count, CTC-DAPI double staining, and scanning electron microscopy. Food Microbiol. 15, 539–550. 10.1006/fmic.1998.0181 [DOI] [Google Scholar]
- FSAI (2015). Raw milk and raw milk filters; Microbial Surveillance Programme (12NS2). Monitoring and surveillance series. Microbiology, Food Safety Authority of Ireland.
- Hilbert F., Scherwitzel M., Paulsen P., Szostak M. P. (2010). Survival of Campylobacter jejuni under conditions of atmospheric oxygen tension with the support of Pseudomonas spp. Appl. Environ. Microbiol. 76, 5911–5917. 10.1128/AEM.01532-10, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey T. J. (1986). Techniques for the optimum recovery of cold injured Campylobacter jejuni from milk or water. J. Appl. Bacteriol. 61, 125–132. 10.1111/j.1365-2672.1986.tb04265.x, PMID: [DOI] [PubMed] [Google Scholar]
- ISO 7218:2014 . Microbiology of food and animal feeding stuffs—general requirements and guidance for microbiological examinations.
- ISO 10272-1:2017 Microbiology of the food chain—horizontal method for detection and enumeration of Campylobacter spp.—part 1: detection method. [DOI] [PubMed]
- ISO 10272-2:2017 Microbiology of the food chain—horizontal method for detection and enumeration of Campylobacter spp.—part 2: colony-count technique. [DOI] [PubMed]
- John A., Connerton P. L., Cummings N., Connerton I. F. (2011). Profound differences in the transcriptome of Campylobacter jejuni grown in two different, widely used, microaerobic atmospheres. Res. Microbiol. 162, 410–418. 10.1016/j.resmic.2011.02.004, PMID: [DOI] [PubMed] [Google Scholar]
- Kaakoush N. O., Castano-Rodriguez N., Mitchell H. M., Man S. M. (2015). Global epidemiology of Campylobacter infection. Clin. Microbiol. Rev. 28, 687–720. 10.1128/CMR.00006-15, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keithlin J., Sargeant J., Thomas M. K., Fazil A. (2014). Systematic review and meta-analysis of the proportion of Campylobacter cases that develop chronic sequelae. BMC Public Health 14:1203. 10.1186/1471-2458-14-1203, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger N. J., Buhler C., Iwobi A. N., Huber I., Ellerbroek L., Appel B., et al. (2014). “Limits of control”—crucial parameters for a reliable quantification of viable Campylobacter by real-time PCR. PLoS One 9:e88108. 10.1371/journal.pone.0088108, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mughini-Gras L., Penny C., Ragimbeau C., Schets F. M., Blaak H., Duim B., et al. (2016). Quantifying potential sources of surface water contamination with Campylobacter jejuni and Campylobacter coli. Water Res. 101, 36–45. 10.1016/j.watres.2016.05.069, PMID: [DOI] [PubMed] [Google Scholar]
- Nocker A., Cheung C. Y., Camper A. K. (2006). Comparison of propidium monoazide with ethidium monoazide for differentiation of live vs. dead bacteria by selective removal of DNA from dead cells. J. Microbiol. Methods 67, 310–320. 10.1016/j.mimet.2006.04.015, PMID: [DOI] [PubMed] [Google Scholar]
- Pacholewicz E., Buhler C., Wulsten I. F., Kraushaar B., Luu H. Q., Iwobi A. N., et al. (2019). Internal sample process control improves cultivation-independent quantification of thermotolerant Campylobacter. Food Microbiol. 78, 53–61. 10.1016/j.fm.2018.09.017, PMID: [DOI] [PubMed] [Google Scholar]
- Razzuoli E., Vencia W., Fedele V., Mignone G., Lazzara F., Rubini D., et al. (2018). Evaluation and validation of an alternative method to detect Campylobacter spp. in dairy products. Ital. J. Food Saf. 7:7180. 10.4081/ijfs.2018.7180, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins D. M., Colwell R. R. (1986). Viable but nonculturable stage of Campylobacter jejuni and its role in survival in the natural aquatic environment. Appl. Environ. Microbiol. 52, 531–538. 10.1128/AEM.52.3.531-538.1986, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosner B. M., Schielke A., Didelot X., Kops F., Breidenbach J., Willrich N., et al. (2017). A combined case-control and molecular source attribution study of human Campylobacter infections in Germany, 2011–2014. Sci. Rep. 7:5139. 10.1038/s41598-017-05227-x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Stel A. X., Boogerd F. C., Huynh S., Parker C. T., Van Dijk L., Van Putten J. P. M., et al. (2017). Generation of the membrane potential and its impact on the motility, ATP production and growth in Campylobacter jejuni. Mol. Microbiol. 105, 637–651. 10.1111/mmi.13723, PMID: [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All datasets generated for this study are included in the article.