Keywords: antibiotics, cholinergic, enteric nervous system, microbiota, weaning
Abstract
The period during and immediately after weaning is an important developmental window when marked shifts in gut microbiota can regulate the maturation of the enteric nervous system (ENS). Because microbiota-derived signals that modulate ENS development are poorly understood, we examined the physiological impact of the broad spectrum of antibiotic, vancomycin-administered postweaning on colonic motility, neurochemistry of enteric neurons, and neuronal excitability. The functional impact of vancomycin on enteric neurons was investigated by Ca2+ imaging in Wnt1-Cre;R26R-GCaMP3 reporter mice to characterize alterations in the submucosal and the myenteric plexus, which contains the neuronal circuitry controlling gut motility. 16S rDNA sequencing of fecal specimens after oral vancomycin demonstrated significant deviations in microbiota abundance, diversity, and community composition. Vancomycin significantly increased the relative family rank abundance of Akkermansiaceae, Lactobacillaceae, and Enterobacteriaceae at the expense of Lachnospiraceae and Bacteroidaceae. In sharp contrast to neonatal vancomycin exposure, microbiota compositional shifts in weaned animals were associated with slower colonic migrating motor complexes (CMMCs) without mucosal serotonin biosynthesis being altered. The slowing of CMMCs is linked to disruptions in the neurochemistry of the underlying enteric circuitry. This included significant reductions in cholinergic and calbindin+ myenteric neurons, neuronal nitric oxide synthase+ submucosal neurons, neurofilament M+ enteric neurons, and increased proportions of cholinergic submucosal neurons. The antibiotic treatment also increased transmission and responsiveness in myenteric and submucosal neurons that may enhance inhibitory motor pathways, leading to slower CMMCs. Differential vancomycin responses during neonatal and weaning periods in mice highlight the developmental-specific impact of antibiotics on colonic enteric circuitry and motility.
INTRODUCTION
The gastrointestinal tract contains its own extensive enteric nervous system (ENS) and harbors the largest microbial ecosystem (microbiota) in the body; both systems control vital gastrointestinal functions and host physiology. Studies have now shown that the myenteric neurons and enteric glia at the mucosa can interact with microbiota in the mature (12, 37, 46, 53, 86) and developing gut (6, 9, 32, 33).
Mice have been central to understanding the physiological significance of host-microbiota interactions and of ENS development. Almost 99% of mouse genes are shared with humans. Moreover, microbiota of mice and humans share many core similarities, including a general predominance of bacterial phyla Firmicutes and Bacteroidetes and their regional compartmentalization (15, 35), and antibiotics cause dysbiosis in both species. However, mice have a shorter and accelerated early life. The period after weaning (from 3 wk of age) in mice corresponds to the period from 6 mo of age for human babies. Yet mice can begin puberty as early as from the time of weaning, and for this reason mice that are ≥3 wk to <8 wk of age are considered adolescents (15, 38). In this study, we refer to the period from postnatal day (P) 20 to P49 in mice as the postweaning period. In humans, major shifts in microbiota still occur during infancy and after weaning due to the transition from mother’s milk to solid food (35). We recently established that the ENS in mice, particularly the submucous plexus, undergoes significant maturation during the period immediately after weaning (55). In this study, we explored the impact of antibiotic exposure on the maturing ENS and microbiota during the postweaning period.
Antibiotics that are widely used to treat bacterial infections significantly impact microbiota communities in the gut. It appears that antibiotics administered during critical developmental windows have greater consequences on host physiology than antibiotic exposure during maturity. Early life antibiotic exposure has received recent attention with reports of increased susceptibility to diseases such as functional gastrointestinal disorders, obesity, metabolic dysfunction, and allergies later in life (10, 14, 49). Vancomycin is used orally to treat clinical infections in pediatric and adult patients (43) and enterocolitis caused by bacterial pathogens such as Staphylococcus aureus (8). Oral vancomycin is also commonly used on its own or in a cocktail with other antibiotics to perturb microbiota in mice, with minimal systemic side effects, as it is poorly absorbed (6, 12, 29, 32, 33).
We have previously shown that oral vancomycin administered to mice during the neonatal period profoundly affects the gut microbiota and ENS function (32). Here, we examined the impact of oral vancomycin exposure on the mouse colon during a later developmental period, immediately after weaning. We demonstrate significant antibiotic-induced shifts in microbiota diversity and communities that are associated with disrupted colonic migrating motor complexes (CMMCs). Furthermore, we demonstrate that vancomycin-induced slowing of CMMCs is linked to disruption of the enteric circuitry underlying this motor pattern, including alterations in the proportions of specific myenteric and submucosal neurochemical subtypes and increased transmission and responsiveness of enteric neurons to electrical stimuli.
MATERIALS AND METHODS
Animals.
Male and female Wnt1-Cre;R26R-GCaMP3 mice in which all neural crest-derived cells, including all enteric neurons and glia, express the genetically encoded calcium indicator GCaMP3 (3, 87) were used. Reporter mice were bred by mating heterozygous Wnt1-Cre male mice (11) with homozygous R26R-GCaMP3 female mice (The Jackson Laboratory). Animals were euthanized by cervical dislocation, as approved by the University of Melbourne Animal Experimentation Ethics Committee. The cecal weight of 6-wk-old mice was measured. Full-length colons from postnatal day (P) 10/11 and 6-wk-old mice were removed and immediately pinned open in a petri dish lined with silicone elastomer (Sylgard 184; Dow Corning, North Ryde, NSW, Australia) containing physiological saline (composition in mM: 118 NaCl, 25 NaHCO3, 11 d-glucose, 4.8 KCl, 2.5 CaCl2, 1.2 MgSO4, and 1.0 NaH2PO4) bubbled with carbogen gas (95% O2-5% CO2) or phosphate-buffered saline (PBS) at room temperature.
Antibiotic treatment.
Mouse pups from each litter were given a dose of sterile water or vancomycin hydrochloride (83 mg · kg−1 · day−1, total volume of 2.0 µL/g body wt; Sigma Aldrich) from P0 to P10/11 by oral feeding daily using a micropipette tip, as previously described (32). Three-week-old mice were removed from their mothers, placed in separate cages from the day of weaning, and treated until 6 wk of age. Six-week-old (P42–49) mice have been defined as being in middle-to-late adolescence (38). Each cage was separated by sex, and sterile water (control) or vancomycin hydrochloride (0.5 g/L) dissolved in sterile drinking water was given ad libitum with twice weekly bottle changes, as previously described (12, 33).
Microbiome and mass spectrometry analysis.
Intestinal segments were cut open along the mesenteric border with the mucosal side up. With the use of sterile microdissection tools, the midcolonic mucosa was sampled for mass spectrometry analysis, whereas fecal content and mucosa were sampled for microbiome analysis or neurotransmitter profiling (mass spectroscopy). Specimens for microbiome and mass spectrometry analysis were immediately snap frozen in liquid nitrogen. The 16S rDNA V4 amplicon data were generated by MiSeq paired-end sequencing (effective read length: 250 bp for V4). The DADA2 package (version 1.8) (5) was used to process de-multiplexed raw sequencing reads using default parameter settings with minor modifications. Specifically, raw sequencing reads were trimmed while maintaining overlap regions for merging paired-end reads; sequencing primers retained in forward and reverse reads were removed. The IdTaxa function in the DECIPHER package (version 2.6.0) (50) and its prebuilt training set (LearnTaxa function) of the SILVA database (release 132) (60) were used for taxonomy assignment (threshold: 0.5) of amplicon sequence variants (ASVs) from the DADA2 output. ASVs that were not classified as bacteria were removed, as were sequences identified as chimeras. Phylogeny of ASVs was generated using DECIPHER for sequence alignment and FastTree (version 2.1.3) (57) for tree construction. Samples with <1,000 reads in the ASV matrix were excluded, and proportional transformation was applied to normalize feature data before downstream analysis. α-Diversity indices, including Shannon and inverse Simpson metric, were calculated using the Phyloseq package (version 1.24.2) (44). β-Diversity analysis was performed using nonmetric multidimensional scaling (NMDS) of weighted UniFrac (40) distance in the Vegan package (version 2.5–5) (54). Benjamini-Hochberg (BH) correction was applied after Mann-Whitney-Wilcoxon test for comparing collapsed taxa at family rank between groups; a BH-adjusted P value of <0.05 was considered significant. Levels of 5-hydroxytryptamine (5-HT, serotonin) biosynthetic intermediates were measured using SRM-MS, as previously described by us (41).
Video imaging of colonic motility patterns.
Gut motility patterns were revealed using spatiotemporal mapping, as previously described (31, 63, 77). Briefly, full-length colon was cannulated in an organ bath continuously superfused with saline and carbogen (95% O2 and 5% CO2) throughout the experiment and heated to 33–35°C. The intraluminal pressures of the colon were maintained between 2 and 4 cmH2O. Tissues were then left to equilibrate for 45 min, followed by 3 × 15 min video recordings. Videos were captured using a Logitech Quickcam Pro 900 camera (I-Tech, Ultimo, NSW, Australia) mounted directly above the organ bath, as described by Swaminathan et al. (77). Recordings were processed using software developed in-house (Scribble 2.0) and a purpose built Matlab (version 2014b) plugin that converts the movement of the gut into a spatiotemporal map, which was then analyzed using the Analyse2 software. A mean value of each parameter was derived from three spatiotemporal maps from each colon. A population mean was then determined by averaging the data obtained per treatment condition.
Neurochemistry of the myenteric and submucosal plexus.
Midcolonic segments (3–4 cm long taken directly below the demarcated striations of the proximal colon) were cut open along the mesenteric border, stretched, and pinned flat with the mucosal side up. Tissues were fixed overnight in 4% formaldehyde in 0.1 M phosphate buffer (pH 7.2) at 4°C. Fixative was cleared the next day with 3 × 10 min PBS washes. Whole mount preparations of the submucosal plexus and myenteric plexus with adherent longitudinal muscle were obtained from the colon via microdissection, as previously described (16, 17, 21). Preparations were permeabilized with 1% Triton X-100/PBS (Proscitech, Thuringowa, QLD, Australia) for 30 min at room temperature, followed by PBS washes (3 × 10 min) before incubation with a combination of primary antisera (Table 1) for 24–72 h at 4°C. Tissue preparations were then washed with PBS (3 × 10 min) and incubated with secondary antisera (Table 1) for 2.15–2.5 h at room temperature. Excess antiserum was removed by washing with PBS (3 × 10 min), and the preparations were mounted on glass slides using fluorescent mounting medium (DAKO, Carpinteria, CA).
Table 1.
Primary and secondary antisera
| Raised In | Dilution | Source | |
|---|---|---|---|
| Primary antibodies | |||
| Hu (ANNA-1) | Human | 1:5000 | Gift from Dr. V. Lennon |
| nNOS | Sheep | 1:1000 | Gift from P. Emson |
| Calbindin | Rabbit | 1:1600 | SWANT |
| ChAT | Goat | 1:100 | Chemicon |
| Calretinin | Goat | 1:1000 | SWANT |
| Medium size neurofilament (145) referred to as NFM | Rabbit | 1:500 | Merck Millipore |
| Synaptophysin | Rabbit | 1:100 | Abcam |
| VIP | Rabbit | 1:1000 | Merck Millipore |
| Secondary antibodies | |||
| Anti-human AF594 | Donkey | 1:500 | Jackson Immuno Laboratories |
| Anti-rabbit AF488 | Donkey | 1:400 | Molecular Probes |
| Anti-rabbit AF594 | Donkey | 1:400 | Molecular Probes |
| Anti-rabbit AF647 | Donkey | 1:400 | Molecular Probes |
| Anti-sheep AF488 | Donkey | 1:400 | Molecular Probes |
| Anti-sheep AF647 | Donkey | 1:500 | Molecular Probes |
ChAT, choline acetyltransferase; NFM, neurofilament-M; nNOS, neuronal nitric oxide synthase; VIP, vasoactive intestinal peptide.
Immunostained myenteric plexus preparations were viewed and imaged using a Zeiss fluorescence microscope (Axio Imager M2), whereas submucosal plexus preparations were viewed and imaged with a Zeiss LSM800 confocal microscope. Images were analyzed using the FIJI software (ImageJ 1.51s; National Institutes of Health). To examine neuron density of the myenteric plexus, two images per preparation of Hu stains were taken using a Plan-Apochromat ×10/0.45 objective and analyzed as previously described (32). The total number of each neuronal subtype [neuronal nitric oxide synthase (nNOS), calbindin, choline acetyltransferase (ChAT), calretinin, neurofilament-M (NFM), and vasoactive intestinal peptide (VIP)] was expressed as a proportion of the total number of Hu+ neurons within the imaged region. At least 140 Hu+ cell bodies and at least 67 Hu+ cell bodies were counted in each myenteric and submucous preparation, respectively. The mean proportion of each neuronal subtype was determined by obtaining averages from at least three animals.
Calcium imaging and analysis.
A segment of midcolon (3–4 cm) from each animal was cut along the mesenteric border and pinned flat, mucosa side up, in an organ bath lined with a silicone elastomer. Submucosal plexus preparations were obtained by removing the mucosa and the underlying smooth muscle layers by microdissection, after which serosa and longitudinal muscle layers were stripped off to produce myenteric plexus/circular muscle preparations. Submucosal and myenteric plexus preparations were stretched over an inox ring and clamped with a rubber O-ring to stabilize the tissue during imaging (16, 21, 39). Preparations were then placed in an organ bath that was constantly superfused (1 mL/min) with 95% O2-5% CO2 bubbled physiological saline at room temperature (between 20 and 24°C) throughout the experiment via a gravity-fed inflow system, as previously described (16, 65). In dissected whole mount preparations, the integrity of cells is better preserved at room temperature, and the analyzed electrically-evoked Ca2+ responses can still provide clear indications of the functionality of the enteric circuitry. Preparations were imaged using a ×20 (NA 1.0) water-dipping objective on an upright Zeiss (Axio Examiner.Z1) microscope with an Axiocam 702 camera (Carl Zeiss Microscopy, North Ryde, NSW, Australia). Images (16 bit) were acquired at 7 Hz with an exposure time of 20 ms. Neurons within enteric ganglia were stimulated electrically via a focal stimulating electrode (tungsten wire; 50 μm diameter) placed on an interganglionic fiber tract leading into the imaged ganglion of choice, where a single pulse and a train of 20 pulses (20 Hz) were elicited to evoke responses corresponding to fast and fast/slow excitatory postsynaptic potentials, respectively (16, 17, 27, 36, 51). Analyses were performed using custom-written directives in IGOR Pro (WaveMetrics, Lake Oswego, OR) (39). Regions of interest were drawn over a selected area of the cytoplasm for each neuron, excluding the nucleus because GCaMP3 is absent from the nuclei (79, 85). The fluorescence intensity was calculated as a fraction of the baseline fluorescence, Fi/F0. Neurons were considered responsive only when they exhibited peaks in intracellular Ca2+ concentration ([Ca2+]i) (calcium transients) with signals that had a minimum increase of five or three (for single pulse-stimulated responses) times the intrinsic noise. The amplitudes of calcium transients were measured as the maximum increase in [Ca2+]i above (ΔFi/F0).
Statistical analysis.
Data are presented as means ± SE, where n is the number of animals examined, unless stated otherwise. Statistical comparisons were performed using unpaired t-tests and Mann-Whitney U tests with GraphPad Prism 5.0 (GraphPad Software, San Diego, CA), where P < 0.05 is considered statistically significant.
RESULTS
Vancomycin exposure has divergent effects on gut microbiota communities during different postnatal developmental periods.
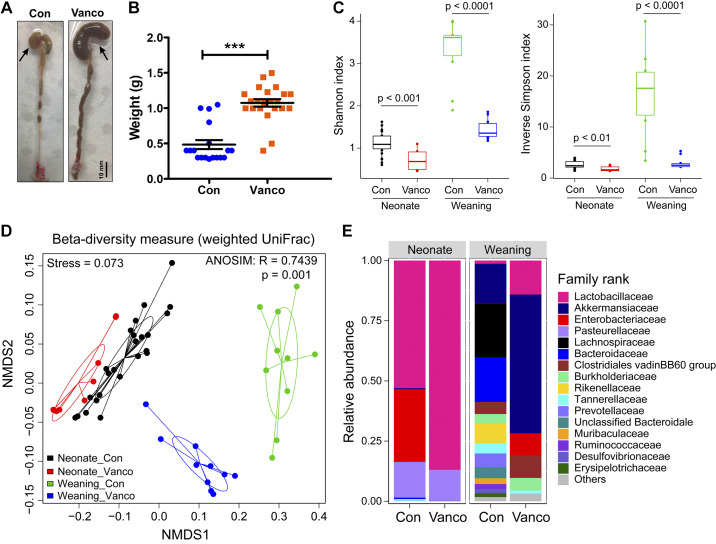
Oral vancomycin significantly increased cecal weights of juvenile mice treated postweaning (Fig. 1, A and B); this is strongly associated with dysbiosis (6, 33). Indeed, when 16S rDNA amplicon reads from 6-wk-old mice were examined, we found that vancomycin exposure significantly decreased microbiome α-diversity and resulted in markedly different community compositions (Fig. 1, C and E).
Fig. 1.
Vancomycin exposure disrupted microbiota. A: representative images of the colons from a control and a mouse that underwent postweaning vancomycin treatment (arrow indicates cecum). B: cecum weight of control- and vancomycin-exposed mice (n = 29–34 mice). Data are shown as means ± SE. Treatment groups were compared statistically using unpaired t tests. ***P < 0.0001. C–E: microbiome analysis performed on fecal samples of control- and vancomycin-treated mice that were treated during the neonatal (n = 10–24 mice) or postweaning period (n = 10 mice). C: α-diversity of fecal samples of control and vancomycin-treated mice that were treated during the neonatal or postweaning period. Data represented as median with 1st and 3rd quartiles; statistical analysis was performed with Mann-Whitney-Wilcoxon test between 2 groups. D: β-diversity represented as nonmetric multidimensional scaling (NMDS) plot of weighted UniFrac distances of the microbiota at each sample point. E: microbiota composition at the family taxonomic level.
We found significant increases in the abundance and communities of microbiota between control P10 and control 6-wk-old mice (Fig. 1, C–E). In control animals, there was family-ranked abundance of Lactobacillaceae, Enterobacteriaceae, and Pasteurellaceae at P10, but by 6 wk of age, there were more types of contributing bacterial families, with Akkermansiaceae, Lachnospiraceae, and Bacteroidaceae being particularly abundant (Fig. 1E). Although vancomycin treatment significantly decreased microbiota diversity in both neonatal and weaned mice, community compositional shifts differed significantly between the distinct developmental stages. Vancomycin treatment resulted in a marked expansion of Lactobacillaceae in the expense of Enterobacteriaceae in the neonates, whereas Akkermansiaceae and Enterobacteriaceae were evident in older animals, with loss of Bacteroidaceae and Lachnospiraceae (Fig. 1E).
Vancomycin exposure postweaning slowed colonic migrating motor complexes.
We examined whether there are any effects on colonic motility patterns when mice were exposed to vancomycin from the day they were weaned. Vancomycin exposure did not affect the frequency or length of all anally propagating contractions (Fig. 2, A–C) in 6-wk-old mice. It also did not affect the frequency of anal short contractions, which are short contractions (<10 mm long) occurring at the anal end of the colon that do not appear to propagate (Fig. 2, A and D). When we focused on colonic migrating motor complexes (CMMCs), which are the orally initiated contractions that propagate anally for at least half the full length of the colonic segment, we found that although vancomycin treatment did not affect CMMC frequency, it significantly decreased their speed of propagation and increased the colonic diameter between CMMCs (Fig. 2, A and E–G).
Fig. 2.
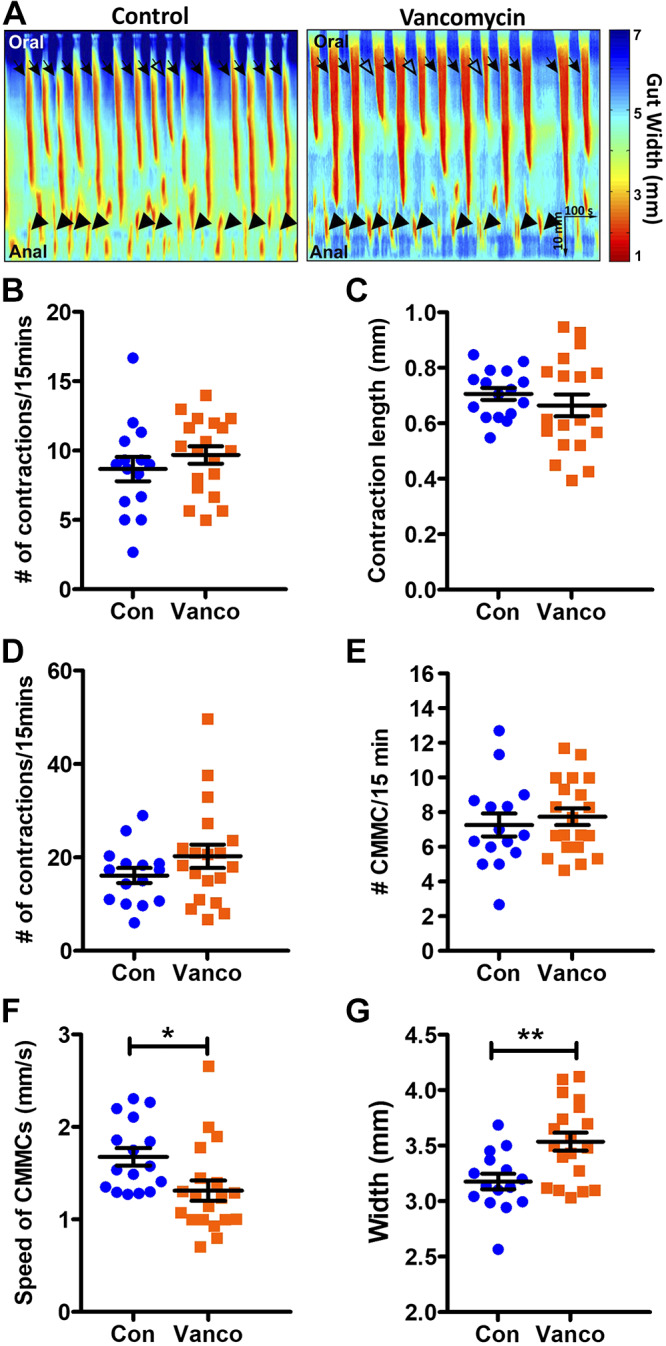
Vancomycin exposure postweaning altered colonic motility. A: representative spatiotemporal maps showing colonic motility patterns in a control and a vancomycin-treated 6-wk-old mouse treated postweaning, with all anally propagating contractions indicated by arrows. Anally propagating patterns that do not propagate past halfway along the length of the colon (open arrows) and those that do [colonic migrating motor complexes (CMMCs) closed arrows] are identified. Short contractions that occur in the anal regions (closed arrowheads) are also denoted. B–F: quantification of the frequency of all anally propagating contractions (B), length of anally propagating contractions (C), frequency of anal short contractions (D), frequency of CMMCs (E), speed of CMMCs (F), and resting gut width (G) of control- (Con) and vancomycin (Vanco)-exposed mice (n = 15–20 mice). Data are represented as means ± SE. Treatment groups were compared statistically using unpaired t tests or Mann-Whitney U tests. *P < 0.05; **P < 0.01.
Vancomycin exposure postweaning did not affect serotonin biosynthesis.
Several studies have found that adult germ-free mice or mice that were depleted of their microbiota via treatment with cocktails of broad-spectrum antibiotics have defective serotonergic metabolism (12, 26, 61, 86). However, in our study, analysis of the serotonin biosynthesis pathway of colonic mucosa via selected-reaction monitored mass spectrometry showed that vancomycin administered postweaning did not affect the levels of serotonin or its metabolite 5-hydroxyindoleacetic acid (5-HIAA) despite significantly increasing the levels of its precursor tryptophan (Fig. 3).
Fig. 3.
Vancomycin (Vanco) exposure postweaning did not affect mucosal serotonin biosynthesis. Mass spectrometry analysis measuring the levels of tryptophan (A), 5-HT (B), and the metabolite 5-hydroxyindoleacetic acid (5-HIAA; C) in the colonic mucosa of control (Con) and Vanco-treated 6-wk-old mice (n = 10 animals). Data are represented as means ± SE. Treatment groups were compared statistically using unpaired t tests and Mann-Whitney U tests. *P < 0.05.
Vancomycin exposure postweaning altered the proportion of myenteric and submucosal neuronal subtypes.
We have previously shown significant neurochemical changes in submucosal but not myenteric neurons during the postweaning period in mice (55). To determine whether vancomycin during this period produces changes in the neurochemistry of enteric neuronal subtypes governing gastrointestinal functions (23, 55, 58, 67), we performed immunohistochemistry on both enteric plexi.
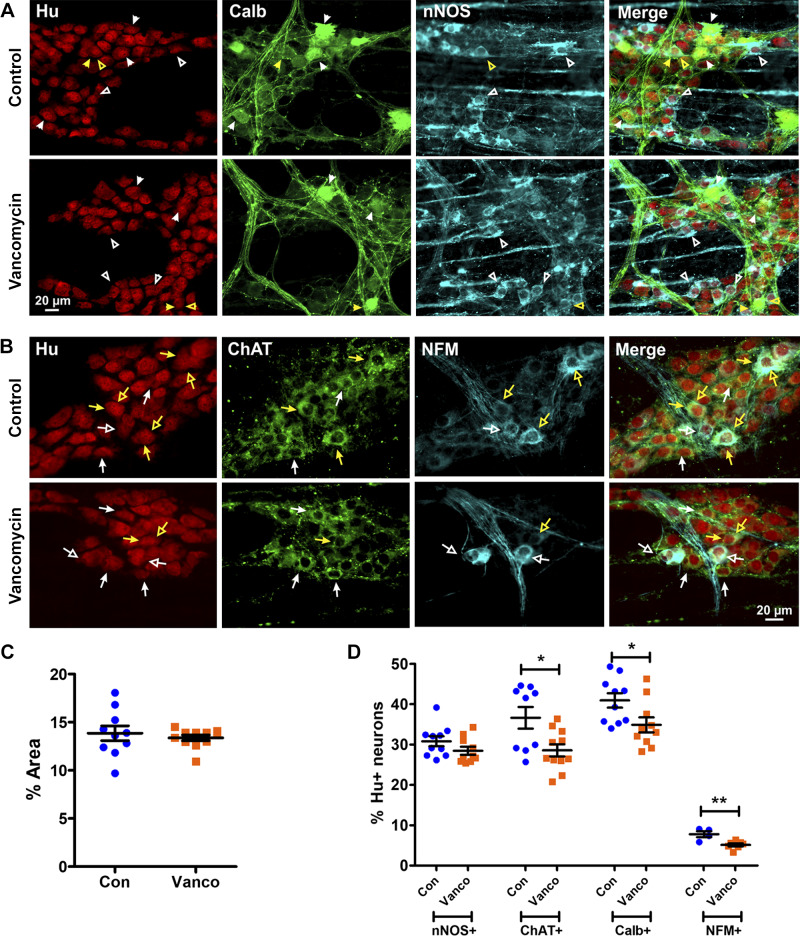
For the myenteric plexus, we examined nNOS (Fig. 4A) and choline acetyltransferase (ChAT; Fig. 4B) immunoreactive neurons, which are functionally distinct subclasses. nNOS is expressed in inhibitory motor neurons and some interneurons within the myenteric ganglia. ChAT is expressed by many myenteric neurons that serve various functions (58, 66, 67, 69). We also examined neurofilament-M (NFM) and calbindin, both markers of intrinsic sensory neurons and some interneurons (Fig. 4, A and B). Although both calbindin and NFM+ neurons are subtypes of cholinergic (ChAT+ neurons), some NFM+ neurons in the colon also express nNOS (24, 30, 55, 58, 66, 70). Vancomycin did not affect Hu+ neuron density (Fig. 4C) or the proportions of nNOS+ neurons (Fig. 4D). However, ChAT+, calbindin+, and NFM+ neurons were significantly reduced in vancomycin-treated mice compared with controls (Fig. 4D).
Fig. 4.
Vancomycin (Vanco) exposure postweaning disrupted myenteric neuronal subtype proportions in the colon. A and B: representative images of myenteric plexus preparations from the midcolon of control- (Con) and Vanco-treated 6-wk-old mice. A: myenteric neurons were stained with the pan-neuronal marker Hu (red), calbindin (green), and neuronal nitric oxide synthase (nNOS; blue). Some calbindin-immunoreactive (closed arrowheads) and neuronal nitric oxide synthase (nNOS)-immunoreactive (open arrowheads) Hu+ neurons are indicated. Hu+ neurons that express both calbindin and nNOS (yellow arrowheads) are rare. B: myenteric neurons are stained for Hu (red), choline acetyltransferase (ChAT; green), and neurofilament-M (NFM; cyan). Some ChAT-immunoreactive (filled arrows) and NFM-immunoreactive (open arrows) Hu+ neurons are indicated. Some Hu+ neurons express both ChAT and NFM (yellow arrows). C: myenteric neuron (Hu+) density of the midcolon (n = 10–11 mice). D: quantification of nNOS+ (n = 10 mice), ChAT+ (n = 9–11 mice), calbindin+ (n = 10 mice) and NFM+ (n = 4–7 mice) neurons relative to Hu+ neurons in the mid colon. Data are represented as means ± SE. Treatment groups were compared statistically using unpaired t tests. *P < 0.05; **P < 0.01.
For the submucosal plexus, we examined ChAT and vasoactive intestinal peptide (VIP) immunoreactivity, which mark distinct classes of secretomotor neurons (Fig. 5A) (19, 48). There is also a small proportion of submucosal neurons that co-express ChAT and VIP (55). NFM neurons were examined (Fig. 5B), although their function in the mouse is unknown. nNOS (Fig. 5C), which is expressed by a subpopulation of VIP+ neurons, and calretinin and calbindin, which stain almost all submucosal neurons, were also examined (19, 55). Vancomycin exposure specifically increased the proportion of ChAT+ neurons that did not coexpress VIP (Fig. 5D) but significantly decreased the proportion of nNOS+ and NFM+ neurons (Fig. 5E)
Fig. 5.
Vancomycin (Vanco) exposure postweaning disrupted submucosal neuronal subtype proportions in the colon. A–C: representative images of submucous plexus preparations from the mid-colon of control- (Con) and Vanco-treated 6-wk-old mice. A: submucosal neurons were stained with the pan-neuronal marker Hu (red), vasoactive intestinal peptide (VIP; green), and choline acetyltransferase (ChAT; blue). VIP-immunoreactive (closed arrowheads) and ChAT-immunoreactive (open arrowheads) Hu+ neurons are indicated. B: submucosal neurons are stained for Hu (red) and neurofilament-M (NFM; green). NFM-immunoreactive (filled arrowheads) Hu+ neurons are indicated. C: submucosal neurons are stained for Hu (red) and neuronal nitric oxide synthase (nNOS; green). nNOS-immunoreactive (filled arrowheads) Hu+ neurons are indicated. D: quantification of VIP+ and or ChAT+ neurons relative to Hu+ neurons in the midcolon (n = 4–5 mice). E: quantification of nNOS+, NFM+, and calbindin (Calb)/calretinin (Calr)+ neurons relative to Hu+ neurons in the mid-colon. Data are represented as means ± SE. Treatment groups were compared statistically using unpaired t tests. *P < 0.05; **P < 0.01.
Vancomycin exposure postweaning increased slow transmission to myenteric neurons and responsiveness of enteric neurons to electrical stimuli.
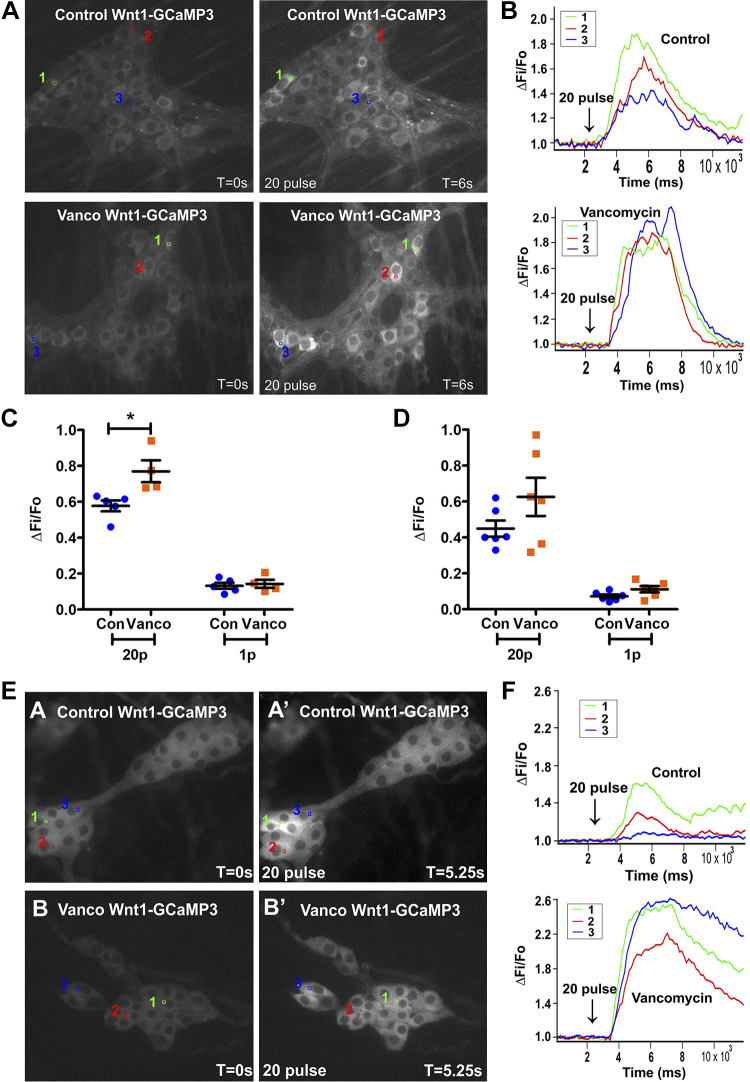
We used the myenteric and submucosal plexus of 6-wk-old mice expressing the Ca2+ indicator GCaMP3 to determine whether the effects of vancomycin on gastrointestinal functions postweaning were associated with changes in Ca2+ transients in different enteric neuronal subtypes. Vancomycin treatment significantly increased the amplitude of 20-pulse train- but not one-pulse-evoked [Ca2+]i responses in myenteric neurons. The 20-pulse train-evoked responses are associated with slow transmission, whereas one-pulse stimuli produce fast excitatory synaptic potentials in enteric neurons (Fig. 6, A–C) (16, 17, 27, 51). Nevertheless, single-pulse stimulation of interganglionic fiber tracts evoked responses in more myenteric neurons after vancomycin treatment (Table 2 and 3). In the submucosal plexus, the amplitudes of electrically evoked [Ca2+]i responses were not significantly affected by vancomycin (Fig. 6, D–F), but antibiotic-exposed submucosal neurons were more responsive to both 20-pulse train- and one-pulse stimulation (Table 2 and 3).
Fig. 6.
Vancomycin (Vanco) exposure postweaning increased slow transmission to myenteric neurons and responsiveness of enteric neurons to electrical stimuli. A: fluorescence micrographs of 20-pulse train (20 Hz)-evoked intracellular Ca2+ concentration ([Ca2+]i) responses in myenteric neurons [GCaMP3 signal at rest (t = 0) and after 20-pulse train stimulation (t = 6 s)] of control (Con)- and Vanco-treated 6-wk-old mice. B: 3 representative responding neurons in each condition are numerically marked (in A), and their color-coded traces show 20-pulse-evoked [Ca2+]i responses. Amplitude of [Ca2+]i transients (ΔFi/F0) from all cells in the myenteric plexus (n = 4–5 mice; C) and submucosal plexus (n = 6 mice; D) stimulated with a 20-pulse train versus a single-pulse stimulus. Graphs are represented as means ± SE. Treatment groups were compared statistically using unpaired t-tests. *P < 0.05. E: fluorescence micrographs of 20-pulse train (20 Hz)-evoked [Ca2+]i responses in submucosal neurons [GCaMP3 signal at rest (t = 0) and after 20 pulse train stimulation (t = 5.25 s)] of Con- and Vanco-treated mice. F: 3 representative responding neurons in each condition are numerically marked (in A), and their color-coded traces show 20-pulse-evoked [Ca2+]i responses.
Table 2.
Treatment induced change in proportion of GCaMP3+ responding neurons
| Electrical Stimulation (Gut Region/Plexus) | Proportion of Responders in Antibiotic Treatment Normalized to Control |
|---|---|
| 1-Pulse | |
| MP | 1.3* |
| SMP | 2.6* |
| 20-Pulse | |
| MP | 1.0 |
| SMP | 1.2* |
MP, myenteric plexus; SMP, submucous plexus.
P < 0.0001 χ2 test.
Table 3.
Number of GCaMP3+ cells that responded to electrical stimulation
| Electrical Stimulation (Treatment) | Gut Region/Plexus | No. of Responders | No. of Nonresponders |
|---|---|---|---|
| 1-Pulse | |||
| Control | MP | 435 | 1,375 |
| Vancomycin* | MP | 888 | 2,053 |
| Control | SMP | 65 | 235 |
| Vancomycin* | SMP | 211 | 165 |
| 20-Pulse | |||
| Control | MP | 646 | 1,164 |
| Vancomycin | MP | 1,097 | 1,844 |
| Control | SMP | 189 | 111 |
| Vancomycin* | SMP | 296 | 80 |
MP, myenteric plexus; SMP, submucous plexus.
P < 0.0001, Fisher’s exact test.
DISCUSSION
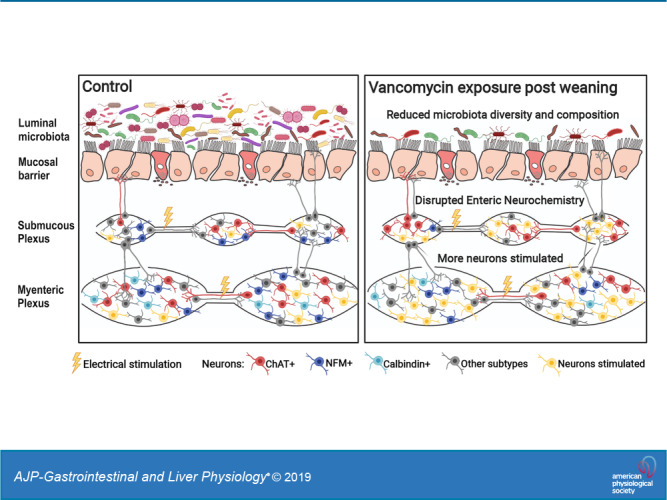
The postweaning period is associated with significant and concurrent maturation of the ENS and microbiota, but the impact of antibiotic exposure on the gut during this time remains unclear. We found that vancomycin exposure during the postweaning period significantly affected both α- and β-diversity of the microbiota as well as community composition. Vancomycin treatment also disrupted the neurochemistry of specific subpopulations of enteric neurons and increased responsiveness of both myenteric and submucosal neurons to electrical stimuli, which could contribute to the reduced speed of CMMCs.
CMMCs are neurally mediated contraction patterns in the mouse colon (63). In the present study, we found that postweaning vancomycin exposure significantly reduced the speed of CMMC propagation. This is consistent with other studies demonstrating slowed colonic transit time in antibiotic-treated animals (1, 6, 26, 84). The slowed CMMCs during the postweaning period contrast with our previous finding that neonatal vancomycin increased incidence and propagation speed of colonic contractions (32), thus demonstrating divergent effects of the antibiotic during different stages of life. The differential effects of vancomycin on colonic contractions may be due to the different effects on microbiota communities in the neonatal compared with the postweaning period, for example, the divergent impact on Enterobacteriaceae.
Intestinal transit time is a key determinant of gut microbiota composition, diversity, metabolism, and clearance, as well as nutrient and water absorption. Slow colonic transit typically causes firm stool consistency as a symptom of constipation. It has also been associated with high gut microbiota richness and diversity (7, 62, 72, 75, 81). However, this was not seen in our study, which found that vancomycin-treated mice had slow colonic transit and reduced microbiota diversity. Slower in vivo gastrointestinal and colonic transit was also reported in pseudo-germ-free adult mice treated antibiotic cocktail that included vancomycin (26). The discrepancies in the effect on gut motility between studies could be due to antibiotics having direct effects on the ENS and/or due to different changes in microbiota composition produced by treatment regimes. Postweaning vancomycin treatment in our study favored expansion of mucinolytic and metabolism-modulating Akkermansiaceae, a feature also reported in malnourished states and in patients with multiple sclerosis (56). Other microbiota phylogenetically associated with Enterobactericaeae and Lactobacillaceae were increased, whereas microbiota communities normally associated with maturity, including Lachnospiraceae and Bacteriodaceae (2, 42), were drastically reduced.
Mucosal serotonin is reported to be modulated by gut microbiota in several studies (12, 32, 61, 86). In contrast, we found that vancomycin exposure postweaning did not alter levels of 5-HT or its metabolite 5-HIAA. It may be that the vancomycin-induced perturbation of gut microbiota in our study was insufficient to affect 5-HT signaling; earlier studies used treatments that effectively abolished the gut microbiota (12, 61, 86). However, our α-diversity metric demonstrated that there was a major reduction in microbiota community richness, and we did find a significant increase in the levels of tryptophan in our vancomycin-treated mice, demonstrating that this pathway was impacted. The metabolic pathway of tryptophan is dominated by the conversion to indoles and kynurenine, which forms kynurenic acid and quiolinic acid signaling intermediates, whereas 5-HT accounts for only a small part of tryptophan metabolism. Thus, our antibiotic treatment may have affected the kynurenine pathway. The metabolites of kynurenine have been extensively studied in the CNS (34, 47), but any roles they may have in the ENS remain unclear and need to be investigated in future studies.
Some enteric neuronal subtypes are susceptible to vancomycin treatment during adolescence, although the total density of neurons was unaltered. The myenteric plexus contains the main neuronal circuitry coordinating gut motility. The total number of cholinergic myenteric neurons (ChAT+), specifically including the NFM+ and calbindin+ subtypes, was significantly reduced in vancomycin-treated postweaning mice. This is consistent with another study that found a reduced density of ChAT+ myenteric neurons in juvenile mice treated with an antibiotic cocktail (6). Some NFM+ neurons express nNOS (55), but vancomycin treatment did not alter the proportion of nNOS+ neurons, suggesting that non-nitrergic NFM+ neurons, which include intrinsic sensory neurons (58), are particularly vulnerable to vancomycin treatment. The antibiotic-induced reduction of ChAT+, calbindin+, and NFM+ neurons without affecting total Hu+ neuron density raises the possibility that myenteric neurons expressing other subtype markers such as 5-HT, calretinin, CGRP, and substance P (6, 58, 82) not examined in the present study may have compensated for the loss of these neurons and should be investigated in future studies.
Effects of postweaning vancomycin on the ENS differed from those produced by neonatal administration, which reduced neuron density, increased the proportion of calbindin+ neurons, and decreased the proportion of nNOS+ neurons (32). In contrast to the weaning period, the neurochemical changes in neonates were accompanied by increased frequency and speed of colonic contractions (32). The motility changes seen as a result of postweaning vancomycin treatment may be explained by the altered neurochemical phenotypes of some of the myenteric neurons and the increased excitability of some neurons. Computational modeling of contractile complexes in the intestine indicates that their propagation occurs via the spread of network activity through recurrent networks of intrinsic sensory neurons, with the speed of propagation being determined by the excitability of the network (78). Reduced numbers of intrinsic sensory neurons, either NFM+ or calbindin+, would reduce the excitatory feedback within the network and hence, slow propagation of complexes, but as frequency of complexes depends on presynaptic mechanisms within these neurons (78), this would be independent of their numbers. On the other hand, the resting diameter of the colon appears to depend on tonic firing of nNOS+ inhibitory motor neurons between contractile complexes (74), which might be expected to be enhanced by the increased excitability detected in our Ca2+ imaging analysis. Other mechanisms are, of course, possible, and a detailed analysis of the relationships between altered excitability and neuronal subtype will be essential for a comprehensive understanding of these mechanisms. For example, the absence of significant shifts in mucosal 5-HT could represent a major physiological variable, as could changes in other metabolites recently reported to regulate ENS maturation and function during development (53).
The submucosal plexus contains the final motor neurons regulating water and electrolyte secretion in the gut but also contributes to the regulation of motility. The proportion of ChAT+ neurons that do not contain VIP was significantly increased after vancomycin treatment during the postweaning period. Because cholinergic submucosal neurons have a secretomotor function (22), the increased proportion of these neurons could cause more water and electrolyte secretion. On the other hand, the proportion of NFM+ and nNOS+ submucosal neurons was significantly reduced by antibiotic treatment. NFM expression has been associated with enteric neurons with a sensory function in the myenteric plexus but is also expressed by some murine submucosal neurons (55), where it has not yet been associated with a particular function. Almost all nNOS+ submucosal neurons also express VIP (19), but the proportion of VIP+ neurons was unaffected by vancomycin, suggesting that the expression of nNOS in the submucosal plexus is vulnerable to antibiotic treatment. As yet the role of submucosal nNOS in mouse colon is unclear, although nitric oxide donors have been shown to modify responses of submucosal neurons in the guinea pig to various synaptic inputs (4). Thus, a reduction in proportion of nNOS in submucosal neurons could lead to less NO released, thereby modifying the response of submucosal neurons to physiological stimuli. The increased excitability of submucosal neurons detected in the Ca2+ imaging could contribute to the slowed colonic transit if they excite the inhibitory motor pathways and/or increase secretomotor activity, which should be investigated in future studies.
The overall increase in excitability of the enteric circuitry may result from vancomycin-induced dysbiosis, although we cannot exclude the possibility of the antibiotic itself activating the ENS. The microbiome has been shown to affect the activity of intrinsic sensory neurons of the ENS. Lactobacillus strains can directly modulate colonic motility by activating calcium-dependent potassium channels in the intrinsic sensory neurons of the ENS (37, 45, 46, 83). Thus an expansion in Lactobacillaceae in our postweaning mice could have direct consequences on enteric neuronal excitability. Furthermore, expansion in Akkermansiaceae and Lactobacillaceae could produce metabolites that can influence ENS function. For example, microbes can break down carbohydrates to produce short-chain fatty acids such as butyrate, which affect the ENS and gut motility (56, 73, 76, 80).
Conclusion.
In conclusion, the postweaning period in mice is a significant time of maturation of the ENS and microbiota in the colon. Vancomycin exposure during this time significantly altered gut microbiota and neuronal activity in both the submucosal and myenteric plexus of the ENS. Changes in the cross-talk between the microbiota and the ENS could contribute to the disruption in CMMCs. The effects of the antibiotic were not mediated by 5-HT signaling and could involve alternative pathways of tryptophan or other metabolic mechanisms.
GRANTS
This research was supported by National Health and Medical Research Council of Australia Project Grants APP1099016 to J. P. P. Foong, J. C. Bornstein, and T. C. Savidge, National Institutes of Health Grants P30-DK-56338 to T. C. Savidge, U01-AI-24290 to T. C. Savidge, and R01-AI-10091401 to T. C. Savidge, and the University of Melbourne International Research and Fee Remission Scholarship to L. Y. Hung.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
T.S. and J.P.P.F. conceived and designed research; L.Y.H., P.P., P.B., Y.W., A.H., R.A.L., and J.P.P.F. performed experiments; L.Y.H., P.P., P.B., Q.W., Y.W., A.H., R.A.L., and J.P.P.F. analyzed data; L.Y.H., P.P., P.B., Q.W., J.C.B., T.S., and J.P.P.F. interpreted results of experiments; L.Y.H., Q.W., and J.P.P.F. prepared figures; L.Y.H., Q.W., J.C.B., T.S., and J.P.P.F. drafted manuscript; L.Y.H., P.P., J.C.B., T.S., and J.P.P.F. edited and revised manuscript; L.Y.H., P.P., P.B., Q.W., Y.W., A.H., R.A.L., J.C.B., T.S., and J.P.P.F. approved final version of manuscript.
REFERENCES
- 1.Anitha M, Vijay-Kumar M, Sitaraman SV, Gewirtz AT, Srinivasan S. Gut microbial products regulate murine gastrointestinal motility via Toll-like receptor 4 signaling. Gastroenterology 143: 1006–1016.e4, 2012. doi: 10.1053/j.gastro.2012.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bergström A, Skov TH, Bahl MI, Roager HM, Christensen LB, Ejlerskov KT, Mølgaard C, Michaelsen KF, Licht TR. Establishment of intestinal microbiota during early life: a longitudinal, explorative study of a large cohort of Danish infants. Appl Environ Microbiol 80: 2889–2900, 2014. doi: 10.1128/AEM.00342-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boesmans W, Martens MA, Weltens N, Hao MM, Tack J, Cirillo C, Vanden Berghe P. Imaging neuron-glia interactions in the enteric nervous system. Front Cell Neurosci 7: 183, 2013. doi: 10.3389/fncel.2013.00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bornstein JC, Marks KA, Foong JP, Gwynne RM, Wang ZH. Nitric oxide enhances inhibitory synaptic transmission and neuronal excitability in guinea-pig submucous plexus. Front Neurosci 4: 30, 2010. doi: 10.3389/fnins.2010.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 13: 581–583, 2016. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caputi V, Marsilio I, Filpa V, Cerantola S, Orso G, Bistoletti M, Paccagnella N, De Martin S, Montopoli M, Dall’Acqua S, Crema F, Di Gangi IM, Galuppini F, Lante I, Bogialli S, Rugge M, Debetto P, Giaroni C, Giron MC. Antibiotic-induced dysbiosis of the microbiota impairs gut neuromuscular function in juvenile mice. Br J Pharmacol 174: 3623–3639, 2017. doi: 10.1111/bph.13965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carroll IM, Chang YH, Park J, Sartor RB, Ringel Y. Luminal and mucosal-associated intestinal microbiota in patients with diarrhea-predominant irritable bowel syndrome. Gut Pathog 2: 19, 2010. doi: 10.1186/1757-4749-2-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cimolai N. Does oral vancomycin use necessitate therapeutic drug monitoring? Infection 48: 173–182, 2020. doi: 10.1007/s15010-019-01374-7. [DOI] [PubMed] [Google Scholar]
- 9.Collins J, Borojevic R, Verdu EF, Huizinga JD, Ratcliffe EM. Intestinal microbiota influence the early postnatal development of the enteric nervous system. Neurogastroenterol Motil 26: 98–107, 2014. doi: 10.1111/nmo.12236. [DOI] [PubMed] [Google Scholar]
- 10.Cox LM, Yamanishi S, Sohn J, Alekseyenko AV, Leung JM, Cho I, Kim SG, Li H, Gao Z, Mahana D, Zárate Rodriguez JG, Rogers AB, Robine N, Loke P, Blaser MJ. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell 158: 705–721, 2014. doi: 10.1016/j.cell.2014.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Danielian PS, Muccino D, Rowitch DH, Michael SK, McMahon AP. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr Biol 8: PS1–PS2, 1998. doi: 10.1016/S0960-9822(07)00562-3. [DOI] [PubMed] [Google Scholar]
- 12.De Vadder F, Grasset E, Mannerås Holm L, Karsenty G, Macpherson AJ, Olofsson LE, Bäckhed F. Gut microbiota regulates maturation of the adult enteric nervous system via enteric serotonin networks. Proc Natl Acad Sci USA 115: 6458–6463, 2018. doi: 10.1073/pnas.1720017115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Vroey B, De Cassan C, Gower-Rousseau C, Colombel JF. Editorial: antibiotics earlier, IBD later? Am J Gastroenterol 105: 2693–2696, 2010. doi: 10.1038/ajg.2010.396. [DOI] [PubMed] [Google Scholar]
- 15.Dutta S, Sengupta P. Men and mice: relating their ages. Life Sci 152: 244–248, 2016. doi: 10.1016/j.lfs.2015.10.025. [DOI] [PubMed] [Google Scholar]
- 16.Foong JP, Hirst CS, Hao MM, McKeown SJ, Boesmans W, Young HM, Bornstein JC, Vanden Berghe P. Changes in nicotinic neurotransmission during enteric nervous system development. J Neurosci 35: 7106–7115, 2015. doi: 10.1523/JNEUROSCI.4175-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foong JP, Nguyen TV, Furness JB, Bornstein JC, Young HM. Myenteric neurons of the mouse small intestine undergo significant electrophysiological and morphological changes during postnatal development. J Physiol 590: 2375–2390, 2012. doi: 10.1113/jphysiol.2011.225938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Foong JP, Tough IR, Cox HM, Bornstein JC. Properties of cholinergic and non-cholinergic submucosal neurons along the mouse colon. J Physiol 592: 777–793, 2014. doi: 10.1113/jphysiol.2013.265686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fung C, Boesmans W, Cirillo C, Foong JPP, Bornstein JC, Vanden Berghe P. VPAC receptor subtypes tune purinergic neuron-to-glia communication in the murine submucosal plexus. Front Cell Neurosci 11: 118, 2017. doi: 10.3389/fncel.2017.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fung C, Unterweger P, Parry LJ, Bornstein JC, Foong JP. VPAC1 receptors regulate intestinal secretion and muscle contractility by activating cholinergic neurons in guinea pig jejunum. Am J Physiol Gastrointest Liver Physiol 306: G748–G758, 2014. doi: 10.1152/ajpgi.00416.2013. [DOI] [PubMed] [Google Scholar]
- 23.Furness JB. The enteric nervous system and neurogastroenterology. Nat Rev Gastroenterol Hepatol 9: 286–294, 2012. doi: 10.1038/nrgastro.2012.32. [DOI] [PubMed] [Google Scholar]
- 24.Furness JB, Robbins HL, Xiao J, Stebbing MJ, Nurgali K. Projections and chemistry of Dogiel type II neurons in the mouse colon. Cell Tissue Res 317: 1–12, 2004. doi: 10.1007/s00441-004-0895-5. [DOI] [PubMed] [Google Scholar]
- 26.Ge X, Ding C, Zhao W, Xu L, Tian H, Gong J, Zhu M, Li J, Li N. Antibiotics-induced depletion of mice microbiota induces changes in host serotonin biosynthesis and intestinal motility. J Transl Med 15: 13, 2017. doi: 10.1186/s12967-016-1105-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gwynne RM, Bornstein JC. Synaptic transmission at functionally identified synapses in the enteric nervous system: roles for both ionotropic and metabotropic receptors. Curr Neuropharmacol 5: 1–17, 2007. doi: 10.2174/157015907780077141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hansen CH, Krych L, Nielsen DS, Vogensen FK, Hansen LH, Sørensen SJ, Buschard K, Hansen AK. Early life treatment with vancomycin propagates Akkermansia muciniphila and reduces diabetes incidence in the NOD mouse. Diabetologia 55: 2285–2294, 2012. doi: 10.1007/s00125-012-2564-7. [DOI] [PubMed] [Google Scholar]
- 30.Hao MM, Young HM. Development of enteric neuron diversity. J Cell Mol Med 13: 1193–1210, 2009. doi: 10.1111/j.1582-4934.2009.00813.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hennig GW, Costa M, Chen BN, Brookes SJ. Quantitative analysis of peristalsis in the guinea-pig small intestine using spatio-temporal maps. J Physiol 517: 575–590, 1999. doi: 10.1111/j.1469-7793.1999.0575t.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hung LY, Boonma P, Unterweger P, Parathan P, Haag A, Luna RA, Bornstein JC, Savidge TC, Foong JPP. Neonatal antibiotics disrupt motility and enteric neural circuits in mouse colon. Cell Mol Gastroenterol Hepatol 8: 298–300.e6, 2019. doi: 10.1016/j.jcmgh.2019.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kabouridis PS, Lasrado R, McCallum S, Chng SH, Snippert HJ, Clevers H, Pettersson S, Pachnis V. Microbiota controls the homeostasis of glial cells in the gut lamina propria. Neuron 85: 289–295, 2015. doi: 10.1016/j.neuron.2014.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kennedy PJ, Cryan JF, Dinan TG, Clarke G. Kynurenine pathway metabolism and the microbiota-gut-brain axis. Neuropharmacology 112: 399–412, 2017. doi: 10.1016/j.neuropharm.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 35.Kostic AD, Howitt MR, Garrett WS. Exploring host-microbiota interactions in animal models and humans. Genes Dev 27: 701–718, 2013. doi: 10.1101/gad.212522.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koussoulas K, Swaminathan M, Fung C, Bornstein JC, Foong JPP. Neurally released GABA acts via GABAC receptors to modulate Ca2+ transients evoked by trains of synaptic inputs, but not responses evoked by single stimuli, in myenteric neurons of mouse ileum. Front Physiol 9: 97, 2018. doi: 10.3389/fphys.2018.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kunze WA, Mao YK, Wang B, Huizinga JD, Ma X, Forsythe P, Bienenstock J. Lactobacillus reuteri enhances excitability of colonic AH neurons by inhibiting calcium-dependent potassium channel opening. J Cell Mol Med 13: 2261–2270, 2009. doi: 10.1111/j.1582-4934.2009.00686.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laviola G, Macrì S, Morley-Fletcher S, Adriani W. Risk-taking behavior in adolescent mice: psychobiological determinants and early epigenetic influence. Neurosci Biobehav Rev 27: 19–31, 2003. doi: 10.1016/S0149-7634(03)00006-X. [DOI] [PubMed] [Google Scholar]
- 39.Li Z, Hao MM, Van den Haute C, Baekelandt V, Boesmans W, Vanden Berghe P. Regional complexity in enteric neuron wiring reflects diversity of motility patterns in the mouse large intestine. eLife 8: e42914, 2019. doi: 10.7554/eLife.42914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71: 8228–8235, 2005. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luna RA, Oezguen N, Balderas M, Venkatachalam A, Runge JK, Versalovic J, Veenstra-VanderWeele J, Anderson GM, Savidge T, Williams KC. Distinct microbiome-neuroimmune signatures correlate with functional abdominal pain in children with autism spectrum disorder. Cell Mol Gastroenterol Hepatol 3: 218–230, 2017. doi: 10.1016/j.jcmgh.2016.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mackie RI, Sghir A, Gaskins HR. Developmental microbial ecology of the neonatal gastrointestinal tract. Am J Clin Nutr 69: 1035S–1045S, 1999. doi: 10.1093/ajcn/69.5.1035s. [DOI] [PubMed] [Google Scholar]
- 43.McFarland LV, Ozen M, Dinleyici EC, Goh S. Comparison of pediatric and adult antibiotic-associated diarrhea and Clostridium difficile infections. World J Gastroenterol 22: 3078–3104, 2016. doi: 10.3748/wjg.v22.i11.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8: e61217, 2013. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McVey Neufeld KA, Mao YK, Bienenstock J, Foster JA, Kunze WA. The microbiome is essential for normal gut intrinsic primary afferent neuron excitability in the mouse. Neurogastroenterol Motil 25: 183–e88, 2013. doi: 10.1111/nmo.12049. [DOI] [PubMed] [Google Scholar]
- 46.McVey Neufeld KA, Perez-Burgos A, Mao YK, Bienenstock J, Kunze WA. The gut microbiome restores intrinsic and extrinsic nerve function in germ-free mice accompanied by changes in calbindin. Neurogastroenterol Motil 27: 627–636, 2015. doi: 10.1111/nmo.12534. [DOI] [PubMed] [Google Scholar]
- 47.Moloney GM, O’Leary OF, Salvo-Romero E, Desbonnet L, Shanahan F, Dinan TG, Clarke G, Cryan JF. Microbial regulation of hippocampal miRNA expression: Implications for transcription of kynurenine pathway enzymes. Behav Brain Res 334: 50–54, 2017. doi: 10.1016/j.bbr.2017.07.026. [DOI] [PubMed] [Google Scholar]
- 48.Mongardi Fantaguzzi C, Thacker M, Chiocchetti R, Furness JB. Identification of neuron types in the submucosal ganglia of the mouse ileum. Cell Tissue Res 336: 179–189, 2009. doi: 10.1007/s00441-009-0773-2. [DOI] [PubMed] [Google Scholar]
- 49.Munyaka PM, Khafipour E, Ghia JE. External influence of early childhood establishment of gut microbiota and subsequent health implications. Front Pediatr 2: 109, 2014. doi: 10.3389/fped.2014.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murali A, Bhargava A, Wright ES. IDTAXA: a novel approach for accurate taxonomic classification of microbiome sequences. Microbiome 6: 140, 2018. doi: 10.1186/s40168-018-0521-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nurgali K, Stebbing MJ, Furness JB. Correlation of electrophysiological and morphological characteristics of enteric neurons in the mouse colon. J Comp Neurol 468: 112–124, 2004. doi: 10.1002/cne.10948. [DOI] [PubMed] [Google Scholar]
- 53.Obata Y, Castaño Á, Boeing S, Bon-Frauches AC, Fung C, Fallesen T, de Agüero MG, Yilmaz B, Lopes R, Huseynova A, Horswell S, Maradana MR, Boesmans W, Vanden Berghe P, Murray AJ, Stockinger B, Macpherson AJ, Pachnis V. Neuronal programming by microbiota regulates intestinal physiology. Nature 578: 284–289, 2020. doi: 10.1038/s41586-020-1975-8. [DOI] [PubMed] [Google Scholar]
- 54.Oksanen J, Kindt R, Legendre P, O’Hara B, Simpson G, Solymos P, Stevens M, Wagner H. The Vegan Package, Version 1.15-1. https://www.researchgate.net/profile/Gavin_Simpson/publication/228339454_The_vegan_Package/links/0912f50be86bc29a7f000000/The-vegan-Package.pdf, 2008.
- 55.Parathan P, Wang Y, Leembruggen AJ, Bornstein JC, Foong JP. The enteric nervous system undergoes significant chemical and synaptic maturation during adolescence in mice. Dev Biol 458: 75–87, 2020. doi: 10.1016/j.ydbio.2019.10.011. [DOI] [PubMed] [Google Scholar]
- 56.Preidis GA, Ajami NJ, Wong MC, Bessard BC, Conner ME, Petrosino JF. Microbial-derived metabolites reflect an altered intestinal microbiota during catch-up growth in undernourished neonatal mice. J Nutr 146: 940–948, 2016. doi: 10.3945/jn.115.229179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26: 1641–1650, 2009. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Qu ZD, Thacker M, Castelucci P, Bagyánszki M, Epstein ML, Furness JB. Immunohistochemical analysis of neuron types in the mouse small intestine. Cell Tissue Res 334: 147–161, 2008. doi: 10.1007/s00441-008-0684-7. [DOI] [PubMed] [Google Scholar]
- 60.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41: D590–D596, 2013. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reigstad CS, Salmonson CE, Rainey JF III, Szurszewski JH, Linden DR, Sonnenburg JL, Farrugia G, Kashyap PC. Gut microbes promote colonic serotonin production through an effect of short-chain fatty acids on enterochromaffin cells. FASEB J 29: 1395–1403, 2015. doi: 10.1096/fj.14-259598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Roager HM, Hansen LB, Bahl MI, Frandsen HL, Carvalho V, Gøbel RJ, Dalgaard MD, Plichta DR, Sparholt MH, Vestergaard H, Hansen T, Sicheritz-Pontén T, Nielsen HB, Pedersen O, Lauritzen L, Kristensen M, Gupta R, Licht TR. Colonic transit time is related to bacterial metabolism and mucosal turnover in the gut. Nat Microbiol 1: 16093, 2016. doi: 10.1038/nmicrobiol.2016.93. [DOI] [PubMed] [Google Scholar]
- 63.Roberts RR, Murphy JF, Young HM, Bornstein JC. Development of colonic motility in the neonatal mouse-studies using spatiotemporal maps. Am J Physiol Gastrointest Liver Physiol 292: G930–G938, 2007. doi: 10.1152/ajpgi.00444.2006. [DOI] [PubMed] [Google Scholar]
- 65.Roosen L, Boesmans W, Dondeyne M, Depoortere I, Tack J, Vanden Berghe P. Specific hunger- and satiety-induced tuning of guinea pig enteric nerve activity. J Physiol 590: 4321–4333, 2012. doi: 10.1113/jphysiol.2012.231134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sang Q, Williamson S, Young HM. Projections of chemically identified myenteric neurons of the small and large intestine of the mouse. J Anat 190: 209–222, 1997. doi: 10.1046/j.1469-7580.1997.19020209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sang Q, Young HM. Chemical coding of neurons in the myenteric plexus and external muscle of the small and large intestine of the mouse. Cell Tissue Res 284: 39–53, 1996. doi: 10.1007/s004410050565. [DOI] [PubMed] [Google Scholar]
- 69.Sang Q, Young HM. The identification and chemical coding of cholinergic neurons in the small and large intestine of the mouse. Anat Rec 251: 185–199, 1998. doi:. [DOI] [PubMed] [Google Scholar]
- 70.Sang Q, Young HM. The identification and chemical coding of cholinergic neurons in the small and large intestine of the mouse. Anat Rec 251: 185–199, 1998. doi:. [DOI] [PubMed] [Google Scholar]
- 72.Sartor RB. Gut microbiota: optimal sampling of the intestinal microbiota for research. Nat Rev Gastroenterol Hepatol 12: 253–254, 2015. doi: 10.1038/nrgastro.2015.46. [DOI] [PubMed] [Google Scholar]
- 73.Soret R, Chevalier J, De Coppet P, Poupeau G, Derkinderen P, Segain JP, Neunlist M. Short-chain fatty acids regulate the enteric neurons and control gastrointestinal motility in rats. Gastroenterology 138: 1772–1782.E4, 2010. doi: 10.1053/j.gastro.2010.01.053. [DOI] [PubMed] [Google Scholar]
- 74.Spencer NJ, Hibberd TJ, Travis L, Wiklendt L, Costa M, Hu H, Brookes SJ, Wattchow DA, Dinning PG, Keating DJ, Sorensen J. Identification of a rhythmic firing pattern in the enteric nervous system that generates rhythmic electrical activity in smooth muscle. J Neurosci 38: 5507–5522, 2018. doi: 10.1523/JNEUROSCI.3489-17.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stephen AM, Wiggins HS, Cummings JH. Effect of changing transit time on colonic microbial metabolism in man. Gut 28: 601–609, 1987. doi: 10.1136/gut.28.5.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Suply E, de Vries P, Soret R, Cossais F, Neunlist M. Butyrate enemas enhance both cholinergic and nitrergic phenotype of myenteric neurons and neuromuscular transmission in newborn rat colon. Am J Physiol Gastrointest Liver Physiol 302: G1373–G1380, 2012. doi: 10.1152/ajpgi.00338.2011. [DOI] [PubMed] [Google Scholar]
- 77.Swaminathan M, Hill-Yardin E, Ellis M, Zygorodimos M, Johnston LA, Gwynne RM, Bornstein JC. Video imaging and spatiotemporal maps to analyze gastrointestinal motility in mice. J Vis Exp: 53828, 2016. doi: 10.3791/53828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Thomas EA, Sjövall H, Bornstein JC. Computational model of the migrating motor complex of the small intestine. Am J Physiol Gastrointest Liver Physiol 286: G564–G572, 2004. doi: 10.1152/ajpgi.00369.2003. [DOI] [PubMed] [Google Scholar]
- 79.Tian L, Hires SA, Mao T, Huber D, Chiappe ME, Chalasani SH, Petreanu L, Akerboom J, McKinney SA, Schreiter ER, Bargmann CI, Jayaraman V, Svoboda K, Looger LL. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat Methods 6: 875–881, 2009. doi: 10.1038/nmeth.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tremaroli V, Bäckhed F. Functional interactions between the gut microbiota and host metabolism. Nature 489: 242–249, 2012. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- 81.Vandeputte D, Falony G, Vieira-Silva S, Tito RY, Joossens M, Raes J. Stool consistency is strongly associated with gut microbiota richness and composition, enterotypes and bacterial growth rates. Gut 65: 57–62, 2016. doi: 10.1136/gutjnl-2015-309618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Verdú EF, Bercik P, Verma-Gandhu M, Huang XX, Blennerhassett P, Jackson W, Mao Y, Wang L, Rochat F, Collins SM. Specific probiotic therapy attenuates antibiotic induced visceral hypersensitivity in mice. Gut 55: 182–190, 2006. doi: 10.1136/gut.2005.066100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang B, Mao YK, Diorio C, Pasyk M, Wu RY, Bienenstock J, Kunze WA. Luminal administration ex vivo of a live Lactobacillus species moderates mouse jejunal motility within minutes. FASEB J 24: 4078–4088, 2010. doi: 10.1096/fj.09-153841. [DOI] [PubMed] [Google Scholar]
- 84.Wichmann A, Allahyar A, Greiner TU, Plovier H, Lundén GO, Larsson T, Drucker DJ, Delzenne NM, Cani PD, Bäckhed F. Microbial modulation of energy availability in the colon regulates intestinal transit. Cell Host Microbe 14: 582–590, 2013. doi: 10.1016/j.chom.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 85.Yamada Y, Mikoshiba K. Quantitative comparison of novel GCaMP-type genetically encoded Ca(2+) indicators in mammalian neurons. Front Cell Neurosci 6: 41, 2012. doi: 10.3389/fncel.2012.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yano JM, Yu K, Donaldson GP, Shastri GG, Ann P, Ma L, Nagler CR, Ismagilov RF, Mazmanian SK, Hsiao EY. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell 161: 264–276, 2015. [Erratum in: Cell 163: 258, 2015.] 10.1016/j.cell.2015.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zariwala HA, Borghuis BG, Hoogland TM, Madisen L, Tian L, De Zeeuw CI, Zeng H, Looger LL, Svoboda K, Chen TW. A Cre-dependent GCaMP3 reporter mouse for neuronal imaging in vivo. J Neurosci 32: 3131–3141, 2012. doi: 10.1523/JNEUROSCI.4469-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]