Abstract
The gut microbe-derived metabolite trimethylamine-N-oxide (TMAO) has recently been linked to cardiovascular disease (CVD) pathogenesis, prompting the development of therapeutic strategies to reduce TMAO. Previous work has shown that experimental alteration of circulating TMAO levels via dietary alterations or inhibition of the host TMAO producing enzyme flavin containing monooxygenase 3 (FMO3) is associated with reorganization of host cholesterol and bile acid metabolism in mice. In this work, we set out to understand whether recently developed nonlethal gut microbe-targeting small molecule choline trimethylamine (TMA) lyase inhibitors also alter host cholesterol and bile acid metabolism. Treatment of mice with the mechanism-based choline TMA lyase inhibitor, iodomethylcholine (IMC), increased fecal neutral sterol loss in the form of coprostanol, a bacteria metabolite of cholesterol. In parallel, IMC treatment resulted in marked reductions in the intestinal sterol transporter Niemann-pick C1-like 1 (NPC1L1) and reorganization of the gut microbial community, primarily reversing choline supplemented diet-induced changes. IMC also prevented diet-driven hepatic cholesterol accumulation, causing both upregulation of the host hepatic bile acid synthetic enzyme CYP7A1 and altering the expression of hepatic genes critical for bile acid feedback regulation. These studies suggest that the gut microbiota-driven TMAO pathway is closely linked to both microbe and host sterol and bile acid metabolism. Collectively, as gut microbe-targeting choline TMA lyase inhibitors move through the drug discovery pipeline from preclinical models to human studies, it will be important to understand how these drugs impact both microbe and host cholesterol and bile acid metabolism.
NEW & NOTEWORTHY The gut microbe-dependent metabolite trimethylamine-N-oxide (TMAO) has been strongly associated with cardiovascular mortality, prompting drug discovery efforts to identify points of therapeutic intervention within the microbe host TMAO pathway. Recently, mechanism-based small molecule inhibitors of the major bacterial trimethylamine (TMA) lyase enzymes have been developed, and these drugs show efficacy as anti-atherothrombotic agents. The novel findings of this study are that small molecule TMA lyase inhibition results in beneficial reorganization of host cholesterol and bile acid metabolism. This study confirms previous observations that the gut microbial TMAO pathway is intimately linked to host cholesterol and bile acid metabolism and provides further rationale for the development of small molecule choline TMA lyase inhibitors for the treatment of cardiometabolic disorders.
Keywords: bile acid, cardiovascular disease, cholesterol, metabolism, microbiome, TMA
INTRODUCTION
There is now unequivocal evidence that microbes resident in the human intestine can directly contribute to the pathogenesis of obesity (2, 7, 22, 30, 35, 39, 50–52), diabetes (25, 61), cardiovascular disease (CVD) (8), chronic kidney disease (26), and cancer (20, 38). Recent technological advances have rapidly facilitated the identification of disease-relevant microbial participants, microbe-derived metabolites, and host dedicated receptor systems that sense microbial products. Gut microbes represent a filter of our greatest environmental exposure: the foods we consume (7). In fact, it is now clear that we each experience a given meal differently, based on our unique gut microbial communities. Biologically active gut microbe-derived metabolites, such as short-chain fatty acids (3, 11), secondary bile acids (29a, 41, 55), and trimethylamine-N-oxide (TMAO) (45, 57), are now recognized as contributors to diseases including obesity, diabetes, CVD, and cancer. However, mechanistic insights into how microbe-derived metabolites promote or ameliorate human disease are only beginning to emerge. Given that mechanistic links are being discovered between microbial metabolites and human disease, there is growing interest in developing gut microbe-targeted therapeutic strategies for the prevention or treatment of diseases in the human host. One of the most mature examples of microbe-targeted drug discovery is the TMAO pathway, where several nonlethal (i.e., as opposed to an antibiotic) small molecule drugs targeting bacterial enzyme systems have recently been developed (37, 47, 56). Initially, the natural product 3,3-Dimethyl-1-butanol (DMB) was identified as a competitive small molecule inhibitor of the gut microbial choline trimethylamine (TMA) lyase system CutC/D both in vitro and in vivo (56). When provided to hyperlipidemic apolipoprotein E (ApoE)-deficient mice, DMB significantly reduced circulating TMAO levels, macrophage foam cell formation, and aortic root atherosclerosis (37, 47, 56), providing one of the first examples of a microbe-targeted nonlethal drug providing benefit to the host. More recently, a family of second-generation mechanism-based CutC/D inhibitors, halomethylcholines, with IC50 in the picomolar to low nanomolar range, have been described as bacterially targeted drugs that very effectively (>10,000-fold more potent than DMB) lower circulating levels of TMA and TMAO in vivo (37). These halomethylcholines (iodomethylcholine, fluoromethylcholine, chloromethylcholine, and bromomethylcholine) choline TMA lyase inhibitors are some of the first-ever described gut microbe-targeted enzyme inhibitors that show antithrombotic effects in preclinical animal models (37). With these powerful pharmacologic tools in hand, there is now a realistic possibility that TMAO-lowering drugs can be advanced from preclinical models into human studies.
Production of TMAO occurs via a metaorganismal metabolic pathway that is initiated when TMA containing nutrients present in high-fat foods or red meat (i.e., phosphatidylcholine and choline) are metabolized by the primary gut microbial choline utilizing enzyme complex CutC/D (16, 17) to generate TMA, which is then further metabolized by the host liver enzyme flavin containing monooxygenase 3 (FMO3) (4) to produce TMAO. Although TMAO has been directly linked to platelet responsiveness and in vivo thrombosis potential via cell autonomous effects on circulating platelets (37, 64, 67, 68), there are several studies that have suggested that the metaorganismal TMAO pathway is also intimately linked to host cholesterol and bile acid metabolism in mice (29, 33, 44, 60), which has additional implications in atherothrombotic diseases. First, dietary supplementation of TMAO significantly reduces intestinal cholesterol absorption and alters the bile acid pool size and composition (29, 60). Using unbiased transcriptional screening approaches, we also discovered that the TMAO-producing enzyme FMO3 is a negative regulator of macrophage reverse cholesterol transport (60). It is important to note that FMO3 is directly transcriptionally regulated by the bile acid sensing nuclear receptor farnesoid X receptor (FXR) (4), providing one potential mechanism by which the TMAO pathway is linked to bile acid and sterol homeostasis. The purpose of this study was to understand whether second generation gut microbe-targeted choline TMA lyase inhibitors also impact host sterol and bile acid homeostasis. Our results show that microbial choline TMA lyase inhibition promotes beneficial reorganization of host bile acid and cholesterol metabolism.
MATERIALS AND METHODS
Mouse studies.
At 8 wk of age, wild-type C57BL/6J male mice were switched from standard rodent chow to one of four experimental synthetic diets containing low (0.02%, wt/wt) or high (0.2%, wt/wt) levels of dietary cholesterol with or without the microbe-targeted choline TMA lyase inhibitor iodomethylcholine (IMC; 0.06%, wt/wt). The base diet used for all four diets was a chemically defined minimal choline chow formulation originally described to provide sufficient levels of dietary choline with 23.5% kcal from protein, 59.3% kcal from carbohydrate, and 17.2% kcal from fat (total kcal/g = 3.1) (57). These experimental diets were synthesized by Envigo-Teklad Diets (Madison, WI), and additional information can be found for each of these diets by referencing the following diet numbers: TD.130104 (low cholesterol diet without IMC), TD.150813 (low cholesterol diet containing IMC), TD.160514 (high-cholesterol diet without IMC), and TD.160515 (high-cholesterol diet containing IMC). Mice were maintained on these diets over a period of 4 consecutive weeks and phenotyped as described below for sterol and bile acid homeostasis. In a confirmatory study, apolipoprotein E-deficient mice (ApoE−/−) were fed a diet containing 1% supplemental choline (Envigo/Teklad no. TD.09041) as previously described for 21 wk (57, 64) before 72-h fecal collection for fecal sterol loss measurements. All mice were maintained in accordance with the Association for the Assessment and Accreditation of Laboratory Animal Care (AAALAC) policies, and all experimental protocols were approved by the institutional animal care and use committee of the Cleveland Clinic.
Standardized necropsy conditions.
To keep results consistent, mice were fasted for 4 h (from 9:00 AM to 1:00 PM) before necropsy. At necropsy, all mice were terminally anesthetized with ketamine-xylazine (100–160 mg/kg ketamine-20–32 mg/kg xylazine), and a midline laparotomy was performed. Blood was collected by heart puncture. After blood collection, a whole body perfusion was conducted by puncturing the inferior vena cava and slowly delivering 10 mL of saline into the heart to remove blood from tissues. Tissues were collected and immediately snap frozen in liquid nitrogen for subsequent biochemical analysis or fixed for morphological analysis.
Quantitation of plasma TMA, TMAO, choline, carnitine, γ-butyrobetaine, and IMC.
Stable isotope dilution high-performance liquid chromatography with online tandem mass spectrometry (LC-MS/MS) was used for quantification of levels of TMAO, TMA, choline, carnitine, and γ-butyrobetaine in plasma, as previously described (28, 37, 58). Their d9(methyl)-isotopologues were used as internal standards. For IMC quantification, synthetic d2-IMC was used as an internal standard (37). LC-MS/MS analyses were performed on a Shimadzu 8050 triple quadrupole mass spectrometer. IMC and d2-IMC, along with other metabolites, were monitored using multiple reaction monitoring of precursor and characteristic product ions as follows: m/z 230.0→58.05 for IMC; m/z 232.0→60.1 for d2-IMC; m/z 76.0→58.1 for TMAO; m/z 85.0→66.25 for d9-TMAO; m/z 60.2→44.2 for TMA; m/z 69.0→49.1 for d9-TMA; m/z 104.0→60.15 for choline; m/z 113.1→69.2 for d9-choline; m/z 118.0→58.1 for betaine; m/z 127.0→66.2 for d9-betaine.
Hepatic lipid analyses.
Extraction of liver lipids and quantification of plasma and hepatic triglycerides, total cholesterol, free cholesterol, and cholesterol esters were conducted using enzymatic assays as described previously (28, 36). Briefly, ∼100 mg of liver was thawed, minced, and weighed in a glass tube. Lipids were extracted in 2:1 CHCl3/methanol at room temperature overnight. The protein was quantitatively separated from the lipid extract, which was then dried down under N2 and redissolved in a measured volume of 2:1 chloroform/methanol. Upper phase was aspirated and dried down; 1% Triton X-100 in CHCl3 was then added, and the solvent was evaporated. Deionized water was then added to each tube and vortexed until the solution was clear. Thereafter, hepatic triacylglycerol levels (Wako Diagnostics), total cholesterol levels (Infinity Cholesterol Reagent, Thermo/Fisher), and free cholesterol (Wako Diagnostics) were quantified enzymatically.
Quantification of fecal neutral sterol excretion.
Quantitative fecal excretion of both plant- and animal-derived sterols were analyzed by gas chromatography coupled with flame ionization detection using methods we have previously described (60). Briefly, after being fed experimental diets for 4 wk (C57BL/6J mice) or 21 wk (ApoE−/− mice), mice were individually housed in a cage with a wire bottom and allowed free access to diet and water for 3 consecutive days (72 h). After a 3-day fecal collection, the mice were weighed, and the feces were collected, dried in a 70°C vacuum oven, weighed, and crushed into a fine powder. A measured mass (50–100 mg) of feces was placed into a glass tube containing 100 μg of 5α-cholestane as an internal standard. The feces were saponified and the neutral lipids were extracted into hexane, and mass analysis of the extracted neutral sterols was conducted by gas chromatography flame ionization detection as described previously (60). Total fecal neutral sterol mass represents the sum of cholesterol and the bacterial metabolite of cholesterol-coprostanol in each sample.
Quantification of plasma bile acid levels.
Total plasma bile acid levels were measured enzymatically using a commercially available enzymatic assay (Mouse Total Bile Acid Assay Kit, Crystal Chem, Inc.). Quantification of individual plasma bile acid species was conducted using a quantitative stable isotope dilution LC-MS/MS analytical method as recently described (15). Briefly, stable isotope-labeled internal standards included were: D4-glycolithocholic acid, D4-glycoursodeoxycholic acid, D4-glycodeoxycholic acid, D4-glycocholic acid, D4-taurolithocholic acid, D4-tauroursodeoxycholic acid, D4-taurochenodeoxycholic acid, D4-taurodeoxycholic acid, and D4-taurocholic acid, D4-lithocholic acid, D4-chenodeoxycholic acid, D4-deoxycholic acid, and D4-cholic acid, and D4-glycochenodeoxycholic acid. Mouse plasma samples were mixed with ice-cold methanolic IS working solution of internal standard, vortexed for 10 min, and centrifuged at 14,000 g for 20 min at 4°C. The supernatant was transferred to glass HPLC vials for LC-MS/MS analysis using a 4000 Q-Trap triple quadrupole tandem mass spectrometer (AB SCIEX, Framingham, MA) equipped with an electrospray ionization source operating in negative ion mode. Mass spectrometry parameters were as follows: ions spray voltage, 4,200 V; ion source heater temperature, 500°C; source gas 1, 35 psi; source gas 2, 45 psi; and curtain gas, 35 psi. Nitrogen gas was used for the nebulizer and curtain and collision gas. Analyses were performed using electrospray ionization in negative-ion mode with multiple reaction monitoring (MRM) of precursor and characteristic product ions specific for each monitored BA. The HPLC system consisted of four binary pumps (LC-20 AD), autosampler operating at 10°C (Nexera X2 SIL-30AC), controller (CBM-20A) (Shimadzu Scientific Instruments, Inc., MD), and a dual column switching valve system Rheodyne (IDEX Health & Science, MA). Chromatographic separations were performed on a reverse-phase column (Kinetix C18, 2.6 µm, 150 mm × 4.6 mm ID; Cat. No. 00F-4462-E0; Phenomenex, Torrance, CA). Mobile phase A was 1 mM ammonium acetate and 0.1% acetic acid in methanol:acetonitrile:water (1:1:3; vol/vol/vol), and mobile phase B was 0.1% acetic acid in methanol:acetonitrile:2-propanol (4.5:4.5:1; vol/vol/vol). Samples were injected onto columns equilibrated in 100% A and separated using a gradient as follows: 0–2 min 0% B; 2–20 min 0–100% B; 20–28.5 min 100% B. Flow rate was programmed as follows: 0.3 mL/min from 0–20 min and 0.5 mL/min from 20–28 min. Samples were introduced to the mass spectrometer for analysis from 9–28 min. To eliminate carry over, an extensive washing step alternating between mobile phase A and B was added at the end of each run as follows: 100% A for 28–35 min, then directly switched to 100% B from 36–46 min, and equilibration step of 100% A from 46–60 min. Calibration curves were built by fitting each analyte concentration (10 different points) to peak area ratios (analyte/internal standard). The limit of detection was defined as the lowest concentration of analyte in sample matrix (e.g., serum) that generated a signal-to-noise ratio of ≥ 3. The limit of quantification was defined as the lowest concentration of analyte in sample matrix that generated a signal-to-noise ratio of ≥ 10.
Real-time PCR analysis of gene expression.
Roughly 20 mg of snap-frozen liver tissue was homogenized in the 1-mL TRIzol reagent (Thermo Fisher Scientific, Cat. No. 15596018). Furthermore, 200 µL of chloroform were added and samples were spun down at 13,000 revolutions/min for 5 min. Upper clear layer were passed through the RNeasy Mini Spin Columns (Cat. No. 74104). DNase treatment (10 U/reaction) was performed according to the Qiagen RNeasy kit (Cat. No. 74104). Concentrations of pure RNA were measured using Nanodrop (Thermo Fisher Scientific, ND-2000). Reverse transcription to generate cDNA was performed using qscript mastermix (QuantaBio Cat. No. 101414-106) as recommended by the manufacturer using 750 ng of RNA template. Resulting cDNA was diluted 10× and used in the real-time PCR reaction using an Applied Biosystems Step One Plus thermocycler. Relative mRNA levels were calculated based on the ΔΔCT method using the Applied Biosystems Step One Plus PCR System as we have previously described (42, 60), using Gapdh as a normalizing gene. Primers used for qPCR are shown in Table 1.
Table 1.
Primers used for real-time qPCR and antibodies used for Western blot analysis
| No. | Gene | Primer |
|---|---|---|
| 1 | Srebp2 | F-tggatgacgcaaaggtcaa R-caggaaggtgaggacacataag |
| 2 | Hmgcs | F- gcatatatagcaatgtctcctgcaa R- gccgtgaactgggtcgaa |
| 3 | Hmgcr | F- cttgtggaatgccttgtgattg R-agcccgaagcagcacatgat |
| 4 | Bsep | F- aaattcaagcaggctcaagacaa R-taccaactggggtgctctttt |
| 5 | Cyp8b1 | F-tgaacgacttcttggcttcc R-cctgtttctgggtcctcttattc |
| 6 | Cyp7a1 | F-tcacaaactccctgtcatacc R-catcacttgggtctatgcttct |
| 7 | Cyp27a1 | F-gcctcacctatgggatcttca R-tcaaagcctgacgcagatg |
| 8 | Abcg8 | F-ctgtacactgctggtccttatt R-caggtttgtcagccagtagat |
| 9 | Abcg5 | F-cttacccacggttcctttca R-actgcctgcttattcctcatta |
| 10 | Abca1 | F-gggtggtgttcttcctcattac R-cacatcctcatcctcgtcattc |
| 11 | Asbt | F-attggatagatggcgacatgg R-gtgtagacgaagaggcaaagag |
| 12 | Shp | F-ctgaagggcacgatcctct R-gcctcctgttgcaggtgt |
| 13 | Npc1l1 | F-tggactggaaggaccatttcc R-gtgccccgtagtcagctat |
| 14 | Ostα | F-gctcacctccctactcttcta R-tacccaaccttgtcgtctttc |
| 15 | Ostβ | F-caggaactgctggaagaaatg R-ccaggaccaggatggaataa |
| 16 | Fxr | F-gggcagaatctggatttgga R-gtagaagcccaggttggaatag |
| 17 | Fgf15 | F-agatggtgcttcatggatctg R-catataccgggctgattcgctac |
| 18 | Ibabp | F-tgagagtgagaagaattacgatgag R-ctgacctctgtgatgatcttg |
| 19 | Gapdh | F-cctcgtcccgtagacaaaatg R- tgaaggggtcgttgatggc |
Abca1, Abcg5, and Abcg8, ATP-binding cassette transporters; Asbt, apical sodium bile acid transporter; Bsep, bile salt export pump; Cyp7a1, Cyp8b1, and Cyp27a1, bile acid modifying cytochrome-P450 family members; Fgf15, fibroblast growth factor 15; Fxr, farnesoid X receptor; Hmgcr, HMG-CoA reductase; Hmgcs, HMG-CoA synthase; Ibabp, ileal bile acid-binding protein; Npc1l1, Niemann-pick C1-like 1; Ostα and Ostβ; organic solute transporter α and β; Shp, small heterodimeric partner; Srebp2, sterol regulatory-binding protein 2. F, forward; R, reverse.
Immunoblotting.
Whole tissue homogenates or membrane preparations were generated as previously described (28), and protein was quantified using the bicinchoninic acid (BCA) assay (Pierce). Proteins were separated by 4–12% SDS-PAGE, transferred to polyvinylidene difluoride (PVDF) membranes, and detected after incubation with specific antibodies as previously described (60). Quantification of blots was performed using ImageJ software (National Institute of Health). Information of antibodies is available in Table 2.
Table 2.
Primary and secondary antibodies
| No. | Antibody | Vendor | Catalog No. |
|---|---|---|---|
| Primary antibody information | |||
| 1 | ABCA1 | Novus-Biological | NB400-105 |
| 2 | NPC1L1 | Gift from Dr. Joyce Repa (Univ. of Texas–Southwestern Medical Center | Not applicable |
| 3 | HMGCOA-reductase | Gift from Dr. Russell DeBose-Boyd (Univ. of Texas–Southwestern Medical Center) | Not applicable |
| 4 | CYP7A1 | Abcam | Ab65596 |
| 5 | Calnexin | Cell Signaling Technologies | (C5C9)#2679 |
| 6 | GAPDH | Cell Signaling Technologies | (D16H11)#8884 |
| Secondary antibody information | |||
| 1 | Anti-rabbit HRP-conjugated | Cell Signaling Technologies | 7074 |
| 2 | Anti-mouse HRP-conjugated | Cell Signaling Technologies | 7076 |
ABCA1, ATP-binding cassette transporter; CYP7A1, bile acid modifying cytochrome-P450 family member; NPC1L1, Niemann-pick C1-like 1.
Cecal microbiome studies and bioinformatic analyses.
Cecal microbial community composition was assessed by sequencing 16S rRNA gene amplicons essentially as we have previously described (21, 28, 57–64). Briefly, 16S rRNA amplicon sequencing was done using Earth Microbiome Project (EMP) standard protocols (https://www.earthmicrobiome.org/protocols-and-standards/16s) (43). Raw 16S amplicon sequence and metadata, were demultiplexed using split_libraries_fastq.py script implemented in QIIME1.9.1 (12). Demultiplexed fastq file was split into sample specific fastq files using split_sequence_file_on_sample_ids.py script from Qiime1.9.1 (12). Individual fastq files without nonbiological nucleotides were processed using Divisive Amplicon Denoising Algorithm (DADA) pipeline (10). We obtained a total of 3,836,268 paired-end reads with an average read length of 151 base pairs. The median sequencing depth per sample was 163,962 reads. After filtering, denoising, and removing chimeras, we retained 3,750,755 (97% of initial) reads. The output of the dada2 pipeline [feature table of amplicon sequence variants (an ASV table)] was processed for α- and β-diversity analysis using phyloseq (46) and microbiomeSeq (https://www.github.com/umerijaz/microbiomeSeq) packages in R. α-Diversity estimates were measured within group categories using estimate_richness function of the phyloseq package (46). Multidimensional scaling (MDS, also known as principal coordinate analysis; PCoA) was performed using Bray-Curtis dissimilarity matrix (32) between groups and visualized by using ggplot2 package (62). We assessed the statistical significance (P < 0.05) throughout, and whenever necessary, we adjusted P values for multiple comparisons according to the Benjamini and Hochberg method to control false discovery rate (FDR) (5) while performing multiple testing on taxa abundance according to sample categories. We performed an analysis of variance (ANOVA) among sample categories while measuring the α-diversity measures using plot_anova_diversity function in microbiomeSeq package (https://www.github.com/umerijaz/microbiomeSeq). Permutational multivariate analysis of variance (PERMANOVA) with 999 permutations was performed on all principal coordinates obtained during PCoA with the ordination function of the microbiomeSeq package. Linear regression (parametric) and Wilcoxon (nonparametric) tests were performed on ASVs abundances against coprostanol levels using their base functions in R (50).
Statistical analysis.
All data were analyzed using either one-way or two-way ANOVA where appropriate, followed by either a Tukey’s or Student’s t test for post hoc analysis. Differences were considered significant at P < 0.05. All mouse data analyses were performed using Graphpad Prism 6 (La Jolla, CA) software.
RESULTS
Iodomethylcholine effectively reduces circulating TMA and TMAO levels in mice.
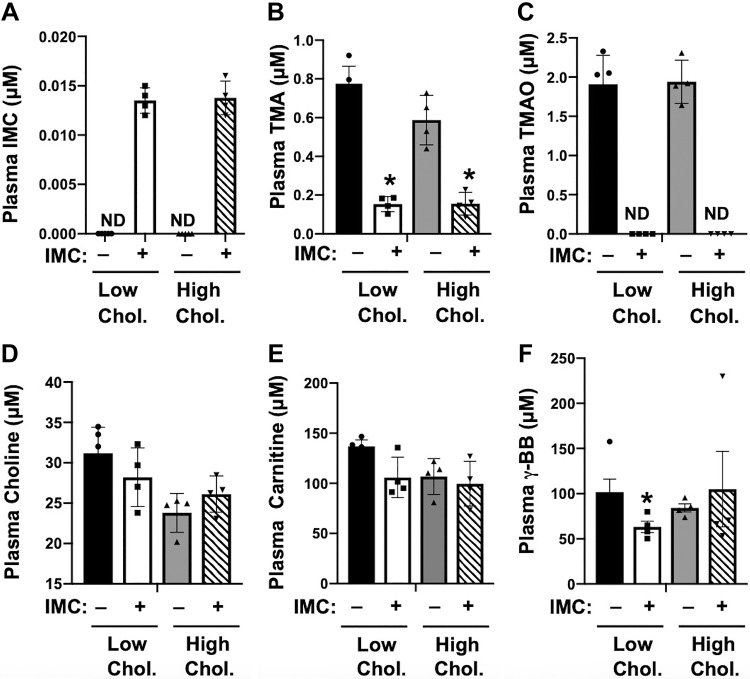
To examine whether small molecule choline TMA lyase inhibitors can alter host cholesterol and bile acid metabolism under normal physiologic conditions, we fed young (8 wk old) wild-type C57BL/6J mice synthetic diets containing either low (0.02%) or high (0.2%) dietary cholesterol in the absence or presence of a highly efficacious second-generation choline TMA lyase inhibitor IMC (37). IMC-treated mice had low nanomolar levels (~13 to 14 nM) of the drug in plasma, and this was similar in both low- and high-cholesterol diet-fed mice (Fig. 1A). It is important to note that compared with this small amount of drug in the circulation in the current study, our previous studies have shown that the vast majority of IMC remains in the cecal and colonic lumen (~5 mM of IMC in lumen), and given the mechanism-based inhibition of CutC/D the vast majority of the drug is disposed of in the feces (37). As expected, IMC treatment resulted in highly significant reductions in circulating TMA and TMAO levels (Fig. 1, B and C), without significantly altering plasma levels of choline or carnitine (Fig. 1, D and E). IMC treatment resulted in a modest reduction in the TMAO-generating precursor γ-butyrobetaine (28), but this only occurred in mice fed a low-cholesterol diet (Fig. 1F). Collectively, these data show that the small molecule CutC/D inhibitor IMC effectively lowers circulating TMA and TMAO levels in mice fed either low or high dietary cholesterol.
Fig. 1.
Small molecule trimethylamine (TMA) lyase inhibitor iodomethylcholine (IMC) suppresses circulating TMA and trimethylamine-N-oxide (TMAO) level. At 8 wk of age, wild-type C57BL/6J male mice were switched from standard rodent chow to 1 of 4 experimental synthetic diets containing low (0.02%, wt/wt) or high (0.2%, wt/wt) levels of dietary cholesterol with or without the microbe-targeted TMA lyase inhibitor IMC (0.06%, wt/wt). Plasma IMC (A), TMA (B), TMAO (C), choline (D), carnitine (E), and γ-butyrobetaine (BB) (F) levels were quantified by liquid chromatography tandem mass spectrometry. Data shown represent the means ± SE for n = 4 to 5 mice per group; *significantly different than the nondrug-treated mice within each diet group. ND, not detected.
Microbial choline TMA lyase inhibition stimulates fecal neutral sterol loss.
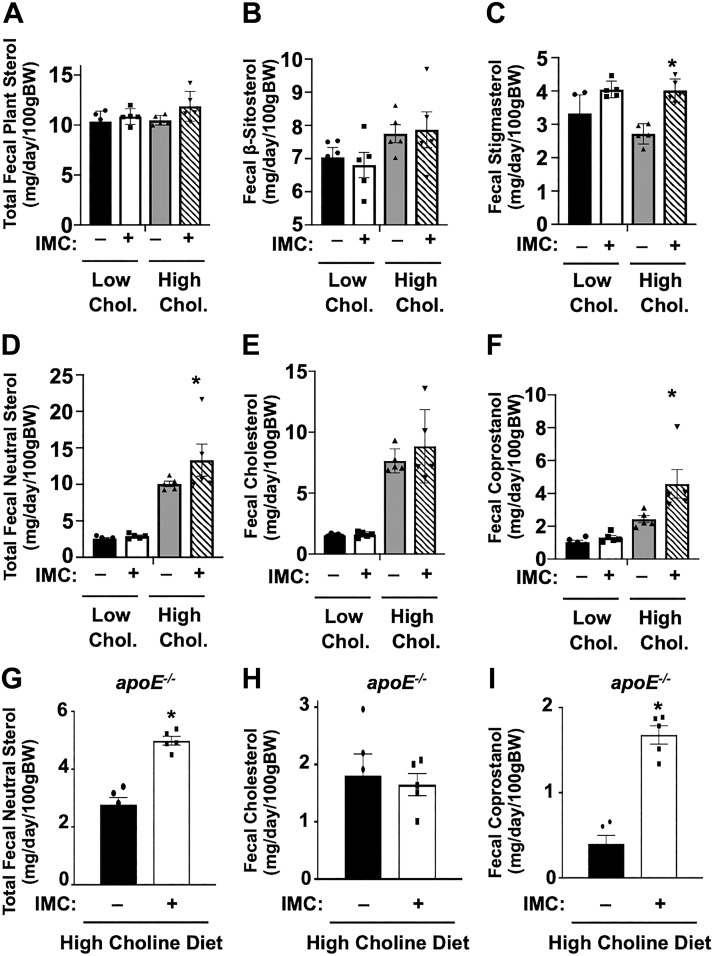
We previously demonstrated that the host TMAO-producing enzyme FMO3 is a negative regulator of macrophage reverse cholesterol transport and mass fecal neutral sterol loss in mice (25). Therefore, we set out to understand whether small molecule TMA lyase inhibitors could likewise influence the fecal disposal of cholesterol or its derivatives. In low-cholesterol diet-fed mice, IMC treatment did not significantly alter the fecal disposal of either plant-derived sterols (β-sitosterol or stigmasterol) or animal-derived sterols (cholesterol and coprostanol) (Fig. 2, A–C). However, when mice were challenged with high (0.2%) dietary cholesterol, IMC-treated mice had a significant increase in total fecal neutral sterol loss (Fig. 2D). Interestingly, the IMC-stimulated increase in total neutral sterol loss in high-cholesterol diet-fed mice could be explained not by increases in fecal cholesterol (Fig. 2E), but instead in a nearly twofold increase in fecal coprostanol loss (Fig. 2F). In contrast, IMC treatment did not significantly alter total fecal plant sterol loss but did cause a modest increase in the fecal disposal of the plant sterol stigmasterol only in high-cholesterol diet-fed mice (Fig. 2C). To confirm our results in a mouse model of atherosclerosis progression, we examined fecal sterol loss in apolipoprotein E-deficient (ApoE−/−) mice fed a high-choline diet (Fig. 2, G–I). Similar to results in wild-type C57Bl/6J mice (Fig. 2, D–F), IMC treatment also significantly increased total fecal neutral sterol, which was due exclusively to an increase in disposal of coprostanol (Fig. 2, G–I). Collectively, these results show that microbial choline TMA lyase inhibitors can selectively promote host fecal disposal of sterol through enhanced excretion of coprostanol, a bacterially derived metabolite of cholesterol (6, 46).
Fig. 2.
Trimethylamine (TMA) lyase inhibition enhances total fecal neutral sterol loss. A–F: at 8 wk of age, wild-type C57BL/6J male mice were switched from standard rodent chow to 1 of 4 experimental synthetic diets containing low (0.02%, wt/wt) or high (0.2%, wt/wt) levels of dietary cholesterol with or without the microbe-targeted TMA lyase inhibitor iodomethylcholine (IMC; 0.06%, wt/wt). After 4 wk on diets, mice were housed on wire bottom cages for a 3-day quantitative fecal collection. G–I: at 4 wk of age, apolipoprotein E-deficient (ApoE−/−) female mice were switched from standard rodent chow to a diet containing 1% supplemental choline with or without the microbe-targeted TMA lyase inhibitor IMC (0.06%, wt/wt) for 21 consecutive weeks. After 21 wk on diets, mice were housed on wire bottom cages for a 3-day quantitative fecal collection. Fecal excretion of total (G), cholesterol (H), and coprostanol (I) animal/microbial sterols (cholesterol and coprostanol) was quantified using gas-liquid chromatography. All data shown represent the means ± SE for n = 5 mice per group; *significantly different than the nondrug-treated mice within each diet group.
Microbial choline TMA lyase inhibition protects against diet-driven hepatic cholesterol accumulation and alters sterol-sensitive transcriptional programs in the liver.
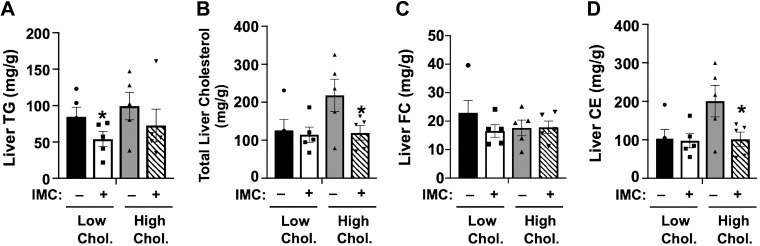
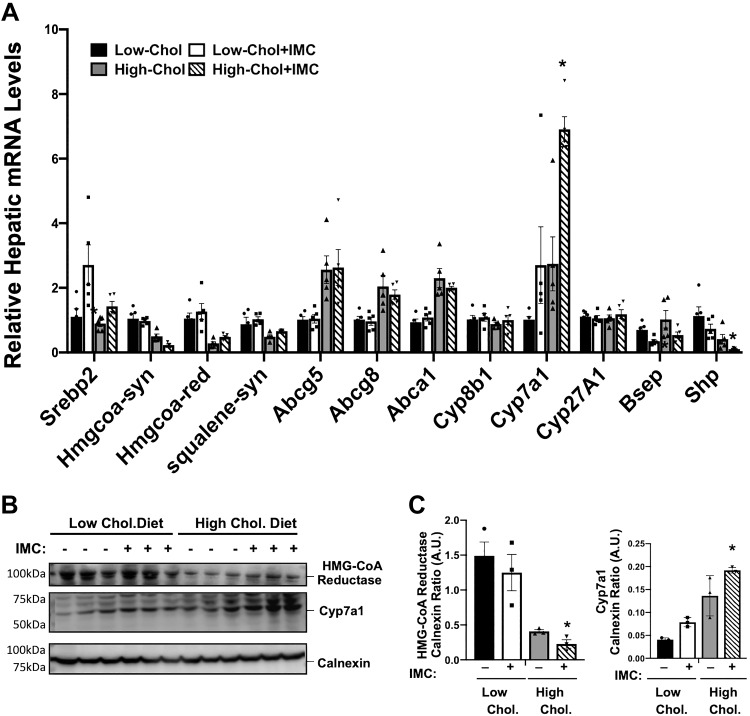
It is well appreciated that excess dietary cholesterol can promote the synthesis of cholesteryl esters (CE) in the liver (34, 65, 66), and excess hepatic CE synthesis under high-dietary cholesterol conditions is a major determinant of atherosclerotic plaque development (24, 25, 54). Therefore, we examined alterations in hepatic cholesterol storage and sterol-sensitive gene expression in the liver of IMC-treated mice as a proxy for atherogenic potential in these wild-type mice. IMC treatment resulted in modest reductions in hepatic triacylglycerol (TG) storage in low-cholesterol diet-fed mice (Fig. 3A). More relevant to atherosclerosis potential than liver TG levels, IMC completely blocked high-cholesterol diet-induced hepatic cholesterol storage (Fig. 3, B and D), which was due exclusively to reductions in cholesteryl esters (Fig. 3D) and not free cholesterol (Fig. 3C). Cellular cholesterol levels are carefully sensed and regulated in the liver by at least two major transcriptional mechanisms involving sterol regulatory element-binding proteins (SREBPs) and liver X receptors (LXRs) (9, 53). Interestingly, IMC treatment caused significant increases in the mRNA levels of sterol regulatory binding protein 2 (Srebp2) itself (~1.5–3.0-fold according to diet) but did not dramatically alter high-cholesterol diet-driven suppression of SREBP2-target genes HMG-CoA synthase, HMG-CoA reductase, or squalene synthase (Fig. 4A). It is important to note that in the high-cholesterol diet setting, IMC did result in significantly lower HMG-CoA synthase mRNA levels and slightly higher HMG-CoA reductase mRNA compared with controls (Fig. 4A). However, protein levels of HMG-CoA reductase were slightly lower in high-cholesterol diet-fed IMC-treated mice compared with controls (Fig. 4C). Altogether, the effects of IMC on SREBP2 target mRNA and protein expression were very modest. Hepatic expression of the canonical LXR target genes encoding ATP-binding cassette transporters Abca1, Abcg5, and Abcg8 were not significantly altered in IMC-treated mice. However, these genes showed the expected high-cholesterol diet-induced induction (Fig. 4A). Unexpectedly, IMC treatment resulted in significantly increased expression (~2 to 3-fold induction) of the bile acid synthetic gene Cyp7a1 under both dietary cholesterol conditions, yet other bile acid synthetic genes including Cyp8b1 and Cyp27a1 were not altered by IMC treatment (Fig. 4A). The IMC-induced increases in Cyp7a1 mRNA levels were also seen at the protein level (Fig. 4, B and C). In addition to alterations in Cyp7a1 expression, IMC treatment resulted in reduced mRNA levels of the bile salt export pump (Bsep), which regulates the enterohepatic circulation of bile salts and small heterodimeric partner (Shp) (a well-described transcriptional corepressor of bile acid synthesis) (Fig. 4). Collectively, these results demonstrate that microbial choline TMA lyase inhibition prevents dietary cholesterol-driven increases in hepatic cholesteryl ester storage and also results in altered expression of genes involved in bile acid metabolism.
Fig. 3.
A–D: trimethylamine (TMA) lyase inhibition blunts high-cholesterol diet-driven hepatic cholesteryl ester storage. At 8 wk of age, wild-type C57BL/6J male mice were switched from standard rodent chow to 1 of 4 experimental synthetic diets containing low (0.02%, wt/wt) or high (0.2%, wt/wt) levels of dietary cholesterol with or without the microbe-targeted TMA lyase inhibitor iodomethylcholine (IMC; 0.06%, wt/wt). After 4 wk on these diets, liver lipids were extracted and the levels of hepatic triacylglycerol (TG; A), total cholesterol (B), free cholesterol (FC; C), and cholesteryl esters (CE; D) were quantified using enzymatic assays. Data shown represent the means ± SE for n = 5 mice per group; *significantly different than the nondrug-treated mice within each diet group.
Fig. 4.
Trimethylamine (TMA) lyase inhibition alters the hepatic expression of key genes involved in sterol and bile acid metabolism. At 8 wk of age, wild-type C57BL/6J male mice were switched from standard rodent chow to 1 of 4 experimental synthetic diets containing low (0.02%, wt/wt) or high (0.2%, wt/wt) levels of dietary cholesterol with or without the microbe-targeted TMA lyase inhibitor iodomethylcholine (IMC; 0.06%, wt/wt). After 4 wk on these diets, liver RNA was extracted and gene expression was measured using quantitative real-time PCR. A: relative mRNA expression of sterol regulatory-binding protein 2 (Srebp2), HMG-CoA reductase (HMG CoA-red), HMG-CoA synthase (HMG CoA-syn), squalene synthase (Squalene-syn), ATP-binding cassette transporters (Abca1, Abcg5, and Abcg8), bile acid modifying cytochrome-P450 family members (Cyp7a1, Cyp8b1, and Cyp27a1), bile salt export pump (Bsep), and small heterodimeric partner (Shp) were quantified using the ΔΔCT method. B and C: Western blot analysis HMG-CoA reductase and Cyp7a1 with densitometric quantification. Data shown in A represent the means ± SE for n = 5 mice per group, whereas data in B and C represent the means ± SE for n = 3 mice per group; *significantly different than the nondrug-treated mice within each diet group. AU, arbitrary units.
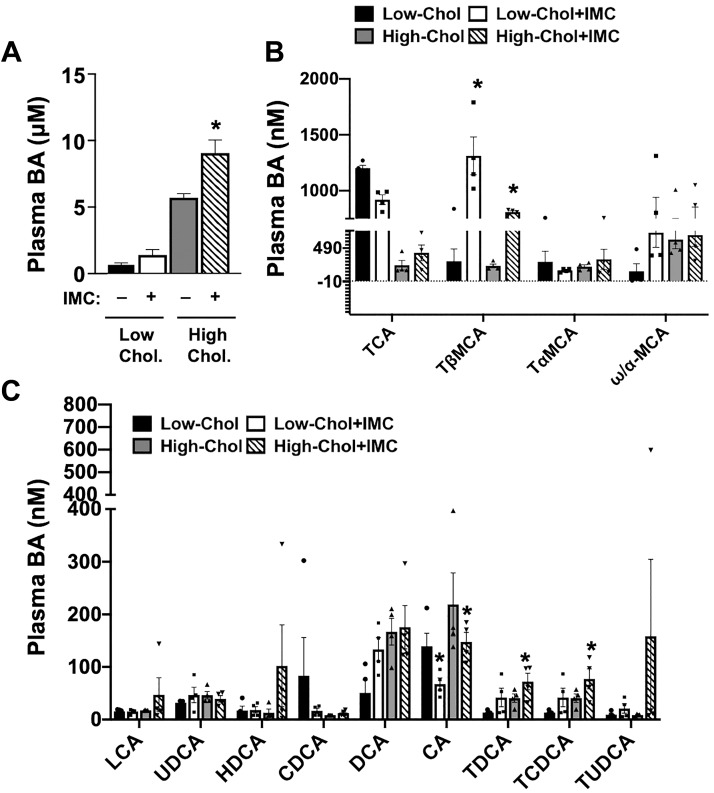
Microbial choline TMA lyase inhibition alters plasma bile acid concentration and composition.
Given the fact that IMC caused striking alterations in the hepatic expression of key genes involved in bile acid synthesis (Cyp7a1), transport (Bsep, also known as Abcb11), and sensing (Shp) (Fig. 4), we next investigated whether IMC treatment impacted the pool and/or composition of circulating bile acids (Fig. 5). IMC treatment in high-cholesterol diet-fed mice significantly increased the total plasma bile acid pool, and a similar trend was seen in low-cholesterol diet-fed mice (Fig. 5A). When we examined the molecular species of circulating bile acid via stable isotope dilution tandem mass spectrometry, a number of compositional changes were apparent (Fig. 5, B and C). IMC treatment resulted in a significant reduction in cholic acid (CA) on both diets (Fig. 5C) and also reduced taurocholic acid (TCA) only in low-cholesterol diet-fed mice (Fig. 5B). Reciprocally, IMC treatment resulted in large increases in tauro-β-muricholic acid (TβMCA) under both low- and high-dietary cholesterol setting (Fig. 5B). These IMC-induced alterations in bile acid composition are anticipated to impact host bile acid receptor signaling and bile acid feedback regulation and could also potentially impact the critical micelle formation function of bile acids, which is discussed below.
Fig. 5.
Trimethylamine (TMA) lyase inhibition alters plasma bile acid levels. At 8 wk of age, wild-type C57BL/6J male mice were switched from standard rodent chow to 1 of 4 experimental synthetic diets containing low (0.02%, wt/wt) or high (0.2%, wt/wt) levels of dietary cholesterol with or without the microbe-targeted TMA lyase inhibitor iodomethylcholine (IMC; 0.06%, wt/wt). After 4 wk on these diets, total plasma bile acids (BAs) were quantified enzymatically (A) or individual species were quantified using stable isotope dilution liquid chromatography tandem mass spectrometry (B and C). Major species of plasma BAs are shown in B, whereas lesser abundant species are shown in C. Data shown represent the means ± SE for n = 4 to 5 mice per group; *significantly different than the nondrug-treated mice within each diet group. CA, cholic acid; CDCA, chenodeoxycholic acid; DCA, deoxycholic acid; HDCA, hyodeoxycholic acid; LCA, lithocholic acid; TαMCA, tauro-α-muricholic acid; TβMCA, tauro-β-muricholic acid; TCA, taurocholic acid; TCDCA, taurochenodeoxycholic acid; TDCA, taurodeoxycholic acid; TUDCA, tauroursodeoxycholic acid; UDCA, ursodeoxycholic acid.
Microbial choline TMA lyase inhibition reorganizes gut microbial communities and host intestinal gene expression in a diet-specific manner.
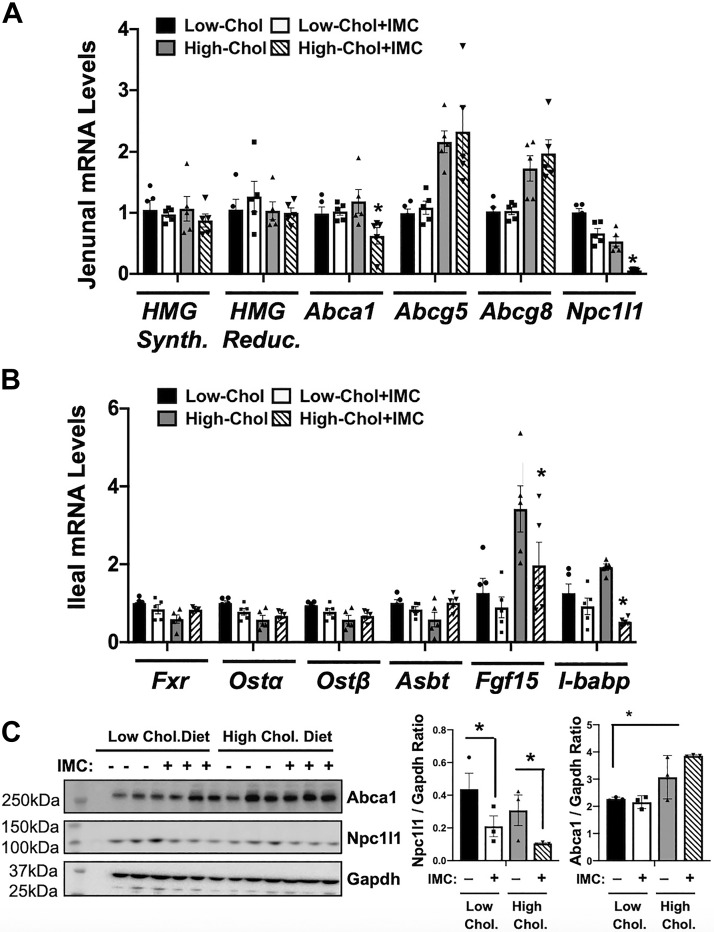
Our previous work demonstrated that small molecule choline TMA lyase inhibitors can induce shifts in the cecal microbiome, particularly in mice fed supplemental choline (37). Furthermore, data shown here demonstrate that TMA lyase inhibition can increase fecal sterol loss and alter circulating bile acid levels, which can serve as a proxy for ileal reabsorption. All of these facts point to the possibility that TMA lyase inhibition alters microbe-host crosstalk in the intestinal microenvironment that then subsequently impacts systemic cholesterol and bile acid balance. Hence, we examined both the cecal microbiome and host intestinal gene expression patterns in IMC-treated mice fed low- or high-cholesterol diets. In the jejunum, where intestinal cholesterol absorption is high but bile acid absorption is low, we found expression of SREBP2 and LXR target genes was unaltered by IMC treatment (Fig. 6, A and C). However, jejunal mRNA expression of the enterocyte sterol transporter Niemann Pick C1-Like 1 (Npc1l1) was markedly suppressed in IMC-treated group under both dietary cholesterol conditions (Fig. 6A). This IMC-induced suppression of Npc1l1 in the jejunum was also seen at the protein level (Fig. 6C). In the ileum, where bile acid reabsorption and FXR-dependent bile acid sensing is most efficient, we found that IMC promoted diet- and gene-specific transcriptional regulation (Fig. 6B). Although Fxr itself, and several other well-characterized FXR-target genes (Asbt, Ostα, and Ostβ), were not altered, two other well-known FXR-target genes [fibroblast growth factor 15 (Fgf15) and the ileal bile acid-binding protein (I-babp)] were significantly reduced in high-cholesterol diet-fed IMC-treated mice (Fig. 6B). These results show that IMC can alter both sterol (Npc1l1) and bile acid (Fgf15 and I-babp) regulatory genes in a highly regional and diet-specific manner in the small intestine.
Fig. 6.
Trimethylamine (TMA) lyase inhibition alters the host intestinal expression of key genes involved in sterol and bile acid transport and metabolism. At 8 wk of age, wild-type C57BL/6J male mice were switched from standard rodent chow to 1 of 4 experimental synthetic diets containing low (0.02%, wt/wt) or high (0.2%, wt/wt) levels of dietary cholesterol with or without the microbe-targeted TMA lyase inhibitor iodomethylcholine (IMC; 0.06%, wt/wt). After 4 wk on these diets, RNA was extracted from either the jejunum (A) or ileum (B), and gene expression was measured using quantitative real-time PCR. The mRNA abundance of HMG-CoA reductase (HMG Reduc.), HMG-CoA synthase (HMG Synth.), ATP-binding cassette transporters (Abca1, Abcg5, and Abcg8), Niemann-pick C1-like 1 (Npc1l1), farnesoid X receptor (Fxr), apical sodium bile acid transporter (Asbt), organic solute transporter α and β (Ostα and Ostβ), fibroblast growth factor 15 (Fgf15), and ileal bile acid-binding protein (I-babp) were quantified using the ΔΔCT method. C: Western blot analysis of Abca1, Npc1l1, and Gapdh with densitometric quantification. Data shown in A represent the means ± SE for n = 5 mice per group, whereas data in B and C represent the means ± SE for n = 3 mice per group; *significantly different than the nondrug-treated mice within each diet group.
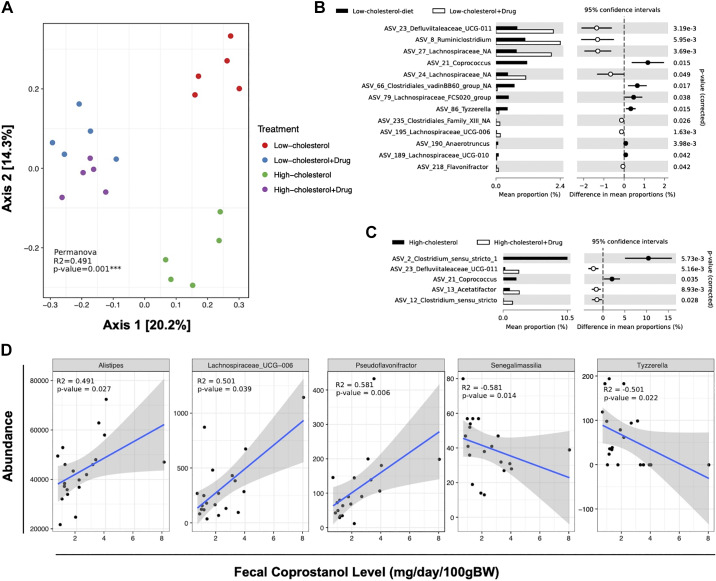
In addition to alteration in host intestinal gene expression, IMC treatment resulted in diet-specific alterations in cecal microbiome communities (Fig. 7). Principal coordinates analysis (PCA) of microbial taxa reveal distinct clusters, indicating that IMC induced cecal microbial community rearrangement in a diet-specific manner (Fig. 7A). When comparing the composition of low-cholesterol microbiomes to that of low-cholesterol samples treated with IMC, we found that a total of 13 amplicon sequence variants (ASVs) significantly differed in abundance (White’s nonparametric t test with Benjamini-Hochberg FDR multiple test correction, adjusted P ≤ 0.01, Fig. 7B). ASVs that increased in abundance in low-cholesterol IMC-treated samples included genera such as Clostridiales_vadin BB60 group, Lachnospiraceae FCS020, Coprococcus, and Tyzzerella, whereas genera that decreased in abundance included Ruminiclostridium and Flavonifractor, among others. Interestingly ASVs from Lachnospiraceae and Defluviitaleaceae UCG-011 dominated the increase in the low-cholesterol diet-fed IMC-treated group but could not be identified at the genus level. Two-group analysis (White’s nonparametric t test with Benjamini-Hochberg FDR multiple test correction, adjusted P ≤ 0.01) revealed five ASVs as statistically significant (Fig. 7C) across high-cholesterol diet-fed groups. ASVs that increased in abundance in high-cholesterol samples included Coprococcus and Clostridium, whereas Acetatifactor was decreased in abundance. Interestingly, different ASVs of Clostridium_sensu_stricto showed an increase in abundance in high-cholesterol and high-cholesterol + IMC samples (Fig. 7C). We also analyzed the correlation between the abundance of ASVs and the fecal concentration of coprostanol (Fig. 7D). The results showed that the abundance of 47 ASVs was significantly correlated with the concentration of coprostanol, 26 showing a negative correlation and 19 a positive correlation. Genera that showed positive correlation with coprostanol value included Lachnospiraceae_UCG-006, Alistipes, and Pseudoflavonifractor, whereas genera that showed negative correlation included Senegalimassilia and Tyzzerella (Fig. 7D). Collectively, these data show that IMC promotes the reorganization of gut microbial communities and alters host intestinal gene expression relevant to cholesterol and bile acid transport and metabolism.
Fig. 7.
Trimethylamine lyase inhibition promotes diet-specific reorganization of cecal microbial communities. Microbiomes were analyzed by sequencing the V4 region of the 16S rRNA using Earth microbiome protocol (EMP) (61). Quality filtered reads were processed into 737 Amplicon sequence variants (ASVs). Multigroup analysis (ANOVA, with Benjamini-Hochberg false discovery rate (FDR) multiple test correction, adjusted P ≤ 0.01) revealed 12 ASVs as differentially abundant across all four groups. A: principal component analyses (PCA). B and C: ASVs significantly different in abundance (White’s nonparametric t test with Benjamini-Hochberg FDR multiple test correction, adjusted P ≤ 0.01). D: correlation analyses comparing the abundance of ASVs and the fecal concentration of coprostanol. Data shown represent the means ± SE for n = 5 individual mice per group.
DISCUSSION
There is a growing appreciation that gut microbes play a contributory role in human health and disease, and the plethora of diverse metabolites produced by bacteria likely play a key role in this metaorganismal communication (7). Although our ability to “drug the microbiome” is still in its infancy, drug discovery in the gut microbiome space has tremendous untapped potential. The choline TMA lyase inhibitors used in this work represent some of the first-ever described mechanism-based small molecule inhibitors targeting a metaorganismal metabolic pathway, and we have previously shown that these drugs have potent anti-atherothrombotic activity (29). The present studies provide new evidence that the choline TMA lyase inhibition can also elicit diverse effects on host cholesterol and bile acid metabolism. The major findings of the current study demonstrate that small molecule microbial choline TMA lyase inhibition: 1) promotes total fecal neutral sterol loss by selectively enhancing the fecal disposal of the bacterially derived cholesterol metabolite coprostanol; 2) prevents high dietary cholesterol-induced hepatic cholesteryl ester storage; 3) alters the hepatic expression of key genes involved in bile acid synthesis and transport; 4) alters the total amount and composition of circulating bile acids; 5) promotes marked reductions in the enterocyte cholesterol transporter Npc1l1 in the jejunum while suppressing only a subset of FXR target genes in the ileum; and 6) results in diet-specific reorganization of cecal microbial communities. Collectively, this study shows for the first time that bacterially targeted choline TMA lyase inhibitors can alter host cholesterol and bile acid metabolism, providing support for the concept that bacterially derived TMA can elicit metaorganismal communication that instructs host sterol metabolism in a diet-related manner.
It has long been appreciated that both gut microbes and host hepatic metabolic pathways play a key role in shaping the size and composition of the enterohepatic pool of bile acids. In fact, the enterohepatic metabolism of bile acids represents one of the most well-studied metaorganismal metabolic pathways to date (23, 36, 55). Bile acids serve an essential role as detergents in the upper gastrointestinal track, facilitating the presentation of hydrophobic compounds (cholesterol, fatty acids, fat soluble vitamins, etc.) for intestinal absorption, and are also recognized to function as diverse signaling molecules that regulate host macronutrient metabolism and energy expenditure via activation of dedicated host receptor systems such as FXR and TGR5 (23, 48, 53, 55). Although much progress has been made to understand the key microbe and host enzymes coordinating bile acid composition, how other metaorganismal metabolic pathways intersect with bile acid metabolism is poorly understood. The present study supports previous studies demonstrating that the gut microbial TMAO pathway is intimately connected to bile acid metabolism (13, 18, 19, 25, 29) and provides new insights into the unique role of microbiota CutC/D-generated TMA. We initially showed that dietary supplementation with TMAO can reduce the total bile acid pool size in apolipoprotein E (ApoE)-deficient mice (29). In parallel, knockdown of the TMAO-producing enzyme FMO3 can also reduce the total bile acid pool, which is associated with marked suppression of the key bile acid synthetic enzymes CYP7A1 and CYP8B1 in the liver (60). It is important to note that the hepatic expression of the murine Fmo3 gene is induced by bile acids given that its promoter in under direct transcriptional control of the nuclear bile acid receptor FXR (4), which provides one potential pathway by which bile acids can control the relative levels of TMA and TMAO. Here we show that small molecule inhibition of the gut microbial choline TMA lyase (CutC/D) alters the size and composition of the circulating bile acid pool, decreasing several bile acid species known to activate FXR (cholic acid and taurocholic acid) and reciprocally increasing the naturally occurring FXR antagonist tauro-β-muricholic acid (TβMCA) (40). Two other recent studies have reported that provision of dietary TMAO can alter the hepatic expression of Cyp7a1 and also alter the relative abundance of FXR agonist versus antagonist bile acid species (13, 18). Although results here are not entirely consistent with these two recent reports in regard to the specific bile acid species altered, this may be due to the fact that we are using small molecule CutC/D inhibitors in contrast to feeding TMAO in the diet (13, 18). One limitation in the current study is that we only examined the bile acid composition in the plasma, and future studies examining the effects of TMA lyase inhibitors on individual bile acid species in the liver, bile, small and large intestine, and feces could provide additional information to better understand the metabolic perturbations induced by IMC and/or inhibition of microbial TMA formation. Collectively, there is a growing body of literature suggesting that bile acids can regulate the TMAO pathway (33) and perturbation of the TMAO pathway can reciprocally alter bile levels (13, 18, 29, 60).
Given that cholesterol is the substrate for primary bile synthesis in the liver, and the fact that bile acids are required for intestinal cholesterol absorption, it is not surprising that we find that fecal sterol loss and hepatic sterol storage are also altered in IMC-treated mice. IMC treatment was associated with increased fecal disposal of the bacterially derived cholesterol metabolite coprostanol (6, 46) and, in parallel, prevented high-cholesterol diet-driven increases in hepatic cholesteryl ester storage. It is tempting to speculate that these interrelated changes in sterol balance may be in part due to striking downregulation of the intestinal cholesterol transporter Npc1l1 (Fig. 6A) coupled with reorganization of cecal microbial composition that may favor cholesterol to coprostanol conversion. Although the transcriptional regulation of Npc1l1 is not completely understood, it is well accepted that activation of the liver X receptor (LXRα) can suppress the expression of Npc1l1 (19). However, in the case of IMC treatment, other LXR target genes such as Abca1, Abcg5, and Abcg8 were unaltered, arguing against a general alteration in LXR activity. Additional work is needed to understand the mechanism(s) by which TMA lyase inhibition promotes a marked decrease in jejunal Npc1l1 expression. As we originally reported (37), IMC treatment does elicit alterations in cecal microbial composition (Fig. 7). Although it is not well understood which murine or human commensals are involved in cholesterol catabolism to coprostanol, we found that the abundance of 47 amplicon sequence variants was significantly correlated with fecal coprostanol concentrations. The genera that were most highly correlated with fecal coprostanol levels were Lachnospiraceae_UCG-006, Alistipes, Pseudoflavonifractor, Senegalimassilia, and Tyzzerella (Fig. 7D). It is interesting to note that one other recent report also found Lachnospiraceae and Alistipes to be correlated with fecal coprostanol levels in humans infected with Clostridium difficile (1), but it will be important to perform in vitro and in vivo studies to confirm that any of these strains can convert cholesterol to coprostanol in the gut. It has been postulated that bacterial conversion to cholesterol to coprostanol could be advantageous to the context of atherosclerosis because coprostanol is poorly absorbed by the intestine (31). Further understanding of how gut microbial TMA production impacts bacterial coprostanol production deserves additional investigation.
Although drug discovery has historically targeted pathways in the human host, a fertile period in biomedical research lies ahead as instead gut microorganisms are targeted to either improve human health or prevent disease. There is tremendous untapped therapeutic potential in developing gut microbe-targeting drugs, but the field is still in its infancy in understanding how selective targeting of microbial enzymes may impact metaorganismal communication. Gut microbe-targeted choline TMA lyase inhibitors represent some of the first-ever described nonantibiotic small molecule inhibitors that irreversibly inhibit bacterial enzymes and accumulate within microbes with limited systemic exposure (37, 56). These drugs have been shown to have potent anti-atherosclerotic and antithrombotic effects in preclinical animal models (26, 27) and hold tremendous therapeutic potential for diverse cardiometabolic disorders. The present results show targeting microbial choline TMA lyase activity should have promise in treatment of diseases associated with altered sterol and bile acid homeostasis, including cardiovascular disease, advanced liver disease, and certain cancers. As small molecule drug discovery advances with the TMAO pathway and beyond, this study provides important reminders that metabolites produced from gut microbes can have profound effects on cholesterol and bile acid homeostasis in the host.
GRANTS
This work was supported by National Institutes of Health (NIH) and Office of Dietary Supplements Grants R01 HL122283 (to J. M. Brown), R01 DK120679 (J. M. Brown), P50 AA024333 (to J. M. Brown), P01 HL147823 (to S. L. Hazen and J. M. Brown), R01 HL130819 (to Z. Wang), and R01 HL103866 (to S. L. Hazen) and the American Heart Association (Postdoctoral Fellowship 17POST3285000 to R. N. Helsley). S. L. Hazen also reports partial support from an award from the Leducq Foundation. Development of lipid mass spectrometry methods reported here were supported in part by generous pilot grants from the Clinical and Translational Science Collaborative of Cleveland (4UL1TR000439) from the National Center for Advancing Translational Sciences (NCATS) component of NIH and the NIH Roadmap for Medical Research, the Case Comprehensive Cancer Center (P30 CA043703), the VeloSano Foundation, and a Cleveland Clinic Research Center of Excellence Award.
DISCLOSURES
Z. Wang and S. L. Hazen report being named as coinventors on pending and issued patents held by the Cleveland Clinic relating to cardiovascular diagnostics and therapeutics. S. L. Hazen reports being a paid consultant for Procter & Gamble, having received research funds from Procter & Gamble and Roche Diagnostics, and being eligible to receive royalty payments for inventions or discoveries related to cardiovascular diagnostics or therapeutics from Cleveland Heart Laboratory and Procter & Gamble. J. C. Garcia-Garcia is an employee of The Procter & Gamble Co. J. A. Buffa reports the right to receive royalties from Procter & Gamble. None of the other authors has any conflicts of interest, financial or otherwise, to disclose.
AUTHOR CONTRIBUTIONS
P.P., R.T., and J.M.B. conceived and designed research; P.P., R.N.H., A.L.B., J.A.B., I.C., I.N., C.B.G., V.G., Z.W., J.C.G.-G., L.C., R.T., N.S., S.L.H., and J.M.B. performed experiments; P.P., R.N.H., A.L.B., J.A.B., I.C., I.N., C.B.G., V.G., Z.W., J.C.G.-G., L.C., R.T., N.S., S.L.H., and J.M.B. analyzed data; P.P., I.C., I.N., C.B.G., Z.W., J.C.G.-G., L.C., R.T., S.L.H., and J.M.B. interpreted results of experiments; P.P., R.N.H., R.T., and J.M.B. prepared figures; P.P., R.N.H., A.L.B., J.A.B., I.C., V.G., J.C.G.-G., L.C., R.T., N.S., S.L.H., and J.M.B. drafted manuscript; P.P., R.N.H., A.L.B., J.A.B., I.C., I.N., C.B.G., V.G., Z.W., J.C.G.-G., L.C., R.T., N.S., S.L.H., and J.M.B. edited and revised manuscript; P.P., R.N.H., A.L.B., J.A.B., I.C., I.N., C.B.G., V.G., Z.W., J.C.G.-G., L.C., R.T., N.S., S.L.H., and J.M.B. approved final version of manuscript.
ACKNOWLEDGMENTS
We thank Dr. Russell DeBose-Boyd (University of Texas–Southwestern Medical Center) for kindly providing the antibody for HMG-CoA reductase.
REFERENCES
- 1.Antharam VC, McEwen DC, Garrett TJ, Dossey AT, Li EC, Kozlov AN, Mesbah Z, Wang GP. An integrated metabolomic and microbiome analysis identified specific gut microbiota associated with fecal cholesterol and coprostanol in Clostridium difficile infection. PLoS One 11: e0148824, 2016. doi: 10.1371/journal.pone.0148824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bäckhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA 101: 15718–15723, 2004. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belizário JE, Faintuch J, Garay-Malpartida M. Gut microbiome dysbiosis and immunometabolism: new frontiers for treatment of metabolic diseases. Mediators Inflamm 2018: 2037838, 2018. doi: 10.1155/2018/2037838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennett BJ, de Aguiar Vallim TQ, Wang Z, Shih DM, Meng Y, Gregory J, Allayee H, Lee R, Graham M, Crooke R, Edwards PA, Hazen SL, Lusis AJ. Trimethylamine-N-oxide, a metabolite associated with atherosclerosis, exhibits complex genetic and dietary regulation. Cell Metab 17: 49–60, 2013. doi: 10.1016/j.cmet.2012.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benjamini Y. Discovering the false discovery rate. J R Stat Soc Series B Stat Methodol 72: 405–416, 2010. doi: 10.1111/j.1467-9868.2010.00746.x. [DOI] [Google Scholar]
- 6.Björkhem I, Gustafsson JA. Mechanism of microbial transformation of cholesterol into coprostanol. Eur J Biochem 21: 428–432, 1971. doi: 10.1111/j.1432-1033.1971.tb01488.x. [DOI] [PubMed] [Google Scholar]
- 7.Brown JM, Hazen SL. The gut microbial endocrine organ: bacterially derived signals driving cardiometabolic diseases. Annu Rev Med 66: 343–359, 2015. doi: 10.1146/annurev-med-060513-093205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown JM, Hazen SL. Microbial modulation of cardiovascular disease. Nat Rev Microbiol 16: 171–181, 2018. doi: 10.1038/nrmicro.2017.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown MS, Goldstein JL. A proteolytic pathway that controls the cholesterol content of membranes, cells, and blood. Proc Natl Acad Sci USA 96: 11041–11048, 1999. doi: 10.1073/pnas.96.20.11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13: 581–583, 2016. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Canfora EE, Jocken JW, Blaak EE. Short-chain fatty acids in control of body weight and insulin sensitivity. Nat Rev Endocrinol 11: 577–591, 2015. doi: 10.1038/nrendo.2015.128. [DOI] [PubMed] [Google Scholar]
- 12.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336, 2010. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen ML, Yi L, Zhang Y, Zhou X, Ran L, Yang J, Zhu JD, Zhang QY, Mi MT. Resveratrol attenuates trimethylamine-N-oxide (TMAO)-induced atherosclerosis by regulating TMAO synthesis and bile acid metabolism via remodeling of the gut microbiota. MBio 7: e02210-15, 2016. doi: 10.1128/mBio.02210-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choucair I, Nemet I, Li L, Cole MA, Skye SM, Kirsop JD, Fischbach MA, Gogonea V, Brown JM, Tang WH, Hazen SL. Quantification of bile acids: a mass spectrometry platform for studying gut microbe connection to metabolic diseases. J Lipid Res 61: 159–177, 2020. doi: 10.1194/jlr.RA119000311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Craciun S, Balskus EP. Microbial conversion of choline to trimethylamine requires a glycyl radical enzyme. Proc Natl Acad Sci USA 109: 21307–21312, 2012. doi: 10.1073/pnas.1215689109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Craciun S, Marks JA, Balskus EP. Characterization of choline trimethylamine-lyase expands the chemistry of glycyl radical enzymes. ACS Chem Biol 9: 1408–1413, 2014. doi: 10.1021/cb500113p. [DOI] [PubMed] [Google Scholar]
- 18.Ding L, Chang M, Guo Y, Zhang L, Xue C, Yanagita T, Zhang T, Wang Y. Trimethylamine-N-oxide (TMAO)-induced atherosclerosis is associated with bile acid metabolism. Lipids Health Dis 17: 286, 2018. doi: 10.1186/s12944-018-0939-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duval C, Touche V, Tailleux A, Fruchart JC, Fievet C, Clavey V, Staels B, Lestavel S. Niemann-Pick C1 like 1 gene expression is down-regulated by LXR activators in the intestine. Biochem Biophys Res Commun 340: 1259–1263, 2006. doi: 10.1016/j.bbrc.2005.12.137. [DOI] [PubMed] [Google Scholar]
- 20.Elinav E, Garrett WS, Trinchieri G, Wargo J. The cancer microbiome. Nat Rev Cancer 19: 371–376, 2019. doi: 10.1038/s41568-019-0155-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gregory JC, Buffa JA, Org E, Wang Z, Levison BS, Zhu W, Wagner MA, Bennett BJ, Li L, DiDonato JA, Lusis AJ, Hazen SL. Transmission of atherosclerosis susceptibility with gut microbial transplantation. J Biol Chem 290: 5647–5660, 2015. doi: 10.1074/jbc.M114.618249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henao-Mejia J, Elinav E, Jin C, Hao L, Mehal WZ, Strowig T, Thaiss CA, Kau AL, Eisenbarth SC, Jurczak MJ, Camporez JP, Shulman GI, Gordon JI, Hoffman HM, Flavell RA. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature 482: 179–185, 2012. doi: 10.1038/nature10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jia W, Xie G, Jia W. Bile acid-microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat Rev Gastroenterol Hepatol 15: 111–128, 2018. doi: 10.1038/nrgastro.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson FL, St Clair RW, Rudel LL. Studies on the production of low density lipoproteins by perfused livers from nonhuman primates. Effect of dietary cholesterol. J Clin Invest 72: 221–236, 1983. doi: 10.1172/JCI110961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khan MT, Nieuwdorp M, Bäckhed F. Microbial modulation of insulin sensitivity. Cell Metab 20: 753–760, 2014. doi: 10.1016/j.cmet.2014.07.006. [DOI] [PubMed] [Google Scholar]
- 26.Knauf F, Brewer JR, Flavell RA. Immunity, microbiota and kidney disease. Nat Rev Nephrol 15: 263–274, 2019. doi: 10.1038/s41581-019-0118-7. [DOI] [PubMed] [Google Scholar]
- 28.Koeth RA, Levison BS, Culley MK, Buffa JA, Wang Z, Gregory JC, Org E, Wu Y, Li L, Smith JD, Tang WH, DiDonato JA, Lusis AJ, Hazen SL. γ-Butyrobetaine is a proatherogenic intermediate in gut microbial metabolism of L-carnitine to TMAO. Cell Metab 20: 799–812, 2014. doi: 10.1016/j.cmet.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, Britt EB, Fu X, Wu Y, Li L, Smith JD, DiDonato JA, Chen J, Li H, Wu GD, Lewis JD, Warrier M, Brown JM, Krauss RM, Tang WH, Bushman FD, Lusis AJ, Hazen SL. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med 19: 576–585, 2013. doi: 10.1038/nm.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29a.Kriaa A, Bourgin M, Potiron A, Mkaouar H, Jablaoui A, Gérard P, Maguin E, Rhimi M. Microbial impact on cholesterol and bile acid metabolism: current status and future prospects. J Lipid Res 60: 323–332, 2019. doi: 10.1194/jlr.R088989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature 444: 1022–1023, 2006. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 31.Lichtenstein AH. Intestinal cholesterol metabolism. Ann Med 22: 49–52, 1990. doi: 10.3109/07853899009147241. [DOI] [PubMed] [Google Scholar]
- 32.McMurdie PJ, Holmes S. Waste not, want not: why rarefying microbiome data is inadmissible. PLOS Comput Biol 10: e1003531, 2014. doi: 10.1371/journal.pcbi.1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miao J, Ling AV, Manthena PV, Gearing ME, Graham MJ, Crooke RM, Croce KJ, Esquejo RM, Clish CB, Vicent D, Biddinger SB; Morbid Obesity Study Group . Flavin-containing monooxygenase 3 as a potential player in diabetes-associated atherosclerosis. Nat Commun 6: 6498, 2015. doi: 10.1038/ncomms7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Repa JJ, Buhman KK, Farese RV Jr, Dietschy JM, Turley SD. ACAT2 deficiency limits cholesterol absorption in the cholesterol-fed mouse: impact on hepatic cholesterol homeostasis. Hepatology 40: 1088–1097, 2004. doi: 10.1002/hep.20439. [DOI] [PubMed] [Google Scholar]
- 35.Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, Griffin NW, Lombard V, Henrissat B, Bain JR, Muehlbauer MJ, Ilkayeva O, Semenkovich CF, Funai K, Hayashi DK, Lyle BJ, Martini MC, Ursell LK, Clemente JC, Van Treuren W, Walters WA, Knight R, Newgard CB, Heath AC, Gordon JI. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 341: 1241214, 2013. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ridlon JM, Harris SC, Bhowmik S, Kang DJ, Hylemon PB. Consequences of bile salt biotransformations by intestinal bacteria. Gut Microbes 7: 22–39, 2016. [Erratum in Gut Microbes 7: 262–280, 2016]. doi: 10.1080/19490976.2015.1127483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roberts AB, Gu X, Buffa JA, Hurd AG, Wang Z, Zhu W, Gupta N, Skye SM, Cody DB, Levison BS, Barrington WT, Russell MW, Reed JM, Duzan A, Lang JM, Fu X, Li L, Myers AJ, Rachakonda S, DiDonato JA, Brown JM, Gogonea V, Lusis AJ, Garcia-Garcia JC, Hazen SL. Development of a gut microbe-targeted nonlethal therapeutic to inhibit thrombosis potential. Nat Med 24: 1407–1417, 2018. doi: 10.1038/s41591-018-0128-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Routy B, Gopalakrishnan V, Daillère R, Zitvogel L, Wargo JA, Kroemer G. The gut microbiota influences anticancer immunosurveillance and general health. Nat Rev Clin Oncol 15: 382–396, 2018. doi: 10.1038/s41571-018-0006-2. [DOI] [PubMed] [Google Scholar]
- 39.Rudel LL, Haines J, Sawyer JK, Shah R, Wilson MS, Carr TP. Hepatic origin of cholesteryl oleate in coronary artery atherosclerosis in African green monkeys. Enrichment by dietary monounsaturated fat. J Clin Invest 100: 74–83, 1997. doi: 10.1172/JCI119524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sayin SI, Wahlström A, Felin J, Jäntti S, Marschall HU, Bamberg K, Angelin B, Hyötyläinen T, Orešič M, Bäckhed F. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab 17: 225–235, 2013. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 41.Schnabl B, Brenner DA. Interactions between the intestinal microbiome and liver diseases. Gastroenterology 146: 1513–1524, 2014. doi: 10.1053/j.gastro.2014.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schugar RC, Shih DM, Warrier M, Helsley RN, Burrows A, Ferguson D, Brown AL, Gromovsky AD, Heine M, Chatterjee A, Li L, Li XS, Wang Z, Willard B, Meng Y, Kim H, Che N, Pan C, Lee RG, Crooke RM, Graham MJ, Morton RE, Langefeld CD, Das SK, Rudel LL, Zein N, McCullough AJ, Dasarathy S, Tang WHW, Erokwu BO, Flask CA, Laakso M, Civelek M, Naga Prasad SV, Heeren J, Lusis AJ, Hazen SL, Brown JM. The TMAO-producing enzyme flavin-containing monooxygenase 3 regulates obesity and the beiging of white adipose tissue. Cell Rep 20: 2451–2461, 2017. [Erratum in Cell Rep 20: 279, 2017]. doi: 10.1016/j.celrep.2017.05.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shalapour S, Lin XJ, Bastian IN, Brain J, Burt AD, Aksenov AA, Vrbanac AF, Li W, Perkins A, Matsutani T, Zhong Z, Dhar D, Navas-Molina JA, Xu J, Loomba R, Downes M, Yu RT, Evans RM, Dorrestein PC, Knight R, Benner C, Anstee QM, Karin M. Inflammation-induced IgA+ cells dismantle anti-liver cancer immunity. Nature 551: 340–345, 2017. [Erratum in Nature 561: E1, 2018]. doi: 10.1038/nature24302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shih DM, Wang Z, Lee R, Meng Y, Che N, Charugundla S, Qi H, Wu J, Pan C, Brown JM, Vallim T, Bennett BJ, Graham M, Hazen SL, Lusis AJ. Flavin containing monooxygenase 3 exerts broad effects on glucose and lipid metabolism and atherosclerosis. J Lipid Res 56: 22–37, 2015. doi: 10.1194/jlr.M051680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Skye SM, Zhu W, Romano KA, Guo CJ, Wang Z, Jia X, Kirsop J, Haag B, Lang JM, DiDonato JA, Tang WH, Lusis AJ, Rey FE, Fischbach MA, Hazen SL. Microbial transplantation with human gut commensals containing CutC is sufficient to transmit enhanced platelet reactivity and thrombosis potential. Circ Res 123: 1164–1176, 2018. doi: 10.1161/CIRCRESAHA.118.313142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Snog-Kjaer A, Prange I, Dam H. Conversion of cholesterol into coprosterol by bacteria in vitro. J Gen Microbiol 14: 256–260, 1956. doi: 10.1099/00221287-14-2-256. [DOI] [PubMed] [Google Scholar]
- 47.Tang WH, Wang Z, Levison BS, Koeth RA, Britt EB, Fu X, Wu Y, Hazen SL. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N Engl J Med 368: 1575–1584, 2013. doi: 10.1056/NEJMoa1109400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thompson LR, Sanders JG, McDonald D, Amir A, Ladau J, Locey KJ, Prill RJ, Tripathi A, Gibbons SM, Ackermann G, Navas-Molina JA, Janssen S, Kopylova E, Vázquez-Baeza Y, González A, Morton JT, Mirarab S, Zech Xu Z, Jiang L, Haroon MF, Kanbar J, Zhu Q, Jin Song S, Kosciolek T, Bokulich NA, Lefler J, Brislawn CJ, Humphrey G, Owens SM, Hampton-Marcell J, Berg-Lyons D, McKenzie V, Fierer N, Fuhrman JA, Clauset A, Stevens RL, Shade A, Pollard KS, Goodwin KD, Jansson JK, Gilbert JA, Knight R; Earth Microbiome Project Consortium . A communal catalogue reveals Earth’s multiscale microbial diversity. Nature 551: 457–463, 2017. doi: 10.1038/nature24621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444: 1027–1031, 2006. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 51.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. A core gut microbiome in obese and lean twins. Nature 457: 480–484, 2009. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Turnbaugh PJ, Bäckhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe 3: 213–223, 2008. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tontonoz P, Mangelsdorf DJ. Liver X receptor signaling pathways in cardiovascular disease. Mol Endocrinol 17: 985–993, 2003. doi: 10.1210/me.2003-0061. [DOI] [PubMed] [Google Scholar]
- 54.Véniant MM, Sullivan MA, Kim SK, Ambroziak P, Chu A, Wilson MD, Hellerstein MK, Rudel LL, Walzem RL, Young SG. Defining the atherogenicity of large and small lipoproteins containing apolipoprotein B100. J Clin Invest 106: 1501–1510, 2000. doi: 10.1172/JCI10695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wahlström A, Sayin SI, Marschall HU, Bäckhed F. Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab 24: 41–50, 2016. doi: 10.1016/j.cmet.2016.05.005. [DOI] [PubMed] [Google Scholar]
- 56.Wang Z, Roberts AB, Buffa JA, Levison BS, Zhu W, Org E, Gu X, Huang Y, Zamanian-Daryoush M, Culley MK, DiDonato AJ, Fu X, Hazen JE, Krajcik D, DiDonato JA, Lusis AJ, Hazen SL. Non-lethal inhibition of gut microbial trimethylamine production for the treatment of atherosclerosis. Cell 163: 1585–1595, 2015. doi: 10.1016/j.cell.2015.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, Feldstein AE, Britt EB, Fu X, Chung YM, Wu Y, Schauer P, Smith JD, Allayee H, Tang WH, DiDonato JA, Lusis AJ, Hazen SL. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 472: 57–63, 2011. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Z, Levison BS, Hazen JE, Donahue L, Li XM, Hazen SL. Measurement of trimethylamine-N-oxide by stable isotope dilution liquid chromatography tandem mass spectrometry. Anal Biochem 455: 35–40, 2014. doi: 10.1016/j.ab.2014.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Warrier M, Shih DM, Burrows AC, Ferguson D, Gromovsky AD, Brown AL, Marshall S, McDaniel A, Schugar RC, Wang Z, Sacks J, Rong X, Vallim TA, Chou J, Ivanova PT, Myers DS, Brown HA, Lee RG, Crooke RM, Graham MJ, Liu X, Parini P, Tontonoz P, Lusis AJ, Hazen SL, Temel RE, Brown JM. The TMAO-generating enzyme flavin monooxygenase 3 is a central regulator of cholesterol balance. Cell Reports 10: 326–338, 2015. doi: 10.1016/j.celrep.2014.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC, Hu C, Wong FS, Szot GL, Bluestone JA, Gordon JI, Chervonsky AV. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature 455: 1109–1113, 2008. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wickham H. ggplot2: Elegant Graphics for Data Analysis. New York: Springer, 2009. doi: 10.1007/978-0-387-98141-3 [DOI] [Google Scholar]
- 63.Yao L, Seaton SC, Ndousse-Fetter S, Adhikari AA, DiBenedetto N, Mina AI, Banks AS, Bry L, Devlin AS. A selective gut bacterial bile salt hydrolase alters host metabolism. eLife 7: e37182, 2018. doi: 10.7554/eLife.37182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhu W, Gregory JC, Org E, Buffa JA, Gupta N, Wang Z, Li L, Fu X, Wu Y, Mehrabian M, Sartor RB, McIntyre TM, Silverstein RL, Tang WHW, DiDonato JA, Brown JM, Lusis AJ, Hazen SL. Gut microbial metabolite TMAO enhances platelet hyperreactivity and thrombosis risk. Cell 165: 111–124, 2016. doi: 10.1016/j.cell.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang J, Kelley KL, Marshall SM, Davis MA, Wilson MD, Sawyer JK, Farese RV Jr, Brown JM, Rudel LL. Tissue-specific knockouts of ACAT2 reveal that intestinal depletion is sufficient to prevent diet-induced cholesterol accumulation in the liver and blood. J Lipid Res 53: 1144–1152, 2012. doi: 10.1194/jlr.M024356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang J, Sawyer JK, Marshall SM, Kelley KL, Davis MA, Wilson MD, Brown JM, Rudel LL. Cholesterol esters (CE) derived from hepatic sterol O-acyltransferase 2 (SOAT2) are associated with more atherosclerosis than CE from intestinal SOAT2. Circ Res 115: 826–833, 2014. doi: 10.1161/CIRCRESAHA.115.304378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhu W, Wang Z, Tang WH, Hazen SL. Gut microbe-generated trimethylamine N-oxide from dietary choline is prothrombotic in subjects. Circulation 135: 1671–1673, 2017. doi: 10.1161/CIRCULATIONAHA.116.025338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhu W, Buffa JA, Wang Z, Warrier M, Schugar R, Shih DM, Gupta N, Gregory JC, Org E, Fu X, Li L, DiDonato JA, Lusis AJ, Brown JM, Hazen SL. Flavin monooxygenase 3, the host hepatic enzyme in the metaorganismal trimethylamine N-oxide-generating pathway, modulates platelet responsiveness and thrombosis risk. J Thromb Haemost 16: 1857–1872, 2018. doi: 10.1111/jth.14234. [DOI] [PMC free article] [PubMed] [Google Scholar]