Abstract
Background and Aim:
As a tick-borne zoonotic pathogen, Ehrlichia canis has already posed a threat to public health and safety. This study aimed to clarify the prevalence and molecular characteristics of E. canis in pet dogs in Xinjiang, China.
Materials and Methods:
A total of 297 blood samples of pet dogs and 709 skin ticks (Rhipicephalus sanguineus sensu lato) were subjected to molecular detection using PCR for E. canis 16S rRNA gene, and then, positive samples were amplified, sequenced, and phylogenetically analyzed for E. canis gp36 gene.
Results:
The PCR detection showed that the positive rate of PCR was 12.12% (36/297) in blood samples and 15.23% (108/709) in tick samples, respectively. Based on the phylogenetic analysis of E. canis gp36 protein, these E. canis strains in different geographical regions of the world can be divided into Genogroup I and Genogroup II. Among them, the Xinjiang epidemic strain XJ-6 and 533, 36, 1055, Kasur1, and Jake strains were clustered into subgroup 1.1 of Genogroup I, while the XJ-2, XJ-21, and XJ-35 strains and the TWN1, TWN4, CM180, and CM196 strains were closely related and belonged to subgroup 2.2 of Genogroup II, displaying high genetic diversity.
Conclusion:
This is the first study focusing on the molecular epidemiology of E. canis infection in pet dogs, which revealed that E. canis infection had been occurred in Xinjiang, China. More importantly, this study confirmed that the substantial variability in immunoreactive protein gp36 from E. canis strains circulating in pet dogs.
Keywords: Ehrlichia canis, genetic characteristics, gp36, pet dog, Rhipicephalus sanguineus sensu lato
Introduction
Ehrlichia canis is the pathogen causing canine monocytic ehrlichiosis (CME). As a member of the strict intracellular parasitic Gram-negative microorganism in the family Anaplasmataceae [1], E. canis is mainly transmitted by ticks (Rhipicephalus sanguineus sensu lato). Dog infected with E. canis is characterized by fever, depression, anorexia, increased ocular and nasal secretions, thrombocytopenia, and anemia, which are harmful to dogs [2,3]. Since E. canis was first detected in Algeria in 1935, it has been found in dogs and ticks in North America, Europe, Asia, and Africa. At present, CME is spreading all over the world [3-7].
Dogs are important companion animal of human beings, which are part of our daily lives. With the increasing number of pet dogs in cities, however, the more frequent exposures and interactions between companion animals and humans increase the infection risks of zoonotic diseases of human beings. In recent years, some important zoonoses (e.g., rabies and rickettsiosis) associated with pet dogs have received more and more attention from medical and veterinary researchers [8,9]. Therefore, as a newly discovered zoonotic pathogen, E. canis infection poses a threat to public health and safety [8]. Meanwhile, E. canis infection is closely related to the distribution and epidemic of Rhipicephalus sanguineus sensu lato. It was also found that the immunogenic protein gp36 of E. canis strains from different geographical regions of the world existed great genetic variation and showed obvious genetic diversity [10-15].
Xinjiang, located in Northwest China, is an area where E. canis vectors, Rhipicephalus sanguineus sensu lato, are populated, and is also one of the important focuses of tick-borne diseases. However, the infection status and molecular characteristics of E. canis in pet dogs are still unclear.
The main purpose of this study was to investigate the infection status and explore the molecular characteristics of E. canis in pet dogs in Xinjiang, China. Here, the blood samples were collected from pet hospitals in Xinjiang and the ticks parasitized on dog surface were detected by PCR, and the genetic evolution of E. canis Xinjiang epidemic strains was analyzed to reveal the genetic evolution relationship between E. canis epidemic strains from Xinjiang, China and other regions over the world, which will provide valuable epidemiological data for the prevention and control of E. canis infection.
Materials and Methods
Ethical approval
The experiments were carried out in accordance with the guidelines issued by the Ethical Committee of Shihezi University.
Source and collection of samples
During March 2016-March 2019, a total of 297 blood samples of dog and 709 samples of ticks (Rhipicephalus sanguineus sensu lato) on the skin surface of pet dogs were collected from pet hospitals in eight different geographical regions of Xinjiang (Tacheng, Yili, Shihezi, Urumqi, Changji, Korla, Aksu, and Kashgar) (Supplementary Figure-1). The collected clinical samples were sealed and placed in an icebox to transport at 4°C to the key laboratory of animal disease prevention and control in Xinjiang.
Supplementary Figure-1:
Geographic locations of the investigated regions in Xinjiang, [Source: www.zhongguolu.com/xinjiang/].
Primer design
Briefly, 16S rRNA and gp36 gene sequences of E. canis deposited in GenBank (http://www.ncbi.nlm.nih.gov/genbank/) were downloaded and compared, and the conservative sequences were selected. The specific primers for 16S rRNA and gp36 gene were designed by Premier 5.0 primer software according to the conservative region, respectively. 16S rRNA FP1-RP1 primer was used for PCR detection of whole blood and tick samples, while gp36 FP2-RP2 primer was used for the amplification of gp36 gene of E. canis epidemic strains in Xinjiang (Table-1). The primers were synthesized by BGI Group (BGI, China).
Table-1.
List of primer sequences used in this study.
| Primer name | Nucleotide sequence (5’®3’) | Target gene | Size of product (bp) |
|---|---|---|---|
| 16S-FP1 | CCTACGTTAGATTAGCTAGTTG | 16S rRNA | 465 |
| 16S-RP1 | CTGGTGTTCCTCCTAATATCTA | ||
| gp36-FP2 | atgctatttatactaatgggttat | gp36 | 845-865 |
| gp36-RP2 | TTAGTACAACCAGTTAGGCATATCAG |
DNA extraction from samples
In brief, DNA was extracted from blood samples of pet dogs using cell genomic DNA extraction kit (TaKaRa, Japan). Tick samples from pet dogs were identified by inverted microscopy and then placed in a grinder (one sample per tick) and grinded with 0.5 mL sterilized saline. The milling fluid was collected in 1.5 mL EP tube and frozen and thawed 3 times. Then, tick’s DNA was extracted according to the instructions of DNA extraction kit (Qiagen, Germany).
Molecular detection of E. canis by PCR
Briefly, PCR detection of E. canis was performed using 16S rRNA FP1-RP1 primers. The PCR reaction was carried out in 50 μl reaction mixture containing H2O2 21 uL, 16S rRNA FP1-RP1 primers 2 uL (0.2 μmol/L), 2×Premix Ex Taq (TaKaRa, Japan) 25 uL, and DNA template 2 uL. The PCR thermocycling conditions were as follows: Pre-denaturation at 95°C for 5 min, denaturation at 95°C for 20 s, annealing at 60°C for 30 s, elongation at 72°C for 30 s, 35 cycles, and followed by final elongation at 72°C for 10 min. Then, PCR product was detected by electrophoresis on 2% agarose gel containing ethidium bromide (TaKaRa, Japan) and was visualized using the Gel Doc XR System 2000 (BioRad, USA). The detection results of PCR were further analyzed statistically.
Amplification of gp36 gene in Xinjiang epidemic strain of E. canis
The samples with positive PCR results were selected, and the next PCR amplification of complete gp36 gene of E. canis was performed using gp36 FP2-RP2 primers. The PCR reaction system is the same as described in 1.4. PCR thermocycling conditions were as follows: Pre-denaturation at 95°C for 5 min, denaturation at 95°C for 20 s, annealing at 60°C for 30 s, extension at 72°C for 50 s; 40 cycles, and final extension at 72°C for 10 min. Then, PCR product was detected by 1.5% agarose gel electrophoresis.
Cloning and sequencing of gp36 gene of E. canis Xinjiang strain
In brief, the PCR product was extracted by DNA Gel Extraction Kit (TaKaRa, Japan) and cloned into pMD18-T vector (TaKaRa, Japan) for sequencing. Three positive clones were selected for each sample and each clone was sequenced (Sangon, China) 3 times. These sequences with completely identical sequencing in positive clone were submitted to GenBank and used for sequence comparison analysis.
Phylogenetic analysis of different geographical strains of E. canis based on amino acid sequences of gp36 protein
DNAStar 7.1 (DNASTAR Inc., USA) and Clustal X 2.1 (http://www.clustal.org/) software were used to compare the nucleotides and deduced amino acid sequences of the Xinjiang strains of E. canis with those of different geographical strains. The identities and genetic variations of the amino acid sequences of gp36 protein were analyzed. Using MEGA 6.0 software (https://www.megasoftware.net/), a phylogenetic tree (Bootstrap value of 1000) was constructed by neighbor-joining method to explore the genetic relationship among different geographical strains.
Statistical analysis
Statistical analysis was performed by SAS software (Version 9.1, SAS Institute, Inc., Cary, NC). The PCR-positive rates in different geographical regions in Xinjiang were compared using Chi-square test. p<0.05 was considered statistically significant, while p<0.01 was considered extremely significant difference.
Results
In 297 pet blood samples and 709 Rhipicephalus sanguineus sensu lato from dogs, the total positive rate of PCR was 12.12% (36/297) and 15.23% (108/709), respectively (Table-2 and Figure-1). The results showed that the positive rate in ticks was higher than that of blood samples, and there were significant differences in the positive rate in different geographical areas, especially in Yili and Shihezi regions (p<0.05).
Table-2.
Molecular detection of E. canis in different geographical regions in Xinjiang, China.
| Region/location | Blood of pet dogs | Ticks from pet dogs | ||||
|---|---|---|---|---|---|---|
| No. of blood samples | No. of positive | Positive rate (%) of E. canis | No. of tick samples | No. of positive | Positive rate (%) of E. canis | |
| Tacheng | 28 | 2 | 7.14 (2/28)a | 89 | 9 | 10.11 (9/89)a |
| Yili | 21 | 5 | 23.81 (5/21)b | 106 | 29 | 27.36 (29/106)b |
| Shihezi | 56 | 11 | 19.64 (11/56)b | 102 | 18 | 17.65 (18/102)b |
| Urumqi | 66 | 6 | 9.09 (6/66)a | 123 | 12 | 9.76 (12/123)a |
| Changji | 32 | 5 | 15.63 (5/32)a | 82 | 9 | 10.98 (9/82)a |
| Korla | 28 | 3 | 10.71 (3/28)a | 71 | 11 | 15.49 (11/71)a |
| Aksu | 39 | 2 | 5.13 (2/39)a | 94 | 13 | 13.83 (13/94)a |
| Kashgar | 27 | 2 | 7.41 (3/27)a | 42 | 7 | 16.67 (7/42)a |
| Total | 297 | 36 | 12.12 (36/297) | 709 | 108 | 15.23 (108/709) |
The different letter in same column means significant difference (p<0.05). E. canis=Ehrlichia canis
Figure-1.

Morphological identification of Rhipicephalus sanguineus sensu lato (a) from pet dogs and molecular detection of Ehrlichia canis 16S rRNA gene (b). (a) Back (♀); (b) ventral (♀); (c) amplification of E. canis 16S rRNA gene. M, DNA marker DL 1000 (1000, 750, 500, 400, 300, 200, and 100); 1, negative samples; 2-4, positive samples.
Four gp36 genes of Xinjiang epidemic strains of E. canis were successfully amplified from 36 positive samples of pet dogs and sequenced (Supplementary Figure-2). The gp36 gene sequences of four strains had been submitted to GenBank under the accession numbers (XJ-2, MN366176; XJ-6, MN366177; XJ-21, MN366178; and XJ-35, MN366179), respectively. By comparing the amino acid sequences of gp36 protein of 42 strains in different geographical regions (Supplementary Table-1 can be obtain from the author), it was found that the genetic variation of gp36 protein was obvious, which shared 65.71-99.26% identities in amino acid sequence (Supplementary Table-2 can be obtain from the author). Among the four Xinjiang epidemic strains, they shared 93.21% and 90.58% identities in nucleotide and amino acid sequence, respectively. However, the strain XJ-2 and XJ-21 contained 13 conserved amino acid tandem repeat units (TRR) (TEDSVSAPA), while the XJ-6 strain lacked one TRR sequence, which only contained 12 TRRs. Notably, XJ-35 strain contained 12 TRRs and a KILFLLQLL mutation sequence (Figure-2). By contrast, the Thailand strain CM180 and CM196 contained 8 and 13 TRRs in amino acid sequences of gp36 protein, respectively, while the Israel strain (strain 611) uniquely possessed TRR of TEDPVSATA.
Supplementary Figure-2.
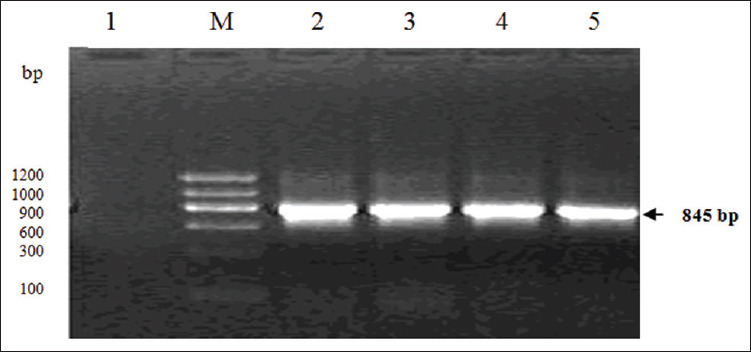
Amplification of gp36 gene of Ehrlichia canis from positive blood samples of pet dogs. (M) DNA marker DL 1200 (1200, 1000, 900, 600, 300, and 100); (1) Negative samples; (2) E. canis XJ-2 strain; (3) E. canis XJ-21 strain; (4) E. canis XJ-35 strain; (5) E. canis XJ-6 strain.
Figure-2.

Alignments of amino acid sequences of gp36 protein among Xinjiang strains of Ehrlichia canis. The tandem repeat region in amino acid sequences of gp36 protein of E. canis was boxed and shadowed.
Phylogenetic analysis based on the amino acid sequence of gp36 protein showed that E. canis strains from different geographical regions could be divided into two gene groups: Genogroup I and Genogroup II (Figure-3), among which the different genogroups could be further classified into different genetic subgroups, showing obvious genetic diversity. The strain XJ-6, together with the Thailand strains (533, 36, and 1055) and the US strain Jake, belonged to subgroup 1.1 of Genogroup I. In contrast, XJ-2, XJ-21, and XJ-35 strains were closely related to the Taiwan strain (TWN1 and TWN4) and the Thailand strain (CM180 and CM196) and belonged to subgroup 2.2 of Genogroup II. The results confirmed that both Genogroup I and Genogroup II epidemic strains of E. canis substantively had been circulating in Xinjiang, China.
Figure-3.

Phylogenetic analysis of different geographical strains of Ehrlichia canis based on amino acid sequences of gp36 protein. The amino acid sequences of gp36 protein of E. canis obtained in this study and available in GenBank were used to construct phylogenetic tree by the neighbor-joining method. Bootstrap values were calculated with 1000 replicates. Filled circle indicates E. canis strain XJ-2, XJ-6, XJ-21, and XJ-35 identified in this study. The GenBank accession numbers of 42 strains of E. canis were as follows: DJ (North Carolina), DQ146153.1; Florida, DQ146152.1; Cameroon 71, DQ146155.1; Sao Paulo, DQ146154.1; Louisiana, DQ146151.1; Demon, DQ085429.1; Oklahoma, DQ085428.1; Jake, DQ085427.1; Londrina, JX312080; 70C, KF233414.1; 56C, KF233413.1; 1055, KT363877.1; 533, KT363876.1; 36, KT363875.1; CM196, MF771085.1; CM180, MF771084.1; 222, KC479021.1; 171, KC479020.1; 105, KC479019.1; Muzaffargarh3, MH608290.1; Kasur2, MH608289.1; Muzaffargarh2, MH549199.1; Muzaffargarh1, MH549198.1; Rawalpindi2, MH549197.1; Rawalpindi1, MH549196.1; Kasur1, MH549195.1; 46, KT357370.1; 37y97, KT357369.1; Ranana, EU118961; 611, EF636663; Nether, KC935387.1; NGR, JN982341.1; TWN1, EF551366.1; TWN3, EF651794.1; TWN17, HQ009756.1; TWN4, EU139491.1; TWN2, EF560599.1; DB63, MG905720.1; B2-15, MG905719.1; B1-17, MG905718.1; B1-7, MG905717.1; B16, MG905716.1; B12, MG905714.1; DB51, MG905713.1; mkk2, MG905711.1; XJ-2, MN366176; XJ-6, MN366177; XJ-21, MN366178; XJ-35, MN366179.
Discussion
According to the difference of 16S rRNA gene, Ehrlichia can be divided into five recognized species: Ehrlichia muris, Ehrlichia chaffeensis, Ehrlichia ewingii, Ehrlichia ruminantium, and E. canis [1]. Among them, E. ruminantium mainly infects ruminants (bovine, sheep, goat, etc.), while E. chaffeensis, E. canis, and E. ewingii can infect humans [8]. Accordingly, as a newly discovered pet source zoonotic pathogen, E. canis has posed a great threat to public health [9].
In recent years, E. canis and its mixed infection with other pathogens have been reported in dogs in many countries and regions around the world [16-19]. Cicuttin et al. [11] performed molecular detection of E. canis on the canine blood samples from Buenos Aires, Argentina, and the PCR-positive rate was 6.7% (15/223). da Costa et al. [15] performed molecular detection of E. canis on Brazilian dogs, and the PCR-positive rate ranged from 14.6% to 42.3%. Daramola et al. [12] detected blood samples from dogs in South West Nigeria used by nested PCR, and the positive rate was 22.9% (47/2015). Malik et al. [13] performed E. canis detection on 151 canine blood samples from three regions of Pakistan using used PCR, with a positive rate of 28% (42/151). In this study, it showed that E. canis infection had occurred in pet dogs and their skin ticks in Xinjiang. Given that, intimate contact with infected pet dogs could enhance the risks of E. canis infection, timely detection, and monitoring of E. canis infection in pet dogs and ticks are necessary for public health.
Based on the genome-wide sequence [1,20], many proteins (such as gp200, gp140, gp36, and gp19) containing an acidic tandem repeat and an anchor protein repeat interacting with host cell were identified and characterized in E. canis [21,22]. gp36, as an important antigen protein inducing immune response, belongs to an acidic serine-rich protein with a major antigen epitope in the tandem repeat region. Doyle et al. found the variability in the number and sequence of tandem repeat units TRR in gp36 protein by comparing the gp36 proteins of the US, Brazilian, and Cameroonian strain of E. canis [21]. However, whether selective immune pressure is responsible for the substantial variability in this immunoreactive protein should be further investigated.
According to the genetic variations, gp36 protein can be used as molecular marker of E. canis. Aguiar et al. classified E. canis into different genotypes: US genotype, Brazil genotype, and the intermediate type [23]. It is worth noting that Bouza-Mora et al. identified a new genotype of E. canis by genetic analysis of gp36 protein in the blood samples from Costa Rica blood donors [9]. Maekawa et al. [7] confirmed significant differences between E. canis strains in the Philippine region and the US and Brazil strains. Furthermore, Nambooppha et al. [14] found two gene clusters in the local strain of E. canis in the local dog population in Thailand, the USA strain, and the Taiwan strain. In this study, we found that E. canis strains from Xinjiang can be divided into two gene groups, which belong to different gene subgroups, respectively, showing substantively genetic diversity.
Conclusion
This study for the 1st time suggested that Genogroup I and Genogroup II strains of E. canis had been circulating in Xinjiang, China, and confirmed that the substantial variability in immunoreactive protein gp36 had occurred, which provided valuable epidemiological data for the traceability and molecular characteristics of E. canis strains from pet dogs.
Authors’ Contributions
QM, MQ, and QJ planned and designed the whole study. WL, LY, and RY carried out the whole work. CK, ZJ, ZZ, YW, and PY collected sample. QM wrote the manuscript. CX and LC helped during manuscript writing and revision. All authors read and approved the final manuscript.
Acknowledgments
We thank the field staff who provided the samples for this study. This work was supported by National Key Research and Development Program, China (No. 2016YFD0501005), young and middle-aged leading science and technology innovation talents plan of Xinjiang Corps, China (No. 2016BC001) and Cooperation project of Urumqi-Chongqing in Science and Technology, China (No. Y161220001).
Competing Interests
The authors declare that they have no competing interests.
Publisher’s Note
Veterinary World remains neutral with regard to jurisdictional claims in published map and institutional affiliation.
References
- 1.Mavromatis K, Doyle C.K, Lykidis A, Ivanova N, Francino M.P, Chain P, Shin M, Malfatti S, Larimer F, Copeland A, Detter J.C, Land M, Richardson P.M, Yu X.J, Walker D.H, McBride J.W, Kyrpides N.C. The genome of the obligately intracellular bacterium Ehrlichia canis reveals themes of complex membrane structure and immune evasion strategies. J. Bacteriol. 2006;188(11):4015–4023. doi: 10.1128/JB.01837-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harrus S, Waner T. Diagnosis of canine monocytotropic ehrlichiosis (Ehrlichia cani s):An overview. Vet. J. 2011;187(3):292–296. doi: 10.1016/j.tvjl.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Paulino P.G, Pires M.S, da Silva C.B, Peckle M, da Costa R.L, Vitari G.V, Vilela J.A.R, de Abreu A.P.M, Massard C.L, Santos H.A. Epidemiology of Ehrlichia canis in healthy dogs from the Southeastern region of the state of Rio de Janeiro, Brazil. Prev. Vet. Med. 2018;159(11):135–142. doi: 10.1016/j.prevetmed.2018.09.012. [DOI] [PubMed] [Google Scholar]
- 4.Piratae S, Pimpjong K, Vaisusuk K, Chatan W. Molecular detection of Ehrlichia canis Hepatozoon canis and Babesia canis vogeli in stray dogs in Maha Sarakham province, Thailand. Ann. Parasitol. 2015;61(3):183–187. doi: 10.17420/ap6103.05. [DOI] [PubMed] [Google Scholar]
- 5.Kaewmongkol G, Lukkana N, Yangtara S, Kaewmongkol S, Thengchaisri N, Sirinarumitr T, Jittapalapong S, Fenwick S.G. Association of Ehrlichia canis Hemotropic Mycoplasma spp Anaplasma platys and severe anemia in dogs in Thailand. Vet. Microbiol. 2017;201(3):195–200. doi: 10.1016/j.vetmic.2017.01.022. [DOI] [PubMed] [Google Scholar]
- 6.Rotondano T.E.F, Krawczak F.D.S, Barbosa W.O, Moraes-Filho J, Bastos F.N, Labruna M.B, Azevedo S.S, Melo M.A, Almeida A.M.P. Ehrlichia canis and Rickettsia spp. In dogs from urban areas in Paraiba state, Northeastern Brazil. Rev. Bras. Parasitol. Vet. 2017;26(2):211–215. doi: 10.1590/S1984-29612017030. [DOI] [PubMed] [Google Scholar]
- 7.Maekawa N, Konnai S, Balbin M.M, Mingala C.N, Gicana K.R.B, Bernando F.A.E, Murata S, Ohashi K. Molecular detection and phylogenetic analysis of Ehrlichia canis in a Philippine dog. Ticks Tick Borne Dis. 2018;9(2):266–269. doi: 10.1016/j.ttbdis.2017.09.012. [DOI] [PubMed] [Google Scholar]
- 8.Conrad M.E. Ehrlichia canis:A tick-borne rickettsial-like infection in humans living in the Southeastern United States. Am. J. Med. Sci. 1989;297(1):35–37. doi: 10.1097/00000441-198901000-00008. [DOI] [PubMed] [Google Scholar]
- 9.Bouza-Mora L, Dolz G, Solórzano-Morales A, Romero-Zuñiga J.J, Salazar-Sánchez L, Labruna M.B, Aguiar D.M. Novel genotype of Ehrlichia canis detected in samples of human blood bank donors in Costa Rica. Ticks Tick Borne Dis. 2017;8(1):36–40. doi: 10.1016/j.ttbdis.2016.09.012. [DOI] [PubMed] [Google Scholar]
- 10.Nazari M, Lim S.Y, Watanabe M, Sharma R.S, Cheng N.A, Watanabe M. Molecular detection of Ehrlichia canis in dogs in Malaysia. PLoS Negl. Trop. Dis. 2013;7(1):e1982. doi: 10.1371/journal.pntd.0001982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cicuttin G.L, De Salvo M.N, Dohmen F.E.G. Molecular characterization of Ehrlichia canis infecting dogs, Buenos Aires. Ticks Tick Borne Dis. 2016;7(5):954–957. doi: 10.1016/j.ttbdis.2016.04.017. [DOI] [PubMed] [Google Scholar]
- 12.Daramola O.O, Takeet M.I, Oyewusi I.K, Oyekunle M.A, Talabi A.O. Detection and molecular characterisation of Ehrlichia canis in naturally infected dogs in southwest Nigeria. Acta Vet. Hung. 2018;66(1):85–95. doi: 10.1556/004.2018.008. [DOI] [PubMed] [Google Scholar]
- 13.Malik M.I, Qamar M, Ain Q, Hussain M.F, Dahmani M, Ayaz M, Mahmood A.K, Davoust B, Shaikh R.S, Iqbal F. Molecular detection of Ehrlichia canis in dogs from three districts in Punjab (Pakistan) Vet. Med. Sci. 2018;4(2):126–132. doi: 10.1002/vms3.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nambooppha B, Rittipornlertrak A, Tattiyapong M, Tangtrongsup S, Tiwananthagorn S, Chung Y.T, Sthitmatee N. Two different genogroups of Ehrlichia canis from dogs in Thailand using immunodominant protein genes. Infect. Genet. Evol. 2018;63(3):116–125. doi: 10.1016/j.meegid.2018.05.027. [DOI] [PubMed] [Google Scholar]
- 15.da Costa R.L, Paulino P.G, da Silva C.B, Vitari G.L.V, Peixoto M.P, de Abreu A.P.M, Santos H.A, Massard C.L. Molecular characterization of Ehrlichia canis from naturally infected dogs from the state of Rio de Janeiro. Braz. J. Microbiol. 2019;50(1):1–12. doi: 10.1007/s42770-018-0020-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baneth G, Harrus S, Gal A, Aroch I. Canine vector-borne co-infections Ehrlichia canis and Hepatozoon canis in the same host monocytes. Vet. Parasitol. 2015;208(1-2):30–34. doi: 10.1016/j.vetpar.2014.12.013. [DOI] [PubMed] [Google Scholar]
- 17.Almazán C, González-Álvarez V.H, de Mera I.G.F, Cabezas-Cruz A, Rodríguez-Martínez R, de la Fuente J. Molecular identification and characterization of Anaplasma platys and Ehrlichia canis in dogs in Mexico. Ticks Tick Borne Dis. 2016;7(2):276–283. [Google Scholar]
- 18.Aroch I, Baneth G, Salant H, Nachum-Biala Y, Berkowitz A, Shamir M, Chai O. Neospora caninum and Ehrlichia canis co-infection in a dog with meningoencephalitis. Vet. Clin. Pathol. 2018;47(2):289–293. doi: 10.1111/vcp.12582. [DOI] [PubMed] [Google Scholar]
- 19.Pesapane R, Foley J, Thomas R, Castro L.R. Molecular detection and characterization of Anaplasma platys and Ehrlichia canis in dogs from northern Colombia. Vet. Microbiol. 2019;233(5):184–189. doi: 10.1016/j.vetmic.2019.05.002. [DOI] [PubMed] [Google Scholar]
- 20.Zhang J, Wang J, Wang C. Complete genome sequence of Ehrlichia canis strain YZ-1, isolated from a beagle with fever and thrombocytopenia. Genome Announc. 2018;6(9):e00133–18. doi: 10.1128/genomeA.00133-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doyle C.K, Cardenas A.M, Aguiar D.M, Labruna M.B, Ndip L.M, Yu X.J, McBride J.W. Molecular characterization of E. canis gp36 and E. chaffeensis gp47 tandem repeats among isolates from different geographic locations. Ann. N. Y. Acad. Sci. 2005;1063(3):433–535. doi: 10.1196/annals.1355.079. [DOI] [PubMed] [Google Scholar]
- 22.Zhang X, Luo T, Keysary A, Baneth G, Miyashiro S, Strenger C, Waner T, McBride J.W. Genetic and antigenic diversities of major immunoreactive proteins in globally distributed Ehrlichia canis strains. Clin. Vaccine Immunol. 2008;15(7):1080–1088. [Google Scholar]
- 23.Aguiar D.M, Zhang X, Melo A.L, Pacheco T.A, Meneses A.M, Zanutto M.S, Horta M.C, Santarém V.A, Camargo L.M, McBride J.W, Labruna M.B. Genetic diversity of Ehrlichia canis in Brazil. Vet. Microbiol. 2013;164(3-4):315–321. doi: 10.1016/j.vetmic.2013.02.015. [DOI] [PubMed] [Google Scholar]


