Summary
The coronavirus disease 2019 (COVID-19) pandemic presents an unprecedented threat to global public health. Herein, we utilized a combination of targeted and untargeted tandem mass spectrometry to analyze the plasma lipidome and metabolome in mild, moderate, and severe COVID-19 patients and healthy controls. A panel of 10 plasma metabolites effectively distinguished COVID-19 patients from healthy controls (AUC = 0.975). Plasma lipidome of COVID-19 resembled that of monosialodihexosyl ganglioside (GM3)-enriched exosomes, with enhanced levels of sphingomyelins (SMs) and GM3s, and reduced diacylglycerols (DAGs). Systems evaluation of metabolic dysregulation in COVID-19 was performed using multiscale embedded differential correlation network analyses. Using exosomes isolated from the same cohort, we demonstrated that exosomes of COVID-19 patients with elevating disease severity were increasingly enriched in GM3s. Our work suggests that GM3-enriched exosomes may partake in pathological processes related to COVID-19 pathogenesis and presents the largest repository on the plasma lipidome and metabolome distinct to COVID-19.
Keywords: COVID-19, lipidomics, metabolomics, exosomes, monosialodihexosyl gangliosides, phosphatidylserines, bis(monoacylglyero)phosphates, biomarker
Graphical Abstract
Highlights
-
•
Quantitative lipidomic and metabolomic profiling of COVID-19 plasma
-
•
Plasma metabolite panel distinguished COVID-19 from healthy controls (AUC = 0.975)
-
•
Differential correlation analyses uncovered metabolic dysregulation in COVID-19
-
•
GM3-enriched exosomes are positively correlated with COVID-19 pathogenesis
Plasma metabolite panel effectively distinguished COVID-19 patients from healthy controls (AUC = 0.975). Plasma monosialodihexosyl gangliosides (GM3s) were negatively correlated with CD4+ T cell count in COVID-19 patients, and GM3-enriched exosomes were positively correlated with disease severity. These observations suggest that GM3-enriched exosomes may participate in pathological processes associated with COVID-19 progression.
Context and Significance
The COVID-19 pandemic presents an unprecedented threat to global public health. Systematic analyses of metabolic alterations during COVID-19 pathogenesis will uncover candidate pathways implicated in disease progression. Here, using a combination of targeted and untargeted tandem mass spectrometry to profile the plasma lipidome and metabolome in COVID-19 patients and healthy controls, Song et al. revealed that the plasma lipidome of COVID-19 bears resemblance to that of monosialodihexosyl ganglioside (GM3)-enriched exosomes. They validated observations made based on plasma by analyzing the lipidome of isolated exosomes and showed that GM3-enriched exosomes are correlated positively with the severity of COVID-19. These results also confer a resource that can be mined to propel future mechanistic investigation of metabolic dysregulation underlying COVID-19 pathogenesis.
Introduction
The novel coronavirus disease 2019 (COVID-19), now declared a pandemic by the WHO, is caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (Wu et al., 2020; Zhou et al., 2020b), which engages angiotensin-converting enzyme 2 (ACE2) as the host entry receptor (Hoffmann et al., 2020; Zhou et al., 2020b). Lung alveolar type II epithelial cells with high ACE2 expression levels are targets for SARS-CoV-2 (Zou et al., 2020). Most COVID-19 cases exhibit mild to moderate symptoms, but some patients with pneumonia suddenly deteriorate into severe respiratory failure and require intubation and mechanical ventilation (Huang et al., 2020; Xu et al., 2020; Zhou et al., 2020a).
Viruses are known to induce profound changes in host cell lipidomes and usurp key energy pathways in their exploitation of host metabolic resources for fueling the different stages of viral infection (Bley et al., 2020). Distinct membrane curvatures and precise lipid compositions can determine the ease of viral entry and replication (Zhang et al., 2019). Disease-associated changes in plasma or serum lipidomes had been previously reported for different viral infections. For example, increases in serum plasmalogen phosphatidylethanolamines (PEps) were observed in Zika virus-infected patients (Queiroz et al., 2019). Reductions in plasma triacylglycerols (TAGs) and free fatty acids (FFAs), and increases in numerous PEps and plasmalogen phosphatidylcholines (PCps) were shown in mice administered with respiratory syncytial virus (RSV) (Shan et al., 2018). For viral infections targeting the respiratory tract and the lungs, the consequential development of acute respiratory distress syndrome (ARDS) may also alter lipids implicated in inflammatory processes. For instance, plasma polyunsaturated phosphatidylcholines (PUFA-PCs), PUFA-PEs, and PUFA-TAGs were observed to decrease in non-survivors of ARDS compared to survivors (Maile et al., 2018), and increases in serum C18-FFAs were associated with ARDS development (Bursten et al., 1996). Bioactive derivatives of PUFAs, such as 13-hydroxyoctadecadienoic acids (13-HODE) and 9-HODE, were identified as mediators that induce and resolve inflammation triggered by influenza infection, respectively (Tam et al., 2013). Reductions in plasma phosphatidylserines (PSs), PEs, diacylglycerols (DAGs), and ceramides (Cer) were observed in fatalities of Ebola virus disease (EVD), while monosialodihexosyl gangliosides (GM3s) d34:0 were found to progressively increase from healthy controls to survivors to fatalities of EVD (Kyle et al., 2019).
Sialic acids are commonly found in cellular secretions and on external surfaces of cells as terminal motifs attached to glycoproteins and glycolipids (i.e., gangliosides) (Schauer, 2009). Sialyation was reported to mediate the binding and spreading of many viruses. Sialyation exhibits dual properties with respect to intercellular communication, either mediating viral binding and recognition, or acting as a “mask” in suppressing immunoreactivity by shielding cellular antigenic sites (Nimmerjahn et al., 2007). The precise effect of sialyation is dependent on specific substituents attached to the sialic acid moiety (Schauer, 2009). In addition to deploying specific lipids in their binding to host cells, viruses may also hijack the host’s endogenous machinery to achieve intercellular spreading. For example, hitherto evidence was present that implicated multivesicular bodies (MVBs)/late endosomes in the extracellular release of hepatitis C virus (HCV) and human immunodeficiency virus (HIV) (Chapuy-Regaud et al., 2013). Retrograde movement of MVBs and subsequent fusion with the plasma membrane release entrapped viral particles into the circulation to infect distant cells. Intracellular accumulation of HIV particles that have escaped lysosomal degradation near the plasma membrane periphery attracts and infects surveying CD4+ T cells via formation of infectious synapse (i.e., trans-infection) (Yu et al., 2014). Coronaviruses, on the other hand, were shown to usurp the trafficking machinery of endoplasmic reticulum-Golgi intermediate compartment (ERGIC), and mature viral particles are released by infected cells in the form of transport vesicles budding from the trans-Golgi network (TGN) (Hong, 2020). In this aspect, the coronavirus murine hepatitis virus strain A59 (MHV-59) was found to exploit the TGN to reach the cell exterior (Tooze et al., 1987), while the Middle East respiratory syndrome (MERS) virus was shown to contain a TGN-localization signal in the C-terminal domain of its M protein (Perrier et al., 2019). Biosynthesis of gangliosides in mammals takes place via stepwise addition of monosaccharides and sialic acid motifs to ceramides at membranes of the Golgi and TGN (Sandhoff and Sandhoff, 2018). Trafficking routes connecting the Golgi apparatus and late endosomes are present (Gruenberg and Stenmark, 2004). For example, the mannonse-6-phosphate receptor (MPR) cycles between the late endosomes and TGN (Gruenberg and Stenmark, 2004; Kobayashi et al., 1998). Disrupting the homeostasis of bis(monoacylglycero)phosphate (BMP), a unique lipid localized to the MVBs, led to mis-sorting of MPR and their accumulation in late endosomes (Kobayashi et al., 1998).
Therefore, viruses are capable of utilizing host-derived lipid membranes in their intercellular transmission to conceal and evade the host’s immune system (Izquierdo-Useros et al., 2010). Bypassing host surveillance allows augmented, unrestrained virus replication during early stages of infection (Channappanavar and Perlman, 2017). Failure of host to curb endogenous transmission at the incipient stage could contribute to aggravated disease severity and adverse outcome, such as lymphopenia leading to sudden episodes of “cytokine storm” (Zhang et al., 2020) and lethal pneumonia (Guan et al., 2020), which are the characteristic features of severe COVID-19 cases. As the primary targets of SARS-CoV-2, the alveolar type II epithelial cells synthesize surfactant phospholipids critical for modulating lung function (Schmitz and Müller, 1991), which are continuously secreted and recycled via the endocytic pathway. Exosomes, which had been isolated from broncho-alveolar lavage fluid, mediate intercellular communication between alveolar macrophages and alveolar epithelial cells that form the frontiers of host defense and immunity against air-borne pathogens (Lee et al., 2018). Therefore, it would be worthwhile to investigate if host-derived exosomes in the circulation were implicated in the pathogenesis of COVID-19.
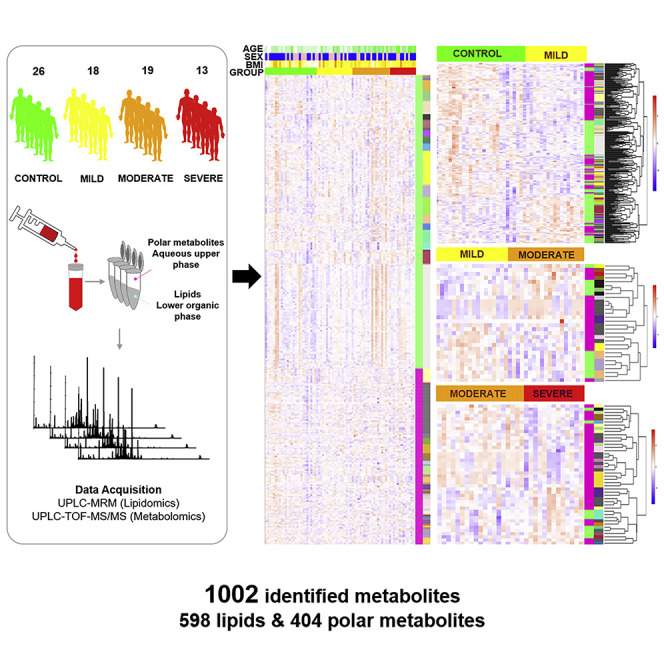
Herein, we present the quantitative serum lipidome and metabolome, comprising 1,002 metabolites (598 lipids and 404 polar small metabolites) quantitated using 71 internal standards (Lam et al., 2017), in a cohort of 76 subjects that included 26 healthy controls and 50 COVID-19 patients of differing disease severity (i.e., mild, moderate, and severe). We provide a systems perspective of COVID-19 pathogenesis using multiscale embedded correlation analyses to unbiasedly reveal pathologically relevant lipid clusters. Plasma lipid alterations associated with COVID-19 pathogenesis resembled exosome-specific lipid profiles, including increases in sphingomyelins (SMs) and GM3s, as well as reductions in DAGs. Based on exosomes isolated from plasma of the same cohort, we validated that exosomes from COVID-19 patients of elevating severity were increasingly enriched in GM3s. We also put forth a plasma-based metabolite panel that distinguished between healthy controls and COVID-19 with an area under the curve (AUC) = 0.975, and present a useful, quantitative omics data repository of COVID-19 plasma and exosomes to propel further mechanistic pursuits.
Results
Blood Routine and Circulating Markers of Systemic Inflammation Indicated Dysregulated Immune Response in COVID-19
Demographic and laboratory findings of the 50 recruited COVID-19 patients (Tables 1 and S1) were in good agreement with published literature on clinical characteristics of COVID-19 in China (Guan et al., 2020). Reductions in lymphocyte count (LC) (p < 0.0001) (Table S1), particularly in the numbers of T lymphocytes and CD4+ T lymphocytes (Figure S1A), were associated with increasing disease severity. Our observations were aligned with previous findings on laboratory-confirmed COVID-19 cases in Wuhan for which particularly drastic reductions in CD4+ T cell counts indicative of dysregulated immune response were noted, especially in severe COVID-19 patients (Qin et al., 2020). Indices of systemic inflammation, including C-reactive protein (CRP) (p = 0.0003), interleukin-6 (IL-6) (p = 0.0958), erythrocyte sedimentation rate (ESR) (p = 0.0008), serum ferritin (SF) (p = 0.0001), and procalcitonin (p = 0.0171), exhibited progressive increases as disease severity increased (Figure S1B; Table S1). We utilized a combination of targeted lipidomics (Lu et al., 2019) and untargeted metabolomics optimized in-house for screening human plasma samples. Our untargeted metabolomics detected an initial pool of 1,552 metabolite peaks with coefficients of variations <20% across quality control samples after subtraction of background noise. After proceeding to structural confirmation based on tandem mass spectrometry (MS/MS) spectra, the consolidated plasma metabolome finally contained 1,002 metabolites (598 lipids and 404 polar metabolites) quantitated using 71 internal standards.
Table 1.
Demographics and Baseline Characteristics of COVID-19 Patients
| Total (n = 50) | Mild (n = 18) | Moderate (n = 19) | Severe (n = 13) | p Value | |
|---|---|---|---|---|---|
| Characteristics | |||||
| Age, years | 43.0 (34.3–53.8) | 32.0 (22.3–40.0) | 45.0 (38.0–53.5) | 50.0 (40.0–78.0) | 0.001 |
| Onset of symptom to hospital admission, days | 5.0 (3.0–8.0) | 4.0 (2.0–5.0) | 6.0 (4.0–7.0) | 8.0 (5.0–10.0) | 0.023 |
| Duration of hospitalization, days | 19.0 (11.0–27.0) | 8.5 (5.3–19.0) | 19.0 (13.0–27.0) | 27.0 (20.0–36.0) | 0.02 |
| Sex | 0.525 | ||||
| Men | 30 (60%) | 9 (50%) | 12 (63%) | 9 (69%) | |
| Women | 20 (40%) | 9 (50%) | 7 (37%) | 4 (31%) | |
| Exposure to Wuhan | 26 (52%) | 13 (72%) | 7 (37%) | 6 (46%) | 0.087 |
| Any comorbidity | 18 (36%) | 3 (17%) | 7 (37%) | 8 (62%) | 0.037 |
| Hypertension | 8 (16%) | 0 | 3 (16%) | 5 (38%) | 0.016 |
| Diabetes | 5 (10%) | 1 (6%) | 2 (11%) | 2 (15%) | 0.664 |
| Malignancy | 1 (2%) | 0 | 0 | 1 (8%) | 0.234 |
| HIV | 1 (2%) | 0 | 0 | 1 (8%) | 0.234 |
| Chronic liver disease | 2 (4%) | 0 | 0 | 2 (15%) | 0.052 |
| Death | 2 (4%) | 0 | 0 | 2 (15%) | 0.052 |
| Signs and Symptoms | |||||
| Fever | 42 (84%) | 13 (72%) | 18 (95%) | 11 (85%) | 0.175 |
| Highest temperature,°C | 0.702 | ||||
| <37.3 | 8 (16%) | 5 (28%) | 1 (5%) | 2 (15%) | |
| 37.3–38.0 | 12 (24%) | 4 (22%) | 5 (26%) | 3 (23%) | |
| 38.1–39.0 | 22 (44%) | 6 (33%) | 10 (53%) | 6 (46%) | |
| >39.0 | 8 (16%) | 3 (17%) | 3 (16%) | 2 (15%) | |
| Cough | 31 (62%) | 8 (44%) | 12 (63%) | 11 (85%) | 0.075 |
| Expectoration | 15 (30%) | 3 (17%) | 5 (26%) | 7 (54%) | 0.076 |
| Rhinorrhoea | 1 (2%) | 1 (6%) | 0 | 0 | 0.404 |
| Myalgia or fatigue | 23 (46%) | 5 (28%) | 7 (37%) | 11 (85%) | 0.004 |
| Nausea and vomiting | 0 | 0 | 0 | 0 | |
| Sore throat | 7 (14%) | 3 (17%) | 2 (11%) | 2 (15%) | 0.853 |
| Shortness of breath | 6 (12%) | 0 | 1 (5%) | 5 (38%) | 0.003 |
| Chest pain | 1 (2%) | 0 | 0 | 1 (8%) | 0.234 |
| Diarrhea | 5 (10%) | 3 (17%) | 2 (11%) | 0 | 0.31 |
Data were median (interquartile range, IQR), or n (%). p values comparing mild, moderate, and severe were computed using the χ2 test (sex, exposure to Wuhan, comorbidities, death, fever, highest temperature, cough, expectoration, rhinorrhea, myalgia or fatigue, nausea and vomiting, sore throat, shortness of breath, chest pain, and diarrhea), one-way ANOVA (onset of symptom to hospital admission, duration of hospitalization, and BMI), or Kruskal-Wallis H test (age). COVID-19, coronavirus disease 2019. See also Figure S1.
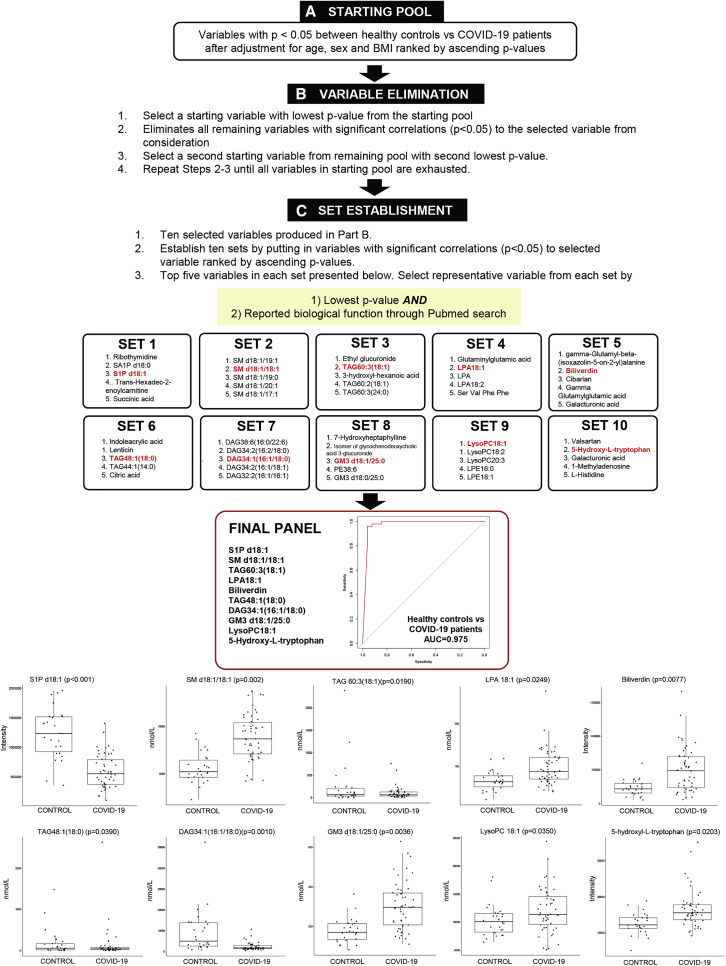
An Integrated Panel of Plasma Lipids and Polar Metabolites Effectively Distinguished COVID-19 Patients from Healthy Controls with AUC = 0.975
To derive a plasma metabolite panel for distinguishing between COVID-19 patients and healthy controls, significant variables (p < 0.05) after adjustment of age, sex, and BMI were included in a starting pool. In an iterative process, ten sets of variables were established from the starting pool (Figure 1 ), and one representative variable was finally selected from each set based on (1) smallest p value and (2) reported biological function from a PubMed search. A final panel of ten metabolites was generated, which separated healthy controls from COVID-19 patients with AUC = 0.975 in a logistic regression model with leave-one-out (LOO) cross validation. Among these metabolites, sphingosine-1-phosphate (S1P) was reduced (p < 0.001) in COVID-19, and its level was raised (p = 0.0065) at hospital discharge relative to admission in a small subset of patients followed longitudinally (Figure S2). S1P generation via sphingosine kinase-2 in monocyte-derived macrophages was recently shown to promote the resolution of inflammation by alveolar macrophages in acute lung injury (Joshi et al., 2020). Biliverdin, the oxidized form of bilirubin, is part of the redox cycle constituting the primary physiologic function of bilirubin as a cytoprotective antioxidant (Baranano et al., 2002). Increases in biliverdin (p = 0.0077) in COVID-19 probably indicated enhanced oxidative stress in disease state, and its level was reduced longitudinally at hospital discharge with marginal significance (p = 0.0558) (Figure S2). Plasma 5-hydroxy-tryptophan was elevated in COVID-19 (p = 0.0203), and its depletion via induction of the indolamine 2, 3-dioxygenase pathway in human alveolar carcinoma type II-like cells was previously reported to suppress the growth of parainfluenza virus type 3 (Rabbani and Barik, 2017). As for lipids, increases in lysophopholipids including lysophosphatidic acid (LPA) 18:1 (p = 0.0249) and lysophosphatidylcholine (LysoPC) 18:1 (p = 0.0350) were observed in COVID-19, while neutral lipids including medium-chain TAG 48:1(18:0) (p = 0.0190), long-chain TAG 60:3(18:1) (p = 0.0390), and DAG 34:1(16:1/18:0) (p = 0.0010) were generally reduced. On the other hand, sphingolipids such as SM d18:1/18:1 (p = 0.002) and GM3 d18:1/25:0 (p = 0.0036) were generally increased with disease.
Figure 1.
Plasma Panel for Differentiating COVID-19 Patients from Heathy Controls
Overview of selection scheme for plasma metabolite panel to differentiate COVID-19 (n = 50) patients from healthy controls (n = 26).
(A) From a total of 1,002 variables measured (598 lipids and 404 polar metabolites), variables with p < 0.05 between healthy controls and COVID-19 patients after adjustment for age, sex, and BMI were sieved out to form a starting pool comprising 322 variables.
(B) A starting variable with the lowest p value was selected, and variables with significant correlations (p < 0.05) to the starting variable selected were removed from consideration. From remaining variables in the starting pool, the next starting variable with the second lowest p value was identified, and the process was repeated in an iterative fashion until all variables in the starting pool were exhausted. This process generated a list of ten variables.
(C) Variables with significant correlations (p < 0.05) to each of the selected variables were added together to form ten established sets. To select a representative variable from each established set, the variable with the smallest p value and with reported biological function from a Pubmed search was chosen. A final panel of ten plasma metabolites, including S1P d18:1, SM d18:1/18:1, TAG60:3(18:1), LPA 18:1, biliverdin, TAG 48:1(18:0), DAG34:1(16:1/18:0), GM3 d18:1/25:0, lysoPC18:1, and 5-hydroxy-L-tryptophan, was generated, which distinguished between healthy controls and COVID-19 patients with an area under the curve (AUC) = 0.975 in a logistic regression model with leave-one-out (LOO) cross-validation. Boxplots for the ten selected metabolites in the final panel were illustrated and p values were indicated on top of each boxplot. Levels of polar metabolites measured using untargeted metabolomics were presented as corrected intensities, and lipids quantitated using targeted lipidomics were presented in nanomoles of lipids per liter (nmol/L) plasma.
See also Figures S2–S4.
Plasma Metabolome Changes Indicated Perturbed Oxidative Pathways of Cellular Energy Production
As baseline characteristics such as age, sex, and BMI were known to significantly influence plasma lipidomes and metabolomes, we constructed logistic regression models with these covariates to search for significant metabolites distinctly associated with different stages of COVID-19 (Figure S3). Several acylcarnitines, such as palmitoylcarnitine, stearoylcarnitine, and oleoylcarnitine, were reduced in COVID-19 (Figure S3, β-oxidation pathway). Reduced circulating levels of acylcarnitines may indicate attenuated entry of fatty acyls into the mitochondria for β-oxidation. Metabolites constituting the tricarboxylic acid (TCA) cycle were generally reduced in COVID-19 (Figure S3, TCA pathway). The reductions in polar metabolites participating in oxidative pathways of energy production (β-oxidation and TCA cycle), particularly in severe patients, may indicate metabolic response to declining lung functions and limiting blood oxygen to lower reliance on oxygen for cellular energy production. Lactate dehydrogenase was found to increase (p = 0.028) with increasing disease severity (Table S1), but plasma lactate was not significantly altered in COVID-19 compared to controls. The overall reductions in these polar metabolites might also reflect a response to change in nutrition, especially in severe patients. Although only 2 out of 13 severe patients were on mechanical ventilation at the point of blood collection (Table S3), a loss in appetite denotes a common general symptom of COVID-19 (Lechien et al., 2020). Interestingly, itaconic acid, a macrophage-specific metabolite derived from cis-aconitate, was progressively reduced with COVID-19 severity (Figure S3, TCA). Itaconate levels were previously reported to positively correlate with the expression of immune-responsive gene 1 (IRG1) in both human and mouse immune cells (Michelucci et al., 2013). Progressive reductions in sulfated steroids were also observed with increasing disease severity (Figure S3, steroid pathway). Numerous amino acids, including tryptophan, valine, proline, citrulline, and isoleucine, were significantly reduced in mild and moderate patients (Figure S3, amino acids). Decreases in specific amino acids were also previously observed in EVD fatalities (Eisfeld et al., 2017).
Plasma Lipidome Distinctly Associated with COVID-19 Resembles Exosomal Membrane Lipid Compositions
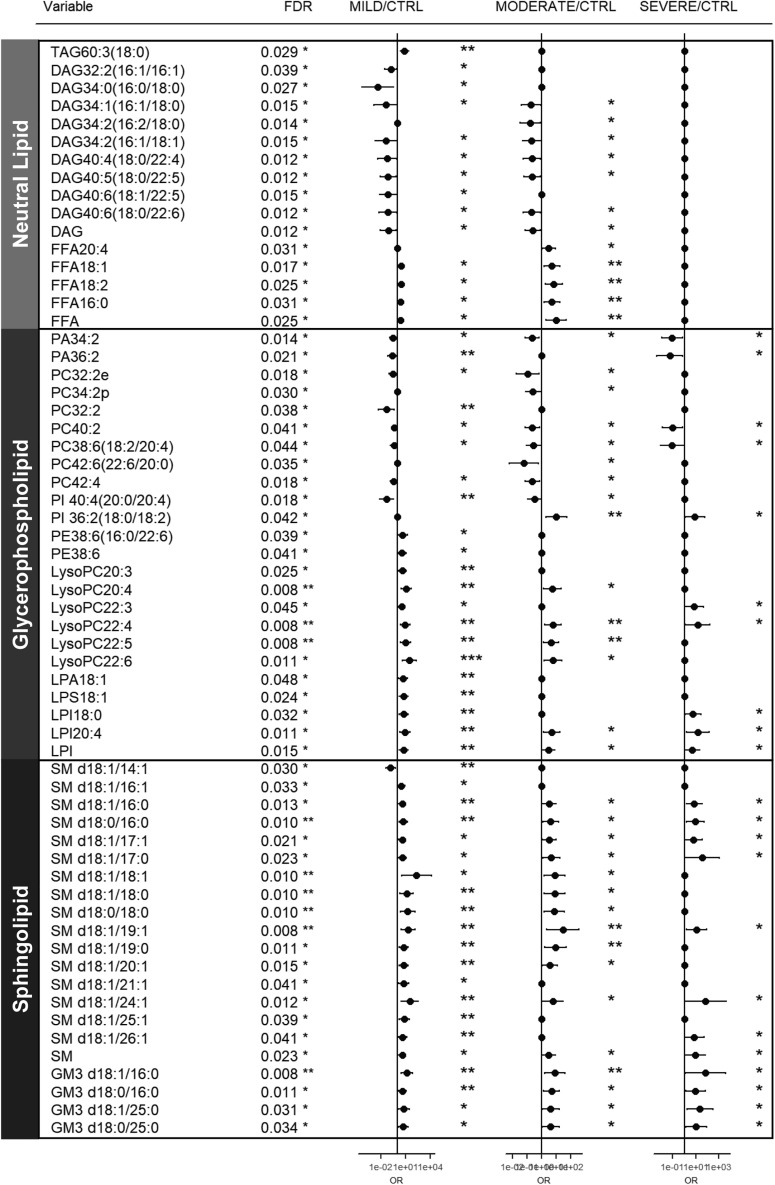
Lipids with overall false discovery rate (FDR) <0.05 were shortlisted and grouped according to major lipid classes for visual clarity in a forest plot (Figure 2 ). We observed reductions in major classes of plasma glycerophospholipids including phosphatidic acids (PAs), phosphatidylinositols (PIs), and PCs, with accompanying increases in their corresponding lysophospholipids (i.e., LPAs, LPIs, and LPCs) that indicate enhanced phospholipase A2 activity in COVID-19 patients. Cytosolic phospholipase A2α (cPLA2α) activation was reported to trigger pulmonary inflammation following pathogen infection (Bhowmick et al., 2017). PUFA-PEs were the only diacyl forms of glycerophospholipids that increased in COVID-19. Changes in phospholipidome (i.e., reductions in PCs and PIs and increases in PEs) observed in COVID-19 corroborated a previous study on plasma lipid alterations in EVD fatalities compared to survivors (Kyle et al., 2019). Among glycerophospholipids, PCs constitute the major membrane components of circulating lipoproteins (Cole et al., 2012), and PC-transfer protein (PC-TP) promotes cellular lipid efflux in nascent high-density lipoprotein (HDL) formation mediated by apolipoprotein A1 (apo-A1) (Baez et al., 2002), the major protein component of HDLs. PI was found to exhibit selective enrichment in plasma HDL fractions and was not detected in plasma lipoprotein-free fractions (Dashti et al., 2011). Changes in phospholipidome therefore suggested reductions in circulating HDLs as COVID-19 progresses, which was in agreement with the observed reductions in apo-A1 (p = 0.0687) as disease severity increases (Figure S1C). As for neutral lipids, marked reductions in several DAGs, with concomitant increases in FFAs (e.g., FFA18:1 and FFA 18:2), were observed in mild and moderate COVID-19 (Figure 2), while TAGs were significantly reduced only in mild cases (Figure S4). Increases in C18-FFAs and diminished TAGs were in agreement with previously reported circulating lipid changes associated with ARDS (Bursten et al., 1996; Maile et al., 2018). In stark contrast to the glycerophospholipid and neutral lipid pathways, sphingolipid classes of SMs and GM3s, e.g., GM3 d18:1/16:0, GM3 d18:0/16:0, GM3 d18:1/25:0, and GM3 d18:0/25:0, displayed progressive increases with increasing severity (Figures 2 and S4), possibly reflecting the augmented secretion of these lipids into the circulation. Of particular interest, lipidomes of exosomal membranes were previously shown to be specifically enriched in SMs (Stoorvogel et al., 2002) with diminished DAGs that cumulatively give rise to enhanced membrane rigidity (Laulagnier et al., 2004). Exosomes from lymphocytes also exhibited cell-type-specific enrichments in GM3s (Wubbolts et al., 2003) and PSs (Brügger et al., 2006). Thus, a gross overview of plasma lipidomic signatures distinctly associated with COVID-19, taking into account baseline cofounders, revealed a close resemblance to that of exosomal lipid compositions (i.e., enriched in SMs and GM3s with reduced DAGs).
Figure 2.
Plasma Lipids Associated with Severity of COVID-19
Logistic regression model with covariates BMI, age, and sex was built with each lipid to search for significant variables that could predict disease severity of subjects (i.e., healthy control, and mild, moderate, and severe COVID-19). Only lipids with false discovery rate (FDR) <0.05 were shortlisted and presented. Forest plots illustrate the magnitude of odds ratios with indicator of significance of the estimate in the model; ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05. For non-significant lipids, the estimates were plotted as zeros. Lipids were broadly classified according to major classes of neutral lipids, glycerophospholipids, and sphingolipids. See also Figure S4.
Association of Plasma Lipids with Pathologically Relevant Clinical Indices
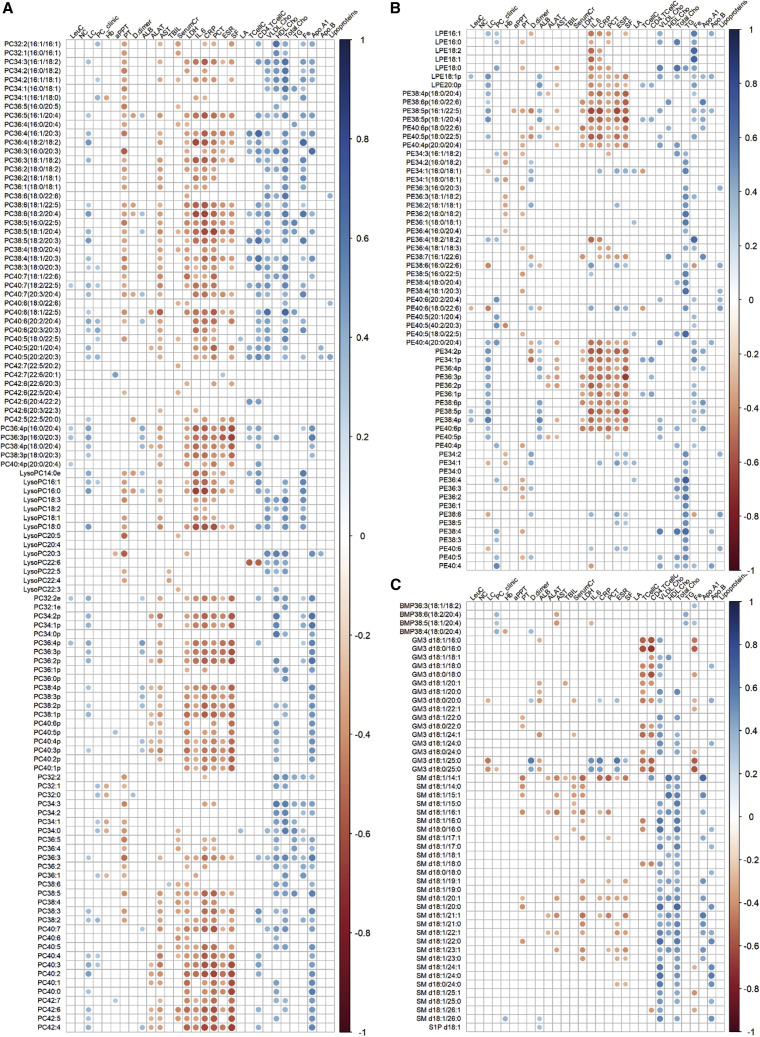
We then evaluated if plasma lipids altered in COVID-19 were significantly correlated with relevant clinical indices. Spearman correlations were performed and only correlations with p < 0.05 were indicated as colored circles on the correlation plots (Figure 3 ). We observed that PCs, particularly PUFA-PCs and PCps, displayed significant negative correlations with clinical indices of systemic inflammation (IL-6, CRP, procalcitonin [PCT], ESR, and SF) (Figure 3A; Table S1). This suggested that reductions in plasma PUFA-PCs and PCps were associated with aggravated systemic inflammation. Corroborating these observations, it was shown in a small cohort of cystic fibrosis patients that serum PUFA-PCs were positive indicators of lung function (measured in terms of predicted forced expiratory volume in 1 s) and negative indicators of systemic inflammation (Grothe et al., 2015). In contrast to PCs, only PEps, but not PUFA-PEs, were significantly and negatively associated with clinical indicators of systemic inflammation (Figure 3B). PCps were also specifically and positively associated with apo-A1 (Figure 3A). Of outstanding interest, we observed that plasma GM3s represented the only pathologically altered lipid class that was strongly and negatively correlated with T cell count and CD4+ T cell count (Figure 3C), which progressively decreased as disease severity increased (Figure S1A). The negative correlations suggest that increases in plasma GM3s were associated with reductions in circulating CD4+ T cell counts in COVID-19. Reductions in circulating CD4+ T cells constitute an important feature of dysregulated immune response reported in COVID-19 (Qin et al., 2020).
Figure 3.
Correlation of Plasma Lipids with Clinical Indices
Correlation plots illustrate spearman correlations between clinical indices with phosphatidylcholines (PC) (A), phosphatidylethanolamines (PE) (B), and multivesicular body-related lipids including bis(monoacylglyero)phosphate (BMPs), monosialodihexosyl gangliosides (GM3), and sphingomyelins (SMs) (C). Only correlations with p < 0.05 were indicated with colored circles. Negative correlations were shown in red and positive correlations were shown in blue, with sizes of circles representing the magnitude of the correlations. LeuC, leukocyte count; NC, neutrophil count; LC, lymphocyte count; PC_clinic, platelet count; Hb, hemoglobin; aPPT, activated partial thromboplastin time; PT, prothrombin time; D.dimer, D-dimer; ALB, albumin; ALAT, alanine aminotransferase; AST, aspartate aminotransferase; TBIL, total bilirubin; Serum Cr, serum creatinine; LDH, lactate dehydrogenase; IL.6, interleukin-6; CRP, C-reactive protein; PCT, procalcitonin; ESR, erythrocyte sedimentation rate; SF, serum ferritin; LA, lactic acid; TCellC, T cell count; CD4.TCellC, CD4+ T cell count; VLDL-Cho, very low-density lipoprotein cholesterol; HDLCho, high-density lipoprotein cholesterol; total cho, total cholesterol; TG, triglycerides; Fe, iron; ApoA1, apolipoprotein A1; ApoB, apolipoprotein B.
Multiscale Embedded Correlation Networks to Uncover Pathologically Relevant Lipid Modules
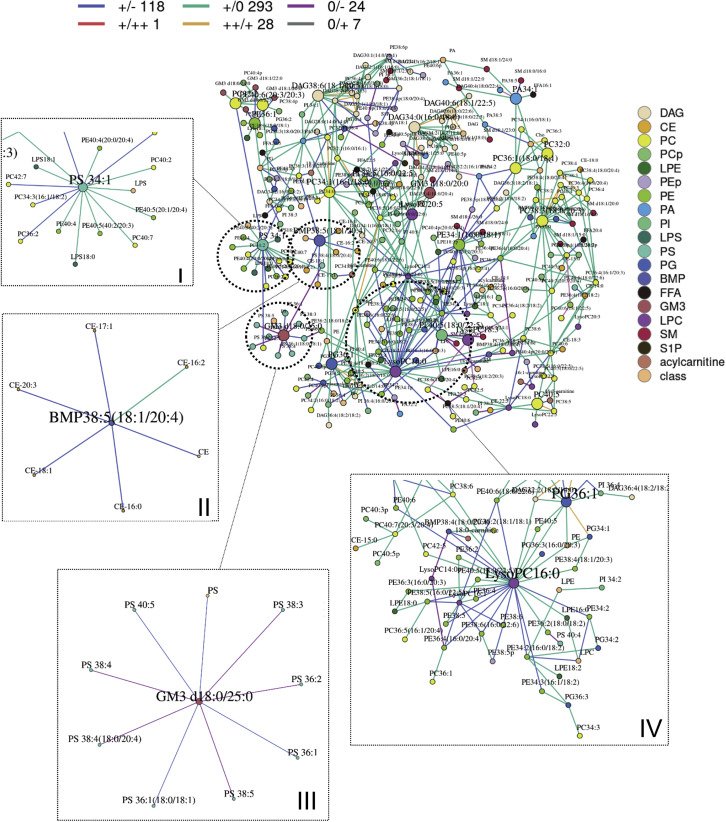
Co-regulated genes often display similar patterns of gene expression, which translates to strong correlations between their gene expression levels (Williams, 2015). Under the same analogy, strong correlations between lipid levels can imply that these lipids lie along a common metabolic pathway and are co-regulated, and changing correlation patterns between lipid-pairs in disease compared to healthy states can potentially indicate pathologically relevant metabolic dysregulation. We had previously shown in a cohort of antecedent diabetes that such a systems approach to interrogate lipidomics data based on differential correlations can sieve out pathway aberrations even before actual changes in metabolite levels set in (Lu et al., 2019). Thus, in order to decipher lipid pathway dysregulation at early stages of infection, we then looked for pathologically relevant lipid modules in mild COVID-19 relative to healthy controls using MEGENA R to construct networks from differentially correlated lipid pairs calculated via the R package DGCA. Only differential correlations with empirical p < 0.05 were displayed (Figure 4 ). Four notable modules in the global network were circled and enlarged for emphasized discussion.
Figure 4.
Differential Correlation Analyses of Plasma Lipids in Mild COVID-19 Relative to Healthy Controls
Multiscale embedded correlation network analysis illustrates the differential correlation of lipids in mild COVID-19 relative to healthy controls to reveal changes in lipid metabolic pathways upon early stage of viral infection. Only lipid pairs with significant differential correlations (empirical p < 0.05) were included. Sign/sign indicates direction and strength of correlation in control/mild COVID-19, and number that follows indicates number of lipid pairs in the global networks exhibiting this pattern of change. For instance, red line +/++ 1 in the upper legend of the global networks indicates that correlation between two connected lipid pairs was positive (+) in controls, and the correlation became even more strongly positive (++) in mild COVID-19 patients, as defined by statistically significant (p < 0.05) increase in correlation coefficients between the lipid pair across the two conditions. A total of 1 lipid pair connected by red lines in the global network displayed this pattern of change (+/++). Blue line +/− : positive in controls → negative in mild COVID-19. Teal line +/0: positive in controls → insignificant in mild COVID-19. Gold line ++/+: strongly positive in controls → weaker positive in mild COVID-19. Purple line 0/−: insignificant correlation in controls → negative correlation in mild COVID-19. Gray line 0/+: insignificant correlation in controls → positive correlation in mild COVID-19. Four modules (I–IV) of biological interest were circled and expanded for better visual clarity. (I) Module with hub PS 34:1 connected to numerous PEs by teal lines, indicating PS-PE positive correlations in healthy controls were lost in mild COVID-19. (II) Module with hub BMP 38:5(18:1/20:4) connected to CEs by blue lines, indicating BMP-CE correlations became negative in mild COVID-19. (III) Module with GM3 d18:0/25:0 as hub connected to several PSs by blue and purple lines, indicating GM3-PS correlations became negative in mild COVID-19. (IV) Module with LysoPC 16:1 as the hub connected to numerous PUFA-PEs by blue lines, indicating lysoPC-PUFA-PE correlations changed from positive in healthy controls to negative in mild COVID-19. PS, phosphatidylserines; PE, phosphatidylethanolamines; BMP, bis(monoacylglycero)phosphates; CE, cholesteryl esters; GM3, monosiaolodihexosyl gangliosides; PUFA-PE, polyunsaturated PEs.
PUFA-PE Accumulation Was Associated with Reductions in PSs in Mild COVID-19
Module I comprises PS 34:1 as the hub connected to numerous PUFA-PEs, i.e., PE 40:4(20:0/20:4) and PE 40:5(20:1/20:4), by green lines (+/0), indicating that the positive association between PS and PUFA-PEs in healthy controls was lost in mild COVID-19. PUFA-PEs were elevated while PSs were reduced in mild COVID-19 relative to controls (Figures 2 and S4). These suggest that PS synthase, which catalyzes the production of PSs from PEs (Han, 2016), may be compromised upon early infection, resulting in a loss of correlation between these lipids.
Correlation between BMPs and CEs Was Altered in Mild COVID-19
Module II comprises BMP 38:5 (18:1/20:4) as the hub connected to numerous cholesteryl esters (CEs) by blue lines (+/−). BMPs represent a class of structurally unique phospholipids enriched in MVBs implicated in cellular cholesterol homeostasis (Hullin-Matsuda et al., 2009). BMPs also exhibit cell-type-specific compositions in their esterified fatty acyl chains, with pulmonary alveolar macrophages exhibiting distinct enrichment in n-6 fatty acids (i.e., linoleic acid 18:2 and arachidonic acid 20:4) (Cochran et al., 1987; Mason et al., 1972). Among the various BMPs analyzed in our study, only BMP 38:5(18:1/20:4) and BMP 38:6(18:2/20:4) were specifically reduced in mild COVID-19 relative to healthy controls (Figure S4), suggesting that these reductions might be specific to pulmonary alveolar macrophages upon infection. BMPs were shown to influence cellular export of cholesterol by controlling cholesterol storage capacity of endosomes (Chevallier et al., 2008), which may explain the positive correlation between BMP 38:5(18:1/20:4) and several CEs in healthy controls. The changes in correlations in mild COVID-19 (i.e., +/− and +/0) suggested that this process might be perturbed, in agreement with the observed reductions in circulating apo-A1 with COVID-19 (Figure S1C). Of interest, we noticed that BMPs enriched in alveolar macrophages, i.e., BMP 38:5(18:1/20:4) (p = 0.0054) and BMP 38:6(18:2/20:4) (p = 0.0004), increased longitudinally with saturated CEs (CE 14:0, p = 0.0017; CE 16:0, p = 0.0092) from hospital admission to discharge in a small subset of patients who recovered from COVID-19 (Figure S2).
Negative Correlation between GM3s and PSs in COVID-19 Pathogenesis
Module III consists of GM3 d18:0/25:0 as the central hub connected to several PSs by purple (0/−) and blue lines (+/−). The connection between PSs and GM3s, which are not directly connected by endogenous lipid biosynthetic pathways, suggested that they might partake in common biological processes in a manner similar to BMPs and CEs as described above. In our study, while PUFA-PSs such as PS 38:4 and PS 40:4 were reduced in mild cases, numerous PS species including PS 40:4, PS 34:1, and PS 36:2 were significantly elevated (p < 0.05) in severe compared to moderate cases (Figure S4). Plasma PSs were also increased (p = 0.0041) during recovery from COVID-19 (Figure S2).
Afflicted PUFA-PC Homeostasis (Enhanced Breakdown or Abated Synthesis) in COVID-19
Module IV is represented by LysoPC 16:0 as the central hub, which became negatively correlated with several PUFA-PEs in mild COVID-19 (blue lines; +/−) relative to healthy controls. Negative correlations between PUFA-PEs and lysoPCs in mild COVID-19 may be explained considering the limiting pool of interconnecting PUFA-PCs that were reduced in mild disease versus controls (Figure 2). Enhanced production of lysoPCs via phospholipase A-mediated cleavage of PUFA-PCs would require increased methylation of PUFA-PEs to generate more PUFA-PCs via phosphatidylethanolamine N-methyltransferase (PEMT). It was demonstrated in a human cohort that PEMT preferentially utilizes PUFA-PEs as substrates, over the more saturated PEs, to selectively produce PUFA-PCs (Grothe et al., 2015).
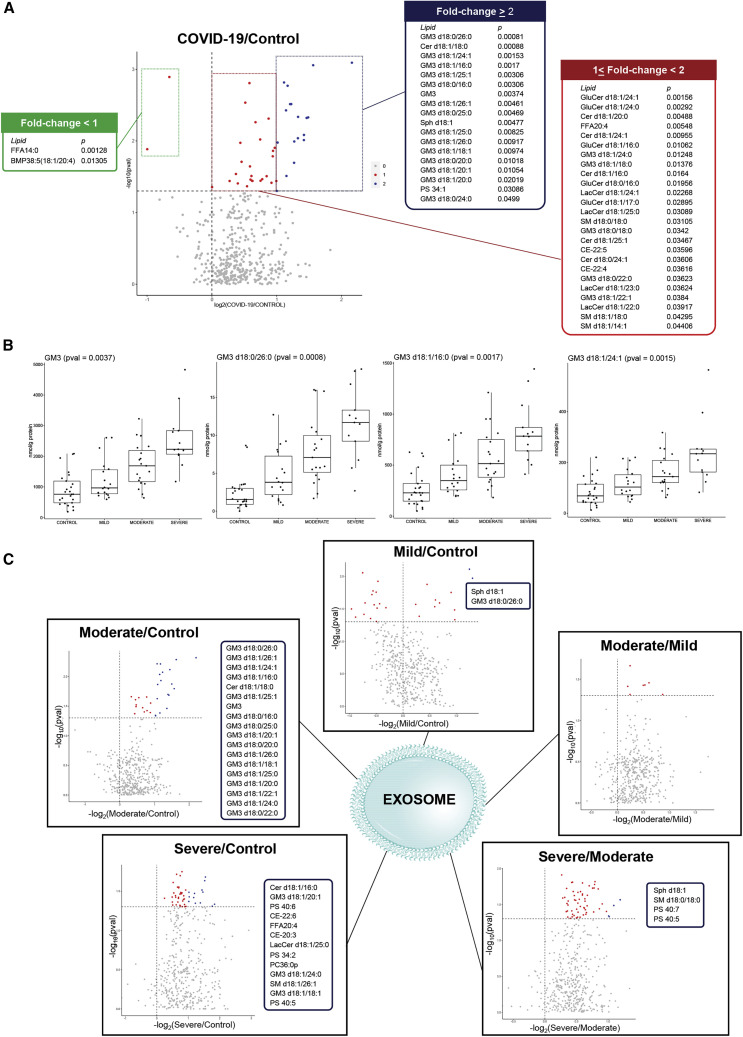
Isolated Exosomes Displayed Increasing Enrichment in GM3s with Elevating Disease Severity
Lipid analyses were conducted on exosomes isolated from the plasma of the same cohort (n = 75). The purity of isolated exosomes was validated by the enrichment in exosome-specific protein marker ALG-2-interacting protein X (Alix) (Théry et al., 2001) (Figure S5A). In line with previous reports on exosome lipid compositions (Subra et al., 2007), the isolated exosomes displayed selective enrichment of raft-associated lipids, including free cholesterol (Cho) and SMs (Figure S5B). Interestingly, we also noted exosome-specific enrichments in 20:4-BMPs, PUFA-PSs, and several GM3s relative to plasma (Figure S5B), suggesting that these lipids might partake in exosome-specific processes. In corroboration with postulations drawn based on plasma lipid profile changes, we found that GM3s were increasingly enriched in the exosomes of COVID-19 patients of elevating disease severity (Figure 5 ). Several GM3s exhibited greater than 2-fold increases in the exosomes (p < 0.05) of COVID-19 patients relative to healthy controls (Figure 5A). Progressive increases with increasing disease severity were similarly noted for GM3s of varying acyl chain lengths, e.g., very long-chain GM3 d18:0/26:0 (p = 0.0008) and GM3 d18:1/24:1 (p = 0.0015), as well as medium-chain such as GM3 d18:1/16:0 (p = 0.0017) (Figure 5B). BMP38:5(18:1/20:4) was reduced in the exosomes of COVID-19 patients compared to controls (Figure 5A). Increasing endosomal BMP content was previously found to impede intercellular transmission of HIV particles (Chapuy-Regaud et al., 2013). PUFA-PSs such as PS 40:7 and PS 40:5 were specifically increased in the exosomes of severe compared to moderate cases (Figure 5C). While plasma changes in sphingolipids, particularly GM3s, were recapitulated in exosomes, many of the changes in phospholipids (with the exception of PSs) were not observed in isolated exosomes. These observations suggested that plasma changes in glycerophospholipids associated with COVID-19 might be attributed to other components, such as lipoproteins, in the circulation.
Figure 5.
Lipid Changes in Exosomes of COVID-19 Patients
(A) Lipid changes in isolated exosomes from plasma of healthy controls (n = 25) and COVID-19 patients (n = 50). Colored dots (blue and red) in volcano plot indicate lipids that were significantly different (p < 0.05) between healthy controls and COVID-19 patients. Blue dots indicate significant lipids with fold change ≥ 2 in COVID-19 patients relative to controls listed in the blue panel, while red dots indicate significant lipids with fold change < 2 in patients relative to controls (listed in red and green panels). Lipids on the right side of vertical line at x = 0 were increased in COVID-19 patients compared to controls (blue and red panels), and lipids on the left side were decreased in COVID-19 patients compared to controls (green panel).
(B) Boxplots illustrate sum of all GM3s (p = 0.0037), and three representatives, GM3 d18:0/26:0 (p = 0.0008), GM3 d18:1/16:0 (p = 0.0017), and GM3 d18:1/24:1 (p = 0.0015), that displayed increasing trends in isolated exosomes from healthy controls to COVID-19 patients of increasing severity. Exosome lipids were expressed in nanomoles of lipids per gram of total protein (nmol/g protein).
(C) Volcano plots illustrate exosome lipids that were significantly different (p < 0.05) in pairwise comparisons indicated on top of each plot. Dots corresponding to significant lipids (p < 0.05) were colored (blue and red), and lipids with fold change ≥ 2 were colored blue and listed in the blue panel accompanying each volcano plot, while lipids with fold change < 2 were colored red.
See also Figure S5.
Discussion
Host-derived membranes can confer disguise against host defense arsenals and greatly facilitate viral exploitation of host cell resources for rapid, unsupervised viral multiplication (Cosset and Dreux, 2014). Indeed, RNA viruses had hitherto been discovered to hijack the exosomal pathway, which normally mediates endogenous intercellular communication, for viral assembly, transmission, and suppression of immune activation (Chahar et al., 2015). Our comprehensive, quantitative analyses of serum lipidome and metabolome in COVID-19 patients and healthy controls revealed that the overall serum lipidomic signatures associated with COVID-19 closely mirrored that of exosomal membrane lipid composition, which comprises enhanced levels of SMs and GM3s (cell-type specific) and reduced amounts of DAGs (Subra et al., 2007), indicating GM3-enriched exosomes might be associated with the pathogenesis of COVID-19. We validated our postulations drawn based on plasma lipidomes using exosomes isolated from the same cohort and demonstrated that exosomes were increasingly enriched in GM3s with elevating disease severity of COVID-19. As GM3s were the only pathologically altered plasma lipids that were negatively correlated with T cell counts and CD4+ T cell counts in COVID-19 patients, we postulate that GM3-enriched exosomes might participate in pathological processes that target CD4+ T cells. Indeed, GD3 gangliosides on the surface of exosomes isolated from fluids of ovarian tumor ascites were causally associated with the functional arrest of T cells, contributing to an immunosuppressive tumor microenvironment. Furthermore, removal of sialic motifs from GD3-coated exosomes obliterated their inhibitory capacity on T cells (Shenoy et al., 2018).
Our global evaluation of metabolic pathway alterations based on differential correlation network analyses also highlighted two lipid modules possibly implicated in COVID-19 pathogenesis, i.e., (1) BMPs and cholesterol homeostasis, and (2) negative correlations between GM3 and PS, which might be related to the secretion and intracellular localization of exosomes upon cellular uptake, respectively. In addition to their roles in cholesterol homeostasis, BMPs also function as stimulators for glycosphingolipid degradative enzymes (Linke et al., 2001). Corroborating with this, we noticed BMP reductions and GM3 elevations in both the plasma and exosomes of COVID-19 patients. Thus, on top of attenuating HDL maturation by impeding cellular export of cholesterol, BMP reduction may also favor the formation of GM3-enriched exosomes. Indeed, it had been proposed that microRNA export via the exosomal pathway and the HDL pathway for intercellular communication is possibly opposing, governed by their differential preferences for endogenous ceramide levels (Vickers et al., 2011). The negative associations between GM3 and PS in mild COVID-19 suggest that these lipids may partake in a common pathological process. Previous studies on artificial viral particles (AVNs) synthesized using nanoparticles coated with different lipid compositions revealed the preferential uptake of GM3-functionalized AVNs by mature dendritic cells, which together with macrophages mediate HIV-1 transmission to T cell targets (Yu et al., 2014, 2015). GM3-functionalized AVNs were sequestered in non-lysosomal compartments near the cell periphery and preserved from lysosomal degradation. The peripheral localization and preservation of viral particles by GM3-functionalized AVNs was highly specific to GM3 content (Xu et al., 2018). Increasing PS content in AVNs was shown to antagonize GM3-mediated compartmentalization, by weakening AVNs binding to macrophages and redirecting AVNs to lysosomal compartment for degradation (Xu et al., 2018). As we observed that both plasma and exosome PSs were significantly increased in severe compared to moderate patients, it remains an interesting possibility if increases in PSs might denote host adaptive response to counteract disease progression, given that plasma PSs were increased longitudinally during the recovery phase and a majority of the severe cases included in this study recovered from the disease (2/13 deaths).
To summarize, our extensive lipidomics and metabolomics analyses of plasma changes associated with COVID-19, in combination with differential correlation network analyses for systems metabolic pathway interrogation, provided evidence that implicated GM3-enriched exosomes in COVID-19 pathogenesis. We validated our postulations drawn based on plasma lipidomes by analyzing lipid changes in isolated exosomes, which demonstrated that exosomes were increasingly enriched in GM3s with elevating disease severity in COVID-19.
Limitations of Study
In this study, lipidomic changes were analyzed in plasma and isolated exosomes, but not in lung tissues or sputum of COVID-19 patients, for which viral counts were expected to be highest. Based on the current study design, it is thus not possible to establish a direct association between the lipidomic changes observed with SARS-CoV-2 infection. Nonetheless, in a previous study on mice infected with RSV that analyzed both lung tissues and plasma lipid changes, multiple correlations between plasma and lung lipids altered by RSV infection were reported (Shan et al., 2018). Metabolite spillage from the lungs into the circulation may also be reasonably expected in cases with compromised alveolar-capillary barrier, such as in moderate and severe COVID-19 patients with pneumonia. While we followed a small group of five patients longitudinally to elucidate plasma metabolites associated with recovery phase (Figure S2), further validation from a larger longitudinal cohort is needed. Our results were based on a single Chinese cohort of COVID-19 patients in China, and future studies in different racial, ethnic, and geographical cohorts will be indispensable for extending our current understanding of lipid metabolic dysregulation in COVID-19 pathogenesis. In addition, while we included age, sex, and BMI as covariates in our logistic regression analyses, the statistically significant age and BMI differences between healthy controls and patients are potential confounders in the current study, due to technical constraints in recruiting age- and BMI-matched healthy controls during the outbreak. Finally, the metabolic dysregulation underlying the observed exosomal enrichment in GM3s awaits further mechanistic validation, although our preliminary evidence suggested that the selective increases in exosomal GM3s might have contributions from non-exosomal plasma components, as indicated by the decreasing GM3 content in exosome-free supernatant with increasing disease severity (Figure S5C).
In conclusion, our work highlighted that GM3-enriched exosomes were correlated with the pathogenesis of COVID-19. We presented the largest, quantitative repository on the plasma lipidome and metabolome distinctly associated with COVID-19, taking into account baseline confounders. Accurate quantitation is sine qua non for facilitating metabolic pathway analysis using multiscale embedded differential correlations, which picks up subtle changes in correlations between quantitated metabolite levels. Using this approach, we uncovered metabolite clusters pathologically relevant to COVID-19 that warrant further mechanistic pursuits.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| ALIX | Abcam | Cat#ab76608; RRID: AB_2042595 |
| Peroxidase-Conjugated Goat anti-Rabbit IgG (H+L) | Beijing Zhongshan Jinqiao Biotechnology Co., Ltd. | Cat#ZB-2301; RRID: AB_2747412 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Chloroform (HPLC grade) | Honeywell | Cat#049-4 |
| Methanol (HPLC grade) | Fisher chemical | Cat#A452-4 |
| Acetonitrile (LCMS grade) | Fisher chemical | Cat#A955-4 |
| Formic acid (98%) | J&K | Cat#299272 |
| RIPA lysis buffer | Meilunbio | Cat#MA0151 |
| Protease inhibitor cocktail | Sigma-Aldrich | Cat#P8340-5ML |
| Ammonium hydroxide solution | Sigma-Aldrich | Cat#05002-1L |
| Ammonium acetate | Sigma-Aldrich | Cat#73594 |
| PC-14:0/14:0 | Avanti Polar Lipids | Cat#850345P |
| d31-PC16:0/18:1 | Avanti Polar Lipids | Cat#860399C |
| PE14:0/14:0 | Avanti Polar Lipids | Cat#850745P |
| d31-PE-16:0/18:1 | Avanti Polar Lipids | Cat#860374C |
| d31-PS-16:0/18:1 | Avanti Polar Lipids | Cat#860403C |
| PA-17:0/17:0 | Avanti Polar Lipids | Cat#830856P |
| PG-14:0/14:0 | Avanti Polar Lipids | Cat#840445P |
| d31-PG-16:0/18:1 | Avanti Polar Lipids | Cat#860384C |
| C14:0-BMP | Avanti Polar Lipids | Cat#857131P |
| d31-PI-16:0/18:1 | Avanti Polar Lipids | Cat#860042P |
| SM-d18:1/12:0 | Avanti Polar Lipids | Cat#860583P |
| LPC-17:0 | Avanti Polar Lipids | Cat#855676P |
| LPE-17:1 | Avanti Polar Lipids | Cat#110699 |
| LPI-17:1 | Avanti Polar Lipids | Cat#850103P |
| LPA-17:0 | Avanti Polar Lipids | Cat#857127P |
| LPS-17:1 | Avanti Polar Lipids | Cat#858141P |
| S1P-d17:1 | Avanti Polar Lipids | Cat#860641P |
| Cer-d18:1/d7-15:0 | Avanti Polar Lipids | Cat#860681P |
| GluCer d18:1/8:0 | Avanti Polar Lipids | Cat#860540P |
| GalCer d18:1/8:0 | Avanti Polar Lipids | Cat#860538P |
| PI-8:0/8:0 | Echelon | Cat#P-0008 |
| d3-GM3 d18:1/18:0 | Matreya LLC | Cat#2052 |
| d3-LacCer d18:1/16:0 | Matreya LLC | Cat#1534 |
| d5-DAG16:0/16:0 | Avanti Polar Lipids | Cat#110537 |
| d5-DAG18:1/18:1 | Avanti Polar Lipids | Cat#110581 |
| TAG(14:0)3-d5 | CDN Isotopes | Cat#D-6958 |
| TAG(16:0)3-d5 | CDN Isotopes | Cat#D-5815 |
| TAG(18:0)3-d5 | CDN Isotopes | Cat#D-5816 |
| d6-cholesterol | CDN Isotopes | Cat#D-2139 |
| d6-CE18:0 | CDN Isotopes | Cat#D-5823 |
| L-Phenylalanine-d8 | Cambridge Isotope Laboratories | Cat# DLM-372-1 |
| L-Tryptophan-d8 | Cambridge Isotope Laboratories | Cat#DLM-6903-0.25 |
| L-Isoleucine-d10 | Cambridge Isotope Laboratories | Cat#DLM-141-0.1 |
| L-Asparagine-13C4 | Cambridge Isotope Laboratories | Cat#CLM-8699-H-0.05 |
| L-Methionine-d3 | Cambridge Isotope Laboratories | Cat#DLM-431-1 |
| L-Valine-d8 | Cambridge Isotope Laboratories | Cat#DLM-311-0.5 |
| L-Proline-d7 | Cambridge Isotope Laboratories | Cat#DLM-487-0.1 |
| L-Alanine-d7 | Cambridge Isotope Laboratories | Cat#DLM-251-PK |
| DL-Serine-d3 | Cambridge Isotope Laboratories | Cat#DLM-1073-1 |
| DL-Glutamic acid-d5 | Cambridge Isotope Laboratories | Cat#DLM-357-0.25 |
| L-Aspartic acid-d3 | Cambridge Isotope Laboratories | Cat#DLM-546-0.1 |
| L-Arginine-d7 | Cambridge Isotope Laboratories | Cat#DLM-541-0.1 |
| L-Glutamine-d5 | Cambridge Isotope Laboratories | Cat#DLM-1826-0.1 |
| L-Lysine-d9 | Cambridge Isotope Laboratories | Cat#DLM-570-0.1 |
| L-Histidine-d5 | Cambridge Isotope Laboratories | Cat#DLM-7855 |
| Taurine-13C 2 | Cambridge Isotope Laboratories | Cat#CLM-6622-0.25 |
| Betaine-d11 | Cambridge Isotope Laboratories | Cat#DLM-407-1 |
| Urea-(13C,15N2) | Cambridge Isotope Laboratories | Cat#CLM-234-0.5 |
| L-lactate-13C3 | Sigma-Aldrich | Cat#485926-500MG |
| Trimethylamine N-oxide-d9 | Cambridge Isotope Laboratories | Cat#DLM-4779-1 |
| Choline-d10 | Cambridge Isotope Laboratories | Cat#DLM-141-0.1 |
| Malic acid-d3 | Cambridge Isotope Laboratories | Cat#DLM-9045-0.1 |
| Citric acid-d4 | Cambridge Isotope Laboratories | Cat#DLM-3487-0.5 |
| Succinic acid-d4 | Cambridge Isotope Laboratories | Cat#DLM-584-1 |
| Fumaric acid-d4 | Cambridge Isotope Laboratories | Cat#DLM-7654-1 |
| Hypoxanthine-d3 | Cambridge Isotope Laboratories | Cat#DLM-2923-0.1 |
| Xanthine-15N2 | Cambridge Isotope Laboratories | Cat#NLM-1698-0.1 |
| Thymidine (13C10,15N2) | Cambridge Isotope Laboratories | Cat#CNLM-3902-25 |
| Inosine-15N4 | Cambridge Isotope Laboratories | Cat#NLM-4264-0.01 |
| Cytidine-13C5 | Cambridge Isotope Laboratories | Cat#CLM-3679-0.05 |
| Uridine-d2 | Cambridge Isotope Laboratories | Cat#DLM-7693-0.05 |
| Methylsuccinic acid-d6 | Cambridge Isotope Laboratories | Cat#DLM-2960-1 |
| Benzoic acid-d5 | Cambridge Isotope Laboratories | Cat#DLM-122-1 |
| Creatine-d3 | Cambridge Isotope Laboratories | Cat#DLM-1302-0.25 |
| Creatinine-d3 | Cambridge Isotope Laboratories | Cat#DLM-3653-0.1 |
| Glutaric acid-d4 | Cambridge Isotope Laboratories | Cat#DLM-3106-5 |
| Glycine-d | Cambridge Isotope Laboratories | Cat# DLM-1674-5 |
| Kynurenic acid-d5 | Cambridge Isotope Laboratories | Cat#DLM-7374-PK |
| L-Citrulline-d4 | Cambridge Isotope Laboratories | Cat#DLM-6039-0.01 |
| L-Threonine-(13C4,15N) | Cambridge Isotope Laboratories | Cat#CNLM-587-0.1 |
| L-Tyrosine-d7 | Cambridge Isotope Laboratories | Cat#DLM-589-0.05 |
| P-cresol sulfate-d7 | Cambridge Isotope Laboratories | Cat#DLM-9786-0.01 |
| Sarcosine-d3 | Cambridge Isotope Laboratories | Cat#DLM-6874-0.1 |
| Trans-4-hydroxy-L-proline-d3 | Cambridge Isotope Laboratories | Cat#DLM-9778-PK |
| Uric acid-(13C; 15N3) | Cambridge Isotope Laboratories | Cat#CNLM-10617-0.001 |
| Critical Commercial Assays | ||
| Invitrogen total exosome isolation kit | Thermo fisher | Cat#4478360 |
| Pierce BCA protein assay kit | Thermo fisher | Cat#23225 |
| BD Multitest CD3/CD8/CD45/CD4 | BD Bioscience | Cat#340499 |
| BD Trucount Tubes | BD Bioscience | Cat#340334 |
| Deposited Data | ||
| Lipidomics and metabolomics raw datasets | This paper | http://dx.doi.org/10.17632/6z5zkzb3hv.3 |
| Software and Algorithms | ||
| RX64-3.6.1 | R Foundation for Statistical Computing | https://www.r-project.org/ |
| Analyst 1.6.3 | Sciex | https://sciex.com/products/software/analyst-software |
| MarkerView 1.3 | Sciex | https://sciex.com/products/software/markerview-software |
| PeakView 2.2 | Sciex | https://sciex.com/products/software/peakview-software |
| Other | ||
| K2 EDTA tube (10 ml) | BD Vacutainer | Cat# 367525 |
| Luna Silica 3 μm column | Phenomenex | Cat#00F-4162-b0 |
| Kinetex-C18 2.6 μm column | Phenomenex | Cat#00D-4462-e0 |
| ACQUITY UPLC HSS T3 1.8 μm column | Waters | Cat#186004680 |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Guanghou Shui (ghshui@genetics.ac.cn).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
These data are available at the Elsevier’s open research data repository website, Mendeley Data (https://data.mendeley.com). The data can be accessed directly via the dataset DOI: http://dx.doi.org/10.17632/6z5zkzb3hv.3.
Experimental Model and Subject Details
Study Participants and Data Collection
We retrospectively recruited a total of 50 patients with COVID-19 from January 22 to February 16, 2020, at the Fifth Medical Center of PLA General Hospital. All enrolled patients were confirmed to be positive for SARS-CoV-2 nucleic acid by real-time polymerase chain reaction. Twenty-six healthy individuals comprising doctors, nurses and researchers also stationed in the same hospital campus during the sample collection period were included as controls. The severity of COVID-19 was judged according to the guidelines for the diagnosis and management of COVID-19 patients (7th edition) by National Health Commission of China. Briefly, mild group (n = 18) included those present mild symptoms without pneumonia; moderate group (n = 19) present with fever, respiratory symptoms, and pneumonia; severe group (n = 13) included severe and critical ill cases. Severe cases are characterized by dyspnea, respiratory frequency ≥30/minute, blood oxygen saturation ≤93%, PaO2/FiO2 ratio < 300, and/or lung infiltrates greater than 50% within 24 to 48 h in pulmonary imaging. Critical ill cases refer to individuals that exhibited respiratory failure and required mechanical ventilation, as well as septic shock, and/or multiple organ dysfunction/failure that required monitoring and treatment in ICU. Our records indicated 2 out of 13 severe patients were on mechanical ventilation at the point of blood sample collection. Demographic, clinical, and laboratory radiological data were extracted from electronic medical records. Baseline characteristics and laboratory findings of COVID-19 patients and healthy controls were summarized in Tables 1 and S1–S3. The admission data of these patients were collected and checked independently by two physicians. The study was performed in accordance with the Declaration of Helsinki principle for ethical research. The study protocol was approved by Ethics Committee of the Fifth Medical Center of PLA General Hospital. Written informed consent was waived by the Ethics Committee of the designated hospital for emerging infectious disease.
Method Details
Plasma Collection and Metabolite Extraction
All blood samples used in this study were collected after an overnight fast. Peripheral blood samples from patients were collected within 24 h upon hospital admission. For patients admitted in the evening, blood samples were taken on the next morning (0500-0600) before breakfast (0700). For patients admitted during daytime, blood samples were collected only on the next morning (0500-0600) before breakfast (0700) (i.e., after one night in the hospital). Throughout the hospitalization period, patients were provided two standard meals per day, scheduled at 0700 and 1700, respectively. Blood was collected in BD Vacutainer (BD 367525). Plasma was separated by centrifugation at 2000 rpm for 10 min. Lipids and metabolites were extracted according to a modified version of the Bligh and Dyer’s protocol (Lu et al., 2019). Plasma (100 μL) for metabolomics and lipidomics analyses was inactivated via the addition of 750 μL of ice-cold chloroform: methanol (1:2) (v/v). Samples were vortexed for 15 s and then incubated for 1 h at 1500 rpm at 4°C. At the end of incubation, 250 μL of ice-cold chloroform and 350 μL of ice-cold MilliQ water were added. Samples were vortexed for 15 s and put on ice for 1 min. This step was repeated once. Samples were then centrifuged at 12 000 rpm for 5 min 4°C to induce phase separation. The lower organic phase was first extracted to a new tube. Then, another 450 μL of ice-cold chloroform was added to the remaining aqueous/methanol phase. Samples were vortexed briefly for 15 s and put on ice for 1 min, and centrifuged at 12 000 rpm for 5 min 4°C. The lower organic phase was extracted and pooled together with the first round organic extract. Double rounds of extraction ensured a better recovery and reduce variations across samples. The remaining aqueous/methanol phase was then centrifuged at 12 000 rpm for 5 min 4°C, and clean supernatant containing polar metabolites were extracted and transferred to new tube. The organic phase was dried in the SpeedVac under OH mode, while aqueous phase was dried under H2O mode. The dried metabolite extracts were shipped on dry ice to the designated laboratory for lipidomics and metabolomics analysis.
Exosome Isolation and Lipid Extraction
Exosomes were isolated from 100 μL of plasma using Invitrogen total exosome isolation kit (Thermofisher Scientific) according to the manufacturer’s protocol. The isolated exosome pellet was resuspended in 100 μL of ice-cold PBS and dispersed completely by repeatedly pipetting up and down. Following this, 750 μL of ice-cold chloroform: methanol (1:2) (v/v) was added to inactivate the samples. Lipid were then extracted in identical steps as described above for plasma samples, and organic extracts pooled from two rounds of extractions were used for lipidomics analysis. The remaining aqueous/methanol phase containing the extracted pellet was dried in the Speedvac under H2O mode. Proteins were extracted from the dried pellet using RIPA lysis buffer with protease inhibitor cocktail (Sigma-Aldrich), and total protein content was determined using Pierce BCA protein assay kit (Thermofisher Scientific) according to the manufacturer’s instructions.
Targeted Lipidomics
Prior to analysis, plasma lipid extracts were resuspended in 100 μL of chloroform: methanol 1:1 (v/v) spiked with appropriate concentrations of internal standards. All lipidomic analyses were carried out on an Exion UPLC coupled with a SCIEX QTRAP 6500 PLUS system as described previously, using an extensive, targeted library tailored for human serum lipidome that confers sufficient lipid coverage to render global lipid pathway analysis (Lam et al., 2014; Lu et al., 2019). All quantification experiments were conducted using internal standard calibration. In brief, polar lipids were separated on a Phenomenex Luna Silica 3 μm column (i.d. 150 × 2.0 mm) under the following chromatographic conditions: mobile phase A (chloroform:methanol:ammonium hydroxide, 89.5:10:0.5) and mobile phase B (chloroform:methanol: ammonium hydroxide: water, 55:39:0.5:5.5) at a flow rate of 270 μL/min and column oven temperature at 25°C. The gradient started with 5% of B and was held for 3 min, which was then increased to 40% of B over 9 min, and was held at 40% for 4 min before further increasing to 70% B over 5 min. The gradient was maintained at 70% B for 15 min before returning to 5% B over 3 min, and was finally equilibrated for 6 min. Individual polar lipid species were quantified by referencing to spiked internal standards of the same lipid class including PC-14:0/14:0, d31-PC16:0/18:1, PE14:0/14:0, d31-PE-16:0/18:1, d31-PS-16:0/18:1, PA-17:0/17:0, PG-14:0/14:0, d31-PG-16:0/18:1, C14:0-BMP, d31-PI-16:0/18:1, SM-d18:1/12:0, LPC-17:0, LPE-17:1, LPI-17:1, LPA-17:0, LPS-17:1, S1P-d17:1, Cer-d18:1/d7-15:0, GluCer d18:1/8:0, GalCer d18:1/8:0 obtained from Avanti Polar Lipids (AL, USA) and PI-8:0/8:0 from Echelon Biosciences, Inc. (UT, USA). d3-GM3 d18:1/18:0 and d3-LacCer d18:1/16:0 were from Matreya LLC (PA, USA). Glycerol lipids including diacylglycerols (DAGs) and triacylglycerols (TAGs) were quantified using a modified version of reverse phase LC/MRM. Separation of neutral lipids were achieved on a Phenomenex Kinetex-C18 2.6 μm column (i.d. 4.6x100 mm) using an isocratic mobile phase containing chloroform:methanol:0.1 M ammonium acetate 100:100:4 (v/v/v) at a flow rate of 300 μL for 10 min. Levels of short-, medium-, and long-chain TAGs were calculated by referencing to spiked internal standards of TAG(14:0)3-d5, TAG(16:0)3-d5 and TAG(18:0)3-d5 obtained from CDN isotopes (Quebec, Canada), respectively. DAGs were quantified using d5-DAG16:0/16:0 and d5-DAG18:1/18:1 as internal standards from Avanti Polar Lipids (Shui et al., 2010). Free cholesterols and cholesteryl esters were analyzed as described previously with d6-cholesterol and d6-CE18:0 cholesteryl ester (CE) (CDN isotopes) as internal standards (Shui et al., 2011). Lipid levels were expressed in nanomoles per L (nmol/L) for plasma, and in nanomoles lipids per g of total protein (nmol/g) for exosomes.
Untargeted Metabolomics
Prior to analysis, aqueous extracts were resuspended in 100 μL of 2% acetonitrile in water. Chromatographic separation was performed on a reversed-phase ACQUITY UPLC HSS T3 1.8 μm column (i.d. 3.0 × 100 mm) (Waters) using an UPLC system (Agilent 1290 Infinity II; Agilent Technologies) as described previously (Tian et al., 2020). MS detection was performed using high-resolution time-of-flight (TOF) mass spectrometry (5600 Triple TOF Plus, Sciex) equipped with an ESI source (Yuan et al., 2012). Data were acquired in TOF full scan method with positive and negative ion modes, respectively. Information-dependent acquisition methods were used for MS/MS analyses of metabolome. The collision energy was set at 35 ± 15 eV. Metabolite identification was compared with standard references, HMDB (https://hmdb.ca/), METLIN (https://metlin.scripps.edu), and literature searches. A total of 45 isotopically-labeled internal standards (IS), purchased from Cambridge Isotope Laboratories, were spiked into the samples for metabolite quantitation, including L-Phenylalanine-d8, L-Tryptophan-d8, L-Isoleucine-d10, L-Asparagine-13C4, L-Methionine-d3, L-Valine-d8, L-Proline-d7, L-Alanine-d7, DL-Serine-d3, DL-Glutamic acid-d5, L-Aspartic acid-d3, L-Arginine-d7, L-Glutamine-d5, L-Lysine-d9, L-Histidine-d5, Taurine-d2, Betaine-d11, Urea-(13C,15N2), L-lactate-13C3, Trimethylamine N-oxide-d9, Choline-d13, Malic acid-d3, Citric acid-d4, Succinic acid-d4, Fumaric acid-d4, Hypoxanthine-d3, Xanthine-15N2, Thymidine (13C10,15N2), Inosine-15N4, Cytidine-13C5, Uridine-d2, Methylsuccinic acid-d6, Benzoic acid-d5, Creatine-d3, Creatinine-d3, Glutaric acid-d4, Glycine-d2, Kynurenic acid-d5, L-Citrulline-d4, L-Threonine-(13C4,15N), L-Tyrosine-d7, P-cresol sulfate-d7, Sarcosine-d3, Trans-4-hydroxy-L-proline-d3, Uric acid-(13C; 15N3). Metabolite levels were normalized according to the following rules (1) ISs were applied to correct peak areas of their corresponding metabolites; (2) When (1) was not feasible due to unavailability of commercial standards, peak areas were corrected with IS of metabolites of the same class, comparable peak intensities, and/or proximity in retention times; (3) Results from Step (2) were evaluated based on relative standard deviation (RSD) values of each metabolite before and after IS correction. Corrected peak areas were adopted if their corresponding RSDs were smaller than that of original areas in quality control samples. For simplicity, all metabolite levels were labeled as “intensity.”
Quantification and Statistical Analysis
Metabolite Panel for Identifying COVID-19
To generate a plasma metabolite panel for differentiating COVID-19 patients from healthy individuals, variables with p < 0.05 between healthy controls and COVIDP19 patients after adjustment for age, sex and BMI were sieved out to form a starting pool. From this pool, a starting variable with lowest p value was added to Set 1, and remaining variables from the starting pool significantly correlated (p < 0.05) with this starting variable were added together to form Set 1. The process then was repeated in an iterative process using starting variable with the second lowest p value, and so on, finally generating a total of ten established sets. Representative metabolite from each established set was chosen based on (1) smallest p value and (2) reported biological function through a PubMed search. The selection process finally created a panel of ten plasma metabolites, and its performance was evaluated in a logistic regression model with leave-one-out (LOO) cross-validation, which distinguished between COVID-19 patients and healthy controls with an area under curve (AUC) = 0.955.
Logistic Regression Analysis
Logistic regression model with covariates BMI, age and sex were built with each variable (lipid/metabolite) to search for significant variables that can predict the conditions (i.e., healthy control, mild, moderate, severe COVID-19) of subjects. The p value of the variable estimate in the model was extracted and those with false discovery rate (fdr) smaller than 0.05 were shortlisted. Forest plots of these significant variables were constructed and the boxplots of individual significant variables were generated. A list of 61 lipids and 89 metabolites were found to be significant (fdr < 0.05) in their respective logistic regression models. Forest plots illustrate the magnitude of estimates on signed log x axis, with indicator of significance of the estimate in the model, with ∗∗∗ representing p < 0.001, ∗∗ representing p < 0.01 and ∗ representing p < 0.05. For non-significant species, the estimates were plotted as zeros.
Spearman Correlation Analysis
Spearman correlation between variables and clinical indicators were performed with data from COVID-19 patients. For each pair of variable and clinical indicator, samples with missing clinical data were omitted from the calculations. Correlation plots for all variables grouped by metabolite class were presented. Only correlations with p < 0.05 were indicated with colored circles. Negative correlations were shown in red and positive correlations were shown in blue, with sizes of circles representing the magnitude of the correlations.
Differential Correlation Analysis
MEGENA R package was used to build correlation networks from differentially correlated lipid pairs in mild COVID-19 relative to healthy controls to reveal changes in lipid co-regulation upon early infection. Differential correlation was calculated using R package DGCA. Only lipid pairs with differential correlation (empirical p < 0.05) were included for analyses.
Acknowledgments
We thank all patients and healthy individuals involved in this study, as well as the dedicated medical and research staff who fight against SARS-CoV-2 worldwide. This work was supported by grants from the Innovative Research Team of the National Natural Science Foundation of China (81721002 to F.-S.W.), and the National Key R&D Program of China (2018YFA0800901 and 2018YFA0506902 to G.S.).
Author Contributions
F.-S.W., G.S., and J.-Y.Z. conceived, designed, and supervised experiments. J.-W.S., X.F., P.X., C.Z., and T.Y. collected clinical samples. W.-J.C., S.-Y.W., F.-P.M., Z.X., J.-L.F., and L.H. collected clinical information. J.-W.S., S.M.L., X.F., H.T., S.Z., R.W., and Z.W. performed the experiments. J.-W.S., W.-J.C., G.H.C., and B.L. performed the statistical analysis. J.-W.S., S.M.L., G.H.C., T.-J.J., M.S., F.-S.W., J.-Y.Z., and G.S. interpreted data. S.M.L. and J.-W.S. wrote the manuscript. All authors read and approved the final manuscript.
Declaration of Interests
S.M.L., G.H.C., and B.L. are employees of LipidALL Technologies.
Published: June 24, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.cmet.2020.06.016.
Contributor Information
Ji-Yuan Zhang, Email: uniquezjy@163.com.
Fu-Sheng Wang, Email: fswang302@163.com.
Guanghou Shui, Email: ghshui@genetics.ac.cn.
Supplemental Information
References
- Baez J.M., Barbour S.E., Cohen D.E. Phosphatidylcholine transfer protein promotes apolipoprotein A-I-mediated lipid efflux in Chinese hamster ovary cells. J. Biol. Chem. 2002;277:6198–6206. doi: 10.1074/jbc.M106799200. [DOI] [PubMed] [Google Scholar]
- Baranano D.E., Rao M., Ferris C.D., Snyder S.H. Biliverdin reductase: a major physiologic cytoprotectant. Proc. Natl. Acad. Sci. USA. 2002;99:16093–16098. doi: 10.1073/pnas.252626999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhowmick R., Clark S., Bonventre J.V., Leong J.M., McCormick B.A. Cytosolic phospholipase A2α promotes pulmonary inflammation and systemic disease during Streptococcus pneumoniae infection. Infect. Immun. 2017;85 doi: 10.1128/IAI.00280-17. e00280-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bley H., Schöbel A., Herker E. Whole lotta lipids-from HCV RNA replication to the mature viral particle. Int. J. Mol. Sci. 2020;21:2888. doi: 10.3390/ijms21082888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brügger B., Glass B., Haberkant P., Leibrecht I., Wieland F.T., Kräusslich H.G. The HIV lipidome: a raft with an unusual composition. Proc. Natl. Acad. Sci. USA. 2006;103:2641–2646. doi: 10.1073/pnas.0511136103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bursten S.L., Federighi D.A., Parsons P., Harris W.E., Abraham E., Moore E.E., Jr., Moore F.A., Bianco J.A., Singer J.W., Repine J.E. An increase in serum C18 unsaturated free fatty acids as a predictor of the development of acute respiratory distress syndrome. Crit. Care Med. 1996;24:1129–1136. doi: 10.1097/00003246-199607000-00011. [DOI] [PubMed] [Google Scholar]
- Chahar H.S., Bao X., Casola A. Exosomes and their role in the life cycle and pathogenesis of RNA viruses. Viruses. 2015;7:3204–3225. doi: 10.3390/v7062770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R., Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin. Immunopathol. 2017;39:529–539. doi: 10.1007/s00281-017-0629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapuy-Regaud S., Subra C., Requena M., de Medina P., Amara S., Delton-Vandenbroucke I., Payre B., Cazabat M., Carriere F., Izopet J. Progesterone and a phospholipase inhibitor increase the endosomal bis(monoacylglycero)phosphate content and block HIV viral particle intercellular transmission. Biochimie. 2013;95:1677–1688. doi: 10.1016/j.biochi.2013.05.019. [DOI] [PubMed] [Google Scholar]
- Chevallier J., Chamoun Z., Jiang G., Prestwich G., Sakai N., Matile S., Parton R.G., Gruenberg J. Lysobisphosphatidic acid controls endosomal cholesterol levels. J. Biol. Chem. 2008;283:27871–27880. doi: 10.1074/jbc.M801463200. [DOI] [PubMed] [Google Scholar]
- Cochran F.R., Roddick V.L., Connor J.R., Thornburg J.T., Waite M. Regulation of arachidonic acid metabolism in resident and BCG-activated alveolar macrophages: role of lyso(bis)phosphatidic acid. J. Immunol. 1987;138:1877–1883. [PubMed] [Google Scholar]
- Cole L.K., Vance J.E., Vance D.E. Phosphatidylcholine biosynthesis and lipoprotein metabolism. Biochim. Biophys. Acta. 2012;1821:754–761. doi: 10.1016/j.bbalip.2011.09.009. [DOI] [PubMed] [Google Scholar]
- Cosset F.L., Dreux M. HCV transmission by hepatic exosomes establishes a productive infection. J. Hepatol. 2014;60:674–675. doi: 10.1016/j.jhep.2013.10.015. [DOI] [PubMed] [Google Scholar]
- Dashti M., Kulik W., Hoek F., Veerman E.C., Peppelenbosch M.P., Rezaee F. A phospholipidomic analysis of all defined human plasma lipoproteins. Sci. Rep. 2011;1:139. doi: 10.1038/srep00139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisfeld A.J., Halfmann P.J., Wendler J.P., Kyle J.E., Burnum-Johnson K.E., Peralta Z., Maemura T., Walters K.B., Watanabe T., Fukuyama S. Multi-platform 'omics analysis of human Ebola virus disease pathogenesis. Cell Host Microbe. 2017;22:817–829.e8. doi: 10.1016/j.chom.2017.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grothe J., Riethmüller J., Tschürtz S.M., Raith M., Pynn C.J., Stoll D., Bernhard W. Plasma phosphatidylcholine alterations in cystic fibrosis patients: impaired metabolism and correlation with lung function and inflammation. Cell. Physiol. Biochem. 2015;35:1437–1453. doi: 10.1159/000373964. [DOI] [PubMed] [Google Scholar]
- Gruenberg J., Stenmark H. The biogenesis of multivesicular endosomes. Nat. Rev. Mol. Cell Biol. 2004;5:317–323. doi: 10.1038/nrm1360. [DOI] [PubMed] [Google Scholar]
- Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X., Liu L., Shan H., Lei C.L., Hui D.S.C., China Medical Treatment Expert Group for Covid-19 Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X. Lipidomics for studying metabolism. Nat. Rev. Endocrinol. 2016;12:668–679. doi: 10.1038/nrendo.2016.98. [DOI] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong W. Combating COVID-19 with chloroquine. J. Mol. Cell Biol. 2020;12:249–250. doi: 10.1093/jmcb/mjaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hullin-Matsuda F., Luquain-Costaz C., Bouvier J., Delton-Vandenbroucke I. Bis(monoacylglycero)phosphate, a peculiar phospholipid to control the fate of cholesterol: implications in pathology. Prostaglandins Leukot. Essent. Fatty Acids. 2009;81:313–324. doi: 10.1016/j.plefa.2009.09.006. [DOI] [PubMed] [Google Scholar]
- Izquierdo-Useros N., Naranjo-Gómez M., Erkizia I., Puertas M.C., Borràs F.E., Blanco J., Martinez-Picado J. HIV and mature dendritic cells: Trojan exosomes riding the Trojan horse? PLoS Pathog. 2010;6:e1000740. doi: 10.1371/journal.ppat.1000740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi J.C., Joshi B., Rochford I., Rayees S., Akhter M.Z., Baweja S., Chava K.R., Tauseef M., Abdelkarim H., Natarajan V. SPHK2-generated S1P in CD11b+ macrophages blocks STING to suppress the inflammatory function of alveolar macrophages. Cell Rep. 2020;30:4096–4109.e5. doi: 10.1016/j.celrep.2020.02.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Stang E., Fang K.S., de Moerloose P., Parton R.G., Gruenberg J. A lipid associated with the antiphospholipid syndrome regulates endosome structure and function. Nature. 1998;392:193–197. doi: 10.1038/32440. [DOI] [PubMed] [Google Scholar]
- Kyle J.E., Burnum-Johnson K.E., Wendler J.P., Eisfeld A.J., Halfmann P.J., Watanabe T., Sahr F., Smith R.D., Kawaoka Y., Waters K.M., Metz T.O. Plasma lipidome reveals critical illness and recovery from human Ebola virus disease. Proc. Natl. Acad. Sci. USA. 2019;116:3919–3928. doi: 10.1073/pnas.1815356116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam S.M., Tong L., Duan X., Petznick A., Wenk M.R., Shui G. Extensive characterization of human tear fluid collected using different techniques unravels the presence of novel lipid amphiphiles. J. Lipid Res. 2014;55:289–298. doi: 10.1194/jlr.M044826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam S.M., Tian H., Shui G. Lipidomics, en route to accurate quantitation. Biochim Biophys Acta Mol Cell Biol Lipids. 2017;1862:752–761. doi: 10.1016/j.bbalip.2017.02.008. [DOI] [PubMed] [Google Scholar]
- Laulagnier K., Motta C., Hamdi S., Roy S., Fauvelle F., Pageaux J.F., Kobayashi T., Salles J.P., Perret B., Bonnerot C., Record M. Mast cell- and dendritic cell-derived exosomes display a specific lipid composition and an unusual membrane organization. Biochem. J. 2004;380:161–171. doi: 10.1042/BJ20031594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechien J.R., Chiesa-Estomba C.M., De Siati D.R., Horoi M., Le Bon S.D., Rodriguez A., Dequanter D., Blecic S., El Afia F., Distinguin L. Olfactory and gustatory dysfunctions as a clinical presentation of mild-to-moderate forms of the coronavirus disease (COVID-19): a multicenter European study. Eur. Arch. Otorhinolaryngol. 2020 doi: 10.1007/s00405-020-05965-1. Published online April 6, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Abston E., Zhang D., Rai A., Jin Y. Extracellular vesicle: an emerging mediator of intercellular crosstalk in lung inflammation and injury. Front. Immunol. 2018;9:924. doi: 10.3389/fimmu.2018.00924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linke T., Wilkening G., Lansmann S., Moczall H., Bartelsen O., Weisgerber J., Sandhoff K. Stimulation of acid sphingomyelinase activity by lysosomal lipids and sphingolipid activator proteins. Biol. Chem. 2001;382:283–290. doi: 10.1515/BC.2001.035. [DOI] [PubMed] [Google Scholar]
- Lu J., Lam S.M., Wan Q., Shi L., Huo Y., Chen L., Tang X., Li B., Wu X., Peng K. High-coverage targeted lipidomics reveals novel serum lipid predictors and lipid pathway dysregulation antecedent to type 2 diabetes onset in normoglycemic Chinese adults. Diabetes Care. 2019;42:2117–2126. doi: 10.2337/dc19-0100. [DOI] [PubMed] [Google Scholar]
- Maile M.D., Standiford T.J., Engoren M.C., Stringer K.A., Jewell E.S., Rajendiran T.M., Soni T., Burant C.F. Associations of the plasma lipidome with mortality in the acute respiratory distress syndrome: a longitudinal cohort study. Respir. Res. 2018;19:60. doi: 10.1186/s12931-018-0758-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason R.J., Stossel T.P., Vaughan M. Lipids of alveolar macrophages, polymorphonuclear leukocytes, and their phagocytic vesicles. J. Clin. Invest. 1972;51:2399–2407. doi: 10.1172/JCI107052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelucci A., Cordes T., Ghelfi J., Pailot A., Reiling N., Goldmann O., Binz T., Wegner A., Tallam A., Rausell A. Immune-responsive gene 1 protein links metabolism to immunity by catalyzing itaconic acid production. Proc. Natl. Acad. Sci. USA. 2013;110:7820–7825. doi: 10.1073/pnas.1218599110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmerjahn F., Anthony R.M., Ravetch J.V. Agalactosylated IgG antibodies depend on cellular Fc receptors for in vivo activity. Proc. Natl. Acad. Sci. USA. 2007;104:8433–8437. doi: 10.1073/pnas.0702936104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrier A., Bonnin A., Desmarets L., Danneels A., Goffard A., Rouillé Y., Dubuisson J., Belouzard S. The C-terminal domain of the MERS coronavirus M protein contains a trans-Golgi network localization signal. J. Biol. Chem. 2019;294:14406–14421. doi: 10.1074/jbc.RA119.008964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin C., Zhou L., Hu Z., Zhang S., Yang S., Tao Y., Xie C., Ma K., Shang K., Wang W., Tian D.S. Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa248. Published online March 12, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queiroz A., Pinto I.F.D., Lima M., Giovanetti M., de Jesus J.G., Xavier J., Barreto F.K., Canuto G.A.B., do Amaral H.R., de Filippis A.M.B. Lipidomic analysis reveals serum alteration of plasmalogens in patients infected with ZIKA virus. Front. Microbiol. 2019;10:753. doi: 10.3389/fmicb.2019.00753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabbani M.A.G., Barik S. 5-hydroxytryptophan, a major product of tryptophan degradation, is essential for optimal replication of human parainfluenza virus. Virology. 2017;503:46–51. doi: 10.1016/j.virol.2017.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhoff R., Sandhoff K. Emerging concepts of ganglioside metabolism. FEBS Lett. 2018;592:3835–3864. doi: 10.1002/1873-3468.13114. [DOI] [PubMed] [Google Scholar]
- Schauer R. Sialic acids as regulators of molecular and cellular interactions. Curr. Opin. Struct. Biol. 2009;19:507–514. doi: 10.1016/j.sbi.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz G., Müller G. Structure and function of lamellar bodies, lipid-protein complexes involved in storage and secretion of cellular lipids. J. Lipid Res. 1991;32:1539–1570. [PubMed] [Google Scholar]
- Shan J., Qian W., Shen C., Lin L., Xie T., Peng L., Xu J., Yang R., Ji J., Zhao X. High-resolution lipidomics reveals dysregulation of lipid metabolism in respiratory syncytial virus pneumonia mice. RSC Advances. 2018;8:29368–29377. doi: 10.1039/c8ra05640d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenoy G.N., Loyall J., Berenson C.S., Kelleher R.J., Jr., Iyer V., Balu-Iyer S.V., Odunsi K., Bankert R.B. Sialic acid-dependent inhibition of T cells by exosomal ganglioside GD3 in ovarian tumor microenvironments. J. Immunol. 2018;201:3750–3758. doi: 10.4049/jimmunol.1801041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shui G., Guan X.L., Low C.P., Chua G.H., Goh J.S., Yang H., Wenk M.R. Toward one step analysis of cellular lipidomes using liquid chromatography coupled with mass spectrometry: application to Saccharomyces cerevisiae and Schizosaccharomyces pombe lipidomics. Mol. Biosyst. 2010;6:1008–1017. doi: 10.1039/b913353d. [DOI] [PubMed] [Google Scholar]
- Shui G., Cheong W.F., Jappar I.A., Hoi A., Xue Y., Fernandis A.Z., Tan B.K., Wenk M.R. Derivatization-independent cholesterol analysis in crude lipid extracts by liquid chromatography/mass spectrometry: applications to a rabbit model for atherosclerosis. J. Chromatogr. A. 2011;1218:4357–4365. doi: 10.1016/j.chroma.2011.05.011. [DOI] [PubMed] [Google Scholar]
- Stoorvogel W., Kleijmeer M.J., Geuze H.J., Raposo G. The biogenesis and functions of exosomes. Traffic. 2002;3:321–330. doi: 10.1034/j.1600-0854.2002.30502.x. [DOI] [PubMed] [Google Scholar]
- Subra C., Laulagnier K., Perret B., Record M. Exosome lipidomics unravels lipid sorting at the level of multivesicular bodies. Biochimie. 2007;89:205–212. doi: 10.1016/j.biochi.2006.10.014. [DOI] [PubMed] [Google Scholar]
- Tam V.C., Quehenberger O., Oshansky C.M., Suen R., Armando A.M., Treuting P.M., Thomas P.G., Dennis E.A., Aderem A. Lipidomic profiling of influenza infection identifies mediators that induce and resolve inflammation. Cell. 2013;154:213–227. doi: 10.1016/j.cell.2013.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry C., Boussac M., Véron P., Ricciardi-Castagnoli P., Raposo G., Garin J., Amigorena S. Proteomic analysis of dendritic cell-derived exosomes: a secreted subcellular compartment distinct from apoptotic vesicles. J. Immunol. 2001;166:7309–7318. doi: 10.4049/jimmunol.166.12.7309. [DOI] [PubMed] [Google Scholar]
- Tian H., Zhou Z., Shui G., Lam S.M. Extensive profiling of polyphenols from two Trollius species using a combination of untargeted and targeted approaches. Metabolites. 2020;10:119. doi: 10.3390/metabo10030119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tooze J., Tooze S.A., Fuller S.D. Sorting of progeny coronavirus from condensed secretory proteins at the exit from the trans-Golgi network of AtT20 cells. J. Cell Biol. 1987;105:1215–1226. doi: 10.1083/jcb.105.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers K.C., Palmisano B.T., Shoucri B.M., Shamburek R.D., Remaley A.T. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 2011;13:423–433. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams G. Database of Gene Co-Regulation (dGCR): a web tool for analysing patterns of gene co-regulation across publicly available expression data. J Genomics. 2015;3:29–35. doi: 10.7150/jgen.10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wubbolts R., Leckie R.S., Veenhuizen P.T., Schwarzmann G., Möbius W., Hoernschemeyer J., Slot J.W., Geuze H.J., Stoorvogel W. Proteomic and biochemical analyses of human B cell-derived exosomes. Potential implications for their function and multivesicular body formation. J. Biol. Chem. 2003;278:10963–10972. doi: 10.1074/jbc.M207550200. [DOI] [PubMed] [Google Scholar]
- Xu F., Bandara A., Akiyama H., Eshaghi B., Stelter D., Keyes T., Straub J.E., Gummuluru S., Reinhard B.M. Membrane-wrapped nanoparticles probe divergent roles of GM3 and phosphatidylserine in lipid-mediated viral entry pathways. Proc. Natl. Acad. Sci. USA. 2018;115:E9041–E9050. doi: 10.1073/pnas.1804292115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C., Liu S., Zhao P., Liu H., Zhu L. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Feizpour A., Ramirez N.G., Wu L., Akiyama H., Xu F., Gummuluru S., Reinhard B.M. Glycosphingolipid-functionalized nanoparticles recapitulate CD169-dependent HIV-1 uptake and trafficking in dendritic cells. Nat. Commun. 2014;5:4136. doi: 10.1038/ncomms5136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Xu F., Ramirez N.G., Kijewski S.D., Akiyama H., Gummuluru S., Reinhard B.M. Dressing up nanoparticles: a membrane wrap to induce formation of the virological synapse. ACS Nano. 2015;9:4182–4192. doi: 10.1021/acsnano.5b00415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Breitkopf S.B., Yang X., Asara J.M. A positive/negative ion-switching, targeted mass spectrometry-based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nat. Protoc. 2012;7:872–881. doi: 10.1038/nprot.2012.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., He G., Filipowicz N.A., Randall G., Belov G.A., Kopek B.G., Wang X. Host lipids in positive-strand RNA virus genome replication. Front. Microbiol. 2019;10:286. doi: 10.3389/fmicb.2019.00286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Zhao Y., Zhang F., Wang Q., Li T., Liu Z., Wang J., Qin Y., Zhang X., Yan X. The use of anti-inflammatory drugs in the treatment of people with severe coronavirus disease 2019 (COVID-19): the perspectives of clinical immunologists from China. Clin. Immunol. 2020;214:108393. doi: 10.1016/j.clim.2020.108393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou X., Chen K., Zou J., Han P., Hao J., Han Z. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front. Med. 2020;14:185–192. doi: 10.1007/s11684-020-0754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
These data are available at the Elsevier’s open research data repository website, Mendeley Data (https://data.mendeley.com). The data can be accessed directly via the dataset DOI: http://dx.doi.org/10.17632/6z5zkzb3hv.3.