Abstract
In silico RNA-RNA interaction prediction is widely applied to identify putative interaction partners and to assess interaction details in base pair resolution. To verify specific interactions, in vitro evidence can be obtained via compensatory mutation experiments. Unfortunately, the selection of compensatory mutations is non-trivial and typically based on subjective ad hoc decisions. To support the decision process, we introduce our COmPensatOry MUtation Selector CopomuS. CopomuS evaluates the effects of mutations on RNA-RNA interaction formation using a set of objective criteria, and outputs a reliable ranking of compensatory mutation candidates. For RNA-RNA interaction assessment, the state-of-the-art IntaRNA prediction tool is applied. We investigate characteristics of successfully verified RNA-RNA interactions from the literature, which guided the design of CopomuS. Finally, we evaluate its performance based on experimentally validated compensatory mutations of prokaryotic sRNAs and their target mRNAs. CopomuS predictions highly agree with known results, making it a valuable tool to support the design of verification experiments for RNA-RNA interactions. It is part of the IntaRNA package and available as stand-alone webserver for ad hoc application.
Keywords: RNA-RNA interaction, compensatory mutation, mutation, design, sRNA
1. Introduction
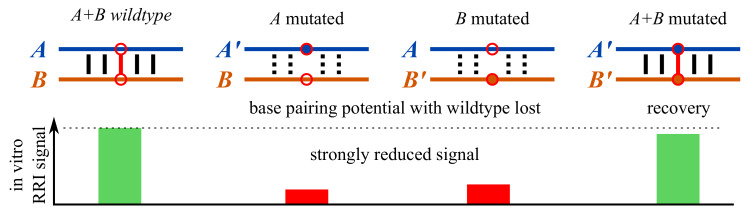
Many non-coding (nc)RNAs, like bacterial small (s)RNAs or eukayotic micro (mi)RNAs, perform their regulatory functions via direct RNA-RNA interaction (RRI) with their target RNAs [1]. This allows the use of in silico RRI prediction approaches to identify potential targets [2,3], which even provide interaction models in base pair resolution. To verify such predictions in vitro, mutation or deletion experiments are conducted [3,4]. Within deletion experiments, whole (potentially interacting) subsequences are removed from one or both RNAs and the in vitro measured interaction potential is compared to results using wildtype sequences. Since deletion of subsequences can have strong side effects, more sophisticated verification experiments are based on compensatory mutations (CoMs). To this end, both potentially interacting RNAs are mutated, such that wildtype-mutant combinations have a reduced base pairing potential within the interaction site that is regained in mutant-only interactions. Often, only a single base pair is mutated, but multiple concurrent CoMs are also used in literature [5]. A depiction of the setup is given in Figure 1. The in vitro RRI potential can be assessed via various experimental protocols [1], e.g., GFP reporter systems [3]. An RRI is considered verified via CoM if the in vitro RRI signal of the wildtype interaction is lost after mutating one sequence, and recovered when both RNAs are mutated (see Figure 1).
Figure 1.
Depiction of an RNA-RNA interaction verification experiment based on compensatory mutations of two RNA sequences A and B. The mutated nucleotides are highlighted by red circles. The lost and regained base pair is given as red line. Black solid lines depict likely formed RRI base pairs, while unlikely base pairs (instable due to reduced RRI) are represented in dotted lines.
The success of a CoM-based RRI verification strongly depends on the mutation selection. If the base-pair-breaking mutation is not weakening the RRI strong enough, no effect is detectable even if the functional RRI is mutated. On the other hand, if the mutation dramatically changes the intra-molecular structure formation of the respective RNA (e.g. when mutating multiple nucleotides at once), reduced RRI signal might be caused by an inaccessibility of the interaction site rather than the loss of base pairing potential. Thus, the design of potent CoMs is typically done manually based on the personal experience of the experimenter.
Here, we introduce CopomuS, a compensatory mutation selector to support this decision process. CopomuS in silico investigates and compares the effect of CoMs on RRI formation to rank candidate CoMs by their verification potential. To this end, IntaRNA [6,7], a state-of-the-art RRI prediction tool [8], is applied. For each CoM, RRI characteristics of the 4 wildtype-mutant sequence combinations are provided. This annotated list of CoMs is sorted by decreasing verification potential (as assessed by CopomuS) and enables an objective subsequent pruning by the experimenter given his/her expert knowledge. That way, CopomuS very much simplifies CoM selection and reliefs the experimenter of the otherwise necessary manual RRI prediction and comparison. Besides the saving of time, users can easily test the effect of different RRI and CoM constraints on the mutations of interest and the results are simple to reproduce. Finally, CopomuS translates the hypothesis behind compensatory mutation experiments into the respective theoretical model. That is, it implements a meta-strategy that checks for and ensures the intended stability difference pattern of wildtype versus mutant combinations.
2. Materials And Methods
2.1. Compensatory Mutations From Literature
To develop and evaluate CopomuS, we first extracted CoMs of successfully verified RRIs from literature, focusing on sRNA-target RRIs where sRNA binding inhibits translation of the target mRNA. The data was extracted from the benchmark set introduced in [9] and comprises both single-nucleotide CoMs (Table A1; 31 RNA pairs) as well as CoMs involving multiple concurrently mutated nucleotides (Table A2; 28 RNA pairs). To model the workflow based on an sRNA target prediction e.g., following [2,3], we use a genomic context around the start codon for each target. All sequences are provided in Table A3, Table A4, Table A5 and Table A6.
2.2. CopomuS Workflow
The central assumptions of CopomuS are that
-
(1)
the (main) regulatory RRI between the RNAs is defined by a single interaction site not interrupted by intra-molecular base pairing and
-
(2)
IntaRNA’s prediction model correctly identifies the important parts of this regulatory RRI.
Only under Assumption-1 we can expect a single CoM to sufficiently alter the RRI potential between the RNAs, since the loss of one site can not be compensated by a concurrently formed second site. A classic example for such a multi-site RRI is the OxyS-fhlA interaction [10]. Furthermore, only under Assumption-2 CopomuS is expected to provide reasonable CoM candidates, since it generates and evaluates them based on the most stable RRIs predicted by IntaRNA.
2.2.1. CoM Generation
Following Assumption-2, CopomuS first computes the most stable RRI that can be formed by the wildtype RNAs using IntaRNA. The (minimal) free energy (MFE) estimate of the RRI computed by IntaRNA provides a stability proxy to assess the RRI potential. That is, the lower the energy the more stable the formed RRI is. Furthermore, the lowest energy RRI is the most likely when assuming a Boltzmann distribution of the energies [11]. The Nearest-Neighbor-model-based energy estimates incorporate both the stability of the inter-molecular base pairing (hybridization energy) and penalty (ED) terms to reflect the accessibility of the interacting subsequences [7]. More precisely, the latter describes the energy needed to break all intra-molecular base pairs that are formed by the RRI’s subsequences [12].
CopomuS supports two modes to select inter-molecular base pairs for CoM candidate generation. Per default, all base pairs of the MFE RRI are considered. In addition, one can extend this set with all base pairs of suboptimal RRIs that can be formed by the subsequences covered by the MFE RRI. This softens the dependency on the accuracy of the reported MFE RRI base-pair pattern if alternative patterns within the same site are possible.
Next, identified base pairs are further filtered given user-defined constraints. For instance, specific base pair types, like AU or GU base pairs, can be filtered. Furthermore, lonely base pairs that can not stack with another base pair on either side can be excluded, since they typically provide only low stability contributions. Similarly, base pairs at putative ends of inter-molecular helices can be removed, since they can only form stackings on one side.
Finally, given the set of base pairs to be considered for mutation, the user can define whether all possible CoMs per base pair are to be considered (i.e., 3 non-compatible mutation alternatives for a given base pair like e.g., GU, GC, AU for UA (Note, CG is omitted since wildtype U can form a base pair with mutant G, which needs to be prevented)) or if the respective CoM candidate is only the ’nucleotide flip’ of the base pair (as often done in literature), i.e., mutating a GC into a CG base pair.
At the end of the CoM generation, CopomuS outputs a list of CoM candidates to be evaluated and ranked.
2.2.2. CoM Characteristics and Ranking
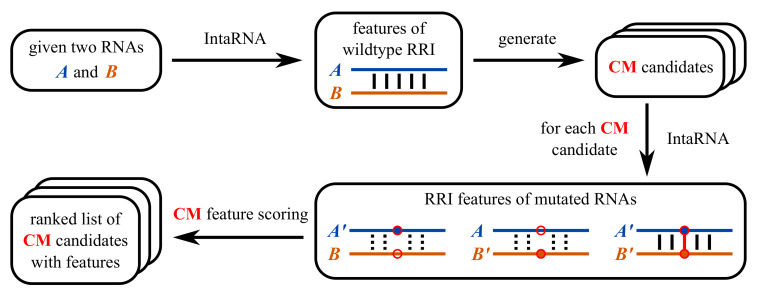
Each CoM candidate is evaluated, using IntaRNA MFE-RRI predictions for all 4 sequence combinations for the current CoM, i.e., wildtype-only, wildtype-mutant as well as mutant-only combinations (see Figure 2). Depending on which ranking the user has specified, various RRI characteristics are aggregated for each combination, e.g., RRI stability in terms of MFE, RRI position, base pairs, or the accessibility of the mutated positions.
Figure 2.
Workflow of CopomuS to generate and rank CoM candidates based on IntaRNA RRI predictions and respective characteristics.
CopomuS implements a hierarchical CoM ranking. To this end, different classification and sorting functions are provided that can be sequentially combined to select for specific CoM characteristics. The most important classifier mfeCover checks whether the CoM is also covered by the mutant-only RRI. If not, the mutant-only RRI was found in a different location or no stable RRI was found at all. Next is the E classifier, which evaluates the desired reduction in RRI potential of wildtype-mutant combinations compared to wildtype-only or mutant-only RRIs. As discussed, the RRI MFE provides a proxy to assess interaction stability. Thus, CopomuS demands that both wildtype-only (ww) and mutant-only (mm) MFE are below zero and both wildtype-mutant MFEs are worse (higher), using two thresholds and , respectively. More formally, MFE(ww) + (MFE(mw),MFE(wm)) and MFE(mm) + (MFE(mw),MFE(wm)) have to be satisfied.
This way, CopomuS can identify CoM candidates that have a high chance to show the expected in vitro RRI signal pattern from Figure 1 needed for RRI verification. The combination and hierarchy of classifier functions already defines a ranking of CoM subsets with decreasing level of constraint satisfaction.
Finally, each subset can be sorted via the minDeltaE function to get a final ranking of CoM candidates. minDeltaE favors CoMs with higher minimal MFE difference between wildtype-mutant combinations to wildtype-only or mutant-only combinations, i.e., it calculates and compares (MFE(mw),MFE(wm)(MFE(ww),MFE(mm))). Therefore, the top-ranked CoM candidate will show the strongest RRI stability reduction for both wildtype-mutant combinations.
2.3. Availability
CopomuS is implemented in Python and part of the IntaRNA package version 3.2.0 or higher (freely available at https://github.com/BackofenLab/IntaRNA/). Due to its modular implementation, it can be easily expanded by further classification or sorting functions. Given its flexible command line interface and the provided CSV-based output, CopomuS can be easily integrated in automated pipelines and workflows. Its webserver (freely available at http://rna.informatik.uni-freiburg.de/CopomuS/) for ad hoc usage is part of the Freiburg RNA tools framework [13] version 4.8.0 or higher.
3. Results
To evaluate the rationals behind CopomuS and to better understand CoM characteristics, we studied RRI characteristics of CoMs from literature. Subsequently, these CoMs were also used to benchmark CopomuS.
3.1. Statistics of CoMs from Literature
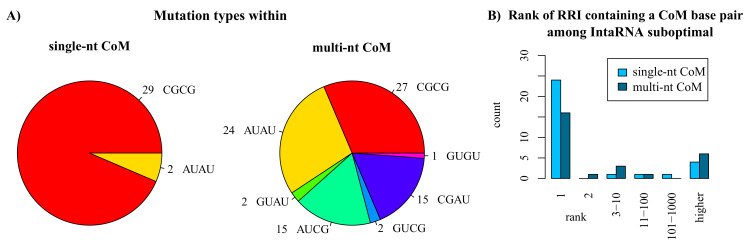
First, the distribution of mutation types was assessed, which is shown in Figure 3A. Within the single-nt CoM data set, mainly GC base pairs (29/31) have been mutated and all base pairs were ’flipped’, i.e., mutated into their wildtype pairing partner. Multi-nt CoMs are also dominated by flipped mutations (52/86) and mutations of GC wildtype base pairs (42/86).
Figure 3.
Characteristics of CoMs known from the literature. (A) Distribution of mutation types where the first two letters encode the wildtype nucleotides and the last two the respective mutated bases (both lex-sorted to reduce classes). For instance, AUCG represents both an AU or a UA wildtype base pair mutated either to CG or GC; (B) Rank of the RRI containing a CoM base pair among IntaRNA’s energy-sorted suboptimal RRI list.
Next, we studied whether or not IntaRNA is able to correctly identify the CoM. We therefore identified the rank of the RRI predicted by IntaRNA (including suboptimals; sorted by energy) that includes at least one of the base pairs of the CoM. Respective statistics are provided in Figure 3B. Most single-nt CoMs are within IntaRNA’s MFE RRI prediction (rank 1; 24/31), while 4 CoMs are not covered by IntaRNA predictions. Similar results are observed for multi-nt CoMs, where 16/27 could be mapped to MFE RRIs and 6/27 are without respective prediction.
All base pairs of the single-nt CoMs can form base pair stackings (to at least one side), i.e., no lonely base pairs have been mutated. Most (24/31) single-nt CoMs are within helices or can stack to both sides.
3.2. Energy Profiling of CoMs from Literature
Next, we evaluated if IntaRNA MFE predictions are a useful proxy for RRI stability in order to identify highly potent CoM candidates based on the RRI stability differences between wildtype-/mutant-only (ww, mm) and wildtype-mutant combinations (wm, mw). We restricted the investigation to the 22 single-nt GC-mutating CoMs found in MFE RRIs. As a background model, we identified all additional GC base pairs (in total 207) from their respective MFE RRIs and treated them as flipped CGCG mutations. For each CoM and wildtype-mutant combination, we computed the respective MFE. Figure 4 visualizes the MFE distributions as well as MFE differences for both the CoMs known from literature as well as the background CoMs.
Figure 4.
(A) Minimum free energy (MFE) distributions of known single-nt GC-mutating CoMs and their background model. The ’Mutations’ data (blue hues) covers 22 CGCG CoMs known from literature, while the ’Background’ data (orange hues) aggregates all remaining 207 CGCG CoMs from the MFE RRIs containing the known CoMs. There are four possible sequence combinations, referring to sRNA-mRNA pairs with respective (w)ildtype/(m)utant annotation. That is, an interaction of wildtype sRNA with the mutated mRNA is denoted by ’wm’, while e.g., ’mm’ refers to the interaction of sRNA and mRNA mutant. Each subplot provides the p-value of the sample t-test comparing the respective distributions. Dotted lines mark mean values, while dashed black lines highlight an energy difference of zero; (B) Pairwise energy differences of mutant combinations compared to wildtype-only ’ww’ or mutant-only ’mm’ MFEs for the CoMs of both the Mutations and Background data set. That is, e.g., ’wm-mm’ refers to the energy difference of a ’wm’ interaction and the respective mutant-only ’mm’ interaction energy. For each data set, p-values of paired sample t-tests that compare the values with the respective reference MFEs (’ww’ or ’mm’) are provided. The minDeltaE feature is defined as (min(MFE(mw),MFE(wm))-max(MFE(ww),MFE(mm))). For further details, see text.
First, we compared the energy distributions of known CoMs and the background shown in Figure 4A. That is, for each wildtype-mutant combination (ww, wm, mw, mm), we compare the set of known CoM MFE values with the set of background CoM MFE values. This is done based on a sample t-test (known CoMs vs. background CoMs, two-tailed, unequal variance, unpaired). Respective p-values are reported in Figure 4A. Here, we only find the mutant-only combinations (mm) to be significantly different.
We also compared for each CoM the MFE of a given RNA combination (e.g., wm) with the respective value of the wildtype-only (ww) or mutant-only (mm) combination from the same data set. Beside visualizing the differences, Figure 4B also shows the p-values of respective paired sample t-tests (combination vs. ww|mm, two-tailed, unequal variance). With regard to wildtype-only (ww) MFE, both the known CoMs as well as the background CoMs show on average higher wildtype-mutant energies (i.e., wm-ww, mw-ww are positive). For the known CoMs, this also holds for mutant-only combinations (mm-ww), while the background CoMs do not show a significant difference between wildtype-only and mutant-only energies (mm-ww). That is, known CoMs show on average a stronger reduction of RRI potential of compensatory mutations (mm-ww) compared to the background. This difference also manifests in the energy differences when relating to the MFE of mutant-only (mm) combinations, i.e., the wildtype-mutant combinations of known CoMs (wm-mm, mw-mm) show in contrast to the background model on average no significant energy change. This is also reflected in the minDeltaE distributions, i.e., the mean minDeltaE difference is about zero for known CoMs while a slightly positive average value is observed for the background.
3.3. CopomuS Parameter Optimization
To find suitable values for the parameters of the MFE-based CoM classification, we performed a parameter sweep in the range given the observations from Figure 4. Here, we used the whole single-nt CoM data set and restricted the CoM candidate generation to the flipping of the MFEs’ GC base pairs. We assessed whether or not the known single-nt CoM was among the valid candidates which fulfilled the MFE-difference constraints for the given values and how many such candidates are left. A low number implies a clear separation of the known CoM from other candidates.
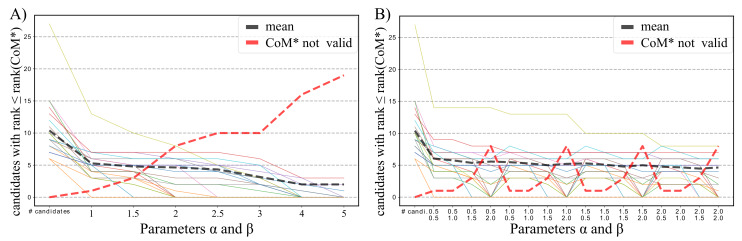
Results are depicted in Figure 5A. An threshold of 1 prunes the candidate list already quite strongly. Increasing the thresholds results in a nearly linear loss of known CoMs among the remaining CoM candidates such that for value 2.5 already 10/31 known CoMs are considered invalid. On the other hand, no significant reduction of the average number of valid candidates can be observed for values between 1 and 2.5.
Figure 5.
Effect of thresholds on MFE-difference-based CoM classification. Each solid line represents the number of valid CoM candidates for an RNA pair with a rank not higher than the known CoM from literature (designated as CoM*; colors differentiate between the single CoMs). The left-most data points represent the overall numbers of CoM candidates in the RNA pair that harbors the respective CoM*. The black dotted line provides the average over all RNA pairs for . The red dotted line depicts the number of RNA pairs for which the known CoM* does not fulfill the energy constraint. (A) Results for equal values of and ; (B) Results for explicit value combinations of and in range [0.5, 2].
We thus performed another parameter screen to investigate non-uniform combinations of and , also summarized in Figure 5B. We observe that the effect of the energy-based candidate pruning is mainly governed by , i.e., the energy difference to the mutant-only MFE. This is in accordance with the MFE difference statistics from Figure 4. As a result, we fix the default values of CopomuS’s E function to and .
3.4. CopomuS Benchmark
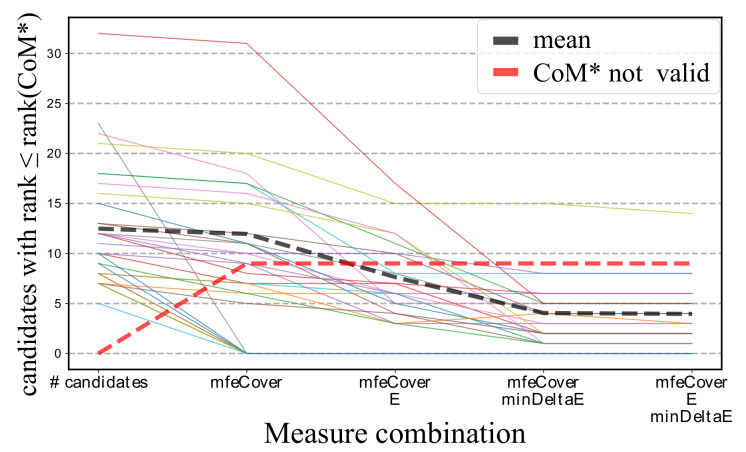
Finally, we evaluated CopomuS’s ranking of known single-nt CoMs among its generated CoM candidates for different combinations of classification and sorting functions. We are interested in combinations for which many known CoMs fulfill all constraints with low (mean) ranking. Given our preliminary studies, we tested the effect of classification based on mfeCover and E (using and ) as well as sorting by minDeltaE. We restrict candidate generation to the flipping of GC and AU base pairs of the MFE RRI that are part of helices (no lonely base pairs or helix ends).
Figure 6 summarizes the results. Classification based on mfeCover has only minor effects, while E-based pruning shows much stronger candidate set reductions without altering the number of known CoMs that are not fulfilling the constraints. minDeltaE sorting alone provides already very good results, which can be slightly enhanced when combined with E classification.
Figure 6.
Effect of different classification and sorting combinations on CoM candidate set sizes. Each solid line represents the number of valid CoM candidates for an RNA pair with a rank not higher than the known CoM from literature (designated as CoM*; colors differentiate between the single CoMs). The left-most data points represent the overall numbers of CoM candidates. The black dotted line provides the average over all RNA pairs. The red dotted line depicts the number of RNA pairs for which the known CoM* does not fulfill the constraints.
4. Discussion
4.1. Statistics of CoMs from Literature
The high abundance of GC mutations is related to the higher base pairing strength of GC base pairs compared to AU or GU base pairs. Thus, preventing the formation of a GC base pair via mutation will have on average a higher impact on RRI stability and thus interaction potential.
Within multi-nt CoMs, GC mutations are not as dominant as in single-nt CoMs. Here, the set of mutations of the multi-nt CoM often aims at the core of a long inter-molecular helix to prevent their formation in wildtype-mutant combinations. Thus, less focus on GC base pairs is possible and even GU base pairs are part of the CoM. If possible, no GU base pair is introduced in the mutant (only 1/86), since GU base pairs have the lowest stability contributions.
Flipping wildtype base pairs in mutants is an easy way to ensure base pair incompatibility in wildtype-mutant combinations and enables (on average) a similar stability of the wildtype-only and mutant-only RRI. Thus, we chose flipping as the standard mode of CoM candidate generation.
The high rate of CoMs found within the IntaRNA MFE predictions supports our choice to build CopomuS’s CoM selection based on IntaRNA RRI predictions. The prediction can fail e.g., if additional interaction partners, like RNA-binding proteins such as Hfq [14], are needed to guide or stabilize the RRI formation. In that case, IntaRNA’s prediction model has to be guided with respective constraints, which can be incorporated in the CopomuS workflow using its optional IntaRNA parameter file. Examples for such constraints are location constraints where RRIs are assumed, structure probing data from e.g., SHAPE experiments [15] or explicit seed interaction information. Another reason for missing a known CoM is that only one base pair pattern of an RRI is investigated, but sometimes slightly different patterns with equal energy within the same boundaries are possible. Since IntaRNA reports only one pattern per RRI boundaries, a known CoM present in an alternative pattern is missed.
The high abundance of stacked base pairs within the data set motivated the introduced CoM candidate filter capabilities of CopomuS, to exclude lonely and helix-end base pairs from the candidate generation.
4.2. Energy Profiling of CoMs from Literature
The pattern of average MFE differences of known CoMs (inversely) follows the desired relation of in vitro RRI signals depicted in Figure 1. That is, wildtype-mutant RRIs show on average lower stability (higher MFEs) compared to wildtype-only or mutant-only interactions. This, in concert with the less prominent pattern within the background model, strongly supports our hypothesis that we can indeed use IntaRNA MFE predictions to select highly potent CoM candidates. Furthermore, the higher mean minDeltaE values of CoMs motivate our final ranking of CoM candidates in CopomuS to identify mutations that show the desired pattern most prominently.
4.3. CopomuS Benchmark
Nine of all known CoMs from literature are not among the remaining valid candidates for all measures and combinations. Seven of these are considered invalid by mfeCover due to a lack of MFE RRI coverage (compare Figure 3B) and two are at helix ends and thus filtered during candidate generation.
As expected, the combination of mfeCover with the E classifier impacts the ranking less than combining mfeCover with minDeltaE; since the latter provides a high-resolution sorting instead. Nevertheless, a combination of all three functions provides the best average rank of about 4 and furthermore instantiates all rationales underlying CopomuS. Thus, we make the combination of mfeCover and E with a final sorting by minDeltaE the default ranking of CopomuS.
5. Conclusions
CopomuS implements an automated, objective evaluation strategy to identify compensatory mutations (CoMs) based on IntaRNA-based RRI stability analyses. That is, top-ranked CoMs show an in silico RRI stability pattern that follows the desired pattern of in vitro RRI potentials. The required scoring functions were derived from characteristics of CoMs used in successful RRI verification experiments known from literature. We could show that the introduced measures efficiently reduce the set of CoM candidates, while the known CoM was found on average within the top-5 candidates. That way, experimenters can easily and reproducibly pick promising candidates from the provided list without the need of time consuming manual RRI prediction and comparison. Therefore, we consider CopomuS a valuable tool to guide the difficult design of highly promising CoM-based RRI verification experiments.
Currently, CopomuS supports only single-nt CoM generation and evaluation. While potent multi-nt CoMs could be derived from top-ranked single-nt CoMs that can stack, no correct ranking of their combination can be done based on the current output. We therefore work on the extension of CopomuS to multi-nt CoM generation and testing, to also provide reliable selection criteria for this setup. Furthermore, we would like to evaluate CopomuS’s ranking order, which is currently not possible due to a lack of experimental data. That is, published CoM data is biased towards “valid” CoMs that were successfully used to verify an RRI. Non-successful CoMs or CoMs with low experimental effect are typically not published. Eventually, an extensive CoM feature and ranking evaluation would require in vitro measurements of multiple CoMs for the same RRI within the same experimental setup to be comparable.
Acknowledgments
We thank Jens Georg for discussions on the project.
Abbreviations
The following abbreviations are used in this manuscript:
| RRI | RNA-RNA interaction |
| CoM | compensatory mutation |
| MFE | minimum free energy |
| nt | nucleotide |
| ww, wm, mw, mm | sRNA-mRNA combinations of (w)ildtype and (m)utated variants |
| AUAU, AUCG, … | CoM classes where the first two letters encode the wildtype and the last two letters the mutated nts, e.g., AUCG encodes the mutation of either an AU or UA RRI base pair into either CG or GC |
Appendix A. Data
Table A1.
Single-nucleotide CoMs from literature used for this study.
| RNA-1 | RNA-2 | Mutation | Source |
|---|---|---|---|
| OxyS_NC_000913 | b2731_NC_000913_-200+100 | G102C&C-13G | [16] |
| CyaR_NC_000913 | b2687_NC_000913_-200+100 | A44U&U-7A | [17] |
| MicA_NC_000913 | b0411_NC_000913_-200+100 | C11G&G-46C | [18] |
| MicA_NC_000913 | b0814_NC_000913_-200+100 | C7G&G17C | [18] |
| RybB_NC_000913 | b0805_NC_000913_-200+100 | C2G&G-71C | [18] |
| RybB_NC_000913 | b2594_NC_000913_-200+100 | C2G&G4C | [18] |
| MicF_NC_000913 | b0889_NC_000913_-200+100 | C2G&G11C | [19] |
| SgrS_NC_000913 | b1101_NC_000913_-200+100 | G178C&C-19G | [20] |
| DsrA_NC_000913 | b2741_NC_000913_-200+100 | C16G&G-103C | [21] |
| RprA_NC_000913 | b2741_NC_000913_-200+100 | C42G&G-103C | [21] |
| ArcZ_NC_000913 | b2741_NC_000913_-200+100 | C70G&G-103C | [21] |
| ArcZ_NC_000913 | b3546_NC_000913_-200+100 | C69G&G-10C | [22] |
| CyaR_NC_000913 | b1824_NC_000913_-200+100 | G32C&C-11G | [3] |
| FnrS_NC_000913 | b2531_NC_000913_-200+100 | C47G&G-3C | [3] |
| RyhB_NC_000913 | b3365_NC_000913_-200+100 | C45G&G5C | [3] |
| RybB_NC_003197 | STM1473_NC_003197_-200+100 | C2G&G19C | [23] |
| MicF_NC_003197 | STM0366_NC_003197_-200+100 | C6G&G-31C | [24] |
| MicF_NC_003197 | STM0959_NC_003197_-200+100 | C6G&G7C | [24] |
| RybB_NC_003197 | STM0413_NC_003197_-200+100 | C2G&G-8C | [25] |
| RybB_NC_003197 | STM0999_NC_003197_-200+100 | C2G&G-39C | [25] |
| RybB_NC_003197 | STM1070_NC_003197_-200+100 | C2G&G31C | [25] |
| RybB_NC_003197 | STM1572_NC_003197_-200+100 | C2G&G25C | [25] |
| RybB_NC_003197 | STM1732_NC_003197_-200+100 | C2G&G19C | [25] |
| RybB_NC_003197 | STM1995_NC_003197_-200+100 | C2G&G19C | [25] |
| RybB_NC_003197 | STM2267_NC_003197_-200+100 | C2G&G-42C | [25] |
| RybB_NC_003197 | STM2391_NC_003197_-200+100 | C2G&G55C | [25] |
| CyaR_NC_003197 | STM0833_NC_003197_-200+100 | A43U&U-5A | [26] |
| SgrS_NC_003197 | STM2945_NC_003197_-200+100 | G176C&C5G | [27] |
| ArcZ_NC_003197 | STM1682_NC_003197_-200+100 | G70C&C22G | [28] |
| ArcZ_NC_003197 | STM2970_NC_003197_-200+100 | G70C&C-12G | [28] |
| MicC_NC_003197 | STM1572_NC_003197_-200+100 | C9G&G69C | [29] |
Table A2.
Multi-nucleotide CoMs from literature used for this study.
| RNA-1 | RNA-2 | Mutation | Source |
|---|---|---|---|
| Spot42_NC_000913 | b2702_NC_000913_-200+100 | U23A&A-4U,C24G&G-5C,U25A&A-6U | [30] |
| Spot42_NC_000913 | b3962_NC_000913_-200+100 | G49C&C21G,U50A&A20U,A51U&U19A | [30] |
| Spot42_NC_000913 | b4311_NC_000913_-200+100 | G5A&C-20U,G6C&U-21G,U7G&A-22C | [30] |
| Spot42_NC_000913 | b1302_NC_000913_-200+100 | G55C&C-33G,G56A&C-34U,A57C&U-35G | [31] |
| Spot42_NC_000913 | b2715_NC_000913_-200+100 | G5A&C-25U,G6C&C-26G,U7G&A-27C | [31] |
| Spot42_NC_000913 | b2801_NC_000913_-200+100 | G49C&C-32G,U50A&A-33U,A51U&U-34A | [31] |
| Spot42_NC_000913 | b3224_NC_000913_-200+100 | G5A&C-54U,G6C&C-55G,U7G&A-56C | [31] |
| Spot42_NC_000913 | b1901_NC_000913_-200+100 | G5A&C-34U,G6C&C-35G,U7G&G-36U | [32] |
| MicA_NC_000913 | b1130_NC_000913_-200+100 | C7G&G7C,G8C&C6G,C9G&G5C,G10C&C4G | [33] |
| GcvB_NC_000913 | b1130_NC_000913_-200+100 | C158G&G-13C,U157A&A-14U,G156C&C-15G ,U155A&A-16U,C154G&G-17C | [34] |
| CyaR_NC_000913 | b1740_NC_000913_-200+100 | A40U&U-3A,G39A&C-2U | [17] |
| CyaR_NC_000913 | b2666_NC_000913_-200+100 | A40U&U7A,G39A&U8U,G38C&C9G | [17] |
| ArcZ_NC_000913 | b1892_NC_000913_-200+100 | U78A&G-60U,U77A&A-59U,G76G&C-58C,U75C&A-57G,G74C&U-56G,G73U&C-55A | [35] |
| OxyS_NC_000913 | b1892_NC_000913_-200+100 | A69U&U-18A,A68A&U-17U,U67C&A-16G ,A66C&U-15G,A65U&U-14A | [35] |
| RybB_NC_000913 | b0721_NC_000913_-200+100 | U12A&A-21U,C13G&G-22C | [36] |
| RyhB_NC_000913 | b0721_NC_000913_-200+100 | U51A&A-15U,A50U&U-14A | [36] |
| Spot42_NC_000913 | b0721_NC_000913_-200+100 | G13C&C-53G,G14C&C-54G | [36] |
| FnrS_NC_000913 | b0755_NC_000913_-200+100 | C47A&G-4U,U48A&A-5U,U49G&A-6C | [5] |
| FnrS_NC_000913 | b1479_NC_000913_-200+100 | U57A&A-13U,U58G&A-14C,U59A&A-15U | [5] |
| FnrS_NC_000913 | b2153_NC_000913_-200+100 | G5U&C-18A,G4C&C-19G | [5] |
| FnrS_NC_000913 | b2303_NC_000913_-200+100 | G5U&C-6A,G4C&C-5G | [5] |
| RyhB_NC_000913 | b1656_NC_000913_-200+100 | C49G&G-6C,C45G&G-3C,G44C&C-2G | [37] |
| RyhB_NC_000913 | b0592_NC_000913_-200+100 | G53U&C-7A,C54U&G-8A,U55C&A-9G | [38] |
| RyhB_NC_000913 | b2155_NC_000913_-200+100 | C47A&G-47U,A48U&U-48A,C49A&G-49U ,A50C&U-50G | [38] |
| Spot42_NC_000913 | b1761_NC_000913_-200+100 | C46G&G86C,C48G&G88C | [3] |
| RybB_NC_003197 | STM0687_NC_003197_-200+100 | C5G&G14C,A4U&U15A | [39] |
| MicA_NC_003197 | STM4231_NC_003197_-200+100 | A22C&U5G,A12G&U14C | [40] |
| GcvB_NC_003197 | STM3930_NC_003197_-200+100 | U84G&A-15C,G85A&C-16U,U86A&A-17U ,U87C&A-18G,U88A&A-19U | [41] |
Table A3.
sRNAs used within this study.
| sRNA_Genome | Sequence |
|---|---|
| ArcZ_NC_000913 | GTGCGGCCTGAAAAACAGTGCTGTGCCCTTGTAACTCATCATAATAATTTACGGCGCAGCCAAGATTTCCCTGGTGTTGGCGCAGTATTCGCGCACCCCGGTCTAGCCGGGGTCATTTTTT |
| ArcZ_NC_003197 | GTGCGGCCTGAAAACAGGACTGCGCCTTTGACATCATCATAATAAGCACGGCGCAGCCACGATTTCCCTGGTGTTGGCGCAGTATTCGCGCACCCCGGTCAAACCGGGGTCATTTTTT |
| CyaR_NC_000913 | GCTGAAAAACATAACCCATAAAATGCTAGCTGTACCAGGAACCACCTCCTTAGCCTGTGTAATCTCCCTTACACGGGCTTATTT |
| CyaR_NC_003197 | GCTGAAAAACATAACCCATAAATGCTAGCTGTACCAGGAACCACCTCCTTGGCCTGCGTAATCTCCCTTACGCAGGCTTATTT |
| DsrA_NC_000913 | AACACATCAGATTTCCTGGTGTAACGAATTTTTTAAGTGCTTCTTGCTTAAGCAAGTTTCATCCCGACCCCCTCAGGGTCGGGATTTTT |
| FnrS_NC_000913 | GCAGGTGAATGCAACGTCAAGCGATGGGCGTTGCGCTCCATATTGTCTTACTTCCTTTTTTGAATTACTGCATAGCACAATTGATTCGTACGACGCCGACTTTGATGAGTCGGCTTTTTTTT |
| GcvB_NC_000913 | ACTTCCTGAGCCGGAACGAAAAGTTTTATCGGAATGCGTGTTCTGGTGAACTTTTGGCTTACGGTTGTGATGTTGTGTTGTTGTGTTTGCAATTGGTCTGCGATTCAGACCATGGTAGCAAAGCTACCTTTTTTCACTTCCTGTACATTTACCCTGTCTGTCCATAGTGATTAATGTAGCACCGCCTAATTGCGGTGCTTT |
| GcvB_NC_003197 | ACTTCCTGAGCCGGAACGAAAAGTTTTATCGGAATGCGTGTTCTGATGGGCTTTTGGCTTACGGTTGTGATGTTGTGTTGTTGTGTTTGCAATTGGTCTGCGATTCAGACCACGGTAGCGAGACTACCCTTTTTCACTTCCTGTACATTTACCCTGTCTGTCCATAGTGATTAATGTAGCACCGCCATATTGCGGTGCTTT |
| MicA_NC_000913 | GAAAGACGCGCATTTGTTATCATCATCCCTGAATTCAGAGATGAAATTTTGGCCACTCACGAGTGGCCTTTT |
| MicA_NC_003197 | GAAAGACGCGCATTTGTTATCATCATCCCTGTTTTCAGCGATGAAATTTTGGCCACTCCGTGAGTGGCCTTTT |
| MicC_NC_003197 | GTTATATGCCTTTATTGTCACATATTCATTTTGTCGCTGGGCCATTGCGTTAACCTTTGCTTTCCAGCGTATAAATTGACAAGCCCGAACGGATGTTCGGGCTTTTTTT |
| MicF_NC_000913 | GCTATCATCATTAACTTTATTTATTACCGTCATTCATTTCTGAATGTCTGTTTACCCCTATTTCAACCGGATGCCTCGCATTCGGTTTTTTTT |
| MicF_NC_003197 | GCTATCATCATTAACTTTATTTATTACCGTCATTCACTTCTGAATGTCTGTTTACCCCTATTTCAACCGGATGCTTCGCATTCGGTTTTTTTT |
| OxyS_NC_000913 | GAAACGGAGCGGCACCTCTTTTAACCCTTGAAGTCACTGCCCGTTTCGAGAGTTTCTCAACTCGAATAACTAAAGCCAACGTGAACTTTTGCGGATCTCCAGGATCCGC |
| RprA_NC_000913 | ACGGTTATAAATCAACATATTGATTTATAAGCATGGAAATCCCCTGAGTGAAACAACGAATTGCTGTGTGTAGTCTTTGCCCATCTCCCACGATGGGCTTTTTTT |
| RybB_NC_000913 | GCCACTGCTTTTCTTTGATGTCCCCATTTTGTGGAGCCCATCAACCCCGCCATTTCGGTTCAAGGTTGATGGGTTTTTT |
| RybB_NC_003197 | GCCACTGCTTTTCTTTGATGTCCCCATTTTGTGGAGCCCATCAACCCCGCCATTTCGGTTCAAGGTTGGTGGGTTTTTT |
| RyhB_NC_000913 | GCGATCAGGAAGACCCTCGCGGAGAACCTGAAAGCACGACATTGCTCACATTGCTTCCAGTATTACTTAGCCAGCCGGGTGCTGGCTTTT |
| SgrS_NC_000913 | GATGAAGCAAGGGGGTGCCCCATGCGTCAGTTTTATCAGCACTATTTTACCGCGACAGCGAAGTTGTGCTGGTTGCGTTGGTTAAGCGTCCCACAACGATTAACCATGCTTGAAGGACTGATGCAGTGGGATGACCGCAATTCTGAAAGTTGACTTGCCTGCATCATGTGTGACTGAGTATTGGTGTAAAATCACCCGCCAGCAGATTATACCTGCTGGTTTTTTTT |
| SgrS_NC_003197 | GATGAAGCAAGAGGAAGAGGTCACTATGCGCCAGTTCTGGTTGAGATATTTTGCCGCGACGGAAAAAACGTCCTGGCTGGCTTGCCTGAGCGCACCGCAGCGCTTAAAAATGCTCGCGGAACTGATGCAGTGGGAGGCGACCGATTGAAGCCAATTGCAGACATCATGTGTGACTGAGTATTGGTGTAGGCGATAGCCTAAAATCACCCGCCAGCAGATAATATCTGCTGGCTTTTTTT |
| Spot42_NC_000913 | GTAGGGTACAGAGGTAAGATGTTCTATCTTTCAGACCTTTTACTTCACGTAATCGGATTTGGCTGAATATTTTAGCCGCCCCAGTCAGTAATGACTGGGGCGTTTTTTA |
Table A4.
genomic subsequences of NC_003197 -200+100 around start codon.
| Locus_Genome_Range | Sequence | Gene |
|---|---|---|
| STM0366_NC_003197_-200+100 | CGTTCATCTTATTAATAGTCAAACCAGATGATTGCGAGTGAGATCACAAAGCAGGGGCGTTTTAATCCGCGTTGTTACGCCGACAGAGCGGGGGCTGACTGGATTTTTCCAGTAATCTACACTACTTATTTAATCAGTCCGAACGGCCTTTTTGTTCTGATAAAGCGATGATGGCGTAATAATAAAACGAGGGTTTTGCTATGAAAACTGGCTACAAGGTTATGCTTGGCGCATTAGCGTTTGTCGTGACAAACGTTTATGCCGCAGAAATCATGAAAAAAACGGACTTTGATAAAGTCG | yahO |
| STM0413_NC_003197_-200+100 | TCATGAAAGATAGTACTGTCGCCGCGTCTAAAATGCGCAAACGTGAACGCAATCGATTACGTAAATGATAGATATGTGAAACAAGACATATTTTTGTGAGCAATGATTTTTATAATAGGCTCCGCAGAAACACGAAATATTTAGAAACGCAAATTGCGTTCTTTTCACTCCCGCAAGGGATTTCAAACAGTGGCATACATATGAAAAAAACTTTACTCGCAGTCAGCGCAGCGCTGGCGCTCACCTCATCTTTTACTGCTAACGCAGCAGAAAATGATCAGCCGCAGTATTTGTCCGACT | tsx |
| STM0687_NC_003197_-200+100 | AGCAGTCGAATGTAACAGAAAGCAATTAAATATGTGCGGTTGCTCATATTATTACATACTGGTTACAGAAAGAGATTGATAATTCGCATCGCGAAAAATAGTCTATTTAACGTAGTAAATGAGGTTTCTCAGCGCTACTTTTTATTTTTTCGCTGTTCGCTTTTGTCGGCAGCAATTTATACGTCAAAGAGGATTAACTTATGCGTACGTTTAGTGGCAAACGTAGTACGCTGGCTCTGGCTATCGCCGGTATCACAGCAATGTCGGGGTGGATCGTTGTTCCGCAGGCGCAAGCCTCCG | ybfM/chiP |
| STM0833_NC_003197_-200+100 | CTGGATGAATGTATCGCGCCGCACGCGCATTATTGGTGCAATAAGCCGGAAAAGTGATGTTAATTGAATAAGATAGCGCGATATGGAAACGTTCTGTTACATGAAAGGCGCCCTTAGACACCGTGAATCGCAAAGAGTTTCCCATTAATTTTTGATATATTTAAAACTTAGGACTTACTTGAAGCACATTTGAGGTGGTTATGAAAAAAATTGCATGTCTTTCAGCACTGGCCGCTGTTCTGGCTTTTTCCGCAGGTACTGCAGTAGCTGCTACTTCTACCGTTACCGGTGGTTACGCTC | ompX |
| STM0959_NC_003197_-200+100 | TGAAATCTACGCATGGCGTGGACAGACGCCATTCGTGATGTCGATAGCTGCCGCGAGGCAACGGTCTTCTCACCATAGACCAGGCATTGCGCGCCGTTAATCCCTCTGGGTTTCGGTCTATCGTGATGGGCAGCGACTCTGAACAGTGATGTGAGTAGAGTCAGGCAGGAGTAGGGAAGGAATACAGAGAGACAATAATAATGGTAGATAGCAAGAAGCGCCCTGGCAAAGATCTCGACCGTATCGATCGTAACATTCTTAATGAACTGCAAAAGGATGGGCGTATTTCCAACGTCGAGC | lrp |
| STM0999_NC_003197_-200+100 | ACATATTATTTCCTTTTGAAACCAAATCTTTATCTTTGTAGCACTTTCACGGTAGCGAAACGTTAGTTTGAATGGAAAGATGCCTGTCAGACACATAAAGACACCAAACTCTCATCAATATTTCTGTAAAGTTTTATTGACGGAATTTATTGACGGCAGTGGCAGGTGTCATATAAAAAAACCAATGAGGGTAATAAATAATGATGAAGCGCAAAATCCTGGCAGCGGTGATCCCTGCCCTGCTGGCTGCTGCAACCGCAAACGCAGCAGAAATTTATAATAAAGATGGTAATAAGCTGG | ompF |
| STM1070_NC_003197_-200+100 | TGTTTTTTTCACATGTCTGACGGAGTTCACACTTGTAAGTTTCCAACTACGTTGTAGACTTTACATCGCCAGGGGTGCTCAGCATAAGCCGTAGATATCGGTAGAGTAACTATTGAGCAGATCCCCCGGTGAAGGATTTAACCGTGTTATCTCGTTGGAGATATTCATGGCGTATTTTGGATGATAACGAGGCGCAAAAAATGAAAAAGACAGCTATCGCGATTGCAGTGGCACTGGCTGGTTTCGCTACCGTAGCGCAGGCCGCTCCGAAAGATAACACCTGGTACGCTGGTGCTAAAC | ompA |
| STM1473_NC_003197_-200+100 | CACATACATGAATACATAATAACAAATATATTCACCATAAATATATGCGTTTCCGATAGTAACTTTTGTATTAATTAATAACATATAAGAAAAGTTAGCATTTGCTGAAATAATATTATTCAGATTAGGATGCCTTTGATTCAACGAATCTGTAGAAGTTCAATCTTTTGCAAATAAGTTAAGTTTTTAAGGATAAAAAAATGAAAAGAAAAGTATTGGCACTTGTCATCCCGGCTCTGCTGGCTGCTGGCGCAGCACACGCCGCTGAAATTTATAACAAAGACGGCAACAAACTGGACC | ompN |
| STM1572_NC_003197_-200+100 | TTAGACAGTCCCTATTTGAATTAATACTCTCAAATGTATTAAGGAGATCTCGATCACACAAATTAAAATAATTTGTAATCTTATGAAACTTATTATTGAACTTATGCCACTCCGTCATTTAAAAATAGTCTGCCATTGACAAACGCCTCGTTTAACAATGGTTGAGGAAACACGCTAAGAAAATTATAAGGATTATTAAAATGAAACTTAAGTTAGTGGCAGTGGCAGTGACTTCCCTGTTGGCAGCAGGCGTTGTAAATGCAGCCGAGGTATATAACAAAGACGGCAATAAACTGGATC | ompD |
| STM1682_NC_003197_-200+100 | CATGCGCTTCGCTTCGGCTACCACGCGCAATAACAAAAGGCGTATGTAACGGCCAGGTTTCCTCATATACCTTAACAGATCTCATCTCTTCCCCTCTGATAGCGCCGGACGCGGCTTGCTACAAATAGTATTTGCCGTGTTAATGAATAATGACTTAAACTGGATTTCGACGTTAACTATAAGTAAATAGGAACATAATTATGTCACAGACTGTACATTTCCAGGGTAACCCGGTCACCGTTGCCAACGTTATTCCGCAGGCTGGTAGCAAAGCACAGGCTTTTACTCTTGTCGCAAAAG | tpx |
| STM1732_NC_003197_-200+100 | GAGCAGACAAATATTTGCATAGCGTGAATATGTCAAAATTGATCTGAATTCCTATAACCAGGATTTTCAATACAAGTTCTAAATTAATCTGGATCAATAAATGTTAAATTATAAGAACAAATGTGATCTGTATTAGATCACTTATTACTTCATTGTGGGTATATTCATCACGCTTTTATAACCATAACGATGGAGCGGGTATGAAAAAATTTACAGTGGCGGCACTGGCGTTAACAACTCTTCTCTCAGGCAGCGCGTTCGCGCACGAAGCCGGAGAATTCTTTATGCGTGCAGGTCCGG | ompW |
| STM1995_NC_003197_-200+100 | GATAATTATAGAATATATATTCTTAGTTACTTATATAGTCTGTATTATAAAAAACCAAACAGAAACAAATTGAAATATTTTAAATACCTTTGTTACATGTTATTTTTTAAATTCCATGAACTTCATAGAATAGTATCAATTTGTAGTTTTGTTGAAGTGGCTACATATTCATATAAATTATTATCATAAGGGAATACATAATGAACAGAAAAGTTCTGGCACTGCTTGTCCCGGCGTTATTAGTGGCAGGCGCAGCAAATGCGGCTGAAGTTTATAATAAAAATGGCAACAAACTCGACC | ompS |
| STM2267_NC_003197_-200+100 | GCTTTAAAAAAGTTCCGTAAAATTCATATTTTGAAACATCTATGTAGATAACTGTAACATCTTAAAAGTTTTAGTATCATATTCGTGTTGGATTATTCTGTATTTTTGCGGAGAATGGACTTGCCGACTGGTTAATGAGGGTTAACCAGTAAGCAGTGGCATAAAAAAGCAATAAAGGCATATAACAGAGGGTTAATAACATGAAAGTTAAAGTACTGTCCCTCCTGGTACCAGCTCTGCTGGTGGCGGGCGCAGCGAATGCGGCTGAAATTTATAATAAAGACGGCAACAAATTAGACC | ompC |
| STM2391_NC_003197_-200+100 | TTAAGCCCGCCGATTTTGCCAGCCAGATCTCGTTTCTAAGATCACAATTGAAAAAACTTATAAACATACTTGCAACATTCTAGCTGGTCAGACCTATACTCTCGCCACTGGTCTGATTTCTAAGTCGTACCGCAGACCCTACACTTCGCGCTCCTGTTACAGCATGTAACATAGTTTGTATAAAAATAATCAATGAGGTTATGGTCATGAGCCAGAAAACCCTGTTTACAAAGTCTGCTCTCGCAGTCGCAGTGGCAATCATCTCCACCCAGGCCTGGTCTGCAGGCTTTCAGTTAAACG | fadL |
| STM2945_NC_003197_-200+100 | AACGACCATTTGCGGCGAATCATCTACCTTTTGTCTGAATTATCGTCACCACAAAGGATTACCAACCATAAATGTGCTGTATTAATAATGTCGTTCAAATTCTCTCCTGTAGTAAACTTTATCTGTTTAATAAAAAAGAGAGAATTGAACGATATATTTTACTCCGGATATTGAATAATATAAATTTGAAGGAAAATATTATGCCAGTCACTTTAAGCTTCGGTAATCATCAAAATTATACGCTTAATGAAAGTCGGCTTGCTCATCTGTTAAGCGCAGATAAAGAAAAAGCAATCCATA | sopD |
| STM2970_NC_003197_-200+100 | TCCATGTAAGAAGCGGATTATTGCATTTGAGATCGGGATCACTGATAGATTCATCACTTAAATGTATCTTTCCGCCCGAAAATTATTACGGCGAAAAATTATATAAAAAGCGTCCCTAAGCAGATTTCATTTTACGATCAGGTCTTTTTTCATTGGATTAGACCAGCAACCTGATTTTTAGCATCCTCCAGGAGAAATAGATGGAAACCACTCAGACCAGCACTATTGCTTCGATTGACTCTCGAAGCGCATGGCGCAAAACGGATACCATGTGGATGCTGGGCCTTTACGGCACGGCTA | sdaC |
| STM3930_NC_003197_-200+100 | CTTCTGATGACTTGAGCAGCGGATTGTGCTTATGGTGCTGCTCATTTACAACATAATCGATGATTTCTTACACAATAAGTGCATTTTTTTAATGCTCCATTTGCCATTTGTCCAAATTTAAGAAAATATTCGCAACAATCGATGTACCCATAACAATAACCGGTACTACCGGAACCGTTGCAAACACGACATGAGGATTTATGGCAGAGAAAAAACCGGAGCTACAGCGTGGGCTGGAAGCTCGTCATATTGAATTGATTGCCCTCGGGGGCACCATCGGCGTCGGACTCTTTATGGGCG | yifK |
| STM4231_NC_003197_-200+100 | GTTGGTAGAAGAGGGCGCCACATTCGCTATCGGCCTGCCGCCAGAGCGCTGTCATCTGTTCCGCGAGGATGGCAGCGCATGTCGTCGTCTGCATCAAGAGCCGGGTGTTTAAGGCCTCCATAAAAAAACGAAACGCAAAACCATTCGCAGTTTTAGAAGGTGGCAGCGTTTAAAGAAAAGCAATGATCTCAGGAGATAGAATGATGATTACTCTGCGCAAACTCCCACTGGCGGTTGCTGTCGCAGCGGGCGTAATGTCCGCTCAGGCAATGGCTGTCGATTTCCACGGTTACGCCCGTT | lamB |
Table A5.
| Locus_Genome_Range | Sequence | Gene |
|---|---|---|
| b0411_NC_000913_-200+100 | AGGGCGAAAGTCAGTACAATCCCCGCCCGAATGTGTGTAAACGTGAACGCAATCGATTACGTAAATGATAGAACTGTGAAACGAAACATATTTTTGTGAGCAATGATTTTTATAATAGGCTCCTCTGTATACGAAATATTTAGAAACGCAATTTGCGCCTTTTTCACTCCCGCAAGGGATTTTCAAACAGTGGCATACATATGAAAAAAACATTACTGGCAGCCGGTGCGGTACTGGCGCTCTCTTCGTCTTTTACTGTCAACGCAGCTGAAAACGACAAACCGCAGTATCTTTCCGACT | tsx |
| b0592_NC_000913_-200+100 | ATAAGCGCAATGTGATGTCCTGCGCCGTTCTGCCCCCTCTCCCTTCCAGGGTGAGGGCTGGGGTGAGGGTTAATGTTCGCACCAGTGCTGGCTGTTCCCCTCACCCTAACCCTCTCCCCAAAGGGGCGAGGGGACGGATTGTGCGCTTTGTCGAATTTGTCATTACGCCCTTAACCTTATTAATAACAGGAAGCTGATTTGTGAGACTCGCCCCGCTCTACCGCAACGCCCTTCTATTAACAGGACTTTTGCTTTCAGGAATAGCCGCAGTTCAGGCCGCTGACTGGCCGCGTCAGATTA | fepB |
| b0721_NC_000913_-200+100 | TCCCGAGCCACCCAGCGTTGTAACGTGTCGTTTTCGCATCTGGAAGCAGTGTTTTGCATGACGCGCAGTTATAGAAAGGACGCTGTCTGACCCGCAAGCAGACCGGAGGAAGGAAATCCCGACGTCTCCAGGTAACAGAAAGTTAACCTCTGTGCCCGTAGTCCCCAGGGAATAATAAGAACAGCATGTGGGCGTTATTCATGATAAGAAATGTGAAAAAACAAAGACCTGTTAATCTGGACCTACAGACCATCCGGTTCCCCATCACGGCGATAGCGTCCATTCTCCATCGCGTTTCCG | sdhC |
| b0755_NC_000913_-200+100 | GTTACGCCCTCGTCATGAGGGCTTTATCTCATATTGTTCAAATCACCAGCAAACACCGACATATTTGCAACTCAATATTCACAACAACCTTACACTGCGCCACTATTTTCGCTATGGTTATGCGTAAGCATTGCTGTTGCTTCGTCGCGGCAATATAATGAGAATTATTATCATTAAAAGATGATTTGAGGAGTAAGTATATGGCTGTAACTAAGCTGGTTCTGGTTCGTCATGGCGAAAGTCAGTGGAACAAAGAAAACCGTTTCACCGGTTGGTACGACGTGGATCTGTCTGAGAAAG | gpmA |
| b0805_NC_000913_-200+100 | TCAAATATGAACTCAATGTAAATAAATGTATTTCTTTTTCGCGCAATGGGTGATAGAAAATCGCTCCAAGTGATAATGCTTATCAAAATTATTATCACTTTCACGAGCACTATCACGGGATTAACAGTGGCATCGCATCCGCAGAGAGGCTTTCTCGTGGCAGTGAAAATTTCAACATATAAGAAAAAGTCACCTGCAAAATGGAAAACAATCGCAATTTCCCTGCCAGACAATTTCATTCGCTCACGTTCTTTGCCGGTCTTTGTATTGGCATCACGCCTGTGGCTCAGGCACTCGCCG | fiu |
| b0814_NC_000913_-200+100 | CTGGATGAATGACAGGGAAAACATGCGTAATACTTACGCAGTTCTCTGAAAAAGTGATTTAAATTTAGATGGATAGCGGTGTATGGAAACGTTCTGTTACATGAAATGGCCCGTTAGACATCACAAATCGCGAAGAGTTTCCCATTAATTTTTGATATATTTAAAACTTAGGACTTATTTGAATCACATTTGAGGTGGTTATGAAAAAAATTGCATGTCTTTCAGCACTGGCCGCAGTTCTGGCTTTCACCGCAGGTACTTCCGTAGCTGCGACTTCTACTGTAACTGGCGGTTACGCAC | ompX |
| b0889_NC_000913_-200+100 | GTGAAATCTACGTATGGCGTGGACAGACGCCATTCGTGATGTCGATAGCTGCCACAAGGCAACGGTCTTCTCACCGTAGACCCAGGCATTGCGCGCCGTGAATCTTCATGATTTCGGTCTATCGTGACGGGTAGCGACTCTGAACAGTGATGTTTCAGGGTCAGACAGGAGTAGGGAAGGAATACAGAGAGACAATAATAATGGTAGATAGCAAGAAGCGCCCTGGCAAAGATCTCGACCGTATCGATCGTAACATTCTTAATGAGTTGCAAAAGGATGGGCGTATTTCTAACGTCGAGC | lrp |
| b1101_NC_000913_-200+100 | CATATGTTTTGTCAAAATGTGCAACTTCTCCAATGATCTGAAGTTGAAACGTGATAGCCGTCAAACAAATTGGCACTGAATTATTTTACTCTGTGTAATAAATAAAGGGCGCTTAGATGCCCTGTACACGGCGAGGCTCTCCCCCCTTGCCACGCGTGAGAACGTAAAAAAAGCACCCATACTCAGGAGCACTCTCAATTATGTTTAAGAATGCATTTGCTAACCTGCAAAAGGTCGGTAAATCGCTGATGCTGCCGGTATCCGTACTGCCTATCGCAGGTATTCTGCTGGGCGTCGGTT | ptsG |
| b1130_NC_000913_-200+100 | GAGCTATCACGATGGTTGATGAGCTGAAATAAACCTCGTATCAGTGCCGGATGGCGATGCTGTCCGGCCTGCTTATTAAGATTATCCGCTTTTTATTTTTTCACTTTACCTCCCCTCCCCGCTGGTTTATTTAATGTTTACCCCCATAACCACATAATCGCGTTACACTATTTTAATAATTAAGACAGGGAGAAATAAAAATGCGCGTACTGGTTGTTGAAGACAATGCGTTGTTACGTCACCACCTTAAAGTTCAGATTCAGGATGCTGGTCATCAGGTCGATGACGCAGAAGATGCCA | phoP |
| b1302_NC_000913_-200+100 | AGCCGGACGTTTGATTGCCGAACTGCTGCGCGGCGACGCCGAACGTTTCGATGCCTTCGCCAATCTGCCGCATTACCCGTTCCCCGGCGGGCGCACGCTGCGTGTGCCGTTTACCGCGATGGGCGCGGCGTATTACAGCCTGCGCGATCGTCTGGGCGTTTAATTTCCGATTAACCGTGAAGAGTCAAAAGGTGTGAAACATGAGCAACAATGAATTCCATCAGCGTCGTCTTTCTGCCACTCCGCGCGGGGTTGGCGTGATGTGTAACTTCTTCGCCCAGTCGGCTGAAAACGCCACGC | puuE |
| b1479_NC_000913_-200+100 | TAGTAAATAACCCAACCGGCAGAAAACGCCCCGCTGAAAAGTAATTCATAACCATCAGTCCTCAATGACGATTAAACACCATTGCCTGCGCAATGGTGTTTTTGTTTTTATCTGCTTTATACTTGAGGCCGACGCCCTGGCGGTAAAGCAAAGACGATAAAAGCCCCCCAGGGATGGATATTCAAAAAAGAGTGAGTGACATGGAACCAAAAACAAAAAAACAGCGTTCGCTTTATATCCCTTACGCTGGCCCTGTACTGCTGGAATTTCCGTTGTTGAATAAAGGCAGTGCCTTCAGCA | maeA |
| b1656_NC_000913_-200+100 | TCTCAGTGAAGACTACTGGCAGCGCCACTATGTTGGCGCTCGTCGGGTAATGACCCCAAAAACACTTCGCTAAAACTTTACCCTGTTGTTACGGCAACAGGGTAAGTTCATCTTTTGTCTCACCTTTTAATTTGCTACCCTATCCATACGCACAATAAGGCTATTGTACGTATGCAAATTAATAATAAAGGAGAGTAGCAATGTCATTCGAATTACCTGCACTACCATATGCTAAAGATGCTCTGGCACCGCACATTTCTGCGGAAACCATCGAGTATCACTACGGCAAGCACCATCAGA | sodB |
| b1740_NC_000913_-200+100 | TTCCGTCCTCTTGTTTATCAGCGTGTTAGATAAGCCTGGAATACATTGGGCGCTTTTTCAAGCCCGTGAACGAAACGGCTCCGCTTTCAGAGGATTCCTGTATGACGTTTTAACCACCATTCAGCCCGCTGTCGCTTGTCGTTTCAGTAGCAACGGGTTAGCTTTAAGGAAGTTTTGTCTTTTCTGTCTGGAGGGGTTCAATGACATTGCAACAACAAATAATAAAGGCGCTGGGCGCAAAACCGCAGATTAATGCTGAAGAGGAAATTCGTCGTAGTGTCGATTTTCTGAAAAGCTACC | nadE |
| b1761_NC_000913_-200+100 | TAACGGTAGCCGGGTGGCAAAACTTTAGCGTCTGAGTTATCGCATTTGGTTATGAGATTACTCTCGTTATTAATTTGCTTTCCTGGGTCATTTTTTTCTTGCTTACCGTCACATTCTTGATGGTATAGTCGAAAACTGCAAAAGCACATGACATAAACAACATAAGCACAATCGTATTAATATATAAGGGTTTTATATCTATGGATCAGACATATTCTCTGGAGTCATTCCTCAACCATGTCCAAAAGCGCGACCCGAATCAAACCGAGTTCGCGCAAGCCGTTCGTGAAGTAATGACCA | gdhA |
| b1824_NC_000913_-200+100 | GCCAGTTTAAGTATCTGCCTGAACTGGCAAGGTTAAGCACAATGATATATCGGCGCGTATTCCGTTGCATAAGTGTGCAAAAAAAGTGGAAGACGTATCGAGATTTGTGCGTCTGATCGAGACATGTTTAAAAATGGCTTGCCATAATTAACGTTGTATGTGATAACAGATTTCGGGTTAAACGAGGTACAGTTCTGTTTATGTGTGGCATTTTCAGTAAAGAAGTCCTGAGTAAACACGTTGACGTTGAATACCGCTTCTCTGCCGAGCCTTATATTGGTGCCTCATGCAGTAATGTGT | yobF |
| b1892_NC_000913_-200+100 | TCGATTTAGGAAAAATCTTAGATAAGTGTAAAGACCCATTTCTATTTGTAAGGACATATTAAACCAAAAAGGTGGTTCTGCTTATTGCAGCTTATCGCAACTATTCTAATGCTAATTATTTTTTACCGGGGCTTCCCGGCGACATCACGGGGTGCGGTGAAACCGCATAAAAATAAAGTTGGTTATTCTGGGTGGGAATAATGCATACCTCCGAGTTGCTGAAACACATTTATGACATCAACTTGTCATATTTACTACTTGCACAGCGTTTGATTGTTCAGGACAAAGCGTCCGCTATGT | flhD |
| b1901_NC_000913_-200+100 | TCCCGCTAAATTTATGCACGTTCTCACTGTAATTCTGCGATGTGATATTGCTCTCCTATGGAGAATTAATTTCTCGCTAAAACTATGTCAACACAGTCACTTATCTTTTAGTTAAAAGGTAATGCTTTGTTTTCCGATTAATTTAACGAATGTCATTCGTTTTTGCCCTACACAAAACGACACTAAAGCTGGAGAGAACCATGCACAAATTTACTAAAGCCCTGGCAGCCATTGGTCTGGCAGCCGTTATGTCACAATCCGCTATGGCGGAGAACCTGAAGCTCGGTTTTCTGGTGAAGC | araF |
| b2153_NC_000913_-200+100 | CTGTGAGTAACTTTCACTTCCGTATTTGCATAACGATGTTTTAACATCTGCTGATGAAAGGCAGCGGCAATTACAATAATTATCGCTGTGAATACTGGATTATGTGCGCCGCCTCACGCACAATAATCAGGCTGTAAATCAGCTTAATAACTTTGCCCCCACGCAGGGCGGAGGCGTCACACCTGCAGGAGAAATCATAAATGCCATCACTCAGTAAAGAAGCGGCCCTGGTTCATGAAGCGTTAGTTGCGCGAGGACTGGAAACACCGCTGCGCCCGCCCGTGCATGAAATGGATAACG | folE |
| b2155_NC_000913_-200+100 | GGATTGATAATTGTTATCGTTTGCATTATCGTTACGCCGCAATCAAAAAAGGCTGACAAATCAGAGGCTGTTCCGGCTTTCTGGGATGATCACCTGCATAAAAAATAAGTCCACCGCGATGCTGCCGTACGCAAGGGGACGTGAAGAAGATGTGAGCGATAACCCATTTTATTTTCGTAGTTACCTCATGGAGATATGGAATGTTTAGGTTGAACCCTTTCGTACGGGTCGGGCTGTGTTTGTCCGCTATTTCTTGTGCATGGCCTGTGTTAGCGGTCGATGATGATGGCGAAACGATGG | cirA |
| b2303_NC_000913_-200+100 | TGGGTTAATGCCTGGACTCGCCAGCGAATTGACCTAGCAATGTATCCGGCAGTCAAGAACTGGCATGAGCGGATCCGTTCGCGCCCTGCCACCGGGCAGGCACTGCTAAAAGCACAACTCGGTGATGAGCGTTCGGATAGTTAACAGAAACAGGTTCTCGTGTATTATTTCATCCTAAGTAAAACAACGGAGAACCTGCAATGGCACAACCTGCCGCTATTATTCGTATAAAGAACCTTCGTTTGCGTACGTTTATCGGAATTAAGGAAGAAGAAATTAACAACCGTCAGGATATTGTTA | folX |
| b2531_NC_000913_-200+100 | ATGGCGTTCACGCCGCATCCGACAACAGGTACAAACGCCACGATAAAAAAATGGCACTGAAGGTTAAATACCCGACTAAATCAGTCAAGTAAATAGTTGACCAATTTACTCGGGAATGTCAGACTTGACCCTGCTATGCAATACCCCCACTTTTACAATAAAAAACCCCGGGCAGGGGCGAGTTTGAGGTGAAGTAAGACATGAGACTGACATCTAAAGGGCGCTATGCCGTGACCGCAATGCTTGACGTTGCGCTCAACTCTGAAGCGGGCCCGGTACCGTTGGCTGATATTTCCGAAC | iscR |
| b2594_NC_000913_-200+100 | CCCCGAGCAACCCGCCAAAAACAGGCTTAGTGTGGCGGCTGCCACCAGATATTTCATGCGCGTCATGACGTTTTGACTTTCCTCAAAATGTAATACGGGAGATTCTCTGTTCCTGCTCCCGGTTAAGACCAGCTACAATAGCACACTATATTAAACGGCAAAGCCGTAAAACCCCAACGATAAACGAAGAAGCAGTATATATGGCACAACGAGTACAGCTCACTGCAACGGTGTCCGAAAACCAACTCGGTCAACGCTTAGATCAGGCTTTGGCCGAAATGTTCCCGGATTATTCACGTT | rluD |
| b2666_NC_000913_-200+100 | TTGCGCGAGTTCAGTCATATTTATTTAAGTATTTTCTAAATTAAGTAAACTCTAAACTAAAAATGCAACATATACCAGCCTCAGCAGCGTAAATGAGAGTAAAAGCGTAAGCTGAAACTGGCAGGCTCCGCTAAAATTACTACGCTTAAGAGATAAAATCTCTTTTTAAACAATGAGTAATTTTCTTATAGGGAGTACATATGGGTTTCTGGAGAATCGTCATCACCATCATTCTGCCGCCGCTCGGCGTGCTGCTCGGTAAAGGGTTCGGTTGGGCGTTCATTATTAATATTCTGTTGA | yqaE |
| b2687_NC_000913_-200+100 | GAAGCCGCTGATACCGAACCGTTTGCGGTGTGGCTGGAAAAACACGCCTGACAGAAAAGAAAAAGGCCACTCGTGAGTGGCCAAAATTTCATCTCTGAATTCAGGGATGATGATAACAAATGCGCGTCTTTCATATACTCAGACTCGCCTGGGAAGAAAGAGTTCAGAAAATTTTTAAAAAAATTACCGGAGGTGGCTAAATGCCGTTGTTAGATAGCTTCACAGTCGATCATACCCGGATGGAAGCGCCTGCAGTTCGGGTGGCGAAAACAATGAACACCCCGCATGGCGACGCAATCA | luxS |
Table A6.
| Locus_Genome_Range | Sequence | Gene |
|---|---|---|
| b2702_NC_000913_-200+100 | CGCACAAGGAAGCGGTAGTCACTGCCCGATACGGACTTTACATAACTCAACTCATTCCCCTCGCTATCCTTTTATTCAAACTTTCAAATTAAAATATTTATCTTTCATTTTGCGATCAAAATAACACTTTTAAATCTTTCAATCTGATTAGATTAGGTTGCCGTTTGGTAATAAAACAATAAATCCTGAAGGAGAGAACAATGATAGAAACCATTACTCATGGTGCAGAGTGGTTTATCGGGCTGTTCCAAAAGGGCGGAGAGGTGTTTACCGGGATGGTGACCGGCATTCTTCCGCTGT | srlA |
| b2715_NC_000913_-200+100 | TGCACAATCGGCGGGAAAAATATTCAGGTGACCGGTTTCACAAATATAAAAAATGAACAATTCACTCTCTTGCTTATTTAGTGACAACTATTCATGATTTTGTGAAACCGGTTTCTTAATTCCGTTTCAGCATCGGCATTTTTCCGTCACGTCGACTGATAACAACTACATCTACCCTACTGATAACAGGATAAAATCCGATGGCCAAAAATTATGCGGCGCTGGCACGCTCGGTGATAGCGGCACTGGGCGGCGTTGATAACATCTCGGCGGTCACGCACTGTATGACGCGGTTGCGCT | ascF |
| b2731_NC_000913_-200+100 | ACTGGGGAAAGACGCGGCGCTGATTGGTGAAGTGGTGGAACGTAAAGGTGTTCGTCTTGCCGGTCTGTATGGCGTGAAACGAACCCTCGATTTACCACACGCCGAACCGCTTCCGCGTATATGCTAATAAAATTCTAAATCTCCTATAGTTAGTCAATGACCTTTTGCACCGCTTTGCGGTGCTTTCCTGGAAGAACAAAATGTCATATACACCGATGAGTGATCTCGGACAACAAGGGTTGTTCGACATCACTCGGACACTATTGCAGCAGCCCGATCTGGCCTCGCTGTGTGAGGCTC | fhlA |
| b2741_NC_000913_-200+100 | CGGGAACAACAAGAAGTTAAGGCGGGGCAAAAAATAGCGACCATGGGTAGCACCGGAACCAGTTCAACACGCTTGCATTTTGAAATTCGTTACAAGGGGAAATCCGTAAACCCGCTGCGTTATTTGCCGCAGCGATAAATCGGCGGAACCAGGCTTTTGCTTGAATGTTCCGTCAAGGGATCACGGGTAGGAGCCACCTTATGAGTCAGAATACGCTGAAAGTTCATGATTTAAATGAAGATGCGGAATTTGATGAGAACGGAGTTGAGGTTTTTGACGAAAAGGCCTTAGTAGAACAGG | rpoS |
| b2801_NC_000913_-200+100 | ATGGTAGTCACATAAAGTCACCTTCTAGCTAATAAGTGTGACCGCCGTCATATTACAGAGCGTTTTTTATTTGAAAATGAATCCATGAGTTCATTTCAGACAGGCAAATATTCACTGATATGAAGCCCGAACTCGCTGGTTTTGCACTTTTGAAAACATAACCGATTACGTGCTTAAGCTTCTGAACCTAAGAGGATGCTATGGGAAACACATCAATACAAACGCAGAGTTACCGTGCGGTAGATAAAGATGCAGGGCAAAGCAGAAGTTACATTATTCCATTCGCGCTGCTGTGCTCAC | fucP |
| b3224_NC_000913_-200+100 | CGCTGTGCCGCAAACCGTTTGGACCGGTAGATGAAAAATATCTGCCAGAACTGAAGGCGCTGGCCCAGCAGTTGATGCAAGAGCGCGGGTGAGTTGTTTCCCCTCGCTCGCCCCTACCGGGTGAGGGGAAATAAACGCATCTGTACCCTACAATTTTCATACCAAAGCGTGTGGGCATCGCCCACCGCGGGAGACTCACAATGAGTACTACAACCCAGAATATCCCGTGGTATCGCCATCTCAACCGTGCACAATGGCGCGCATTTTCCGCTGCCTGGTTGGGATATCTGCTTGACGGTT | nanT |
| b3365_NC_000913_-200+100 | ATCTATTTCTATAAACCCGCTCATTTTGTCTATTTTTTGCACAAACATGAAATATCAGACAATTCCGTGACTTAAGAAAATTTATACAAATCAGCAATATACCCATTAAGGAGTATATAAAGGTGAATTTGATTTACATCAATAAGCGGGGTTGCTGAATCGTTAAGGTAGGCGGTAATAGAAAAGAAATCGAGGCAAAAATGAGCAAAGTCAGACTCGCAATTATCGGTAACGGTATGGTCGGCCATCGCTTTATCGAAGATCTTCTTGATAAATCTGATGCGGCCAACTTTGATATTA | nirB |
| b3546_NC_000913_-200+100 | CTAAAGTCTCTTTTCAAACTTGCATTTTTGTAAATTTGTGCTTCATGCACACTCTTTCCCCACACTTTTTCCCTTTGCTGTGGTCTACTTATTCGCGCGTGTAGATTTTACTTATCTGACTACCTCCGCACTTTTTCCCTGCCGGGCCTGAAAAGCCACTAAGCAGGGTGTTATCACCTGTTTGTCCAGGGTTTGTTTGCATGAGATACATCAAATCGATTACACAGCAGAAGCTGAGCTTTTTGCTTGCAATCTATATTGGCCTTTTTATGAATGGCGCGGTTTTTTACCGCCGCTTCG | eptB |
| b3962_NC_000913_-200+100 | TACGTACAGCGGAAACCTGCCGCTTAAACGGAGAGTATCGTCGATAAAAATCCAATAAAACGTCAGGGCAAAAGTAAGAAACAGACAAAGCAAAGGCCGCTCAGGATATAGCCAGATAAATGACGGGGATCAATTGGCTTACCCGCGATAAAATGTTACCATTCTGTTGCTTTTATGTATAAGAACAGGTAAGCCCTACCATGCCACATTCCTACGATTACGATGCCATAGTAATAGGTTCCGGCCCCGGCGGCGAAGGCGCTGCAATGGGCCTGGTTAAGCAAGGTGCGCGCGTCGCAG | sthA |
| b4311_NC_000913_-200+100 | GTATTTAATCTGGATCTCTGTTTATTTAAATAATGTGAAAAGAGATTTTTCACAGGAGACCTTATACAAAAAAATATAAAATACAGCTACCGGTTGCCAAAGACACTATAAGCCTGGCAAAAAAATATTACACAACATAAATGCTAATTGTTTATGCGGGCTTTGTATTGCTTTCTGTATCCTACAAATGAGTGAAATTTATGAAAAAGGCTAAAATACTTTCTGGCGTATTATTACTGTGCTTTTCGTCCCCATTAATTTCTCAGGCTGCGACACTGGACGTACGTGGTGGATATCGTA | nanC |
Author Contributions
Conceptualization, M.R.; Methodology, M.R., M.U. and F.G.; Software, F.G.; Validation, M.R. and F.G.; Writing–original draft preparation, M.R.; Writing–review and editing, M.R., F.G., M.U. and R.B.; Funding acquisition, R.B., M.R. All authors have read and agreed to the published version of the manuscript.
Funding
Supported by the German Research Foundation (Deutsche Forschungsgemeinschaft DFG) [BA2168/16-1, BA2168/21-1 and BA2168/3-3] and under Germany’s Excellence Strategy (CIBSS-EXC-2189-Project ID 390939984). The article processing charge was funded by the Baden-Wuerttemberg Ministry of Science, Research and Art and the University of Freiburg in the funding programme Open Access Publishing.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- 1.Gong J., Ju Y., Shao D., Zhang Q. Advances and challenges towards the study of RNA-RNA interactions in a transcriptome-wide scale. Quant. Biol. 2018;6 doi: 10.1007/s40484-018-0146-5. [DOI] [Google Scholar]
- 2.Wright P.R., Georg J. Workflow for a Computational Analysis of an sRNA Candidate in Bacteria. Methods Mol. Biol. 2018;1737:3–30. doi: 10.1007/978-1-4939-7634-8_1. [DOI] [PubMed] [Google Scholar]
- 3.Wright P.R., Richter A.S., Papenfort K., Mann M., Vogel J., Hess W.R., Backofen R., Georg J. Comparative genomics boosts target prediction for bacterial small RNAs. Proc. Natl. Acad. Sci. USA. 2013;110:3487–3496. doi: 10.1073/pnas.1303248110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharma C.M., Papenfort K., Pernitzsch S.R., Mollenkopf H.J., Hinton J.C.D., Vogel J. Pervasive post-transcriptional control of genes involved in amino acid metabolism by the Hfq-dependent GcvB small RNA. Mol. Microbiol. 2011;81:1144–1165. doi: 10.1111/j.1365-2958.2011.07751.x. [DOI] [PubMed] [Google Scholar]
- 5.Durand S., Storz G. Reprogramming of anaerobic metabolism by the FnrS small RNA. Mol. Microbiol. 2010;75:1215–1231. doi: 10.1111/j.1365-2958.2010.07044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mann M., Wright P.R., Backofen R. IntaRNA 2.0: Enhanced and customizable prediction of RNA-RNA interactions. Nucleic Acids Res. 2017;45:W435–W439. doi: 10.1093/nar/gkx279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Busch A., Richter A.S., Backofen R. IntaRNA: Efficient prediction of bacterial sRNA targets incorporating target site accessibility and seed regions. Bioinformatics. 2008;24:2849–2856. doi: 10.1093/bioinformatics/btn544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Umu S.U., Gardner P.P. A comprehensive benchmark of RNA–RNA interaction prediction tools for all domains of life. Bioinformatics. 2017;33:988–996. doi: 10.1093/bioinformatics/btw728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gelhausen R., Will S., Hofacker I.L., Backofen R., Raden M. IntaRNAhelix—Composing RNA-RNA interactions from stable inter-molecular helices boosts bacterial sRNA target prediction. J. Bioinform. Comput. Biol. 2019;17:1940009. doi: 10.1142/S0219720019400092. [DOI] [PubMed] [Google Scholar]
- 10.Argaman L., Altuvia S. fhlA repression by OxyS RNA: Kissing complex formation at two sites results in a stable antisense-target RNA complex. J. Mol. Biol. 2000;300:1101–1112. doi: 10.1006/jmbi.2000.3942. [DOI] [PubMed] [Google Scholar]
- 11.Wright P.R., Mann M., Backofen R. Structure and interaction prediction in prokaryotic RNA biology. Microbiol. Spectr. 2018;6 doi: 10.1128/microbiolspec.RWR-0001-2017. [DOI] [PubMed] [Google Scholar]
- 12.Mückstein U., Tafer H., Hackermüller J., Bernhart S.H., Stadler P.F., Hofacker I.L. Thermodynamics of RNA–RNA binding. Bioinformatics. 2006;22:1177–1182. doi: 10.1093/bioinformatics/btl024. [DOI] [PubMed] [Google Scholar]
- 13.Raden M., Ali S.M., Alkhnbashi O.S., Busch A., Costa F., Davis J.A., Eggenhofer F., Gelhausen R., Georg J., Heyne S., et al. Freiburg RNA tools: A central online resource for RNA-focused research and teaching. Nucleic Acids Res. 2018;46:W25–W29. doi: 10.1093/nar/gky329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moller T., Franch T., Hojrup P., Keene D.R., Bächinger H.P., Brennan R.G., Valentin-Hansen P. Hfq: A Bacterial Sm-like Protein that Mediates RNA-RNA Interaction. Mol. Cell. 2002;9:23–30. doi: 10.1016/S1097-2765(01)00436-1. [DOI] [PubMed] [Google Scholar]
- 15.Miladi M., Montaseri S., Backofen R., Raden M. Integration of accessibility data from structure probing into RNA-RNA interaction prediction. Bioinformatics. 2019;35:2862–2864. doi: 10.1093/bioinformatics/bty1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Altuvia S., Zhang A., Argaman L., Tiwari A., Storz G. The Escherichia coli OxyS regulatory RNA represses fhlA translation by blocking ribosome binding. EMBO J. 1998;17:6069–6075. doi: 10.1093/emboj/17.20.6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Lay N., Gottesman S. The Crp-activated small noncoding regulatory RNA CyaR (RyeE) links nutritional status to group behavior. J. Bacteriol. 2009;191:461–476. doi: 10.1128/JB.01157-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gogol E.B., Rhodius V.A., Papenfort K., Vogel J., Gross C.A. Small RNAs endow a transcriptional activator with essential repressor functions for single-tier control of a global stress regulon. Proc. Natl. Acad. Sci. USA. 2011;108:12875–12880. doi: 10.1073/pnas.1109379108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holmqvist E., Unoson C., Reimegard J., Wagner E.G.H. A mixed double negative feedback loop between the sRNA MicF and the global regulator Lrp. Mol. Microbiol. 2012;84:414–427. doi: 10.1111/j.1365-2958.2012.07994.x. [DOI] [PubMed] [Google Scholar]
- 20.Kawamoto H., Koide Y., Morita T., Aiba H. Base-pairing requirement for RNA silencing by a bacterial small RNA and acceleration of duplex formation by Hfq. Mol. Microbiol. 2006;61:1013–1022. doi: 10.1111/j.1365-2958.2006.05288.x. [DOI] [PubMed] [Google Scholar]
- 21.Mandin P., Gottesman S. Integrating anaerobic/aerobic sensing and the general stress response through the ArcZ small RNA. EMBO J. 2010;29:3094–3107. doi: 10.1038/emboj.2010.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moon K., Six D.A., Lee H.J., Raetz C.R.H., Gottesman S. Complex transcriptional and post-transcriptional regulation of an enzyme for lipopolysaccharide modification. Mol. Microbiol. 2013;89:52–64. doi: 10.1111/mmi.12257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bouvier M., Sharma C.M., Mika F., Nierhaus K.H., Vogel J. Small RNA binding to 5’ mRNA coding region inhibits translational initiation. Mol. Cell. 2008;32:827–837. doi: 10.1016/j.molcel.2008.10.027. [DOI] [PubMed] [Google Scholar]
- 24.Corcoran C.P., Podkaminski D., Papenfort K., Urban J.H., Hinton J.C.D., Vogel J. Superfolder GFP reporters validate diverse new mRNA targets of the classic porin regulator, MicF RNA. Mol. Microbiol. 2012;84:428–445. doi: 10.1111/j.1365-2958.2012.08031.x. [DOI] [PubMed] [Google Scholar]
- 25.Papenfort K., Bouvier M., Mika F., Sharma C.M., Vogel J. Evidence for an autonomous 5’ target recognition domain in an Hfq-associated small RNA. Proc. Natl. Acad. Sci. USA. 2010;107:20435–20440. doi: 10.1073/pnas.1009784107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Papenfort K., Pfeiffer V., Lucchini S., Sonawane A., Hinton J.C.D., Vogel J. Systematic deletion of Salmonella small RNA genes identifies CyaR, a conserved CRP-dependent riboregulator of OmpX synthesis. Mol. Microbiol. 2008;68:890–906. doi: 10.1111/j.1365-2958.2008.06189.x. [DOI] [PubMed] [Google Scholar]
- 27.Papenfort K., Podkaminski D., Hinton J.C.D., Vogel J. The ancestral SgrS RNA discriminates horizontally acquired Salmonella mRNAs through a single G-U wobble pair. Proc. Natl. Acad. Sci. USA. 2012;109:757–764. doi: 10.1073/pnas.1119414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Papenfort K., Said N., Welsink T., Lucchini S., Hinton J.C.D., Vogel J. Specific and pleiotropic patterns of mRNA regulation by ArcZ, a conserved, Hfq-dependent small RNA. Mol. Microbiol. 2009;74:139–158. doi: 10.1111/j.1365-2958.2009.06857.x. [DOI] [PubMed] [Google Scholar]
- 29.Pfeiffer V., Papenfort K., Lucchini S., Hinton J.C.D., Vogel J. Coding sequence targeting by MicC RNA reveals bacterial mRNA silencing downstream of translational initiation. Nat. Struct. Mol. Biol. 2009;16:840–846. doi: 10.1038/nsmb.1631. [DOI] [PubMed] [Google Scholar]
- 30.Beisel C.L., Storz G. The base-pairing RNA Spot 42 participates in a multioutput feedforward loop to help enact catabolite repression in Escherichia coli. Mol. Cell. 2011;41:286–297. doi: 10.1016/j.molcel.2010.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beisel C.L., Updegrove T.B., Janson B.J., Storz G. Multiple factors dictate target selection by Hfq-binding small RNAs. EMBO J. 2012;31:1961–1974. doi: 10.1038/emboj.2012.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen J., Gottesman S. Spot 42 sRNA regulates arabinose-inducible araBAD promoter activity by repressing synthesis of the high-affinity low-capacity arabinose transporter. J. Bacteriol. 2016 doi: 10.1128/JB.00691-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coornaert A., Lu A., Mandin P., Springer M., Gottesman S., Guillier M. MicA sRNA links the PhoP regulon to cell envelope stress. Mol. Microbiol. 2010;76:467–479. doi: 10.1111/j.1365-2958.2010.07115.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coornaert A., Chiaruttini C., Springer M., Guillier M. Post-transcriptional control of the Escherichia coli PhoQ-PhoP two-component system by multiple sRNAs involves a novel pairing region of GcvB. PLoS Genet. 2013;9:e1003156. doi: 10.1371/journal.pgen.1003156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Lay N., Gottesman S. A complex network of small non-coding RNAs regulate motility in Escherichia coli. Mol. Microbiol. 2012;86:524–538. doi: 10.1111/j.1365-2958.2012.08209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Desnoyers G., Masse E. Noncanonical repression of translation initiation through small RNA recruitment of the RNA chaperone Hfq. Genes Dev. 2012;26:726–739. doi: 10.1101/gad.182493.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vecerek B., Moll I., Afonyushkin T., Kaberdin V., Blasi U. Interaction of the RNA chaperone Hfq with mRNAs: Direct and indirect roles of Hfq in iron metabolism of Escherichia coli. Mol. Microbiol. 2003;50:897–909. doi: 10.1046/j.1365-2958.2003.03727.x. [DOI] [PubMed] [Google Scholar]
- 38.Wang J., Rennie W., Liu C., Carmack C.S., Prevost K., Caron M.P., Masse E., Ding Y., Wade J.T. Identification of bacterial sRNA regulatory targets using ribosome profiling. Nucleic Acids Res. 2015;43:10308–10320. doi: 10.1093/nar/gkv1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Balbontín R., Fiorini F., Figueroa-Bossi N., Casadesús J., Bossi L. Recognition of heptameric seed sequence underlies multi-target regulation by RybB small RNA in Salmonella enterica. Mol. Microbiol. 2010;78:380–394. doi: 10.1111/j.1365-2958.2010.07342.x. [DOI] [PubMed] [Google Scholar]
- 40.Bossi L., Figueroa-Bossi N. A small RNA downregulates LamB maltoporin in Salmonella. Mol. Microbiol. 2007;65:799–810. doi: 10.1111/j.1365-2958.2007.05829.x. [DOI] [PubMed] [Google Scholar]
- 41.Yang Q., Figueroa-Bossi N., Bossi L. Translation enhancing ACA motifs and their silencing by a bacterial small regulatory RNA. PLoS Genet. 2014;10:e1004026. doi: 10.1371/journal.pgen.1004026. [DOI] [PMC free article] [PubMed] [Google Scholar]