Abstract
Background
Strategies to prevent Mycobacterium tuberculosis (Mtb) infection are urgently required. In this study, we aimed to identify correlates of protection against Mtb infection.
Methods
Two groups of Mtb-exposed contacts of tuberculosis (TB) patients were recruited and classified according to their Mtb infection status using the tuberculin skin test (TST; cohort 1) or QuantiFERON (QFT; cohort 2). A negative reading at baseline with a positive reading at follow-up classified TST or QFT converters and a negative reading at both time points classified TST or QFT nonconverters. Ribonucleic acid sequencing, Mtb proteome arrays, and metabolic profiling were performed.
Results
Several genes were found to be differentially expressed at baseline between converters and nonconverters. Gene set enrichment analysis revealed a distinct B-cell gene signature in TST nonconverters compared to converters. When infection status was defined by QFT, enrichment of type I interferon was observed. A remarkable area under the curve (AUC) of 1.0 was observed for IgA reactivity to Rv0134 and an AUC of 0.98 for IgA reactivity to both Rv0629c and Rv2188c. IgG reactivity to Rv3223c resulted in an AUC of 0.96 and was markedly higher compared to TST nonconverters. We also identified several differences in metabolite profiles, including changes in biomarkers of inflammation, fatty acid metabolism, and bile acids. Pantothenate (vitamin B5) was significantly increased in TST nonconverters compared to converters at baseline (q = 0.0060).
Conclusions
These data provide new insights into the early protective response to Mtb infection and possible avenues to interfere with Mtb infection, including vitamin B5 supplementation.
Analysis of blood from highly exposed household contacts from The Gambia who never develop latent Mycobacterium tuberculosis infection shows distinct transcriptomic, antibody, and metabolomic profiles compared to those who develop latent tuberculosis infection but prior to any signs of infection.
Keywords: tuberculosis, immunology, RNA sequencing, mtb proteome arrays, mtb resisters
It is estimated that more than 2 billion people are latently infected with Mycobacterium tuberculosis (Mtb) worldwide, resulting in 10 million new cases and 1.6 million deaths each year [1]. Strategies to prevent Mtb infection prior to establishment of latency are urgently needed. A recent study showed the rate of sustained QuantiFERON (QFT) conversion was reduced by 45.4% with a booster BCG vaccination of adolescents in a high-transmission setting [2]. Evidence for natural resistance to Mtb infection has been shown in some healthcare workers [3], South African miners [4], and sailors [5] who all were highly exposed but never showed signs of latent Mtb infection (LTBI). A proportion of highly Mtb-exposed household contacts also shows no evidence of infection [6], and these individuals have a much lower rate of progression to tuberculosis (TB) disease than those with LTBI [7]. Recent studies from a Ugandan long-term cohort of Mtb “resisters” showed evidence of higher immunoglobulin (Ig) M antibody reactivity in the resisters, which suggests that the definition of infection may not be accurate using interferon (IFN)-γ responses to Mtb antigens alone [8]. Another recent article from an Indonesian cohort looking at early infection conversions (within 14 weeks of exposure) found BCG vaccination provided some protection, which decreased with increasing exposure [9].
Several cell subsets have been proposed to be involved in resistance to Mtb infection, including both innate (ie, macrophages) and adaptive (ie, B or T cells) subsets [10]. Genome-wide association analysis has also identified loci that are associated with innate or adaptive resistance to Mtb infection [11]. A recent study has shown that early clearance of Mtb is associated with enhanced heterologous (ie, trained) innate immune responses [12].
Our aim in this study was to conduct an unbiased profiling of the global immune space in Mtb infection converters and nonconverters in The Gambia.
METHODS
Study Participants
This study was nested within a larger study of household contacts at Medical Research Council Unit The Gambia (MRCG). Household contacts of confirmed TB cases were consecutively recruited to this study within 2 weeks of diagnosis of the TB index case (Supplementary Figure 1). All participants were symptom screened to rule out active TB disease, and infection status was determined using either the tuberculin skin test (TST) or QFT (see the Supplementary Methods for details). Whole blood RNA was stabilized in Paxgene RNA blood tubes and stored at −80°C until analysis. Heparinized blood was centrifuged (600gmax, 10 minutes), and the plasma collected and stored at −80oC until analysis.
RNA Sequencing
RNA was extracted using an RNeasy mini kit (Qiagen, Germany) according to the manufacturer’s instructions and shipped to the Beijing Genome Institute (Hong Kong) (see Supplementary Methods for further details).
IgG and IgA Mtb Proteome Arrays
IgG and IgA serum antibody reactivity to 4000 Mtb antigens was analyzed using a full proteome microarray by Antigen Discovery Inc. (USA) as previously described [13, 14] (see the Supplementary Methods for further details).
Metabolomic Profiling
Sample preparation was carried out as described previously [15] at Metabolon, Inc (see the Supplementary Methods for further details).
Statistical Analyses
See the Supplementary Methods for a detailed description of the analysis pipeline. Multiple testing was corrected using the Benjamini-Hochberg procedure for all analyses.
RESULTS
Participant Demographics
Analysis of TST at baseline and 3 months defined 2 participant groups: cohort 1, TST converters (those who were negative [0 mm] at baseline and converted to positive [>10 mm] by 3 months) and TST nonconverters (ie, those who remained at 0 mm at both time points; Supplementary Figure 1). Similarly, analysis of QFT at baseline and 6 months defined QFT converters and nonconverters (cohort 2). An exposure score was determined based on the smear grade of the index case (all index cases had a smear grade of 2 or above) and sleeping proximity of the contact to the index case (Supplementary Figure 1). For metabolic profiling and Mtb proteome arrays, paired plasma samples from 40 converters and 38 nonconverters (adults from cohort 1 only) at baseline and 3 months were sent to Metabolon and to Antigen Discovery for metabolomics and antibody arrays, respectively. There was no significant difference in age or sex between the groups, but there was a significantly higher proportion of converters living in the same room as the index case (ie, closest proximity; P = .0340) compared to nonconverters (Table 1). For RNA sequencing, 2 batches of analysis were performed based on TST (cohort [batch] 1) or QFT (cohort [batch] 2). For batch 1, 100 baseline RNA samples were sequenced. Quality control prior to library preparation revealed that only 46/100 samples met the quality standards and were appropriate for sequencing (RNA Integrity Number [RIN] ≥ 7.0), presumably due to the long storage duration (up to 13 years). Of these 46 samples, 35 were TST nonconverters and 11 were TST converters (Table 1). There was a similar proportion of males in both groups, but a significant difference in age (median [interquartile range] of nonconverters = 10 [5–17] compared to 20 [16–32] for the converters; P = .0005). There was no significant difference in proximity or BCG vaccination status between the groups. For cohort 2, samples from 16 QFT nonconverters and 11 QFT converters were sequenced (Table 1). No significant difference in age, sex, BCG vaccination status, or proximity was seen between the groups. Progression to active disease was assessed in all participants for a 24-month period with only 1 QFT converter progressing after 12 months and none of the nonconverters progressing. No progressors were seen in the TST converter/nonconverter group.
Table 1.
Participant Demographics
| Demographic | Nonconverters | Converters | P Value |
|---|---|---|---|
| Plasma | n = 40 | n = 38 | |
| Age, median (IQR) | 22 (18–34) | 26 (20–37) | ns |
| Male, n (%) | 14 (35) | 14 (37) | ns |
| Proximity, n (%) | |||
| Different house | 15 (38) | 10 (26) | ns |
| Different room | 21 (53) | 17 (45) | ns |
| Same room | 4 (10) | 11 (29) | .0340 |
| RNA tuberculin skin test | n = 35 | n = 11 | |
| Age, median (IQR) | 10 (5–17) | 20 (16–32) | .0005 |
| Male, n (%) | 18 (51) | 5 (45) | ns |
| BCG vaccine, n (%) | 15 (38) | 4 (40) | ns |
| Proximity, n (%) | |||
| Different house | 19 (54) | 6 (55) | ns |
| Different room | 14 (40) | 5 (45) | ns |
| Same room | 3 (9) | 2 (18) | ns |
| RNA QuantiFERON | n = 16 | n = 11 | |
| Age, median (IQR) | 29 (19–47) | 18 (17–34) | ns |
| Male, n (%) | 8 (50) | 4 (36) | ns |
| BCG vaccine, n (%) | 9 (59) | 8 (71) | ns |
| Proximity, n (%) | |||
| Different house | 0 | 0 | ns |
| Different room | 0 | 0 | ns |
| Same room | 16 (100) | 11 (100) | ns |
Abbreviations: BCG, Bacillus Calmette–Guérin; IQR, interquartile range; ns, not significant.
RNA Sequencing
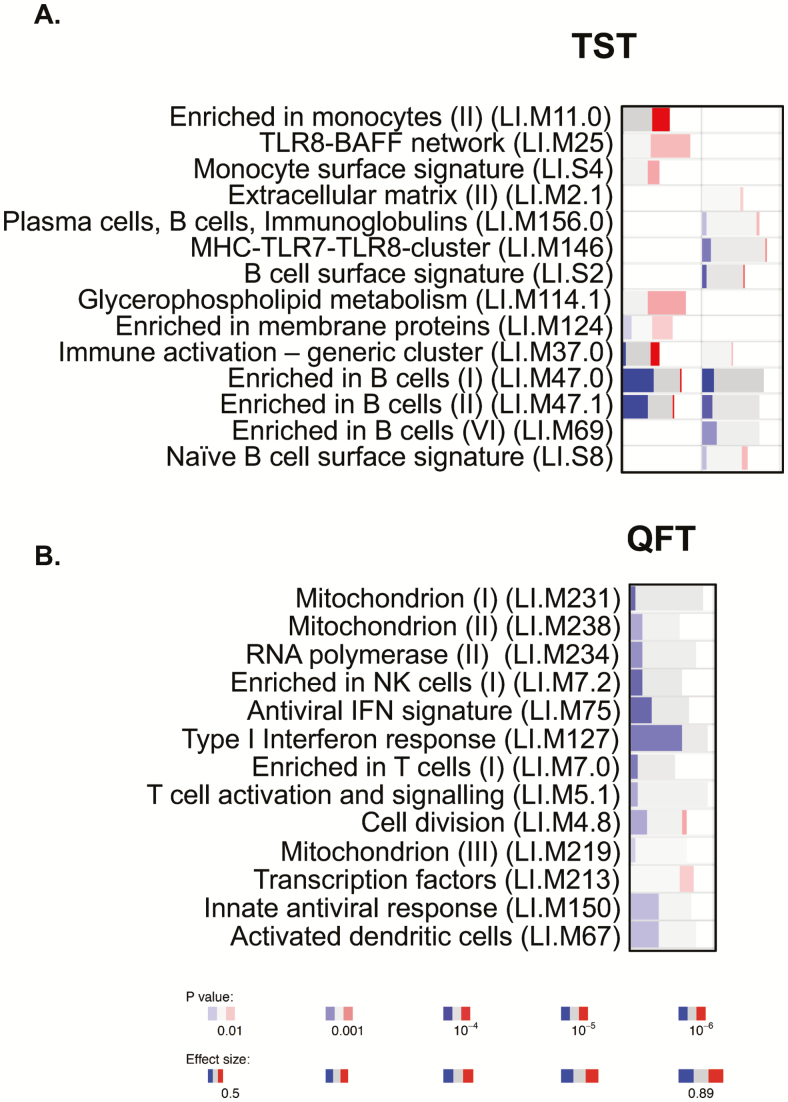
Several differentially expressed genes were observed between converters and nonconverters at baseline (Table 2). For the TST-defined group, the most differentially expressed genes included TMEM56 (P = 1.25E-08, q < 10E-03), RP11-364L4.1 (P = 4.52E-08, q < 10E-03), LCN2 (P = 1.81E-07, q < 10E-03), CLIC2 (P = 3.50E-07, q < 10E-02), and WASHC3 (P = 3.54E-07, q < 10E-02; Table 2). For the QFT-defined group, the most differentially expressed genes included RPS9 (P = 3.14E-10), MDM4 (P = 6.09E-07), HSFX3 (7.6E-07), and IGF1R (P = 5.25E-06), all of which were upregulated in the converters (Table 3, Supplementary Table 1). Due to the significant difference in age between the TST converters and nonconverters, we also performed filtered analysis of adults only (Table 4). Interestingly, the top genes in this group included several immune-regulatory genes such as CXCL10 (P = 3.10E-05), HLA-DQB1 (P = 8.53E-04), and CD22 (P = 1.58E-03). However, significance was lost after adjusting for the false discovery rate (FDR), most likely due to low numbers in both groups. Next, we performed gene set enrichment analysis using the R-package tmod. In the total TST-defined group, there was a strong and statistically significant enrichment in B cell–related genes in the TST nonconverters (Figure 1A, left), which was still evident in the filtered subgroup (Figure 1A, right). TST converters had significant enrichment for TLR8-BAFF and monocyte-related genes (Figure 1A), although this was lost when filtering was performed. The responses in the QFT nonconverters were dominated by IFN type I and antiviral gene signatures (LI.M75 and LI.M127; Figure 1B). The enriched B cell–related modules included, among others, the genes BLK, CD19, CD22, CD24, CD72, CD79, CD200, CXCR5, CR2, and FCR1/2. The genes in the enriched IFN-related modules included TAP1, IFIH1, IRF7, PARP9, STAT1, PLSCR1, IFITM1, HERC5, DDX60, USP18, RSAD2, and IFIT1. It is important to note that there were no significant differences in white blood cell counts between converters and nonconverters (data not shown).
Table 2.
Differentially Expressed Genes in the Tuberculin Skin Test Group With a False Discovery Rate ≤0.01
| Gene | Log-fold Change | P Value | False Discovery Rate | Description |
|---|---|---|---|---|
| TMEM56 | 2.29 | 1.25E-08 | 1.95E-04 | Transmembrane protein 56; protein coding |
| RP11-364L4.1 | 2.83 | 4.52E-08 | 3.54E-04 | Pseudogene |
| LCN2 | 2.42 | 1.81E-07 | 9.43E-04 | Lipocalin 2; iron sequestering; innate immunity |
| CLIC2 | 1.22 | 3.50E-07 | 1.11E-03 | Chloride intracellular channel; regulates cell processes |
| WASHC3 | 0.60 | 3.54E-07 | 1.11E-03 | Endocytosis |
| AC010642.1 | −1.50 | 8.25E-07 | 2.15E-03 | Zinc finger protein pseudogene |
| CCDC153 | 1.43 | 1.25E-06 | 2.79E-03 | Coiled-coil domain-containing protein 153 |
| HNMT | 1.28 | 1.60E-06 | 3.01E-03 | Inactivates histamine by N-methylation |
| FAM114A1 | 1.59 | 1.73E-06 | 3.01E-03 | Protein coding gene; neuronal cell development |
| MED20 | 0.92 | 2.03E-06 | 3.18E-03 | Transcriptional coactivator complex |
| RAB10 | 0.80 | 3.17E-06 | 4.36E-03 | Regulates intracellular vesicle trafficking |
| TCF4 | −0.89 | 3.52E-06 | 4.36E-03 | Transcription factor 4; extracellular signal regulated kinase signaling |
| CXCL9 | 2.39 | 3.62E-06 | 4.36E-03 | T-cell trafficking; antimicrobial |
| SLC14A1 | 1.80 | 3.95E-06 | 4.42E-03 | Mediates urea transport in erythrocytes |
| ATG3 | 0.67 | 4.56E-06 | 4.76E-03 | Regulation of autophagy during cell death |
| ANAPC13 | 0.79 | 4.93E-06 | 4.82E-03 | Class I major histocompatibility complex antigen processing and presentation |
| TRIQK | 0.75 | 5.89E-06 | 5.20E-03 | Cell growth and maintenance of cell morphology |
| GLRX | 1.10 | 5.98E-06 | 5.20E-03 | Member of glutaredoxin family; antioxidant defense |
| NCOA4 | 0.98 | 8.41E-06 | 6.92E-03 | Androgen receptor binding |
| BNIP3L | 1.26 | 9.52E-06 | 7.45E-03 | Pro-apoptotic subfamily within B-cell lymphoma-2 family of proteins |
| LINC00662 | 1.23 | 1.05E-05 | 7.68E-03 | Noncoding RNA |
| DPCD | 0.95 | 1.08E-05 | 7.68E-03 | Possible role in the formation/function of ciliated cells |
| ARV1 | 1.09 | 1.15E-05 | 7.84E-03 | Endoplasmic reticulum cholesterol and bile acid homeostasis |
| ARL6IP5 | 0.73 | 1.35E-05 | 8.27E-03 | Regulates intracellular concentrations of taurine and glutamate; expression affected by vitamin A |
| FBXO9 | 0.78 | 1.37E-05 | 8.27E-03 | F-box protein family; phosphorylation-dependent ubiquitination |
| DENND5B | −1.24 | 1.37E-05 | 8.27E-03 | Promotes the exchange of guanosine diphosphate to guanosine triphosphate |
| TBC1D27 | −1.14 | 1.47E-05 | 8.37E-03 | Pseudogene |
| OLFM4 | 3.14 | 1.50E-05 | 8.37E-03 | Olfactomedin 4; antiapoptotic factor that promotes tumor growth |
| TMEM55A | 1.02 | 1.57E-05 | 8.49E-03 | Superpathway of inositol phosphate compounds |
| EVPL | 1.42 | 1.81E-05 | 9.44E-03 | Member of the plakin family of proteins; component of desmosomes and epidermal cornified envelope |
| IGFBP7 | 0.97 | 1.98E-05 | 9.79E-03 | Member of the insulin-like growth factor–binding protein family |
| DCAF6 | 0.72 | 2.03E-05 | 9.79E-03 | Ligand-dependent coactivator of nuclear receptors |
| FAM26F | 1.32 | 2.07E-05 | 9.79E-03 | Calcium homeostasis modulator family member 6 |
Table 3.
Differentially Expressed Genes in the QuantiFERON Group With a False Discovery Rate ≤0.05
| Gene | Log-fold Change | P Value | False Discovery Rate | Description |
|---|---|---|---|---|
| RPS9 | 5.39 | 3.14E-10 | 4.87E-06 | Ribosomal protein S9 |
| MDM4 | 0.73 | 6.09E-07 | 3.97E-03 | Mouse double minute 4 (MDM4), p53 regulator |
| HSFX3 | 6.04 | 7.66E-07 | 3.97E-03 | Heat shock transcription factor family |
| RP60S | −0.92 | 4.10E-06 | 1.59E-02 | 60S ribosomal protein pseudogene |
| IGF1R | 0.72 | 5.25E-06 | 1.63E-02 | Insulin-like growth factor 1 receptor |
| STRN3 | 0.80 | 9.23E-06 | 1.80E-02 | Striatin 3; calmodulin binding protein |
| RPL39P3 | −0.85 | 9.27E-06 | 1.80E-02 | Ribosomal protein L39 pseudogene 3 |
| PCNA | −0.63 | 9.60E-06 | 1.80E-02 | Proliferating cell nuclear antigen |
| PCK2 | −0.62 | 1.18E-05 | 1.80E-02 | Mitochondrial |
| POLR2L | −0.63 | 1.26E-05 | 1.80E-02 | RNA polymerase II subunit L |
| TFP1 | 5.47 | 1.31E-05 | 1.80E-02 | Transferrin pseudogene 1 |
| SYNE3 | 0.59 | 1.39E-05 | 1.80E-02 | Actin binding |
| LAMTOR2 | −0.68 | 1.71E-05 | 1.82E-02 | Mitogen activated protein kinases and mammalian target of rapamycin activator 2 |
| RNF181 | −0.60 | 1.73E-05 | 1.82E-02 | Ring finger protein 181 |
| NKTR | 0.73 | 1.82E-05 | 1.82E-02 | Natural killer cell triggering receptor |
| PMVK | −0.60 | 1.87E-05 | 1.82E-02 | Phosphomevalonate kinase |
| C19orf70 | −0.60 | 2.07E-05 | 1.89E-02 | Chromosome 19 open reading frame 70 |
| DPM3 | −0.67 | 2.18E-05 | 1.89E-02 | Dolichyl-phosphate mannosyltransferase |
| HLA-DMA | −6.98 | 2.51E-05 | 2.06E-02 | MHC class II, DM alpha |
| HLA-B | −6.14 | 2.90E-05 | 2.13E-02 | MHC class I, B |
| HNRNPH1 | 0.65 | 2.96E-05 | 2.13E-02 | Heterogeneous nuclear ribonucleoprotein H1 |
| RARRES3 | −0.67 | 3.02E-05 | 2.13E-02 | Retinoic acid receptor responder 3 |
| RBM25 | 0.58 | 4.35E-05 | 2.74E-02 | RNA binding motif protein 25 |
| LILRB3 | −3.44 | 4.46E-05 | 2.74E-02 | Leukocyte immunoglobulin like receptor B3 |
| TMSB10 | −0.59 | 4.54E-05 | 2.74E-02 | Thymosin beta 10 |
| STX16 | 0.48 | 4.61E-05 | 2.74E-02 | Syntaxin 16 |
| FBXO6 | −0.80 | 4.77E-05 | 2.74E-02 | F-box protein 6 |
| MALAT1 | 0.69 | 5.26E-05 | 2.90E-02 | Metastasis-associated lung adenocarcinoma |
| 0.87 | 5.47E-05 | 2.90E-02 | Novel transcript, sense intronic to suppressor of tumorigenicity | |
| RPL29P11 | 2.33 | 5.60E-05 | 2.90E-02 | Ribosomal protein L29 pseudogene 11 |
| LMBR1L | 0.47 | 6.41E-05 | 3.20E-02 | Limb development membrane protein 1 like |
| TMEM170B | 0.80 | 6.59E-05 | 3.20E-02 | Transmembrane protein 170B |
| LTB | 5.44 | 7.12E-05 | 3.35E-02 | Lymphotoxin beta |
| PLEKHF1 | −0.69 | 7.33E-05 | 3.35E-02 | Triggers caspase-independent apoptosis |
| SERPING1 | −1.76 | 7.85E-05 | 3.49E-02 | Complement component 1 inhibitor |
| LINC01089 | 0.67 | 8.31E-05 | 3.59E-02 | Long intergenic nonprotein coding RNA 1089 |
| 0.96 | 9.11E-05 | 3.83E-02 | Uncharacterized LOC100288123 | |
| SF3B1 | 0.49 | 9.67E-05 | 3.87E-02 | Splicing factor 3b subunit 1 |
| NDUFA4 | −0.50 | 9.71E-05 | 3.87E-02 | NDUFA4, mitochondrial complex associated |
| NKG7 | −0.85 | 1.05E-04 | 4.01E-02 | Natural killer cell granule protein 7 |
| ULK1 | 0.53 | 1.06E-04 | 4.01E-02 | unc-51 like autophagy activating kinase 1 |
| SRSF11 | 0.57 | 1.15E-04 | 4.14E-02 | Serine and arginine rich splicing factor 11 |
| EPSTI1 | −1.18 | 1.17E-04 | 4.14E-02 | Epithelial stromal interaction 1 |
| EIF4A1 | 0.98 | 1.17E-04 | 4.14E-02 | Eukaryotic translation initiation factor 4A1 |
| ATP11A | 0.47 | 1.23E-04 | 4.24E-02 | ATPase phospholipid transporting 11A |
| PSMG3 | −0.57 | 1.52E-04 | 4.91E-02 | Proteasome assembly chaperone 3 |
| OAS1 | −1.46 | 1.53E-04 | 4.91E-02 | 2’-5’-oligoadenylate synthetase 1 |
| SERINC5 | 0.69 | 1.54E-04 | 4.91E-02 | Serine incorporator 5 |
| BANF1 | −0.57 | 1.58E-04 | 4.91E-02 | Barrier to autointegration factor 1 |
| FOXK1 | 0.55 | 1.59E-04 | 4.91E-02 | Forkhead box K1 |
| AQR | 0.67 | 1.61E-04 | 4.91E-02 | Aquarius intron-binding spliceosomal factor |
| PAN3 | 0.45 | 1.66E-04 | 4.93E-02 | Poly(A)-specific ribonuclease subunit PAN3 |
| ECI1 | −0.55 | 1.68E-04 | 4.93E-02 | Enoyl-CoA delta isomerase 1 |
| ZMYND15 | 1.10 | 1.73E-04 | 4.98E-02 | Zinc finger myeloid, Nervy, and DEAF-1-type containing 15 |
Abbreviations; CoA, co-enzyme A; MHC, major histocompatibility complex.
Table 4.
Differentially Expressed Genes in the Tuberculin Skin Test Group Aged ≥18 Years
| Gene | Log-fold Change | P Value | False Discovery Rate |
|---|---|---|---|
| CXCL10 | 3.97 | 3.10E-05 | ns |
| MTRNR2L1 | 4.19 | 3.23E-05 | ns |
| LILRB2 | 3.61 | 4.67E-04 | ns |
| HNMT | 1.40 | 4.89E-04 | ns |
| EEF1A1P5 | 1.50 | 6.91E-04 | ns |
| ADAMTS1 | 1.51 | 7.54E-04 | ns |
| HLA-DQB1 | −5.65 | 8.53E-04 | ns |
| SRGAP3 | −1.10 | 9.82E-04 | ns |
| HOPX | 0.85 | 1.12E-03 | ns |
| CD22 | −0.84 | 1.58E-03 | ns |
| CLIC2 | 1.16 | 1.65E-03 | ns |
| RP11-394B2.5 | −0.85 | 1.70E-03 | ns |
| FAM26F | 1.60 | 1.71E-03 | ns |
| CCDC114 | −1.22 | 1.74E-03 | ns |
| GSKIP | 0.67 | 1.94E-03 | ns |
| MRPS35 | 0.60 | 2.08E-03 | ns |
| RP11-374P20.4 | −1.44 | 2.19E-03 | ns |
| LCN2 | 2.07 | 2.43E-03 | ns |
| AC020951.1 | −1.17 | 2.68E-03 | ns |
| KIF19 | −1.02 | 2.74E-03 | ns |
Abbreviation: ns, not significant.
Figure 1.
Modular analysis of RNA sequencing data. A, Analysis of tuberculin skin test converters compared to nonconverters. Left column is unfiltered data, and right column is only participants aged ≥18 years. B, Analysis of QuantiFERON converters compared to nonconverters (all aged ≥18 years). RNA sequencing data were analyzed using Tmod analysis (R-statistics). Depth of color indicates significance level, and width indicates effect size (area under the curve). Blue indicates higher in nonconverters, and red indicates higher in converters. Abbreviations: IFN, interferon; NK, natural killer; QFT, QuantiFERON; TST, tuberculin skin test.
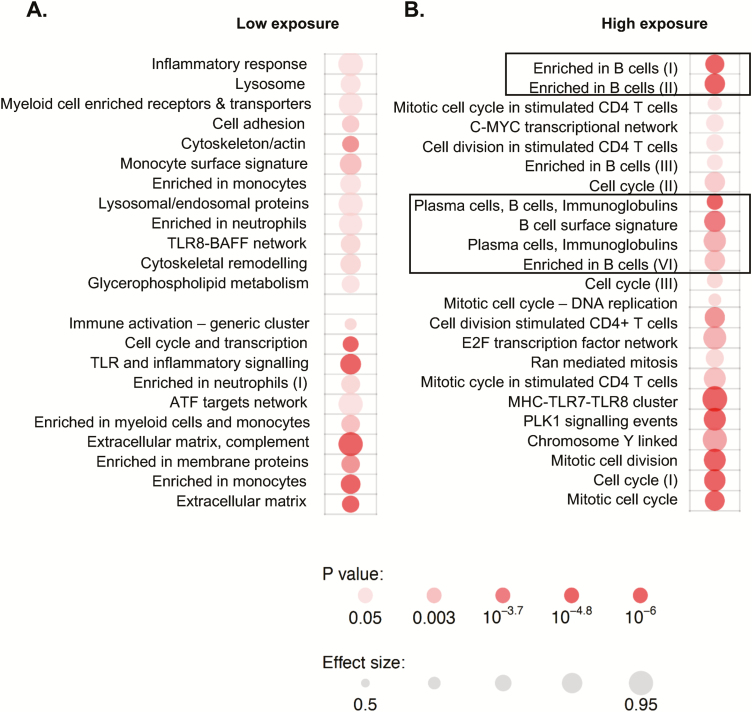
Given the differences in exposure of each participant to the index TB patient, we next assessed how this affected gene expression profiles (Figure 2). In the participants with the lowest exposure, the enriched gene modules were related to innate immune cells, including neutrophils, monocytes, and Toll-like receptor (TLR) pathways. The B-cell signature enrichment was only evident in those with the highest exposure to the index TB case (Figure 2).
Figure 2.
Exposure level influences gene expression profiles. A, Analysis of tuberculin skin test (TST) converters vs nonconverters with low exposure. B, Analysis of TST converters vs nonconverters with high exposure. Exposure was defined based on smear grade of the index case and sleeping proximity of the contact to the index case. Only the high-exposure group showed the B-cell signature seen when all samples were analyzed (rectangles). Depth of color indicates significance level, and size indicates effect. Abbreviations: ATF, activating transcription factor; C-MYC, c-Master Regulator of Cell Cycle Entry and Proliferative Metabolism; TLR, Toll-like receptor.
Mtb Antibody Arrays
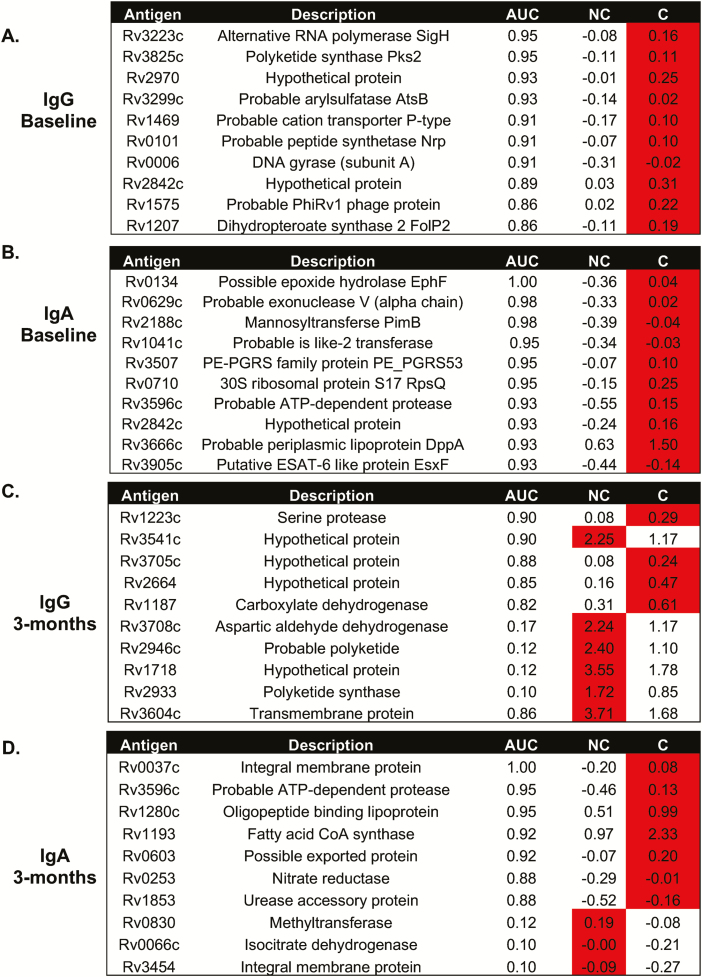
Analysis of both IgG and IgA reactivity to the Mtb proteome showed similar results, with extensive differences in reactivity evident (Supplementary Table 2). The most differential responses were seen in those with the highest exposure (proximity 3). Analysis of IgG responses at baseline to Rv2131c, Rv0363c, and Rv3223c resulted in an area under the curve (AUC) of 0.73, 0.83, and 0.96, respectively (Figure 3A). With IgA reactivity to Rv0134 at the closest proximity, the AUC was 1.00 (Figure 3B). The reactivity was higher in the converters compared to the nonconverters at baseline (Figure 3A and 3B). At 3 months, differential responses were still evident with an AUC of 0.90 seen for Rv1223-IgG levels and an AUC of 1.00 for Rv0037c-IgA levels (both higher in the converters). IgG levels to Rv3541c were higher in the nonconverters compared to converters at 3 months, with an AUC of 0.90. Interestingly, the only antigen to elicit a similar response at 3 months compared to baseline was Rv3596c-IgA, which was higher in the converters at both time points and gave an AUC of 0.93 at baseline and 0.95 at 3 months.
Figure 3.
Differential antibody responses to the Mycobacterium tuberculosis (Mtb) proteome. Analysis of IgG (A and C) and IgA (B and D) antibody responses to Mtb antigens at baseline (A and B) and 3 months (C and D). The most differentially expressed antibodies are shown. Red indicates the group with the highest response. Abbreviations: AUC, area under the curve; C, converters; Ig, immunoglobulin; NC, nonconverters.
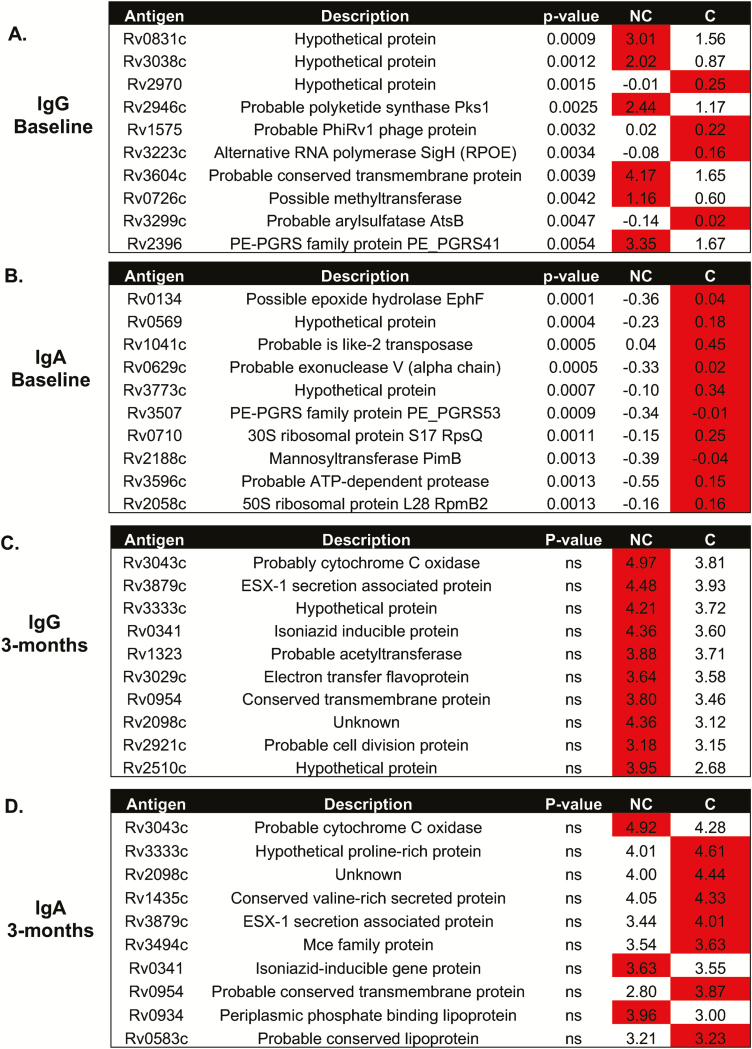
We also analyzed antigens that gave the greatest reactivity (ie, highest antibody levels). For the nonconverters at baseline, the highest IgG responses were seen for Rv0831c (log-fold change [LFC], 1.93), Rv3038c (LFC, 2.32), Rv2946c (LFC, 2.09), Rv3604c (LFC, 2.53), Rv0726c (LFC, 1.93), and Rv2396 (LFC, 2.00; Figure 4A), whereas IgA responses were always lower in the TST nonconverters (Figure 4B). By 3 months, there were no significant differences between the 2 groups for either IgG or IgA responses (Figures 4C and 4D), with high levels in both converters and nonconverters. IgG responses were all elevated in the nonconverters by 3 months (Figure 4C), but the majority of IgA responses were increased in the converters at both time points (Figure 4D).
Figure 4.
Mycobacterium tuberculosis (Mtb) antigens with the highest reactivity. Analysis of IgG (A and C) and IgA (B and D) antibody responses to Mtb antigens at baseline (A and B) and 3 months (C and D). The antibodies showing the highest responses are shown. Red indicates the group with the highest response. Abbreviations: AUC, area under the curve; C, converters; Ig, immunoglobulin; NA, not applicable; NC, nonconverters.
Next, we analyzed reactivity to ESAT-6, dominant secretory antigens from Mtb. Only individuals in the closest proximity to the index case showed any differential antibody reactivity to ESAT-6 antigens, with the most significant being IgG reactivity to Rv3875 (P = .0008). IgA reactivity to Rv3872 (P = .025) and Rv3871 (P = .030) were both increased in converters compared to nonconverters at baseline (Supplementary Table 2).
Metabolomic Profiling
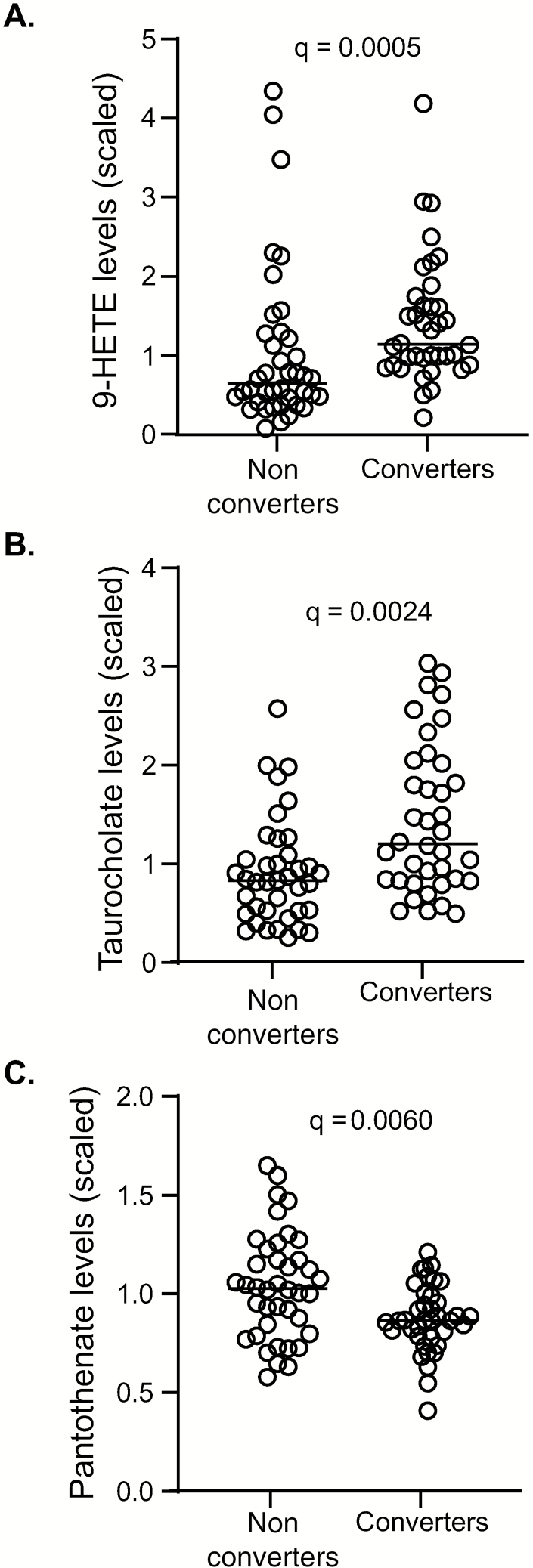
TST converters showed a relative increase in inflammatory biochemicals at baseline compared to nonconverters such as 9-HETE (q = 0.0005; Figure 5A). Primary bile acids such as glycochenodeoxycholate sulfate and taurocholenate were also increased in TST converters at baseline (q = 0.0024 for both; Figure 5B [taurocholenate shown]). Kynurenine was elevated in converters compared to nonconverters at 3 months, but differences were not significant after correcting for FDR (Supplementary Table 3). In nonconverters, increases were observed for pimelate, suberate, azelate, and undecanedioate at 3 months compared to baseline, with trends toward an increase in dodecanedioate and eicosanodioate, consistent with changes in omega-oxidation. Significantly higher levels in biochemicals related to beta-oxidation (eg, medium-chain or long-chain fatty acids, acylcarnitines, and the ketone body 3-hydroxybutyrate) and in arginine and homoarginine were seen in converters compared to nonconverters at both baseline and 3 months (Supplementary Table 3). There was also a significantly lower relative abundance in alpha-ketoglutarate and a higher relative abundance in homocitrulline in converters compared to nonconverters at baseline only (Supplementary Table 3). One of the few metabolites that was higher in TST nonconverters compared to converters was pantothenate (vitamin B5), with a normalized median (interquartile range) of 1.03 (0.81–1.22) compared to 0.87 (0.78–0.99) for the converters (q = 0.0060; Figure 5C).
Figure 5.
Metabolomic profiling of tuberculin skin test (TST) converters and nonconverters. Metabolite analysis of plasma from TST converters and nonconverters was performed. At baseline, increases in (A) 9-HETE and (B) taurocholenate levels in converters was seen. (C) Pantothenate levels were increased in nonconverters at baseline. Data were analyzed using the Mann–Whitney U test. Line indicates median.
DISCUSSION
We characterized host transcriptomic, metabolomic, and antibody responses to Mtb in TST nonconverters and converters prior to any signs of infection as determined by current methods. We have previously shown higher levels of soluble interleukin (IL)-17 in nonconverters at baseline [21], suggesting they have encountered Mtb and mounted an immune response and are not simply anergic to antigens in the current tests and that Mtb was not blocked by physiological barriers. In this study, we identified a distinct antigen-specific antibody signature reflected by differential antibody responses in TST converters and nonconverters to multiple Mtb antigens. Intriguingly, defining infection based on QFT rather than TST revealed a distinct type I IFN/antiviral gene signature.
Analysis of RNA from TST converters and nonconverters at baseline highlighted several differentially expressed genes. These included CXCL9 and BNIP3L, which were both upregulated in converters. CXCL9 is a T-cell chemoattractant induced by IFN-γ, which has been shown to exacerbate pathology in severe TB disease [22]. BNIP3L (BCL2 interacting protein 3 like) is a proapoptotic protein whose substrate is PARK2, which in turn is important for the ubiquitination and clearance of Mtb [23]. We also observed upregulation of SLC14A1, which is involved in urea transport, and of LCN2, which is involved in iron sequestration and innate immunity in converters prior to conversion. Polymorphisms in SLC14A1 have been shown to be associated with susceptibility to TB [24], while LCN2 (lipocalin 2) has been shown to confer a growth advantage on Mtb in the early stages of infection by supplying a source of iron to Mtb in infected macrophages [25]. When modular analysis was performed, we observed a significant enrichment of B-cell genes in TST nonconverters compared to converters, while converters had enrichment of monocyte genes and TLR8-BAFF, which is important in the early airway epithelial response to Mtb [26]. The B-cell genes of interest included CD72, which is thought to mediate B cell–T cell interaction, and CD19, CD22, CD24, and CD200, which are all phenotypic/maturation markers. High-affinity FCR genes are enriched in active TB compared to latent TB [27] and were recently shown to comprise prognostic markers of progression to active TB in Gambian household contacts [28].
Interestingly, different gene signatures were identified when QFT was used to diagnose infection status. While different mechanisms will be inevitable since TST is based on a delayed-type hypersensitivity reaction of 48–72 hours and QFT is based on overnight in vitro antigen stimulation of T cells, the upregulation of these signatures in the uninfected groups shows that they are likely to play a role in resistance of Mtb. Increased production of type I IFNs (IFN-α/-β) as part of the antiviral response inhibits the downstream effects of type II IFN (IFN-γ) responses known to be critical for Mtb control [29]. Thus, the increase in these genes in the QFT nonconverters was surprising. However, evidence has been presented that in the absence of a response to IFN-γ, type I IFN can play a nonredundant protective role against TB in mice [30] and is required for the production of IL-12 and tumor necrosis factor in humans [31]. This suggests that low abundance of type I IFNs may be protective in the context of a low burden of Mtb. It has also been shown in experimental models that type I IFN may play a protective role in the context of BCG-induced immunity and could be targeted to improve preventive vaccination against TB [32, 33]. B cells have gained recognition recently, with several studies highlighting the immunoregulatory role of B cells in controlling Mtb infections, which extends beyond their direct antibody-mediated effector functions [34, 35]. Furthermore, a previous study showed preferential IgA VH3‐23, D3‐3, JH4 gene usage in TST-negative nurses with high and repeated exposure to Mtb [36]. B cells have also been shown to be a source of type I IFN [37]. Thus, it is possible that the differences seen in our TST- and QFT-defined signatures (ie, B cells vs type I IFN) are actually reflecting related immunologic pathways to protect against early Mtb infection.
Because the enrichment of B-cell genes was unexpected, we analyzed IgG and IgA plasma antibody reactivity to all 4000 Mtb proteins from these individuals. TST converters had significantly higher levels of both IgG and IgA antibodies to multiple Mtb antigens at baseline, which allowed excellent discrimination between converters and nonconverters. This was particularly evident when proximity was accounted for; exposed contacts in closest proximity to the index case who later converted their TST had higher antibody levels at baseline, with levels increasing for both converters and nonconverters by 3 months. While this may relate to the length of exposure, which is difficult to ascertain in this cohort, our data suggest that antibodies are playing a role in the response to Mtb infection but are not necessarily protecting the host from becoming infected (although a few antigens elicited significantly higher IgG responses in the nonconverters). Since we identified a distinct B-cell signature in the nonconverters, we conclude that other antibody-independent functions of B cells are driving protection against Mtb infection, and this is currently being assessed in our laboratory.
We observed several differences in the relative abundances of circulating plasma metabolites, including changes in biomarkers of inflammation, fatty acid metabolism, and bile acids. Increases in inflammation-related metabolites in converters could indicate increased immune activation, with 1 previous study correlating IL-1β secretion with increased susceptibility to infection [38]. Interestingly, 1 of the only metabolites to be at statistically higher abundance in nonconverters compared to converters was pantothenate (vitamin B5). Vitamin B5 has recently been shown to significantly inhibit the growth of Mtb in mice by regulating innate immunity and adaptive immunity [39]. Together with our findings, this suggests that vitamin B5 could be harnessed for host-directed intervention to enhance management of TB.
A recent study from Uganda has shown an enhanced antibody reactivity to Mtb antigens in long-term Mtb resisters [8]. In addition, exposed but uninfected Chinese healthcare workers have been shown to have circulating protective antibodies [3], suggesting that these individuals have encountered antigen and have since cleared it. This supports the findings in our study and in another recent study from Indonesia [9] that show that enhanced antibody reactivities persist to clear the pathogen. The INFECT study in Indonesia also showed 45% protection from primary infection by BCG in the early clearer group [9], while we observed no influence of BCG. The level of protection by BCG in INFECT was strongest in those with low exposure, which may explain this difference.
In conclusion, despite being clinically healthy and showing no signs of latent infection by canonical diagnostic tests, we were able to demonstrate differential immune responses in patients who would later convert to a positive TST compared to those who remained TST negative. A potential shortcoming of our study is the relatively small number of evaluated participants and the unbalanced design (especially in the TST group), which may have had a negative impact on the robustness of our findings. An independent future validation of our study is necessary to confirm our predictions.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. The authors thank the National Tuberculosis (TB) Control Program, participants, and their families. The authors also thank the Medical Research Council Unit The Gambia TB Clinic staff and TB immunology and TB microbiology laboratory staff.
Financial support. The clinical cohort enrollment and follow-ups were funded through Medical Research Council (United Kingdom) core support for the TB Platform at the Medical Research Council Unit, The Gambia. All experimental analyses were funded by the European Union’s Horizon 2020 Research and Innovation Programme under project TBVAC2020 with grant agreement 643381, with subawards to J. S. S. and S. H. E. K.
Potential conflicts of interest. All authors report no potential conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. World Health Organization. Global Tuberculosis Report 2018. Available at: http://www.who.int/tb/publications/global_report/en/. Accessed 27 July 2019.
- 2. Nemes E, Geldenhuys H, Rozot V, et al. ; C-040-404 Study Team Prevention of M. tuberculosis infection with H4:IC31 vaccine or BCG revaccination. N Engl J Med 2018; 379:138–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Li H, Wang XX, Wang B, et al. . Latently and uninfected healthcare workers exposed to TB make protective antibodies against Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 2017; 114:5023–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hanifa Y, Grant AD, Lewis J, Corbett EL, Fielding K, Churchyard G. Prevalence of latent tuberculosis infection among gold miners in South Africa. Int J Tuberc Lung Dis 2009; 13:39–46. [PubMed] [Google Scholar]
- 5. Houk VN. Spread of tuberculosis via recirculated air in a naval vessel: the Byrd study. Ann N Y Acad Sci 1980; 353:10–24. [DOI] [PubMed] [Google Scholar]
- 6. Morrison J, Pai M, Hopewell PC. Tuberculosis and latent tuberculosis infection in close contacts of people with pulmonary tuberculosis in low-income and middle-income countries: a systematic review and meta-analysis. Lancet Infect Dis 2008; 8:359–68. [DOI] [PubMed] [Google Scholar]
- 7. Hill PC, Brookes RH, Fox A, et al. . Longitudinal assessment of an ELISPOT test for Mycobacterium tuberculosis infection. PLoS Med 2007; 4:e192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lu LL, Smith MT, Yu KKQ, et al. . IFN-γ-independent immune markers of Mycobacterium tuberculosis exposure. Nat Med 2019; 25:977–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Verrall AJ, Alisjahbana B, Apriani L, et al. . Early clearance of Mycobacterium tuberculosis: the INFECT case contact cohort study in Indonesia. J Infect Dis 2019. [epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 10. Simmons JD, Stein CM, Seshadri C, et al. . Immunological mechanisms of human resistance to persistent Mycobacterium tuberculosis infection. Nat Rev Immunol 2018; 18:575–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cobat A, Gallant CJ, Simkin L, et al. . Two loci control tuberculin skin test reactivity in an area hyperendemic for tuberculosis. J Exp Med 2009; 206:2583–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Verrall AJ, Schneider M, Alisjahbana B, et al. . Early clearance of Mycobacterium tuberculosis is associated with increased innate immune responses. J Infect Dis 2019. Epub ahead of print [DOI] [PubMed] [Google Scholar]
- 13. Davies DH, Liang X, Hernandez JE, et al. . Profiling the humoral immune response to infection by using proteome microarrays: high-throughput vaccine and diagnostic antigen discovery. Proc Natl Acad Sci U S A 2005; 102:547–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kunnath-Velayudhan S, Salamon H, Wang HY, et al. . Dynamic antibody responses to the Mycobacterium tuberculosis proteome. Proc Natl Acad Sci U S A 2010; 107:14703–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Weiner J 3rd, Maertzdorf J, Sutherland JS, et al. ; GC6-74 Consortium Metabolite changes in blood predict the onset of tuberculosis. Nat Commun 2018; 9:5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010; 26:139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-seq experiments with respect to biological variation. Nucleic Acids Res 2012; 40:4288–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Domaszewska T, Scheuermann L, Hahnke K, et al. . Concordant and discordant gene expression patterns in mouse strains identify best-fit animal model for human tuberculosis. Sci Rep 2017; 7:12094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zyla J, Marczyk M, Domaszewska T, Kaufmann SHE, Polanska J, Weiner J. Gene set enrichment for reproducible science: comparison of CERNO and eight other algorithms. Bioinformatics 2019pii: btz447. doi: 10.1093/bioinformatics/btz447. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Statistical Society 1995; 57: 289–300 [Google Scholar]
- 21. Coulter F, Parrish A, Manning D, et al. . IL-17 production from T helper 17, mucosal-associated invariant T, and γδ cells in tuberculosis infection and disease. Front Immunol 2017; 8:1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hasan Z, Jamil B, Khan J, et al. . Relationship between circulating levels of IFN-gamma, IL-10, CXCL9 and CCL2 in pulmonary and extrapulmonary tuberculosis is dependent on disease severity. Scand J Immunol 2009; 69:259–67. [DOI] [PubMed] [Google Scholar]
- 23. Manzanillo PS, Ayres JS, Watson RO, et al. . The ubiquitin ligase parkin mediates resistance to intracellular pathogens. Nature 2013; 501:512–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cheepsattayakorn A, Cheepsattayakorn R. Human genetic influence on susceptibility of tuberculosis: from infection to disease. J Med Assoc Thai 2009; 92:136–41. [PubMed] [Google Scholar]
- 25. Dahl SL, Woodworth JS, Lerche CJ, et al. . Lipocalin-2 functions as inhibitor of innate resistance to Mycobacterium tuberculosis. Front Immunol 2018; 9:2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. McClure R, Massari P. TLR-dependent human mucosal epithelial cell responses to microbial pathogens. Front Immunol 2014; 5:386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Singhania A, Verma R, Graham CM, et al. . A modular transcriptional signature identifies phenotypic heterogeneity of human tuberculosis infection. Nat Commun 2018; 9:2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Suliman S, Thompson E, Sutherland J, et al. ; and the GC6-74 and ACS Cohort Study Groups. Four-gene Pan-African blood signature predicts progression to tuberculosis. Am J Respir Crit Care Med 2018. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McNab F, Mayer-Barber K, Sher A, Wack A, O’Garra A. Type I interferons in infectious disease. Nat Rev Immunol 2015; 15:87–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Desvignes L, Wolf AJ, Ernst JD. Dynamic roles of type I and type II IFNs in early infection with Mycobacterium tuberculosis. J Immunol 2012; 188:6205–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rivas-Santiago CE, Guerrero GG. IFN-α boosting of Mycobacterium bovis bacillus Calmette Güerin-vaccine promoted Th1 type cellular response and protection against M. tuberculosis infection. Biomed Res Int 2017; 2017:8796760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gröschel MI, Sayes F, Shin SJ, et al. . Recombinant BCG expressing ESX-1 of Mycobacterium marinum combines low virulence with cytosolic immune signaling and improved TB protection. Cell Rep 2017; 18:2752–65. [DOI] [PubMed] [Google Scholar]
- 33. Joosten SA, van Meijgaarden KE, Del Nonno F, et al. . Patients with tuberculosis have a dysfunctional circulating B-cell compartment, which normalizes following successful treatment. PLoS Pathog 2016; 12:e1005687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Phuah J, Wong EA, Gideon HP, et al. . Effects of B cell depletion on early Mycobacterium tuberculosis infection in cynomolgus macaques. Infect Immun 2016; 84:1301–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chin ST, Ignatius J, Suraiya S, et al. . Comparative study of IgA VH 3 gene usage in healthy TST(-) and TST(+) population exposed to tuberculosis: deep sequencing analysis. Immunology 2015; 144:302–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Akkaya M, Akkaya B, Miozzo P, et al. . B cells produce type 1 IFNs in response to the TLR9 agonist CpG-A conjugated to cationic lipids. J Immunol 2017; 199:931–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jenum S, Grewal HM, Hokey DA, et al. ; TB Trials Study Group The frequencies of IFNγ+IL2+TNFα+ PPD-specific CD4+CD45RO+ T-cells correlate with the magnitude of the QuantiFERON® gold in-tube response in a prospective study of healthy Indian adolescents. PLoS One 2014; 9:e101224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang G, Zhou B, Li S, et al. . Allele-specific induction of IL-1β expression by C/EBPβ and PU.1 contributes to increased tuberculosis susceptibility. PLoS Pathog 2014; 10:e1004426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. He W, Hu S, Du X, et al. . Vitamin B5 reduces bacterial growth via regulating innate immunity and adaptive immunity in mice infected with Mycobacterium tuberculosis. Front Immunol 2018; 9:365. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.