Abstract
Coronaviruses are contagious pathogens primarily responsible for respiratory and intestinal infections. Research efforts to develop antiviral agents against coronavirus demonstrated the main protease (Mpro) protein may represent effective drug target. X-ray crystallographic structure of the SARS-CoV2 Mpro protein demonstrated the significance of Glu166, Cys141, and His41 residues involved in protein dimerization and its catalytic function. We performed in silico screening of compounds from Curcuma longa L. (Zingiberaceae family) against Mpro protein inhibition. Employing a combination of molecular docking, scoring functions, and molecular dynamics simulations, 267 compounds were screened by docking on Mpro crystallographic structure. Docking score and interaction profile analysis exhibited strong binding on the Mpro catalytic domain with compounds C1 (1E,6E)-1,2,6,7-tetrahydroxy-1,7-bis(4-hydroxy-3-methoxyphenyl)hepta-1,6-diene-3,5-dione) and C2 (4Z,6E)‐1,5‐dihydroxy‐1,7‐bis(4‐hydroxyphenyl)hepta‐4,6‐dien‐3‐one as lead agents. Compound C1 and C2 showed minimum binding score (–9.08 and –8.07 kcal/mole) against Mpro protein in comparison to shikonin and lopinavir (≈ −5.4 kcal/mole) a standard Mpro inhibitor. Furthermore, principal component analysis, free energy landscape and protein-ligand energy calculation studies revealed that these two compounds strongly bind to the catalytic core of the Mpro protein with higher efficacy than lopinavir, a standard antiretroviral of the protease inhibitor class. Taken together, this structure based optimization has provided lead on two natural Mpro inhibitors for further testing and development as therapeutics against human coronavirus.
Communicated by Ramaswamy H. Sarma
Keywords: SARS-CoV-2, Curcuma longa, main protease, inhibitor, MD simulation
Introduction
Coronaviruses belong to the family of Coronaviridae, a diverse group of envelop-positive-strand RNA virus. In December 2019, a new coronavirus (SARS-CoV2) caused an outbreak in China has shown to cause a number of respiratory and enteric diseases. The illness caused by SARS-CoV2 is known as COVID-19. SARS-CoV2 is highly contagious virus responsible for the global outbreak of a life threatening respiratory illness, intestinal infection, congestive heart and renal failures. According to an estimate of May 21, 2020, more than 5 million cases of COVID-19 have been reported in 200 countries and territories, resulting in more than 323,256 deaths. Efforts such as drug repurposing and vaccine development are underway for the treatment of COVID-19, as such development of effective and targeted inhibitors is exceedingly desirable.
The + ssRNA coronaviruses have genetic material that can function both as a genome and as messenger RNA encode nonstructural protein 5 (Nsp5). The Nsp5 protein has three domains D1, D2 and a unique domain D3. D1 and D2 domain contributes to 3-chymotrypsin like protease activity (3CLpro). 3CLpro of Nsp5 known as Mpro is one of the best characterized drug targets. In SARS-CoV2, the special protease Mpro operates on eleven maturation cleavage sites at the carboxy terminus of the large polyprotein 1ab (Stobart et al., 2012). Auto cleavage of Nsp 5 protein results in a mature product with the 3-chymotrypsin like 2 catalytic activity (3CLpro) which further splits to form the eleven non-structural mature products known as Nsp4-Nsp16 proteins (Stobart et al., 2013). ORF 1ab gene of SARS-CoV2 encodes a replicase PP1ab. Mpro cleaved products of PP1ab are essential for the transcription and replication of SARS-CoV2 viral RNA (Wu et al., 2020). This proteolytic cleavage of PP1ab is crucial for the viral life cycle and is an attractive target for anti-SARS-CoV2 drug discovery.
Mpro molecule comprise of three domains, domain I and II (six-stranded antiparallel β barrels) and domain III (a globular cluster of five helices). Substrate binding site resides in a cleft between domain I and II, while domain III is involved in catalytic function. Dimerization of the protein is required for its catalytic regulation. Glu166 residue is a key amino acid involve in the dimerization of Mpro and creation of substrate binding pocket. Further Cys141 and His41 residue forms a catalytic dyad on the active site of the protein essential for its catalytic function (Anand et al., 2003). Recently studies have been reported on targeting SARS-CoV-2 Mpro protein using computer based drug designing approach (Das et al., 2020; Islam et al., 2020; Mittal et al., 2020; Umesh et al., 2020).
High-throughput in silico based screening of compounds against biological targets has recently gained much attention. As such, in silico screening tools have gained considerable importance in meeting the special challenges of antiviral drug discovery. In this process, natural or synthetic compound libraries are filtered by different screening methods such as docking and ligand-based similar searches, computer-aided prediction of properties limiting to a smaller set of promising candidates for their biologic testing. This rational approach makes the drug discovery process highly efficient, goal-oriented and cost-effective. Screening of natural compound libraries have led to few lead molecules which are safe drug candidates against several diseases. In the present study, we screened a small library of 267 phytochemicals from Curcuma longa L. (Zingiberaceae family) against Mpro protein inhibition.
Materials and methods
Curcuma longa phytochemical retrieval and preparation
A total of 267 compounds present in Curcuma longa were obtained from the various literature and search engine platforms such as PubMed, Google Scholar, Web of Science, Science Direct, Scopus, Semantic Scholar, Medline and PubMed Central (Lu, 2011). The structures of phytochemicals were prepared using Marvin Sketch software (https://chemaxon.com/products/marvin). The 3 D or 2 D structures of other reference compounds against targeted protein was retrieved from the NCBI PubChem in .sdf format (Xie, 2010). Open label molecule format converter (O’Boyle et al., 2011) performed conversion of 2 D to 3 D conformation, conversion from .sdf to .mol file. Ligands energy was minimized by applying mmff94 force field and conjugate gradients optimization algorithm using PyRx-Python prescription 0.8 for 200 steps (Chitrala & Yeguvapalli, 2014).
Protein retrieval and preparation
Three dimensional structure of SARS-CoV2 main protease (Mpro, 3CLpro) (PDB ID: 5RFS) protein was obtained from Protein Data Bank (https://www.rcsb.org/) (Berman et al., 2000). The resolution of the retrieved structure was 1.70 Å. The 3 D structure of Mpro protein was loaded into UCSF Chimera for molecular docking preparation (Pettersen et al., 2004). Protein model was cleaned and optimized by removing the ligands as well other heteroatoms including water. After that energy minimization of protein structures was performed by steepest descent method has 100 steps (step size 0.02 Å), and conjugate gradient method has ten steps (step size0.02 Å) by using UCSF Chimera.
Molecular docking studies
For docking experiments, the protein and the ligands were loaded into Auto Dock Tools 1.5.6 (ADT) (Trott & Olson, 2010). Gestgeiger partial charges assigned after merging non polar hydrogen and torsions applied to the ligands by rotating all rotatable bonds. Docking calculations were performed on the protein model. Polar hydrogen atoms, Kollman charges, and solvation parameters were added with the aid of Auto Dock tools. Auto Dock 4.2 offers the option of three search algorithms to explore the space of active binding with different efficacy. We used the Lamarckian Genetic Algorithm in this study. The grid box includes the entire binding site of the particular proteins and provides enough space for the ligands translational and rotational walk. For each of the 30 independent runs, a maximum number of 27,000 GA operations generated on a single population of 150 individuals. Operator weights for the rate of crossover, rate of gene mutation, and elitism was defaulting parameters (0.80, 0.02, and 1, respectively). Thereafter, LigPlot+ (v.1.4.5) and UCSF Chimera (v.1.10.2) used for visualization of the interaction pattern in the protein-ligand complex (Laskowski & Swindells, 2011).
MM-GBSA analysis
Prime MM-GBSA (Molecular Mechanics/Generalized Born Model and Solvent Accessibility) was used to evaluate the protein and protein-ligand binding energies, which includes the VSGB solvent model, OPLS_2005 force field, and rotamer search algorithms. The Prime MM-GBSA simulation was performed by using the Glide pose viewer file to compute the total free energy of binding. The MM/GBSA calculations were attained to evaluate the relative binding affinity of protein and protein-ligand complex (reported in kcal/mol). As the MM/GBSA binding energies are approximate free energies of binding, a more negative value indicates stronger binding (Kalirajan et al., 2019).
Prediction of inhibitory concentration
The prediction of IC50 concentration of lead compounds were performed having binding affinity below –6.00 kcal/mol. Standard inhibitors IC50 including shikonin (15.75 µM), tideglusib (1.55 µM), carmofur (1.82 µM), PX-12 (21.39 µM) and disulfiram (9.35 µM) were retrieved from in vitro experiments performed elsewhere (Jin et al., 2020). Linear regression analysis was performed through GraphPad Prism software to generate the pIC50.
Molecular dynamics simulation
Lead Curcuma longa compounds as predictors of Mpro protease inhibitor of SARS CoV2 were identified from docking study subjected to molecular dynamics simulation and evaluate their conformational space and inhibitory potential. Molecular dynamics simulation was performed using GROningen MAchine for Chemical Simulations (GROMACS) version 5.1.1. Protein parameters were generated using gromos43a1 force field. Ligand parameters for same force field were generated using PRODRG server (Schüttelkopf & van Aalten, 2004). Gmxeditconf tool was used to generate dodecahedron simulation box. Solvation was performed with SPC water model using gmx solvate tool. Gmxgenion tool was utilized to electro-neutralize the system. Following neutralization, energy minimization was performed to remove steric clashes and optimize of structure. After energy minimization, system was equilibrated in two steps. In first step of 100 picoseconds of NVT equilibration, system was heated up to 300 K to stabilize the temperature of the system. In second step of 100 picoseconds of NPT ensemble, pressure and density of system was stabilized. Barostat was maintained using Parrinello-Rahman barostat (Parrinello & Rahman, 1981). Bonds length were kept conserved using linear constraint solver (LINCS) algorithm (Hess et al., 1997). Long range interaction were handled using particle mesh Ewald (PME) summation method (Darden et al., 1993). This well equilibrated system with desired temperature and pressure was used to compute trajectory of 30 nanoseconds on a Linux machine with Intel core i-7 processor (32 GB RAM).
Trajectory analysis and protein-ligand interaction energy analysis
Trajectory analysis was performed using various GROMACS analysis tools. The root mean square deviation (RMSD) and root mean square fluctuations (RMSF) of protein were calculated using gmx rms, and gmxrmsf tools respectively. Solvent accessible surface area and radius of gyration were computed by gmxsasa and gmx gyrate tools respectively. Various energy related parameters were estimated using gmx energy tool. Secondary structure estimation was done by gmxdodssp tool. Hydrogen bonds were analyzed using gmxhbond tool. VMD (Humphrey et al., 1996) and PyMol (DeLano, 2002) was used for the visualization and plots were prepared using Grace Software.
Quantification of interaction energy between lopinavir, C1 and C2 was performed by calculating strength of the interaction between ligands and protease active site amino acid residue; it is also useful to estimate the non-bonded interaction energy between target protein and ligand. Tools like gmx grompp and gmx energy were used to calculate the interaction energy between the target protein and ligand. Interaction energy between two interacting species constitutes two interactions, interactions between charged components and non-charged charged components called as Coulombic short range (SR) interactions and Lennard-Jones-SR interaction energies.
Principal component analysis and free energy landscape
Principal component analysis (PCA) is a widely used analytical technique to illustrate the slow and functional motions for bio-molecules (Bahar et al., 1998). The principal components of the protein were obtained by diagonalizing and solving the eigenvalue and eigenvectors for the covariance matrix. The eigenvectors are a representation of the direction of the motion whereas eigenvalues represent the magnitude of motion along with the direction. The covariance matrix for illustration of PCA was calculated using GROMACS analysis tool gmx cover. gmx cover builds and also diagonalizes the covariance matrix. Another GROMACS analysis tool gmxanaeig was utilized to calculate the overlap between principal components and coordinates of the trajectory.
Free energy landscape (FEL) is a representation of possible conformations taken by a protein in molecular dynamics simulation along with the Gibbs free energy. FEL represents two variables that reflect specific properties of the system and measure conformational variability. FEL was calculated using probability distribution from the essential plane composed of first two eigenvectors. gmx sham tool was used for construction of FEL.
Results and discussion
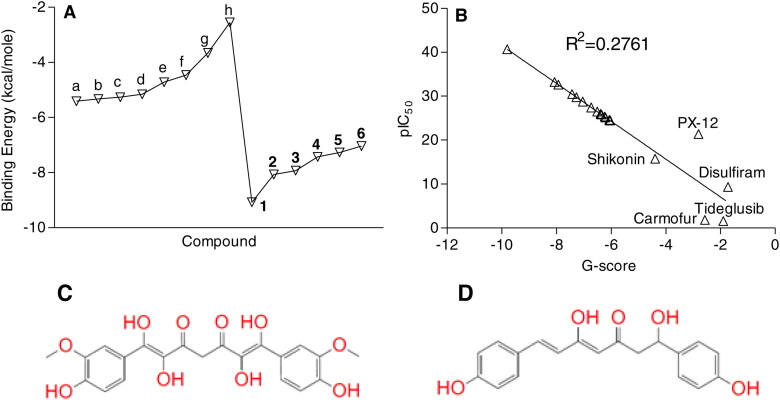
Docking screening of 267 Curcuma longa phytochemicals against Mpro protein revealed six lead compounds exhibiting better binding potential than standard/reference compounds. These phytochemicals showed tight binding energy ranging from –9.08 to –7.04 kcal/mole to Mpro protein, compared to the standard inhibitors viz. shikonin, lopinavir, alpha ketomide, tideglusib, 1,3-Bis(p-anilinophenoxy)-2-propanol (N3), carmofur, 2-(Secbutyldisulfanyl)-1h-imidazole (PX-12), and disulfiram ranging from –5.41 to –2.55 kcal/mole (Figure 1(A)). The nomenclature, structure, and molecular weight of these six compounds (cutoff less than –5.00 kcal/mole binding energy) are described in the Supplementary Table 1. Compound C1 (1E,6E)-1,2,6,7-tetrahydroxy-1,7-bis(4-hydroxy-3-methoxyphenyl)hepta-1,6-diene-3,5-dione) and C2 (4Z,6E)‐1,5‐dihydroxy‐1,7‐bis(4‐hydroxyphenyl)hepta‐4,6‐dien‐3‐one showed minimum binding score of –9.08 and –8.07 kcal/mole against Mpro protein, compared to shikonin (–5.41 kcal/mole) a standard Mpro inhibitor. Type of amino acid residue involved in the protein-ligand interaction such as hydrogen bonding, and hydrophobic interaction are summarized in Supplementary Table 2. In addition, on the basis of reported 50% inhibitory concentration (IC50) of standard/reference compounds against Mpro protein, we attempted to predict the IC50 of the lead phytochemicals. The predicted IC50 values for the lead compounds were in the range of 40.6 to 24.4 µM which were comparatively higher from the IC50 of standard compounds that range between 15.7 to 21.3 µM (Figure 1(B)). A relatively weak positive correlation (R2 = 0.2761) was noted among the binding energy and pIC50 of the test compounds (Figure 1(B)). MM-GBSA analysis of the lead (C1 and C2) and standard inhibitor (lopinavir) bound Mpro protein and unbound protein was performed and the results are shown in Supplementary Table 3. Free energy of unbound protein; lead inhibitor, C1, and C2 bound Mpro protein complex was –12755.75, –2831.23, –12663.69, and –12903.46 respectively in MM-GBSA analysis.
Figure 1.
Docking score of lead Curcuma longa phytochemicals against SARS-CoV2 main protease Mpro and predicted IC50 concentration. (A) Docking score of lead phytochemicals (–7.04 kcal/mol ≤) against SARS-CoV2 main protease Mpro. (B) Predicted IC50 concentrations of the lead molecules. (C) and (D) Structure of lead Curcuma longa phytochemicals C1 (1E,6E)-1,2,6,7-tetrahydroxy-1,7-bis(4-hydroxy-3-methoxyphenyl)hepta-1,6-diene-3,5-dione) and C2 (4Z,6E)‐1,5‐dihydroxy‐1,7‐bis(4‐hydroxyphenyl)hepta‐4,6‐dien‐3‐one. Abbreviations: a = Shikonin; b = Lopinavir; c = Alpha ketomide; d = Tideglusib; e = 1,3-Bis(p-anilinophenoxy)-2-propanol (N3); f = Carmofur; g = 2-(Secbutyldisulfanyl)-1h-imidazole (PX-12); h = Disulfiram.
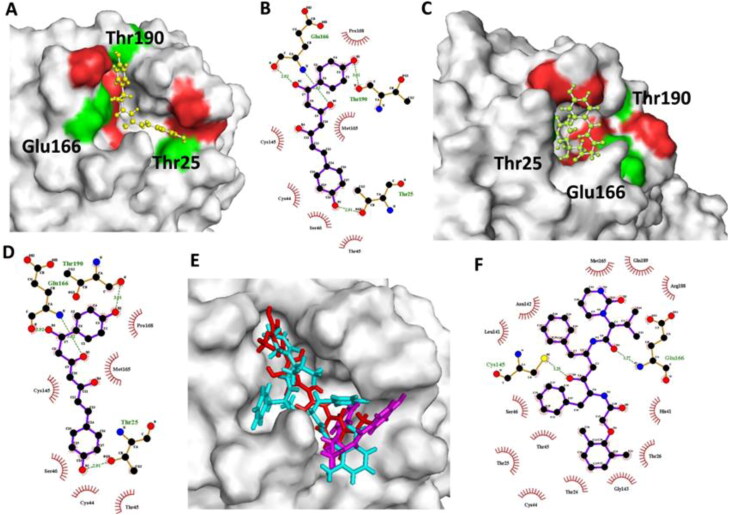
Next we determined the surface structure of two lead phytochemicals (C1 and C2), their binding with Mpro protein and the type of interaction of amino acid residues. The binding pocket of Mpro protein docked with the standard inhibitor, lopinavir and compounds C1 and C2 are shown in docking pose (Figure 2(A)). Amino acid Thr190, Thr25, and Glu166 residues were involved in hydrogen bonding with compounds C1 and C2. Glu166 interacted with O5, Thr190 and Thr25 interacted with O1 and O2 of the C1 and C2 with 3.02, 3.01 and 2.91 Å bond length. Thr45, Cys44, Ser46, Cys145, Pro168 and Met165 amino acid residues interacted with the Mpro protein by hydrophobic interaction (Figure 2). Importantly, C1 and C2 utilized virtually all amino acid residues (Thr45, Cys44, Met165, Glu166, Cys145, and Ser46) in their interaction with the Mpro protein, compared to the standard inhibitors and Mpro protein interaction (Supplementary Table 2). Compounds C1 and C2 showed interaction with amino acids Glu166, Cys145, and Pro168 whose involvement is recently reported with compound13b-Mpro interaction (Zhang et al., 2020). Compound C1 demonstrated unique interaction with Pro168 and Thr190 residue in comparison to lopinavir that might be responsible for its minimum binding energy against Mpro protein.
Figure 2.
Curcuma longa lead phytochemical interaction with SARS-CoV2 Mpro protein. (A) Surface structure of Mpro protein interacted with lead compound C1 (1E,6E)-1,2,6,7-tetrahydroxy-1,7-bis(4-hydroxy-3-methoxyphenyl)hepta-1,6-diene-3,5-dione). Green color represents the mentioned amino acid involved in hydrogen bonding. (B) Ligplot representation of lead compound C1 and Mpro protein (C) Curcuma longa lead phytochemical C2 (4Z,6E)‐1,5‐dihydroxy‐1,7‐bis(4‐hydroxyphenyl)hepta‐4,6‐dien‐3‐one) bound with SARS-CoV2 main protease Mpro. Green color represents the mentioned amino acid involved in hydrogen bonding. (D) Ligplot representation of compound C2 and Mpro protein. (E) Surface structure with superimposed Curcuma longa phytochemicals C1 (red color) and C2 (magenta color) and standard inhibitor lopinavir (cyan color). (F) Ligplot representation of standard inhibitor lopinavir with SARS-CoV2 main protease Mpro.
The enzyme SARS-CoV-2 Mpro along with other proteases is involved in the generation of various nonstructural proteins by cleaving the polyproteins (translated from viral RNA) at specific sites. It has been reported that Cys-His dyad along with Ala, Gly, Gln, Leu and Ser amino acid residues marks the Mpro cleavage site (Zhang et al., 2020). Similarly, our in silico docking studies revealed that phytochemicals C1 and C2 binds with Ser46 and Cys44 residues which might disrupt the Cys-His dyad formation crucial for Mpro protease activity. Compound 1, 2, 6, and 7 formed hydrogen bond with Mpro amino acid residues His41, Glu166, Cys44, Thr25 Thr26 and Thr190 using oxygen atoms on carbon 3, 7, and 18; carbon 3, 7, 18; carbon 3 and 18; and carbon 3, 7, 18, respectively. Oxygen atom at C12 and C14 of compound 4 and 7; and at C10 and 18 of compound 8 were involved in hydrogen bonding with Asn119 and Asn142 amino acid residues. Other lead compounds 3, 4, 5, and 6 showed better tight binding to Mpro active site in comparison to standard inhibitors (Figure 1(A)). Besides hydrogen bond interaction, compound 3, 4, 5, 6 and 7 showed hydrophobic interaction with His41 and Met49 (Supplementary Table 2). Zhang et al. (2020) showed the involvement of His41 catalytic and Met49 other than catalytic site residues in the interaction with potential Mpro inhibitor site (Zhang et al., 2020). Our results indicate that the oxygen atoms in the lead structure play an important role in Mpro active site binding and stabilizing the complex through hydrogen bond interaction. Moreover we also superimposed the Mpro free and Mpro ligand bound protein structure to determine the effect of ligand binding on the 3 D conformational change in the protein structure. Minor deviation in RMSD value (0.341 Å) among the superimposed structure showed that the ligand binding did not affect the overall 3 D structure of the protein (Figure 3(B)).
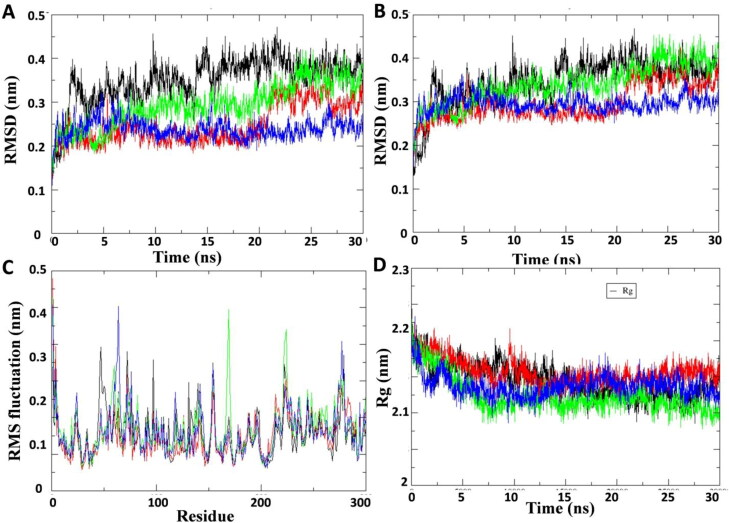
Figure 3.
Plot of molecular dynamic simulation trajectories of COVID-19 Mpro protein and protein-ligand complex during 30 ns simulation. (A) The root mean square deviation (RMSD) of solvated SARS-CoV2 Mpro protein backbone and in complex with Curcuma longa compounds C1, C2 and standard protease inhibitor lopinavir during 30 ns molecular dynamics simulation. (B) The root mean square deviation (RMSD) of solvated SARS-CoV2 Mpro whole protein and in complex with compounds C1, C2 and lopinavir during 30 ns molecular dynamics simulation. (C) The root mean square fluctuation (RMSF) values of solvated SARS-CoV2 Mpro protein and in complex with compounds C1, C2 and lopinavir plotted against residue numbers. (D) Plot of radius of gyration (Rg) during 30 ns molecular dynamics simulation of SARS-CoV2 Mpro protein and in complex with compounds C1, C2 and lopinavir. Unbound protein parameters are depicted in black color. Protein-ligand complex C1, C2 parameters are shown in green and blue color respectively. Protein-lopinavir complex is shown in red color.
We utilized the X-ray crystal structure of recently published COVID-19 Mpro protein (PDB ID: XYZ) to dock the Curcuma longa phytochemicals and the standard inhibitor lopinavir. These virtual structures were subjected to 30 ns MD simulations to study the comparative conformation dynamics of the unbound and bound ligand forms. Energy, temperature, pressure and density of the unbound protein of compounds C1, C2 and standard inhibitor lopinavir were stabilized during the 30 ns simulation (Supplementary Figure 1). Further potential energy of the protein and protein ligand complexes are provided in Supplementary Figure 2. Root-mean-square deviation (RMSD) analysis computes the average distance between the atoms of test protein during simulation. The analysis provides insights into protein conformation, stability and equilibrium of the system during simulation (Gohlke et al., 2003).The average backbone RMSD for unbound and compounds C1, C2 and standard lopinavir bound Mpro protein were found to vary between 0.23 to 0.27 nm and remained stable throughout the entire MD simulation period (Figure 3(A,B)). Unbound and compound C1 bound Mpro protein backbone simulation was stable for last 6 ns of the simulation (Figure 3(A)). It should be noted that compound C2 and Mpro protein complex backbone showed stable RMSD (between 0.2 and 0.25 Å) till the termination of the simulation (Figure 3(A)). The whole protein and protein-ligand complex RMSD showed comparative more stability than the protein backbone RMSD during 30 ns simulation. RMSD of compound C2 bound Mpro protein complex was more stable (≈0.3 Å) than Mpro-lopinavir complex throughout the 30 ns simulation (Figure 3(B)). These results endorsed that simulations are perfect for further computational analysis.
Next we analyzed the effect of Curcuma longa phytochemicals compound C1 and C2 binding on internal dynamics in comparison to lopinavir binding by calculating the root mean square fluctuation (RMSF) of the Cα atoms (Figure 3). Average RMSF value for protein, protein-lead compound C1 and C2 were 0.20, 0.21, and 0.20 nm respectively. Result showed that binding of C1 and C2 to Mpro protein mostly decreased the RMSF values. It should be noted that compound C1 and C2 binding generated substantial decrease of RMSF values in some regions which involve residue 40–55 and 80–115 (Figure 3(C)). Moreover the result showed that the binding of compound C2 did not exhibit fluctuation at 150–200 amino acid residues in comparison to unbound protein. Amino acid residue of this region plays an important role in binding of the standard inhibitor and phytochemicals from Curcuma longa (Supplementary Table 2). These results indicate that the key amino acid residues in 40–55; 80–115; and 150–200 regions may generate strong interaction with the phytochemicals.
Radius of gyration (Rg) is a parameter to assess the folding of regular secondary structures into 3-dimensional protein structure. Rg indicates change in protein structure compactness and its overall dimension. The effect of compounds C1 and C2 binding on Rg value of the protein was computed and compared with lopinavir binding to Mpro (Figure 3(D)). Binding of C1 and C2 initially (≈5–10 ns) decreased the Rg value, however, after that up to 30 ns the values was relatively in the range of 2.1 to 2.15 nm for both unbound and ligand bound protein. Average Rg value for unbound protein, protein-lead compound C1 and C2 were 2.14, 2.12 and 2.13, respectively during 30 ns simulation. Secondary structures in the unbound protein were not much affected due to the binding of these compounds to the Mpro protein during the 30 ns simulation (Supplementary Figure 3). The result suggested tight packing of the protein after ligand binding thus making a stable complex.
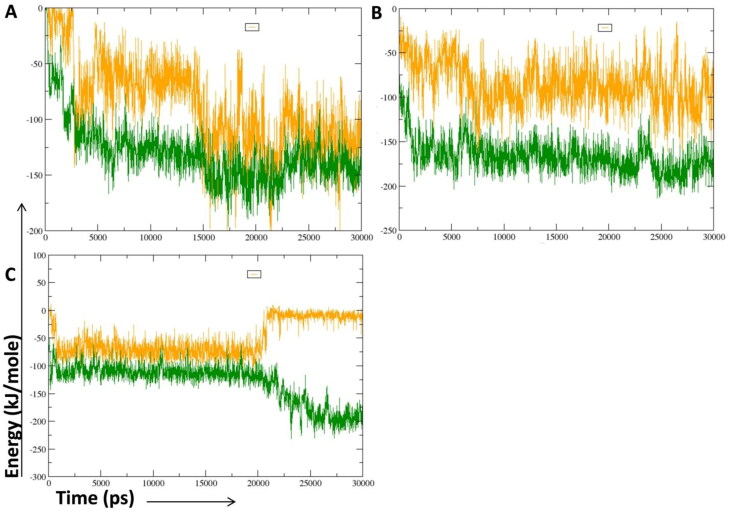
Non bonded interaction involved in lead compound (C1 and C2) and lopinavir bound Mpro protein active site was computed in terms of short range (SR) Coulomb and Lennard-Jones energies during 30 ns MD simulation (Figure 4(A–C)). The total interaction energy of protein-ligand complex was also calculated. The average Coulomb-SR energy for C. longa lead phytochemical C1, C2 and lopinavir interacting with amino acid at active site of Mpro protein was –87.0000 ± 15, –85.1703 ± 6.7, and –51.266 ± 12 kJ/mole respectively. The average Lennard-Jones-SR energy for C1, C2 and lopinavir interacting with the Mpro amino acid residue was –130.396 ± 9.7, –166.697 ± 4.5, and –130.529 ± 15 kJ/mole respectively. The result showed that lead C2-Mpro interaction at active site possesses significantly lower Lennard-Jones-SR energy than lopinavir. Moreover, the total interaction energy of C2-Mpro interaction (–251.8673 ± 5.6 kJ/mole) at active was significantly lower than, C1-Mpro, and lopinavir-Mpro interactions (–217.396 ± 12.35 and –181.795 ± 13.5 kJ/mole respectively). Over all, the result showed that due to significantly lower total interaction energy, C2 might act as potent Mpro inhibitor in comparison to lopinavir.
Figure 4.
Short range energy evaluation between the protein and the hit molecules. (A) The short range Coulombic and Lennard-Jones interaction energy of C1 interacting with amino acid residue at Mpro active site. (B) The short range Coulombic and Lennard-Jones interaction energy of C2 interacting with amino acid residue at Mpro active site. (C) The short range Coulombic and Lennard-Jones interaction energy of lopinavir interacting with amino acid residue at Mpro active site. Coulombic and Lennard-Jones short range (SR) interaction energy are shown in orange and green color.
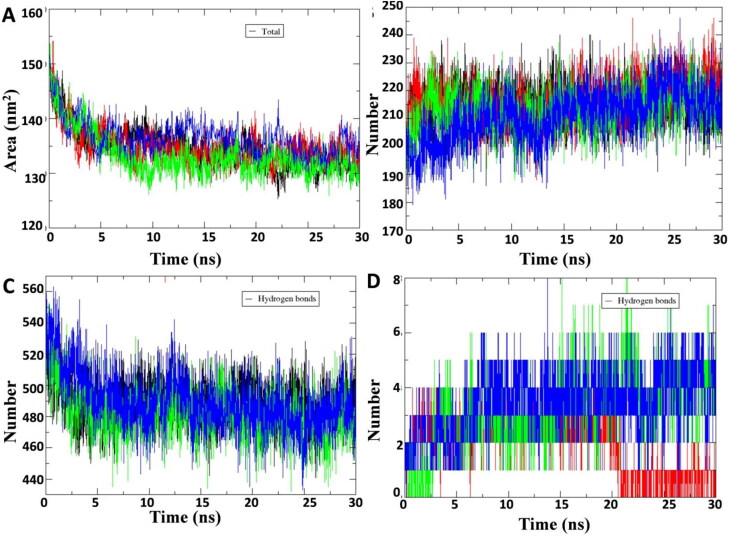
Solvent accessibility surface area (SASA) determines the bimolecular surface area assessable to surrounding solvent molecules. The change in SASA for unbound protein and protein-lead compound C1 and C2 complex were analyzed and compared with protein-lopinavir complex (Figure 5(A)). A significant change in SASA was observed due to binding of C1 and C2 at the active site of the protein. Average SASA value of unbound protein and protein-lead compound C1 and C2 complex during the 30 ns simulation were 135, 133, and 137 nm2, respectively. Hydrogen bond formation plays an important role in the stabilization of protein and protein–ligand complex structures by minimizing the energy of the system. Intra molecular, protein-water and protein-ligand hydrogen bonding pattern were studied in unbound and ligand bound Mpro protein (Figure 5(B,C)). Average value of intra molecular and protein-water hydrogen bonding in unbound protein C1 and C2 bound protein complex were 215, 214 and 211; and 489, 483 and 486 during simulation. Moreover the average H-bonding in protein-lead compound 1 and 2 complex was 3.11 and 3.51 during the 30 ns simulation (Figure 5(D)). Overall, the H-bonding pattern in protein-lead compound C1 and C2 interaction showed the energetically favorable and stable complex formation.
Figure 5.
Plot of solvent accessible surface (SASA) region and hydrogen bond formation during 30 ns MD simulation. (A) Plot of solvent accessible surface area (SASA) during 30 ns molecular dynamics simulation of SARS-CoV2 Mpro protein and in complex with Curcuma longa compounds C1, C2 and standard protease inhibitor lopinavir. (B) Plot of number of hydrogen bond formation with in the SARS-CoV2 Mpro protein, SARS-CoV2 Mpro protein complex with compound C1, C2 and lopinavir. (C) Plot of number of hydrogen bond formation between water and SARS-CoV2 Mpro protein, SARS-CoV2 Mpro protein complex with compound C1, C2 and lopinavir. (D) Plot of number of hydrogen bond formation between SARS-CoV2 Mpro protein and compound C1, C2 and lopinavir. Unbound protein parameters are depicted in black color. Protein-ligand complex C1 and C2 parameters are shown in green and blue color. Protein-lopinavir complex is shown in red color.
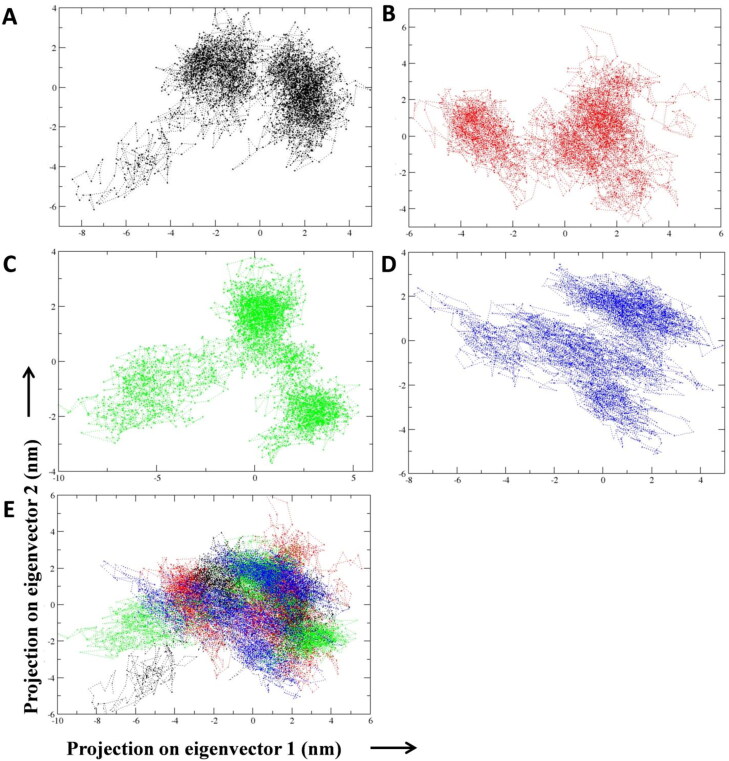
Next we studied the collective motion of the unbound and bound compounds C1 and C2 with Mpro protein from the molecular dynamics trajectories using principal component analysis (PCA). The analysis is based on the strenuous motion of Cα atom of the protein through eigenvectors (overall direction of motion of the atoms) and eigenvalues (atomic contribution of motion) (David & Jacobs, 2014). MD trajectories of unbound, lead compound as well as lopinavir bound protein were examined with the PC for better understanding of the structural and conformational changes in Mpro protein due to ligand binding. PCA analysis indicated that lead compound 2 bound Mpro protein complex depicted lesser collective motion of the protein in comparison to unbound and lead compound 1 bound protein (Figure 6(A–D)). As a result of lesser flexibility, conformational space covered by lead 2-protein complex was narrower than he unbound protein. These results conclude that compound C2 bound Mpro protein was more stable than the unbound and lopinavir bound protein.
Figure 6.
Projection of protein atoms in phase space along the first two principal eigenvectors. (A) COVID-19 Mpro protein (B) COVID-19 Mpro protein complexed with standard protease inhibitor lopinavir. (C) COVID-19 Mpro protein complexed with Curcuma longa compound C1. (D) COVID-19 Mpro protein complexed with Curcuma longa compound C2. (E) Superimposed plot showing COVID-19 Mpro unbound protein and protein complexed with compound C1, C2 and lopinavir.
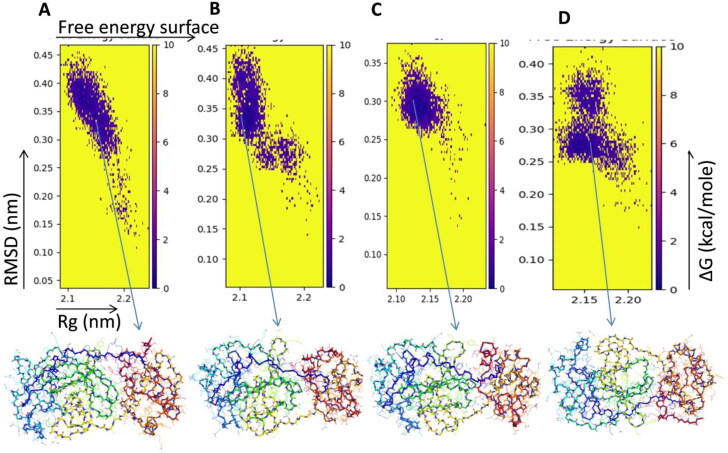
To visualize the energy minima landscape of unbound, C1, C2 and lopinavir bound Mpro protein, we studied the free energy landscape (FEL) against first two principal components PC1 (radius of gyration-Rg) and PC2 (root-mean-square deviation-RMSD) which revealed ΔG value 0 to 10 kJ/mol (Figure 7(A–C)). The shape and size of the minimal energy area (shown in blue) indicate the stability of the protein and protein-lead compound complex. Smaller and more centralized blue areas suggest the stability of the protein and their corresponding complex. Figure 7 indicates that compound C2 is more stable than unbound protein as well as lead compound C1 and lopinavir bound Mpro protein. Thus these compounds have the potential to induce Mpro protein to enter the local energy minimal state.
Figure 7.
Free energy landscape of the first two principal components for (A) COVID-19 Mpro protein. (B) COVID-19 Mpro protein-C1 complex. (C) COVID-19 Mpro protein-C2 complex. (D) COVID-19 Mpro protein-lopinavir complex.
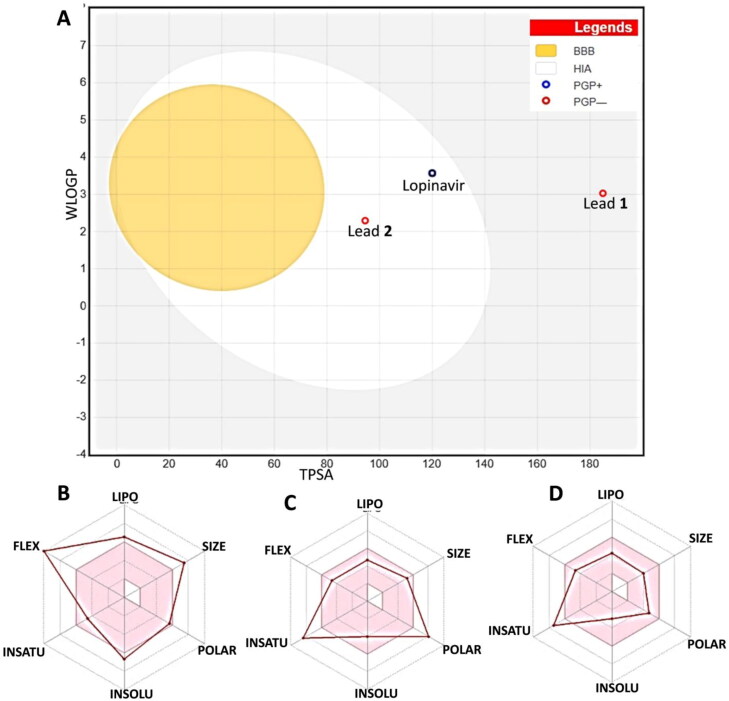
The predictions on drug-likeness and toxicity were performed. Compound C1 is predicted as not absorbed and not brain penetrant as it located outside the Egg (white circle) whereas compound C2 and lopinavir were predicted as well-absorbed based on their position inside the Egg (Figure 8(A–D)). The lead compound C2 exhibited inaccessibility (out of Egg yolk) to blood brain barrier. It is noteworthy that compound C2 showed high probability of passive absorption by the gastrointestinal tract. Our results further showed that standard inhibitor lopinavir is predicted to be actively effluxed by P-glycoprotein (blue dot) while compound C1 and C2 are predicted as non-substrate of P-gp (red dot) (Daina et al., 2017). Bioavailability radar analysis showed that lopinavir and C1 are too flexible and polar predicting that these compounds may not be orally bioavailable. Compound C2 in contrast, showed flexibility and polarity in the optimal region (pink color) of drug-likeness (Figure 8(D)).
Figure 8.
Boiled egg diagram and bioavailability radar map of Curcuma longa compounds C1, C2 and standard protease inhibitor lopinavir. (A) Boiled egg diagram of lopinavir and compound C1 and C2. Bioavailability radar map of (B) Lopinavir (C) compound C1 and (D) compound C2 depicting the LIPO (lipophilicity), SIZE (molecular weight), POLAR (polarity), INSOLU (insolubility) INSATU (insaturation) and FLEX (rotatable bond flexibility) parameters.
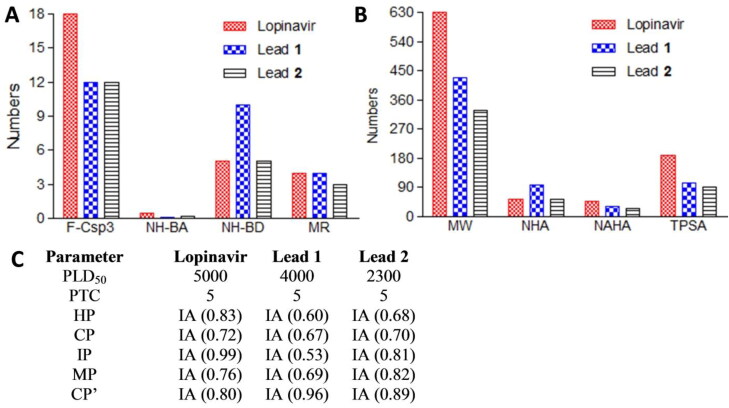
Physiochemical properties such as fraction Csp3 (FCsp3), H-bond acceptor count (NH-BA), H-bond donor count (NH-BD), molar refractivity (MR), molecular weight (MW), number of heavy atoms (NHA), number of aromatic heavy atoms (NAHA) and topological polar surface area (TPSA) of lopinavir and compounds C1 and C2 are depicted in Figure 9(A,B). Compound C1 and C2 possess most of the physiochemical properties in the range of drug-likeness properties. Toxicological profile of lopinavir and C1 and C2 are shown in Figure 9(C). The predicted LD50 of compound C2 was less than half of the standard inhibitor. Hepatotoxicity, carcinogenicity, immunotoxicity, mutagenicity and cytotoxicity prediction depicted inactive scores for all the test compounds. It should be noted that all test compounds were designated to the category of toxicity class (Figure 9(C)). Overall toxicity prediction showed C2 to possess low toxicity profile than compound C1.
Figure 9.
Physiochemical properties and toxicological parameters of COVID-19 Mpro standard inhibitor lopinavir, compounds C1 and C2. (A) F-Csp3, NH-BA, NH-BD and MR physiochemical parameters of standard and compounds C1 and C2. (B) MW, NHA, NAHA, and TPSA physiochemical parameters of standard and compounds C1 and C2. (C) Toxicological profile of standard and C1 and C2 compounds. F-Csp3, Fraction of sp3 carbon; NH-BA, Number of hydrogen bond acceptor; NH-BD, Number of hydrogen bond donor; MR, Molar refractivity; MW, Molecular weight; NHA, Number of heavy atoms; NAHA, Number of aromatic heavy atoms; and TPSA, Topological polar surface area; PLD50, Predicted LD50 [mg/kg body weight]; PTC, Predicted toxicity class; HP, Hepatotoxicity Probability; CP, Carcinogenicity Probability; IP, Immunotoxicity Probability; MP, Mutagenicity Probability; CP’, Cytotoxicity Probability; IA, Inactive.
Conclusions
The present investigation identified some lead compounds obtained from Curcuma longa in the inhibition of COVID-19 Mpro. Molecular docking study confirmed the binding potential of compounds C1 and C2 at the active site of the enzyme. The 30 ns simulation revealed that the protein-lead compound complex possess stable conformation and lower protein-ligand interaction energy. Compound C2 exhibited decent oral bioavailability and lower predicted LD50 value than the standard Mpro inhibitor lopinavir in computational investigation. Other physiochemical and toxicological property prediction of the compound C1 and C2 necessitates further in vitro and in vivo validation of COVID-19 antiviral activity. Approximately five Curcuma longa compounds showed tight binding at Mpro active site demonstrating the presence of more than one inhibitor in a single natural product indicates its potential against COVID-19 virulence. Overall, the study concludes that Curcuma longa possess COVID-19 Mpro inhibitory potential for further testing and development as therapeutics against human coronavirus.
Supplementary Material
Funding Statement
Efforts are supported by the Department of Defense Grants W81XWH-19-1-0720 and W81XWH-18-1-0618 and VA Merit Review 1I01BX002494 to SG. PPK acknowledges financial support from University Grants Commission, India in the form of CSIR-UGC Senior Research fellowship. SK acknowledges University Grants Commission, India and Department of Science and Technology, India for providing financial support in the form of UGC-BSR Research Start-Up-Grant [No. F.30–372/2017 (BSR)] and DST-SERB Grant [EEQ/2016/000350] respectively. SK acknowledges Central University of Punjab, Bathinda, India for providing Research Seed Money Grant [GP-25]. AKS, KSP, and MS acknowledge CSIR-India, DBT-India and DST-India funding agencies respectively for providing financial assistance in the form of Junior Research Fellowship.
Disclosure statement
The authors declare no conflict of interest.
References
- Anand K., Ziebuhr J., Wadhwani P., Mesters J. R., & Hilgenfeld R. (2003). Coronavirus main proteinase (3CLpro) structure: Basis for design of anti-SARS drugs. Science (New York, N.Y.), 300(5626), 1763–1767. 10.1126/science.1085658 [DOI] [PubMed] [Google Scholar]
- Bahar I., Atilgan A. R., Demirel M. C., & Erman B. (1998). Vibrational dynamics of folded proteins: Significance of slow and fast motions in relation to function and stability. Physical Review Letters, 80(12), 2733–2736. 10.1103/PhysRevLett.80.2733 [DOI] [Google Scholar]
- Berman H. M., Westbrook J., Feng Z., Gilliland G., Bhat T. N., Weissig H., Shindyalov I. N., & Bourne P. E. (2000). The Protein Data Bank. Nucleic Acids Research, 28(1), 235–242. 10.1093/nar/28.1.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitrala K. N., & Yeguvapalli S. (2014). Computational prediction and analysis of breast cancer targets for 6-methyl-1, 3, 8-trichlorodibenzofuran. PLoS One, 9(11), e109185 10.1371/journal.pone.0109185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daina A., Michielin O., & Zoete V. (2017). SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Scientific Reports, 7, 42717 10.1038/srep42717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darden T., York D., & Pedersen L. (1993). Particle mesh Ewald: An N⋅ log (N) method for Ewald sums in large systems. The Journal of Chemical Physics, 98(12), 10089–10092. 10.1063/1.464397 [DOI] [Google Scholar]
- Das S., Sarmah S., Lyndem S., & Singha Roy A. (2020). An investigation into the identification of potential inhibitors of SARS-CoV-2 main protease using molecular docking study. Journal of Biomolecular Structure & Dynamics, 1–11. Advance online publication. 10.1080/07391102.2020.1763201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David C. C., & Jacobs D. J. (2014). Principal component analysis: A method for determining the essential dynamics of proteins . Methods in Molecular Biology, 1084, 193–226. 10.1007/978-1-62703-658-0_11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano W. L. (2002). PyMOL. DeLano Scientific. [Google Scholar]
- Gohlke H., Kiel C., & Case D. A. (2003). Insights into protein-protein binding by binding free energy calculation and free energy decomposition for the Ras-Raf and Ras-RalGDS complexes. Journal of Molecular Biology, 330(4), 891–913. 10.1016/S0022-2836(03)00610-7 [DOI] [PubMed] [Google Scholar]
- Hess B., Bekker H., Berendsen H. J., & Fraaije J. G. (1997). LINCS: A linear constraint solver for molecular simulations. Journal of Computational Chemistry, 18(12), 1463–1472. [DOI] [Google Scholar]
- Humphrey W., Dalke A., & Schulten K. (1996). VMD: Visual molecular dynamics. Journal of Molecular Graphics, 14(1), 28–33. 10.1016/0263-7855 [DOI] [PubMed] [Google Scholar]
- Islam R., Parves M. R., Paul A. S., Uddin N., Rahman M. S., Mamun A. A., Hossain M. N., Ali M. A., & Halim M. A. (2020). A molecular modeling approach to identify effective antiviral phytochemicals against the main protease of SARS-CoV-2. Journal of Biomolecular Structure & Dynamics, 1–12. Advance online publication. 10.1080/07391102.2020.1761883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., … Yang H. (2020). Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature,1–6 . Advance online publication. 10.1038/s41586-020-2223-y [DOI] [PubMed] [Google Scholar]
- Kalirajan R., Pandiselvi A., Gowramma B., & Balachandran P. (2019). In-silico design, ADMET Screening, MM-GBSA binding free energy of some novel isoxazole substituted 9-anilinoacridines as HER2 inhibitors targeting breast cancer. Current Drug Research Reviews, 11(2), 118–128. 10.2174/2589977511666190912154817 [DOI] [PubMed] [Google Scholar]
- Laskowski R. A., & Swindells M. B. (2011). LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery. Journal of Chemical Information and Modeling, 51(10), 2778–2786. 10.1021/ci200227u [DOI] [PubMed] [Google Scholar]
- Lu Z. (2011). PubMed and beyond: A survey of web tools for searching biomedical literature. Database: The Journal of Biological Databases and Curation, 2011, baq036 10.1093/database/baq036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal L., Kumari A., Srivastava M., Singh M., & Asthana S. (2020). Identification of potential molecules against COVID-19 main protease through structure-guided virtual screening approach. Journal of Biomolecular Structure & Dynamics, 1–19. Advance online publication. 10.1080/07391102.2020.1768151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Boyle N. M., Banck M., James C. A., Morley C., Vandermeersch T., & Hutchison G. R. (2011). Open Babel: An open chemical toolbox. Journal of Cheminformatics, 3, 33 10.1186/1758-2946-3-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrinello M., & Rahman A. (1981). Polymorphic transitions in single crystals: A new molecular dynamics. Journal of Applied Physics, 52(12), 7182–7190. 10.1063/1.328693 [DOI] [Google Scholar]
- Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., & Ferrin T. E. (2004). UCSF Chimera - A visualization system for exploratory research and analysis. Journal of Computational Chemistry, 25(13), 1605–1612. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- Schüttelkopf A. W., & van Aalten D. M. (2004). PRODRG: A tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallographica. Section D, Biological Crystallography, 60(Pt 8), 1355–1363. 10.1107/S0907444904011679 [DOI] [PubMed] [Google Scholar]
- Stobart C. C., Lee A. S., Lu X., & Denison M. R. (2012). Temperature-sensitive mutants and revertants in the coronavirus nonstructural protein 5 protease (3CLpro) define residues involved in long-distance communication and regulation of protease activity. Journal of Virology, 86(9), 4801–4810. 10.1128/JVI.06754-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stobart C. C., Sexton N. R., Munjal H., Lu X., Molland K. L., Tomar S., Mesecar A. D., & Denison M. R. (2013). Chimeric exchange of coronavirus nsp5 proteases (3CLpro) identifies common and divergent regulatory determinants of protease activity. Journal of Virology, 87(23), 12611–12618. 10.1128/JVI.02050-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O., & Olson A. J. (2010). AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry, 31(2), 455–461. 10.1002/jcc.21334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umesh Kundu, D., Selvaraj C., Singh S. K., & Dubey V. K. (2020). Identification of new anti-nCoV drug chemical compounds from Indian spices exploiting SARS-CoV-2 main protease as target. Journal of Biomolecular Structure & Dynamics, 1–9. Advance online publication. 10.1080/07391102.2020.1763202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Liu Y., Yang Y., Zhang P., Zhong W., Wang Y., Wang Q., Xu Y., Li M., Li X., Zheng M., Chen L., & Li H. (2020). Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharmaceutica Sinica B, 10(5), 766–788. Advance Online Publication, 10.1016/j.apsb.2020.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X. Q. (2010). Exploiting PubChem for virtual screening. Expert Opinion on Drug Discovery, 5(12), 1205–1220. 10.1517/17460441.2010.524924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., & Hilgenfeld R. (2020). Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science (New York, N.Y.), 368(6489), 409–412. 10.1126/science.abb3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.