Abstract
Porphyromonas gingivalis produces proteases capable of degrading cytokines, host heme proteins and some antimicrobial peptides. In this study, we show that P. gingivalis culture supernatants fully or partially degrade human neutrophil peptide α-defensins and human β-defensins after 30 min. This observation suggests that proteases from P. gingivalis degrade defensins and this activity could abrogate defensin-related innate immune functions.
Key Words: Proteases, Gingipains, Defensins, Porphyromonas gingivalis
Introduction
Periodontitis is a common inflammatory disease of the supporting structures of the teeth caused by the host response to specific microorganisms or groups of microorganisms [1]. Initially, periodontitis is characterized by inflammation of the gingiva, loss of soft tissue attachment and loss of bone. Ultimately, periodontitis results in progressive destruction of the periodontal ligament and alveolar bone, which can lead to tooth loss in adults.
The decline of periodontal health and the onset of periodontal disease is associated with alterations in the microflora in the gingival crevice. Periodontitis sites contain elevated levels of Gram-negative anaerobic bacteria [2, 3, 4]. One of these species, Porphyromonas gingivalis, is a late colonizer in the oral biofilm and thought to be a dominant etiologic agent.
P. gingivalis produces numerous virulence factors that contribute to its pathogenicity [1, 5, 6]. Attachment and invasion of P. gingivalis is facilitated by adhesins that include hemagglutinins and 3 distinct fimbriae: major fimbriae, minor fimbriae and Pg-II fimbriae. Local tissue damage and evasion of host defense mechanisms are facilitated by capsular polysaccharides and lipopolysaccharide.
P. gingivalis also produces proteases [7, 8, 9, 10]. These include: gingipain HRgpA, gingipain RgpB and gingipain K, which belong to the cysteine and serine catalytic class of peptidases; gene products TPR and PRTT (periodontain), which belong to papain- and streptopain-like catalytic classes of peptidases; serine endopeptidase; glycyl-prolyl peptidase (dipeptidyl peptidase IV); prolyl tripeptidyl peptidase. All these proteases have been implicated in the pathogenesis of periodontal disease [7, 11]. They can degrade pro-inflammatory cytokines [12], cell adhesion molecules like PECAM-1 [13], host heme proteins [14] and some nonhuman antimicrobial peptides [15]. However, little is known about their ability to degrade human neutrophil peptide α-defensins (HNP-1) and human β-defensins (HBD1, HBD2 and HBD3).
HNPs and HBDs are expressed in oral tissues, salivary glands and gingival tissue [16, 17, 18]. They are also present in salivary secretions and gingival crevicular fluid. For example, HBD2 and HBD3 are present in saliva [19], while HNP-1, HNP-2, HNP-3, HNP-4, HBD1 and HBD2 are present in gingival crevicular fluid [20, 21].
Recently, we observed that defensins can bind to adhesins of P. gingivalis strain 381 [22]. It is possible that binding of defensins to these adhesins may inhibit microbial adherence to tissues, attenuate pro-inflammatory cytokine responses [22] and facilitate delivery of bound antigen to antigen-presenting cells with defensin receptors [23]. Here, we assess whether proteases in culture supernatants from P. gingivalis degrade human α- and β-defensins and thus have the potential to interfere with these important innate immune functions.
Materials and Methods
P. gingivalis strain 381 (obtained from Ann Progulske-Fox, Department of Oral Biology, University of Florida, Gainesville, Fla., USA) was grown in tryptic soy broth (Difco Laboratories, Detroit, Mich., USA) supplemented with 5 µg/ml hemin (Sigma, St. Louis, Mo., USA) and vitamin K (Sigma) for 72 h at 37°C in an atmosphere that contained 85% N2-10% H2-5% CO2 [24]. Bacterial cells were pelleted from the culture by centrifugation at 7,232 g for 15 min at 4°C. Ten milliliters of the culture supernatant was filtered (0.22 µm filter, Millex GV; Millipore, Billerica, Mass., USA), dialyzed for 2 days at 4°C against distilled water to remove smaller media components and salts, and lyophilized. The lyophilized material was then reconstituted to 1 ml in distilled water. This preparation contained 8.6 mg/ml protein and contained numerous bands on SDS-PAGE (gel not shown).
Stock solutions (200 µg/ml) of HNP-1, HBD1, HBD2 and HBD3 (PeproTech Inc., Rocky Hill, N.J., USA) were prepared in 0.01 M sodium phosphate buffer with 0.14 M sodium chloride, pH 7.2 (0.01 M PBS, pH 7.2).
The control and test solutions were prepared and included: (1) 0.01 M PBS, pH 7.2 alone as a control solution, (2) a mixture of HNP-1, HBD1, HBD2 and HBD3 in PBS, (3) the P. gingivalis culture supernatant in PBS and (4) the P. gingivalis culture supernatant and the 4-defensin mixture in PBS (table 1). For this, 40 µl of 0.01 M PBS pH 7.2 (treatments 1 and 3) or defensin solutions (10 µl each of HNP-1, HBD1, HBD2 and HBD3 in 0.01 M PBS pH 7.2; treatments 2 and 4) were put into microcentrifuge tubes and dried for 1 h under vacuum by rotary evaporation. Distilled water (treatments 1 and 2) or dialyzed P. gingivalis culture supernatant (treatments 3 and 4) were then added and incubated for 30 min at 37°C.
Table 1.
Protocol for preparation of samples to assess degradation of human α- and β-defensins by culture supernatants of P. gingivalis strain 381
| Ingredient | Treatment 1: PBS | Treatment 2: defensin mixture in PBS | Treatment 3: culture supernatant in PBS | Treatment 4: defensin mixture in culture supernatant |
|---|---|---|---|---|
| 0.01 M PBS, pH 7.21 | 40 µl | 40 µl | ||
| HNP-12 | 10 µl | 10 µl | ||
| HBD1 | 10 µl | 10 µl | ||
| HBD2 | 10 µl | 10 µl | ||
| HBD3 | 10 µl | 10 µl | ||
| The above solutions for each treatment (total volume of 40 µl each) were put into microcentrifuge tubes and then dried for 1 h under vacuum by rotary evaporation. | ||||
| Distilled water | 40 µl | 40 × l | ||
| Culture supernatant3 | 40 × l | 40 × l | ||
0.01 M sodium phosphate buffer with 0.14 M sodium chloride, pH 7.2.
Solutions (200 µg/ml) of HBD1, HBD2, HBD3 and HNP-1 in 0.01 M PBS, pH 7.2.
P. gingivalis culture supernatant was dialyzed against distilled water.
Cyano-4-hydroxycinnamic acid (CHCA; 20 mg/ml) was dissolved in a mixture (1:1) of isopropanol:acetone containing cellulose nitrate (10 mg/ml). Somatostatin (3,147 Da; 1 pm/µl) and bovine insulin (5,734.5 Da; 5 pm/µl) were used as standards to calibrate the instrument around the masses of human α- and β-defensins. The CHCA mixture was added to both the standards and the test samples, mixed, and 1 µl was ‘spotted’ on the MALDI plate. Spots were air dried and analyzed in a Biflex III MALDI-TOF (Bruker Daltonics Inc., Billerica, Mass., USA). The mass range was gated from 0 to 7,500 m/z.
Results
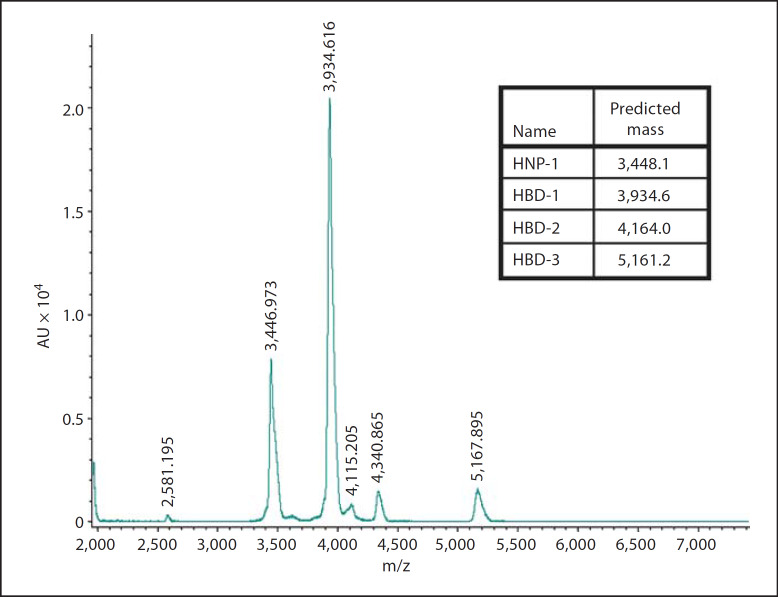
Distinct peaks were seen in treatment 1 containing HNP-1, HBD1, HBD2 and HBD3 (fig. 1). Predicted masses were 3,448.1 Da (HNP-1), 3,934.6 Da (HBD1), 4,164.0 Da (HBD2) and 5,161.2 Da (HBD3). Distinct peaks were seen at 3,446.973 m/z, 3,934.616 m/z, 4,115.205 m/z and 5,167.895 m/z. Proteins with masses of 2,867.81 Da and 5,734.61 Da were used as internal calibration standards. Each defensin could be individually detected and thus this mixture could be used as a detection system to assess defensin degradation simultaneously.
Fig. 1.
MALDI-TOF mass spectrometry showing the masses of human α-defensin HNP-1 and human β-defensins HBD1, HBD2 and HBD3. Predicted masses were 3,448.1 Da for HNP-1, 3,934.6 Da for HBD1, 4,164.0 Da for HBD2 and 5,161.2 Da for HBD3. Distinct peaks were seen at 3,446.973 m/z, 3,934.616 m/z, 4,340.865 m/z and 5,167.895 m/z.
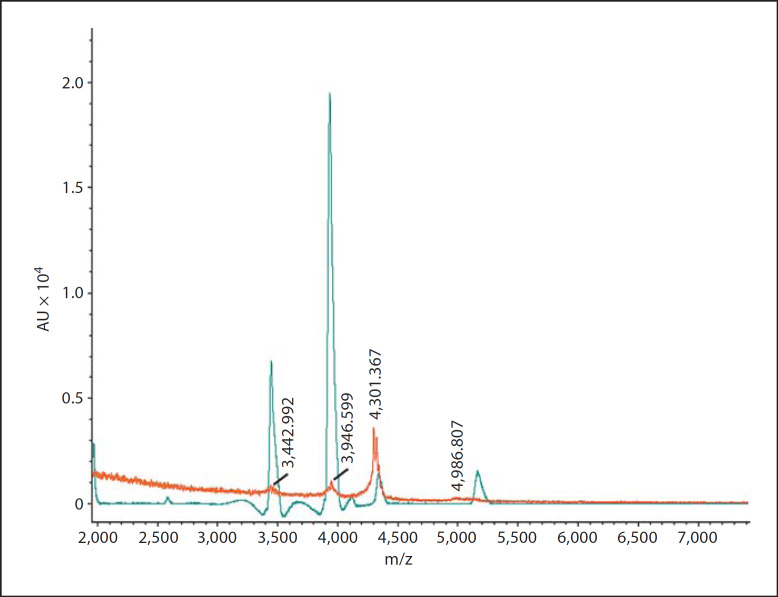
No peaks were seen in 0.01 M PBS, pH 7.2 (treatment 1) or the dialyzed culture supernatant of P. gingivalis (treatment 3; range 0–7,500 m/z; results not shown). Small peaks for HNP-1 (3,442.992 m/z) and HBD1 (3,946.599 m/z) were seen in the defensin mixture incubated with the culture supernatant of P. gingivalis (treatment 4). Signals of 4,301.367 m/z and 4,986.807 m/z may represent fragments of other defensins. Overlay spectrograms of the defensin mixture and the culture supernatant of P. gingivalis incubated with the defensin mixture show that defensins were degraded and nearly absent in the latter mixture (fig. 2).
Fig. 2.
Overlays of spectrograms after MALDI-TOF mass spectrometry (range 0–7,500 m/z). Shown is the spectrogram of the defensin mixture and the spectrogram of the culture supernatant of P. gingivalis incubated with the defensin mixture. Note that the defensins were degraded and nearly absent in the latter mixture.
Discussion
Proteases from P. gingivalis alter host receptors, degrade cytokines and antimicrobial peptides, and activate coagulation, complement and kallikrein/kinin pathways [25]. In this study, we used MALDI-TOF mass spectrometry to show that proteases in culture supernatants from P. gingivalis degrade human α- and β-defensins. Whether this activity is unique to P. gingivalis strain 381 or common among other P. gingivalis is not yet known.
P. gingivalis proteases have different cleavage sites [7, 8, 9, 10, 25, 26]. Whether degradation of defensins occurs via 1 specific protease of P. gingivalis strain 381 or via the simultaneous activity of multiple proteases is also not known. Clearly, defensins are rich in arginine and lysine, often target sites of these proteases.
The minimum inhibitory concentrations of defensins for P. gingivalis vary greatly from 34.6 to >250 µg/ml [24, 27, 28]. Of course, it is tempting to speculate that susceptible strains of P. gingivalis may produce narrower spectrums and lower concentrations of proteases and resistant strains produce wider spectrums and higher concentrations of proteases. However, the resistance of P. gingivalis to direct killing by nondefensin antimicrobial peptides [Dhvar4, a congener of histatin 5; K4-s4(1–15), a shorter derivative of dermaseptin S4; LL-37, a human cathelicidin] was found to be protease independent and more likely related to a low affinity of antimicrobial peptides to P. gingivalis [29].
It is clear that bacteria that produce proteases capable of degrading antimicrobial peptides are pathogenic. Metalloprotease ZapA of uropathogenic Proteus mirabilis cleaves HBD1 and LL-37 [30]. Proteolysis of HBD1 resulted in 6 peptides, while proteolysis of LL-37 resulted in 9 or more peptides. The antimicrobial activity of HBD1 and LL-37 was significantly reduced following ZapA treatment, suggesting that proteolysis results in inactivation of these peptides. The data suggest that a function of ZapA during urinary tract infections is the proteolysis of antimicrobial peptides associated with the innate immune response.
Metalloproteinase (aureolysin) and a glutamylendo-peptidase (V8 protease) of Staphylococcus aureus cleaved and inactivated LL-37 in a time- and concentration-dependent manner [31]. S. aureus strains that produce significant amounts of aureolysin were found to be less susceptible to fragment LL-17–37 than strains expressing no aureolysin activity, suggesting that aureolysin production by S. aureus contributes to its resistance to LL-37. In the case of P. gingivalis, it is not known if the production of protease and the degradation of defensins correlates with the resistance of strains to defensins.
The degradation of defensins may have other implications to the innate immune system. Human α- and β-defensins regulate innate immunity and enhance adaptive immunity [32, 33, 34, 35]. In addition to their direct antimicrobial activity, they have chemotactic effects on phagocytic and mast cells, induce inflammatory mediators, regulate the functions of phagocytes and the complement system, interact with G protein-coupled regulators on immature dendritic cells, stimulate dendritic cell maturation, have direct effects on T cells and enhance antigen-specific immune responses in vivo [23]. They also bind to bacterial membranes, lipopolysaccharides and some bacterial toxins. Inactivation of HNP and HBD composition and concentration after production could inhibit defensin-induced receptor-mediated internalization of microbial antigens to immature dendritic cells [23] or not attenuate antigen-induced pro-inflammatory cytokine responses [22].
In summary, supernatants for P. gingivalis degrade human α-defensin HNP-1 and human β-defensins HBD1, HBD2 and HBD3. Determining the spectrum of protease activity of many strains of P. gingivalis on defensins as well as determining the activity of individual proteases of a single strain of P. gingivalis on defensins would help characterize the role of proteases in the ability of P. gingivalis in evading hosting defenses, colonizing host tissues and inducing inflammatory periodontal diseases.
Acknowledgments
We are grateful to James D. Herd for preparation of the figures and to Elizabeth A. Schmitt, Cairn Communications, Mahtomedi, Minn., USA for critically reading the manuscript. This work was supported by funds from NIH/NIDCR R01 DE014390.
References
- 1.Offenbacher S. Periodontal diseases: pathogenesis. Ann Periodontol. 1996;1:821–878. doi: 10.1902/annals.1996.1.1.821. [DOI] [PubMed] [Google Scholar]
- 2.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–144. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 3.Ledder RG, Gilbert P, Huws SA, Aarons L, Ashley MP, Hull PS, McBain AJ. Molecular analysis of the subgingival microbiota in health and disease. Appl Environ Microbiol. 2007;73:516–523. doi: 10.1128/AEM.01419-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paster BJ, Boches SK, Galvin JL, Ericson RE, Lau CN, Levanos VA, Sahasrabudhe A, Dewhirst FE. Bacterial diversity in human subgingival plaque. J Bacteriol. 2001;183:3770–3783. doi: 10.1128/JB.183.12.3770-3783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lamont RJ, Jenkinson HF. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol Mol Biol Rev. 1998;62:1244–1263. doi: 10.1128/mmbr.62.4.1244-1263.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Progulske-Fox A, Kozarov E, Dorn B, Dunn W Jr., Burks J, Wu Y. Porphyromonas gingivalis virulence factors and invasion of cells of the cardiovascular system. J Periodontal Res. 1999;34:393–399. doi: 10.1111/j.1600-0765.1999.tb02272.x. [DOI] [PubMed] [Google Scholar]
- 7.Potempa J, Travis J. Porphyromonas gingivalis proteinases in periodontitis, a review. Acta Biochim Pol. 1996;43:455–465. [PubMed] [Google Scholar]
- 8.Banbula A, Mak P, Bugno M, Silberring J, Dubin A, Nelson D, Travis J, Potempa J. Prolyl tripeptidyl peptidase from Porphyromonas gingivalis: a novel enzyme with possible pathological implications for the development of periodontitis. J Biol Chem. 1999;274:9246–9252. doi: 10.1074/jbc.274.14.9246. [DOI] [PubMed] [Google Scholar]
- 9.Banbula A, Bugno M, Goldstein J, Yen J, Nelson D, Travis J, Potempa J. Emerging family of proline-specific peptidases of Porphyromonas gingivalis: purification and characterization of serine dipeptidyl peptidase, a structural and functional homologue of mammalian prolyl dipeptidyl peptidase IV. Infect Immun. 2000;68:1176–1182. doi: 10.1128/iai.68.3.1176-1182.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Travis J, Pike R, Imamura T, Potempa J. Porphyromonas gingivalis proteinases as virulence factors in the development of periodontitis. J Periodontal Res. 1997;32:120–125. doi: 10.1111/j.1600-0765.1997.tb01392.x. [DOI] [PubMed] [Google Scholar]
- 11.Imamura T. The role of gingipains in the pathogenesis of periodontal disease. J Periodontol. 2003;74:111–118. doi: 10.1902/jop.2003.74.1.111. [DOI] [PubMed] [Google Scholar]
- 12.Mezyk-Kopec R, Bzowska M, Potempa J, Bzowska M, Jura N, Sroka A, Black RA, Bereta J. Inactivation of membrane tumor necrosis factor α by gingipains from Porphyromonas gingivalis. Infect Immun. 2005;73:1506–1514. doi: 10.1128/IAI.73.3.1506-1514.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yun PL, Decarlo AA, Chapple CC, Hunter N. Functional implication of the hydrolysis of platelet endothelial cell adhesion molecule 1 (CD31) by gingipains of Porphyromonas gingivalis for the pathology of periodontal disease. Infect Immun. 2005;73:1386–1398. doi: 10.1128/IAI.73.3.1386-1398.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sroka A, Sztukowska M, Potempa J, Travis J, Genco CA. Degradation of host heme proteins by lysine- and arginine-specific cysteine proteinases (gingipains) of Porphyromonas gingivalis. J Bacteriol. 2001;183:5609–5616. doi: 10.1128/JB.183.19.5609-5616.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Devine DA, Marsh PD, Percival RS, Rangarajan M, Curtis MA. Modulation of antibacterial peptide activity by products of Porphyromonas gingivalis and Prevotella spp. Microbiology. 1999;145:965–971. doi: 10.1099/13500872-145-4-965. [DOI] [PubMed] [Google Scholar]
- 16.Bissell J, Joly S, Johnson GK, Organ CC, Dawson D, B. McCray PB, Jr, Guthmiller JM. Expression of β-defensins in gingival health and in periodontal disease. J Oral Pathol Med. 2004;33:278–285. doi: 10.1111/j.0904-2512.2004.00143.x. [DOI] [PubMed] [Google Scholar]
- 17.Dale BA, Kimball JR, Krisanaprakornkit S, Roberts F, Robinovitch M, O'Neal R, Valore EV, Ganz T, Anderson GM, Weinberg A. Localized antimicrobial peptide expression in human gingiva. J Periodontal Res. 2001;36:285–294. doi: 10.1034/j.1600-0765.2001.360503.x. [DOI] [PubMed] [Google Scholar]
- 18.Dunsche A, Acil Y, Dommisch H, Siebert R, Schroder JM, Jepsen S. The novel human β-defensin-3 is widely expressed in oral tissues. Eur J Oral Sci. 2002;110:121–124. doi: 10.1034/j.1600-0722.2002.11186.x. [DOI] [PubMed] [Google Scholar]
- 19.Ghosh SK, Gerken TA, Schneider KM, Feng Z, McCormick TS, Weinberg A. Quantification of human beta-defensin-2 and −3 in body fluids: application for studies of innate immunity. Clin Chem. 2007;53:757–765. doi: 10.1373/clinchem.2006.081430. [DOI] [PubMed] [Google Scholar]
- 20.Diamond DL, Kimball JR, Krisanaprakornkit S, Ganz T, Dale BA. Detection of β-defensins secreted by human oral epithelial cells. J Immunol Methods. 2001;256:65–76. doi: 10.1016/s0022-1759(01)00442-2. [DOI] [PubMed] [Google Scholar]
- 21.Pisano E, Cabras T, Montaldo C, Piras V, Inzitari R, Olmi C, Castagnola M, Messana I. Peptides of human gingival crevicular fluid determined by HPLC-ESI-MS. Eur J Oral Sci. 2005;113:462–468. doi: 10.1111/j.1600-0722.2005.00246.x. [DOI] [PubMed] [Google Scholar]
- 22.Pingel LC, Kohlgraf KG, Hansen CJ, Eastman CG, Dietrich DE, Burnell KK, Srikantha RN, Xiao X, Bélanger M, Progulske-Fox A, Cavanaugh JE, Guthmiller JM, Johnson GK, Joly S, Kurago ZB, Dawson DV, Brogden KA. Human β-defensin 3 binds to hemagglutinin B (rHagB), a non-fimbrial adhesin from Porphyromonas gingivalis, and attenuates a pro-inflammatory cytokine response. Immunol Cell Biol. 2008 doi: 10.1038/icb.2008.56. E-pub ahead of print. [DOI] [PubMed] [Google Scholar]
- 23.Yang D, Biragyn A, Hoover DM, Lubkowski J, Oppenheim JJ. Multiple roles of antimicrobial defensins, cathelicidins, and eosinophil-derived neurotoxin in host defense. Annu Rev Immunol. 2004;22:181–215. doi: 10.1146/annurev.immunol.22.012703.104603. [DOI] [PubMed] [Google Scholar]
- 24.Joly S, Maze C, McCray PB, Jr, Guthmiller JM. Human β-defensins 2 and 3 demonstrate strain-selective activity against oral microorganisms. J Clin Microbiol. 2004;42:1024–1029. doi: 10.1128/JCM.42.3.1024-1029.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Travis J, Banbula A, Potempa J. The role of bacterial and host proteinases in periodontal disease. Adv Exp Med Biol. 2000;477:455–465. doi: 10.1007/0-306-46826-3_46. [DOI] [PubMed] [Google Scholar]
- 26.Curtis MA, Aduse-Opoku J, Rangarajan M. Cysteine proteases of Porphyromonas gingivalis. Crit Rev Oral Biol Med. 2001;12:192–216. doi: 10.1177/10454411010120030101. [DOI] [PubMed] [Google Scholar]
- 27.Ji S, Hyun J, Park E, Lee BL, Kim KK, Choi Y. Susceptibility of various oral bacteria to antimicrobial peptides and to phagocytosis by neutrophils. J Periodontal Res. 2007;42:410–419. doi: 10.1111/j.1600-0765.2006.00962.x. [DOI] [PubMed] [Google Scholar]
- 28.Ouhara K, Komatsuzawa H, Yamada S, Shiba H, Fujiwara T, Ohara M, Sayama K, Hashimoto K, Kurihara H, Sugai M. Susceptibilities of periodontopathogenic and cariogenic bacteria to antibacterial peptides, β-defensins and LL37, produced by human epithelial cells. J Antimicrob Chemother. 2005;55:888–896. doi: 10.1093/jac/dki103. [DOI] [PubMed] [Google Scholar]
- 29.Bachrach G, Altman H, Kolenbrander PE, Chalmers NI, Gabai-Gutner M, Mor A, Friedman M, Steinberg D. Resistance of Porphyromonas gingivalis ATCC 33277 to direct killing by antimicrobial peptides is protease independent. Antimicrob Agents Chemother. 2008;52:638–642. doi: 10.1128/AAC.01271-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Belas R, Manos J, Suvanasuthi R. Proteus mirabilis ZapA metalloprotease degrades a broad spectrum of substrates, including antimicrobial peptides. Infect Immun. 2004;72:5159–5167. doi: 10.1128/IAI.72.9.5159-5167.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sieprawska-Lupa M, Mydel P, Krawczyk K, Wojcik K, Puklo M, Lupa B, Suder P, Silberring J, Reed M, Pohl J, Shafer W, McAleese F, Foster T, Travis J, Potempa J. Degradation of human antimicrobial peptide LL-37 by Staphylococcus aureus-derived proteinases. Antimicrob Agents Chemother. 2004;48:4673–4679. doi: 10.1128/AAC.48.12.4673-4679.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ganz T. Defensins: antimicrobial peptides of innate immunity. Nat Rev Immunol. 2003;3:710–720. doi: 10.1038/nri1180. [DOI] [PubMed] [Google Scholar]
- 33.Lehrer RI. Primate defensins. Nat Rev Microbiol. 2004;2:727–738. doi: 10.1038/nrmicro976. [DOI] [PubMed] [Google Scholar]
- 34.Brogden KA. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol. 2005;3:238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 35.Yang D, Chertov O, Oppenheim JJ. The role of mammalian antimicrobial peptides and proteins in awakening of innate host defenses and adaptive immunity. Cell Mol Life Sci. 2001;58:978–989. doi: 10.1007/PL00000914. [DOI] [PMC free article] [PubMed] [Google Scholar]