Abstract
In Gram-negative bacterial infection, lipopolysaccharide (LPS) readily overwhelms the host innate immune system, which could result in inflammation and sepsis in severe cases. Therefore, developing anti-LPS molecules would confer an efficient antibacterial strategy. We used SELEX (Systemic Evolution of Ligands by EXponential enrichment) to isolate single-stranded DNA (ssDNA) aptamers. By immobilizing and exposing different orientations of the LPS molecule on hydrophobic and hydrophilic surfaces, two populations of aptamers were captured from a library of 1014–15 ssDNA oligonucleotides. Progressive SELEX enriched the aptamers towards thymidine residues. The more hydrophobic aptamers with T-rich loops showed strong molecular recognition for the lipid A moiety of LPS, binding at affinity of up to KD of 10–9M, and eliciting 95% neutralization of endotoxicity. The longer ssDNAs exhibited greater avidity for LPS and conferred more efficacious antagonism against LPS. The nucleotide composition imposes subtle influence on the aptamer folding and affinity for LPS.
Key Words: Endotoxin neutralization, ssDNA aptamers, Oligonucleotides, Molecular recognition, Binding kinetics
Introduction
Sepsis is a clinical syndrome associated with severe consequences such as multiple organ failure/multiple organ dysfunction syndrome (MOF/MODS) and hence it is correlated with high mortality rate [1]. Bacterial endotoxin or lipopolysaccharide (LPS) is a major component of the outer membrane of Gram-negative bacteria [2] that is acutely detected by the host immune system. It is responsible for 45–60% of sepsis [3]. Once released from the bacterial cell wall during infection, it interacts with LPS-binding protein (LBP) in the plasma [4]. LBP binds with high affinity to LPS at its lipid A domain [5]. LPS, in complex with LBP, is presented to membrane-bound CD14, which elicits signaling through TLR4 in the macrophages [6]. Activation of macrophages leads to cytokine release, expression of adhesion molecules, phagocytosis and free radical production [7, 8, 9]. Overexpression and dysregulation of these potent mediators, particularly tumor necrosis factor-α (TNF-α), interleukin (IL) and interferon-γ (IFN-γ), contribute to the clinical manifestations of sepsis.
Several approaches have been explored to prevent the cascade of LPS-induced events in humans. To provide a broad cross-protection against LPS from various bacteria, monoclonal antibodies directed against the conserved lipid A region have been developed. However, clinical studies documented a lack of benefit with the monoclonal antibodies [10, 11]. The ineffectiveness of these antibodies could be attributable to the inaccessibility of the biologically active lipid A region of LPS to the antibodies. In the bloodstream, LPS is found either as aggregates or associated with circulating lipoproteins. Other anti-sepsis strategies include the use of natural inhibitor of cytokine production, such as anti-inflammatory cytokines, IL-4 and IL-10, and the development of anti-cytokine antibodies such as anti-TNF-α antibody [12, 13]. However, none of these clinical trials have yielded satisfactory results, possibly due to the complex and interdependent roles played by cytokines in vivo. Nevertheless, complete removal of cytokines like TNF-α and IL-1 during infection is probably disadvantageous, because the physiological concentration of TNF-α orchestrates the inflammatory reaction of the infected host. Hence, there is a need to search for an effective therapy. Development of anti-endotoxin is a worthwhile prophylactic approach to prevent the activation of the downstream inflammatory mediators, hence effectively blocking sepsis.
Recently, oligoaptamers with exceptionally high affinity for their cognate ligands have been derived from combinatorial libraries, by iterative rounds of in vitro selection and amplification process, called Systematic Evolution of Ligands by EXponential enrichment (SELEX) [14, 15, 16]. Aptamers can be RNA, modified RNA, single-stranded DNA (ssDNA) or double-stranded DNA. Hitherto, targets of aptamers can be varied from simple ions, low-molecular-weight ligands and proteins to whole cells. The concept of SELEX relies on the ability of aptamers to fold into unique three-dimensional structures in the presence of their respective targets, and bind with high affinity and specificity, being able to discriminate closely related targets/ligands. Furthermore, aptamers offer advantages over protein-based affinity reagents due to their stability, ease of regeneration and simple modification for detection and immobilization [17]. Moreover, some aptamers have shown higher affinity and specificity towards target molecules than those of antibodies [18]. Aptamers have the advantage of not triggering an immune response, which is one of the major limitations of antibodies for in vivo use. These features make aptamers emerge as a class of molecules that rival antibodies in both therapeutic and diagnostic applications.
Here, we explored the possibility of selecting for ssDNA that binds LPS specifically. Dwarakanath et al. [19] reported a quantum dot-labeled aptamer able to bind to bacteria and to cause a blue shift in fluorescence emission. However, specific information was lacking on the aptamer sequence/structure and their binding affinity to LPS, or to which part of the LPS molecule the aptamer was targeting. Here, we used SELEX to select for oligoaptamers from an ssDNA library comprising 1014–15 different sequences to specifically target the LPS. This is based on the premise that selected oligoaptamers are able to bind to LPS and neutralize the endotoxicity. Each of the oligoaptamers contains 40 random bases flanked by defined primer binding sites containing 23 bases at the 5’ end and 30 bases at the 3’ end. The 5’ and 3’ end primers contain BamHI and HindIII sites for subsequent cloning. Thus, each oligoaptamer totals 93 nucleotides in length. A comparison between two specific SELEX-selected 93-mer ssDNAs (M7-2 and M7-5) and a synthetic 93-mer Poly-C sequence, and further analysis of a series of ssDNAs of increasing lengths from 18 to 93 bases showed that ssDNAs bound the lipid A moiety of LPS in a length-dependent manner to neutralize the endotoxicity. The longer aptamers achieved greater binding affinity for LPS. The sequence specificity or base composition of the ssDNA seems to exert a subtle effect on the secondary structure folding of the ssDNAs and hence their efficacy in LPS binding and neutralization.
Materials and Methods
Materials
A library containing 1014–15 ssDNAs of 93 nucleotides each (Millennium Science, Singapore, Pte. Ltd) was employed in SELEX. Each ssDNA member contained 40 randomized nucleotides flanked by defined primer-binding sites (5’-CC GGA TCC GTT GAT ATA AAA TTC (N40) TTG ATT GTG GTG TTG GCT CCC AAG CTT CGG-3’). The forward and reverse primers containing BamHI and HindIII restriction enzyme sites (underlined), respectively, were used in PCR for the synthesis of dsDNA and ssDNA. Different lengths of single-stranded oligonucleotides of random sequences were synthesized by 1st Base Pte Ltd, Singapore. LPS from Escherichia coli 055:B5 (Sigma) was used as the ligand for the selection of oligonucleotides while E. coli lipid A (F583, Rd mutant, Sigma) and ReLPS from Salmonella minnesota R595 (Re mutant, List Biological Laboratories, Inc., USA) were used in functional studies of the selected aptamers. A PyroGene kit for measuring LPS was from Lonza, Inc., USA. Human TNF-α BD OptEIA was purchased from BD Biosciences Pharmingen (San Diego, Calif., USA) and CellTiter96 Aqueous One Solution Cell Proliferation Assay was from Promega (Madison, Wisc., USA). Pyrogen-free water for making buffers was from Baxter (Morton Grove, Ill., USA).
Selection of LPS-Specific Aptamers
The specific activity of the E. coli LPS (Sigma) was calibrated against a reference standard LPS from Lonza, Inc., USA. Both PolySorpTM and MaxiSorpTM 96-well plates (Nunc) were first coated with 4 µg/ml (˜1 µM containing 400 EU/ml) of the E. coli LPS diluted in phosphate-buffered saline (PBS) [20]. The library of ssDNAs in PBS, pH 7.4, was heat-denatured at 95°C for 10 min and cooled immediately on ice. An aliquot of 1 µM of the ssDNA library, referred to as ‘apt lib’, which contained all the 1014–15 members, was used in the first round of selection. The ssDNAs were allowed to interact with the immobilized LPS at 37°C for 2 h. The non-specifically bound ssDNAs were washed off with PBS containing 0.05% Tween-20. Bound aptamers were eluted with 100 mM NaOH and precipitated overnight at −20°C in 3 M NaOAc and 95% ethanol. The selected ssDNA aptamers were used as templates in performing pilot PCR with varying number of cycles to produce dsDNA templates with few aberrant products.
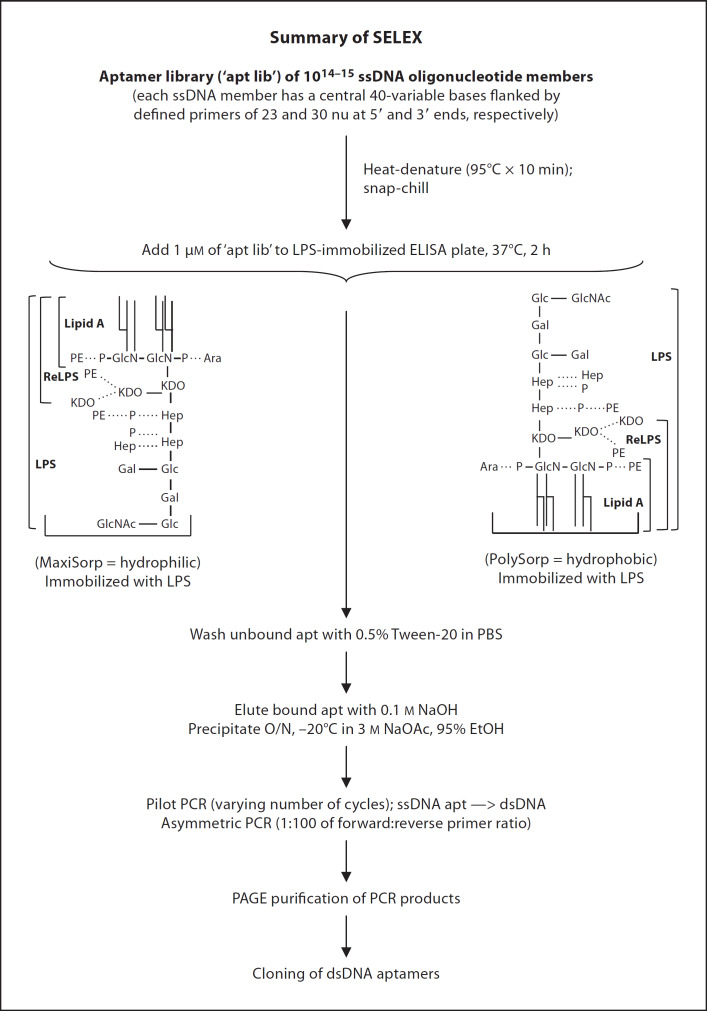
The random dsDNA was used as template in asymmetric PCR with primer ratio of 1:100 of reverse:forward primers at different amplification cycle numbers. Large-scale asymmetric PCR was performed with the desired cycle numbers attained. The PCR products were purified and eluted from 9% polyacrylamide gel using elution buffer containing 0.5 M NH4Ac, 10 mM Mg(Ac)2, 1 mM EDTA (pH 8.0) and 0.1% SDS. As the aptamers became purer and more specific for the target, the amount of LPS and the incubation period between aptamers and LPS were gradually reduced in progressive rounds of selection to increase the stringency of selection. The progress of SELEX was evaluated and seven rounds of selection were accomplished. Figure 1 summarizes the SELEX process.
Fig. 1.
Schematic summary of SELEX procedure. The PolySorpTM and MaxiSorpTM 96-well plates (Nunc) were coated with 4 µg/ml (˜1 µM containing 400 EU/ml) of LPS from E. coli. The ssDNA aptamer library (‘apt lib’ containing 1014–15 members) was heat-denatured at 95°C for 10 min and cooled immediately on ice. An aliquot of 1 µM of the ‘apt lib’ was used in the first round of selection. The ssDNAs were incubated with the immobilized LPS at 37°C for 2 h. The non-specifically bound ssDNAs were washed off with PBS containing 0.05% Tween-20. Bound aptamers were eluted with 100 mM NaOH and precipitated overnight (O/N) at −20°C in 3 M NaOAc and 95% ethanol. The selected ssDNA aptamers were used as templates in pilot PCR with varying number of cycles to produce dsDNA templates with few aberrant products. The random dsDNA was used as template in asymmetric PCR (primer ratio of 1:100 of reverse:forward) at different amplification cycle numbers followed by large-scale asymmetric PCR with the desired cycle number. The PCR products were PAGE-purified. As the aptamers became purer and more specific for the target, the amount of LPS and the incubation period between aptamers and LPS were gradually reduced in progressive rounds to increase the stringency of selection.
Cloning and Sequencing of Selected Oligoaptamers
Aptamers obtained from different rounds of selection were converted to dsDNA before being cloned into pGEM-T Easy Vector (Promega, USA). Plasmids were purified with Wizard Plus SV Miniprep DNA purification system (Promega, USA). Sequencing reaction was carried out with the BigDyeTM Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) using SP6 vector primer. DNA sequencing was performed using ABI Prism 7000 automated sequencer.
Functional Assay of Aptamers – Competition for LPS in the Pyrogene rFC Inhibition Assay
In the PyroGene assay, LPS binds the recombinant factor C (rFC), which is activated to catalyze the hydrolysis of a synthetic substrate to release a fluorescent product that is quantifiable at excitation and emission wavelengths of 380 and 440 nm, respectively. To analyze the functionality of the oligoaptamers, their ability to bind LPS in competition against rFC activity was tested. The inhibition of the LPS-induced rFC activity by the oligoaptamers was assayed by preincubating with 1 EU/ml of E. coli LPS at 37°C for 30 min in a PolySorp 96-well plate (FluoronuncTM, Nunc) prior to addition of the rFC and substrate. The fluorescence of the product was measured immediately and subsequently at 15-min intervals, using a spectrofluorimeter (LS-50B, PerkinElmer).
In a separate assay the inhibition of lipid A and ReLPS by the selected aptamers and other random oligonucleotides of variable lengths was studied. Here, 1 EU/ml of E. coli lipid A or ReLPS from S. minnesota R595 was used in place of the E. coli LPS to activate the rFC enzyme activity. The rationale for this modification in the competition assay was to examine the ability of the aptamers at targeting not just the overall LPS molecule, but also (a) lipid A, the biologically potent moiety of LPS, and (b) ReLPS, which represents the basic core component that exists in all forms of LPS from Gram-negative bacteria. Prior to the assay with aptamers, standard curves were established to calibrate the lipid A and ReLPS against a reference standard LPS from Lonza, Inc., USA.
Surface Plasmon Resonance Analysis of Real-Time Biointeraction between Aptamer and Lipid A
The real-time interaction between different concentrations of the aptamers and lipid A was performed with Biacore 2000 instrument. An HPA sensor chip was precoated with 0.25 mg/ml of E. coli lipid A in PBS. In all experiments, 100 mM pyrogen-free Hepes, pH 9.0, containing 100 mM NaCl, 10 mM MgCl2 and 1.5% glycerol was used as the running buffer at a flow rate of 30 µl/min. The HPA chip was regenerated with 100 mM NaOH until the baseline was achieved. The affinity constant of interaction between the oligoaptamers and lipid A was calculated using BIAevaluation software 3.2.
TNF-α Assay
The inhibitory effect of the aptamers on LPS-induced TNF-α release was assayed with THP-1 cells. The cells were grown at 37°C in a humidified environment in the presence of 5% CO2 with RPMI 1640 medium supplemented with 10% FBS, penicillin (100 U/ml), and streptomycin (0.1 mg/ml). THP-1 monocytes were transformed into macrophages by the addition of a concentrated stock solution of phorbol myristic acid (PMA; Sigma; 0.3 mg/ml in dimethyl sulfoxide) to a final concentration of 30 ng/ml PMA and 0.01% dimethyl sulfoxide. PMA-treated cell suspension was immediately plated into a sterile 96-well microtiter plate (NunclonTM surface, Nunc) at a density of 1 × 106 cell/ml and allowed to differentiate for 48 h at 37°C. Immediately before stimulation by 10 ng/ml LPS or LPS preincubated with the aptamers, the culture medium was removed and the cells were washed twice with serum-free RPMI 1640. Reaction mixtures of LPS with or without aptamers were incubated at 37°C for 30 min prior to their addition into a 96-well plate coated with cells. After 6 h incubation at 37°C, the culture supernatant was collected and the concentration of human TNF-α was assayed by ELISA according to the manufacturer's instructions.
Cytotoxicity Assay
To analyze the potential cytotoxicity of the aptamers, 50 µl of human monocytes, THP-1, at a density of 2 × 106 cells/ml in RPMI 1640 were incubated with 50 µl of twofold serial dilutions of aptamers in a 96-well plate for 24 h at 37°C. Then, 20 µl of CellTiter 96 Aqueous One Solution Reagent was added and further incubated for 90 min at 37°C. MTS tetrazolium compound [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium] is normally bioreduced by living cells into a colored formazan product that is soluble in tissue culture medium. The absorbance at 490 nM was measured using an ELISA plate reader. But, cells undergoing acute cytotoxicity are unable to metabolize MTS.
Results
Selection of ssDNA Oligoaptamers That Bind LPS
Several parameters and conditions were considered with the aim to optimize and establish a SELEX approach that was suitable for selecting LPS-specific aptamers from a library of 1 × 1014–15 oligonucleotide sequences each containing 93 bases within which 40 are random: (i) the number of PCR cycles was optimized to avoid overamplification, which would be evidenced by mis-annealed products; (ii) the incubation time of the oligoaptamers with LPS was progressively decreased over seven iterative rounds of selection with a view to increasing the stringency and specificity for LPS; (iii) the method of recovery of the ssDNA sequences, where efficiency of elution from PAGE gel and affinity chromatography by streptavidin-biotin aptamers was compared. The affinity chromatography approach incurred drastic loss of aptamers due to high avidity of the ssDNA to the streptavidin beads, and inefficiency in regeneration of the resin; (iv) screening and selection for LPS-binding aptamers via a solid-phase method whereby LPS was immobilized on 96-well plates, and (v) whether the orientation of LPS on the 96-well plates would impose any effect on the selection of LPS-specific oligoaptamers.
In consideration of these parameters and the conditions of SELEX, MaxiSorp and PolySorp plates were tested for their suitability as LPS-binding platforms upon which the oligoaptamers were selected. When immobilized on the hydrophilic surface of the MaxiSorp plate, it is anticipated that the hydrophilic oligosaccharide chain of LPS oriented downwards whilst the hydrophobic lipid A tails would be exposed on the surface for interaction with the ssDNA. Conversely, on the hydrophobic PolySorp plate surface, the acyl chains of lipid A were immobilized downwards and the hydrophilic sugar residues of the LPS would be exposed for interaction with ssDNA. The round 3 selected oligoaptamers from the PolySorp plate (referred to as P3 aptamers) were used as the starting point for further selection on the MaxiSorp plate. The P3 aptamers contained a sufficient degree of heterogeneity of oligonucleotide sequences to afford such a transition of selection. A further four rounds of selection were also accomplished for LPS-coated on the MaxiSorp plate.
Selected 93-mer Oligoaptamers Bind and Neutralize the Endotoxicity of LPS
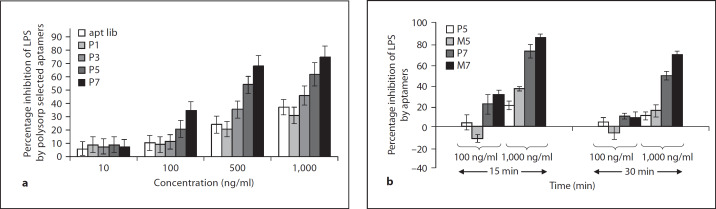
The LPS neutralization ability of the oligoaptamers was measured by using the PyroGene rFC-based assay [21], which is highly sensitive to endotoxin. Oligoaptamers selected on the PolySorp plate (P1-P7) were found to attenuate LPS-induced rFC activity in a dose-dependent manner. This inhibition was specific and the efficiency apparently increased with progressive rounds of selection (fig. 2a), where P7 oligoaptamers were found to inhibit rFC activity by as much as 75%. Moreover, comparison of the aptamers obtained between the two selection platforms (MaxiSorp and PolySorp) showed that the M5 and M7 aptamers exerted 20% greater inhibition of LPS-induced rFC activity than the P5 and P7 counterparts (fig. 2b). This suggests that different orientations of the LPS immobilized on the hydrophobic or hydrophilic plate surface selected dissimilar populations of aptamers, which elicit different efficacy and functional specificity. It is conceivable that exposure of the lipid A moiety of LPS immobilized on the hydrophilic surface of the MaxiSorp plate resulted in the selection for oligoaptamers which exhibited greater LPS-antagonistic potency.
Fig. 2. a.
Inhibition of LPS-induced rFC activity by oligoaptamers selected from different rounds using PolySorp plate. In the presence of the aptamers which specifically bind LPS/lipid A, the rFC activity was attenuated. This reduction in rFC activity measured in fluorescence unit (FU), which is dose-dependent on the aptamers, reflects the efficacy of LPS neutralization by the aptamers. The percentage inhibition of LPS-induced rFC activity was calculated by subtracting averaged relative fluorescence unit (RFU) of each sample against an averaged baseline RFU value of LPS alone, and divided by the average RFU of LPS. The standard errors of at least three independent experiments are presented. b Different orientations of LPS impose an effect on the activity of aptamers selected. rFC assay was performed on the PolySorp- and MaxiSorp-selected aptamers, to compare their anti-LPS activity. P and M annotate PolySorp and MaxiSorp plate-selected aptamers, respectively.
Primary Sequence Analysis and Secondary Structure Prediction of Selected Oligoaptamers
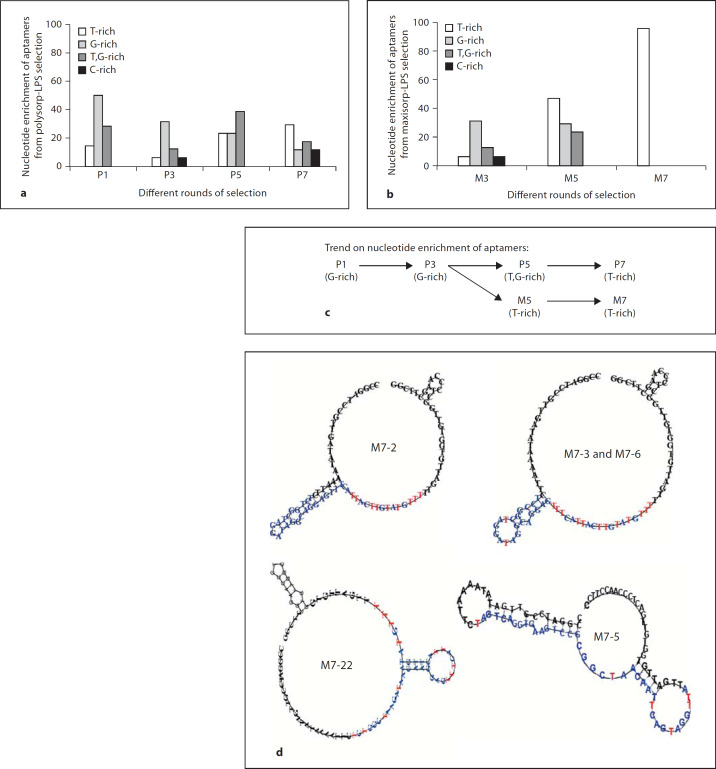
In order to assess the enrichment of the DNA sequence through progressive rounds of SELEX, the LPS-specific oligoaptamers from rounds 1, 3, 5, and 7 obtained from both the PolySorp-LPS and MaxiSorp-LPS selection methods were cloned and sequenced. The DNA sequences showed progressive enrichment towards thymidine (T) residues in both situations (fig. 3a, b). Figure 3c shows that the MaxiSorp-LPS selection yielded T-rich aptamers (M5) at two rounds earlier than those selected from the
Fig. 3. a, b.
Nucleotide enrichment of aptamers derived from both (a) PolySorp-LPS and (b) MaxiSorp-LPS selections. The percentage of oligoaptamers derived from different rounds of (a) PolySorp-LPS, P; (b) MaxiSorp-LPS, M, selections in which they are categorized according to the nucleotide content in the variable region. c Summary of the predominant groups of aptamers obtained in each selection method at each iterative round of SELEX. d Predicted secondary structure of M7 oligoaptamers from SELEX, which contain T-rich ring. In contrast, M7-5 shows a secondary stem-loop which contains more A,T residues. The variable regions of the aptamers are highlighted in blue while the T residues in the variable region that are exposed are marked red.
PolySorp-LPS platform (P7). Interestingly, 70% of M7 aptamers contained identical bases (represented by M7-2) while another 26% of the population displayed only slight differences in the 40-base variable region (table 1). All the primary sequences of M7 aptamers were subjected to secondary structure prediction using the Vienna Secondary Structure Prediction Program (http://rna.tbi.univie.ac.at/). The highly homogeneous sequences of M7 aptamers
Table 1.
Primary sequence of the 40 variable bases (40Ns) of the M7 oligoaptamers
| apt lib | CCGGATCCGTTGATATAAAATTC(40Ns)TTGATTGTGGTGTTGGCTCCCAAGCTTCGG |
|---|---|
| M7-2 (70%) | TCTGGCTAGGATAGGCAGGAGTTTCATTACTTGTATGTTT |
| M7-1 | TCTGGCTAGGATAGGCAGGAGTTTCATTACTTATATGTTT |
| M7-3/M7-6 | TCCGGCTAGGATAGGCAGGAGTTTCATTACTTGTATGTTT |
| M7-13 | TCTGGCTAGGATAGGCAGGTGTTTCATTACTTGTATGTTT |
| M7-14 | TCTGGCTAGGATAGGCAGGAGCTTCATTATTTGTATGTTT |
| M7-22 | TCTGGCTAAGATAAACAAGGAGTTTCATTACTTGTATGTTT |
| M7-5 | TAGTCAGGTGAAGTCCGCGGCTAACAATTCAGTAGGTTA |
The ‘apt lib’ is the complete aptamer library of 1014–15 members, each containing 40 variable bases (40Ns) flanked by the forward and reverse primers on the left and right.
Nucleotides that deviate from the sequences of the identical clones are in bold.
M7-22 and M7-5 harbor 41 and 39 variable residues (Ns), respectively at their centers.
70% of the M7 clones share similar base sequence, as represented by the M7-2 clone.
showed a propensity to fold into a T-rich ring where the thymine side chains were exposed and clustered at one locus in the ring (fig. 3d). In contrast, the M7-5 oligoaptamer folded into a secondary structure which is totally distinct from the rest of M7 members.
Endotoxin-Antagonistic Activity Is Dependent on the Length and Secondary Structure of Oligoaptamers
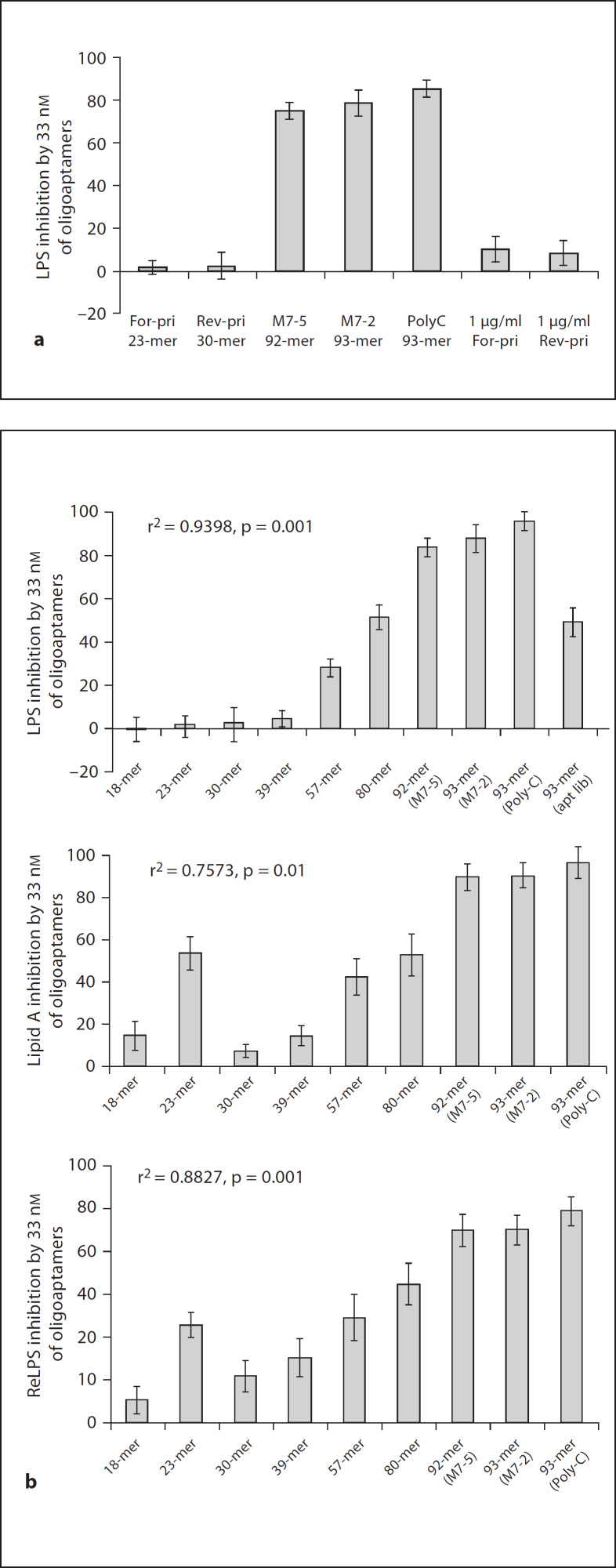
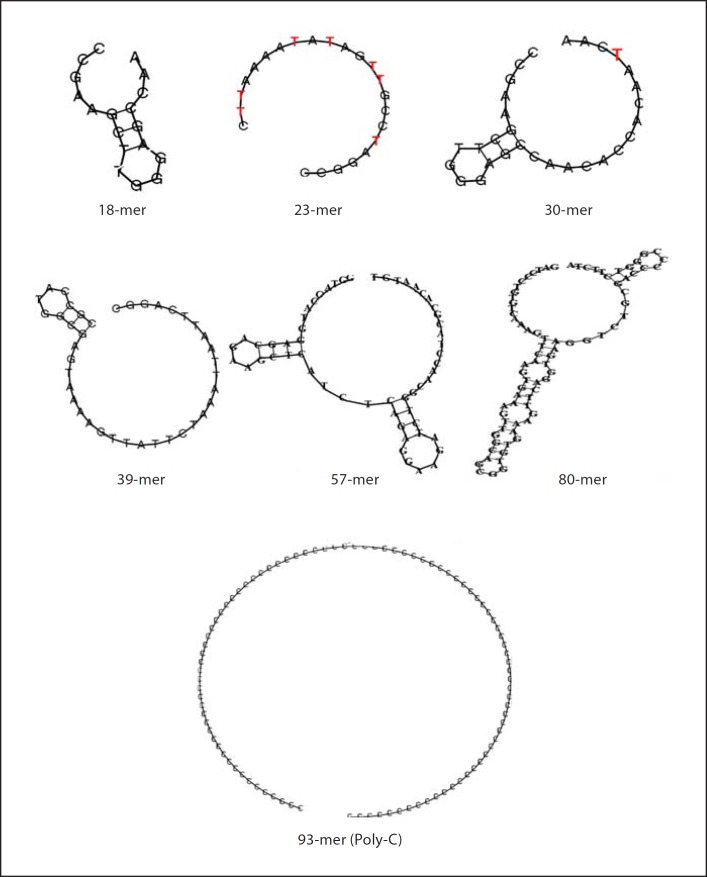
We further investigated the anti-LPS activity of 33 nM of the single clones, M7-2 (93 bases) and M7-5 (92 bases) in comparison to synthetic oligonucleotides of: (i) 93’-mer Poly-C; (ii) 23-mer forward primer and the 30-mer reverse primer which flank each of the 40 random bases of the original oligoaptamer library. Interestingly, the forward and reverse primers exhibited only 10% anti-LPS activity, while the three 92–93 bases long oligos (M7-2, M7-5 and Poly-C) exerted up to 95% anti-LPS activity, regardless of their nucleotide sequence (fig. 4a). This provided us with the first clue that the high LPS inhibition activity of the 93-mer oligoaptamers is mainly attributable to the length. In order to confirm the length dependency of the oligonucleotides in recognizing and neutralizing the endotoxicity of LPS, we tested a series of random ssDNAs of increasing lengths, ranging from 18 to 80 bases in comparison with the 93-mers (table 2). Indeed, we demonstrated that the LPS inhibition ability correlates well with the length of the oligoaptamers (fig. 4b).
Fig. 4. a.
Comparison of LPS inhibition ability of long oligoaptamers (92-, 93-mer) and short oligos (23-, 30-mer). The oligoaptamers were compared in equimolar (33 nM) and equal amounts (33 nM = 1 µg/ml). M7-2, M7-5, which are single clones of 93- and 92- mer from the M7 population; the synthetic 93-mer Poly-C, and the forward and reverse primers (23- and 30-mer, respectively) were compared for their ability to inhibit LPS-induced rFC activity. b Relationship between oligonucleotide length and their LPS-, ReLPS- and lipid A binding and neutralization efficacy. Functional comparison of different length of oligoaptamers (18–93- mer) was performed using rFC assay. Endotoxin at 1 EU/ml derived from E. coli LPS, ReLPS of S. minnesota and E. coli lipid A were used to activate rFC activity, and 33 nM of each of the oligoaptamers were used to determine their ability to compete against rFC for LPS/ReLPS/lipid A.
Table 2.
Primary sequence of nine oligoaptamers of variable lengths tested
| ssDNA | Sequences |
|---|---|
| 18-mer | 5’-ccgaagcttgggagccaa-3’ |
| 23-mer | 5’-ccggatccgttgatataaaattc-3’ |
| 30-mer | 5’-ccgaagcttgggagccaacaccacaatcaa-3’ |
| 39-mer | 5’-cgccatggcgagtaaaagttattctaaattaattcaggc-3’ |
| 57-mer | 5’-ggtaccatggagcagaagctcatctcagaggaagatctggcaacctacgcacaatgt-3’ |
| 80-mer | 5’-gatccctgtgtcaaagttcaagtgaaagttggcagcggtgtgaaagttcaggtgaaggtctgcgacccccgggtcttcta-3’ |
| 92-mer (M7-5) | 5’-ccggatccgttgatataaaattctagtcaggtgaagtccgcggctaacaattcagtaggttattgattgtggtgttggctcccaagcttcgg-3’ |
| 93-mer (M7-2) | 5’-ccggatccgttgatataaaattctctggctaggataggcaggagtttcattacttgtatgtttttgattgtggtgttggctcccaagcttcgg-3’ |
| 93-mer (Poly-C) | 5’-ccccccccccccccccccccccccccccccccccccccccccccccccccccccccccccccccccccccccccccccccccccccccccccc-3’ |
Since lipid A is the endotoxic principle conserved in LPS molecules from different strains of Gram-negative bacteria [27], the ability of the 92–93 base oligoaptamers to exert strong molecular recognition of the lipid A moiety, and to effectively neutralize the endotoxicity, demonstrate the potential of developing these aptamers into a powerful LPS antagonist. At the same time, we noticed that compared to the 30- and 39-mer oligoaptamers, the 23-mer (previously used as the forward primer for PCR of the SELEX library) seems to exhibit a substantially higher inhibition of ReLPS and lipid A, indicating that the 23-mer may contain certain bases or propensity to fold and fit better into the lipid A domain. To ascertain this, we tabulated the nucleotide composition of the ssDNA oligoaptamers (table 3), which showed that the 23-mer is A,T-rich, being partially similar to the T-rich M7-2 and M7-5 aptamers, but different from the rest of the sequences that are either A- or G-rich and not T-rich. Secondary structure prediction (fig. 5) shows that the 23- and 93-mer Poly-C both folded into rings. Furthermore, the 23-mer has more T residues exposed, compared to only one T residue in the ring of the 30-mer. Taken together with the secondary structure predictions of M7-2 and M7-5 (fig. 3d), the T residues seem to play an important role in the oligoaptamer-LPS interaction. However, it appears that the influence from the T residues is less pronounced than that of the length of the oligoaptamers. This is corroborated by the anti-endotoxic potency of the 93-mer Poly-C, which exerted a similar efficacy of inhibition of lipid A and ReLPS as did the M7-2 and M7-5.
Table 3.
Nucleotide profile of oligoaptamers of different lengths
| Oligos | A, % | T, % | C, % | G, % |
|---|---|---|---|---|
| 18-mer | 27.8 | 11.1 | 27.8 | 33.3 |
| 23-mer | 30.4 | 30.4 | 21.7 | 17.4 |
| 30-mer | 36.7 | 10 | 33.3 | 20 |
| 39-mer | 33.3 | 28.5 | 17.9 | 20.5 |
| 57-mer | 31.6 | 17.5 | 24.6 | 26.3 |
| 80-mer | 22.5 | 25 | 21.3 | 31.3 |
| 92-mer (M7,-5) | 22.8 | 30.4 | 19.6 | 27.2 |
| 93-mer (M7,-2) | 19.4 | 36.6 | 17.2 | 26.9 |
| 93-mer (Poly-C) | 100 |
The composition of each nucleotide is tabulated in percentage and the highest percentages are in bold.
Fig. 5.
Predicted secondary structure of the synthetic non-SELEX oligoaptamers. T residues in 23- and 30-mer that are exposed are marked red.
Comparison of the Binding Affinity (KD) of Different Length Oligoaptamers to LPS
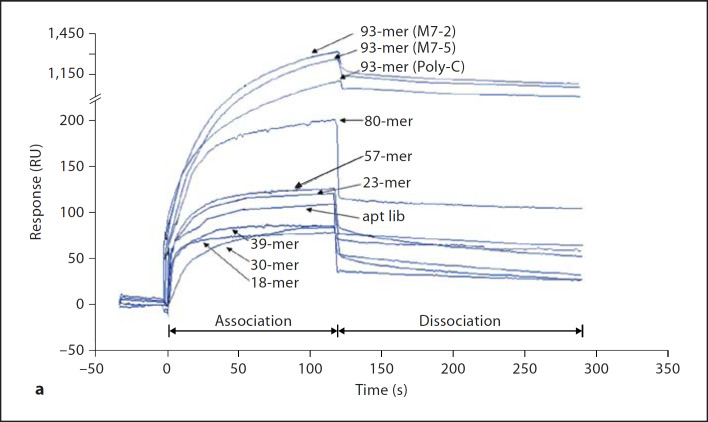
Using surface plasmon resonance analysis, the binding affinity of the nine oligonucleotides to lipid A was measured and compared. Figure 6a shows a series of sensorgrams obtained by injecting 0.4 µM of each oligoaptamer into an HPA flow cell immobilized with lipid A. The corresponding binding affinities are presented in figure 6b. The kinetic constant (KD) was calculated from the sensorgrams by fitting with a 1:1 Langmuir binding model. This real-time biointeraction analysis revealed increasing affinity with increasing length of the oligoaptamers ranging from 10–6 to 10–9 M, except for the 23-mer, which exerted higher than expected affinity (10–8 M) compared to its 30- and 39-mer counterparts. Consistent with earlier finding that the 23-mer has a higher endotoxin neutralization effect (PyroGene rFC assay) against lipid A and ReLPS, the real-time biointeraction analysis here shows that the 23-mer has a lower KD value (higher affinity) for lipid A than those of the 30- and 39-mers. Nevertheless, it must be noted that on one hand, the SELEX procedure has apparently resulted in the selection of the M7-2 and M7-5 with gain-of-affinity for LPS when compared to the mixed pool of ssDNAs in the original aptamer library (‘apt lib’, fig. 6), thus indicating that indeed the T-rich loop of the selected aptamers contributes to the LPS binding. Yet, a dichotomy is observed with the 93-mer Poly-C sequence which shares equally high anti-LPS efficacy even though without T residue, suggesting that on balance, the length rather than the sequence composition of the oligoaptamer is the predominant determinant of LPS binding and neutralization.
Fig. 6. a.
Sensorgrams depicting the real-time interaction between oligoaptamers of different lengths towards lipid A immobilized on a Biacore HPA chip. 60 µl aliquots of each of the nine oligoaptamers as well as aptamer library (‘apt lib’) at a series of different concentrations, ranging from 1, 0.4, 0.16 to 0.064 µM, were injected into the HPA chip at a flow rate of 30 µl/min with 100 mM Hepes, pH 9.0, containing 100 mM NaCl, 10 mM MgCl2 and 1.5% glycerol as the running buffer. During the dissociation phase, the running buffer was introduced for 120 s. For clarity, only sensorgrams of 0.4 µM of oligoaptamers are shown here. b The kinetic constant (KD values) of the oligoaptamers indicate the affinity of interaction between the oligoaptamers (of 18–93-mer) and lipid A, which was calculated using BIAevaluation software with a 1:1 Langmuir binding model.
Aptamers Exhibit Minimal Cytotoxicity and Inhibit LPS-Induced TNF-α Secretion Regardless of Their Lengths
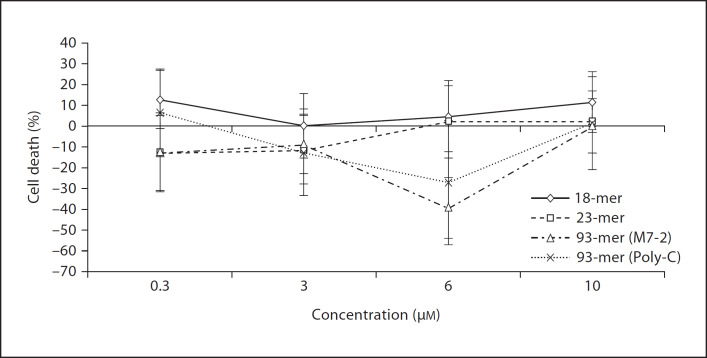
The 18-mer, 23-mer and both 93-mers (M7-2 and Poly-C) were tested for their ability to inhibit LPS-induced TNF-α release from THP-1 cells. Irrespective of their lengths, all four oligoaptamers elicited up to ˜40% inhibition of LPS-induced TNF-α production, probably through the binding and neutralization of the LPS. All the aptamers tested over the wide range of concentrations showed negligible cytotoxicity and have no effect on the cell viability (fig. 7).
Fig. 7.
Aptamers show a minimal adverse effect on cell viability. Four different concentrations, 0.3, 3, 6, 10 µM, of the aptamers were examined for their cytotoxicity/antiproliferative potential against the THP-1 cells. The resulting cell viability was determined by measuring A490nm and the percentage of cell death was calculated after subtraction of the control values.
Discussion
LPS has been strongly linked to the pathophysiology of sepsis [22], where it is associated with mortality rate of 60–70% [3] and remains the main cause of death in intensive care units [23]. Thus, abrogation of the inflammatory response with therapy early in infection may render the patient less susceptible to sepsis. Many therapeutic strategies have been attempted for several decades, but have resulted in limited success. The use of oligonucleotide aptamers offers advantages over that of antibodies in anti-endotoxin strategies because these aptamers can easily be produced with a high degree of accuracy, reproducibility, and purity. They are not temperature-sensitive and do not undergo reversible denaturation, thus having a much longer shelf life [17, 24]. We have procured oligoaptamers that block LPS. The LPS neutralization ability of the aptamers seemed to increase progressively with rounds of SELEX, and function in a dose-dependent manner. A similar functional enrichment was observed in the selection of tubulin-specific DNA aptamers [25] and selection of aptamers that bind differentiated but not parental cells [26]. In our case, the M7 aptamers which probably target the core of LPS and the lipid A moiety, achieved ˜95% efficacy in attenuating endotoxicity (fig. 2, 4). In contrast, the P7 aptamers which probably target the sugar residues of LPS appeared slightly less efficient in neutralizing endotoxicity. By molecular weight, lipid A constitutes around 40% of the whole LPS structure [27]. It is conceivable that when LPS was immobilized on the hydrophobic PolySorp surface where the lipid A moiety faced downwards, the bulk of the exposed polysaccharide chains of LPS could possibly hinder the accessibility of the aptamers to the lipid A moiety. Since lipid A is the endotoxic principle of LPS [28], an oligoaptamer that targets this moiety is expected to be highly efficacious in neutralizing the endotoxicity. This explains why M7 aptamers were more antagonistic of LPS than the P7 aptamers. Furthermore, solution assay with LPS or ReLPS or lipid A in the liquid phase also showed that the M7 aptamers were more efficacious.
Cloning and sequencing of the selected oligonucleotides showed that within each of the 93-mer ssDNAs, the central domain of 40 variable bases actually ranged in length from 39 to 41 nucleotides compared to the designated 40 nucleotides in the randomized library (table 1). Such differences in the lengths of the variable region have been reported for many SELEX experiments [29–31] and are presumably caused by random insertions and deletions during multiple rounds of PCR amplification [32] or limitation of nucleotide coupling efficiency during the chemical synthesis of the randomized library. An examination of the primary sequence of the oligoaptamers selected from the two different platforms (MaxiSorp/PolySorp) revealed a strong tendency to enrich from G- to T-rich residues amongst the majority of round 7 oligoaptamers, which contained more than 35% T residues in their sequences. Sequence enrichment towards a specific nucleotide after iterative rounds of selection has been also reported [25, 33, 34, 35]. Notably, MaxiSorp-LPS selection achieved T-enrichment two rounds earlier than PolySorp-LPS selection, suggesting that the former platform offered more stringent selection. The observed convergence of nucleotide enrichment towards T residue is not outlandish as work of Schneider et al. [36] on E. coli rho factor has shown that the final aptamers are not affected by the stringency of selection. It was noted that even though high stringency can enhance the selection of aptamers, those with similar sequences can be obtained from both high and low stringency selections, suggesting a degree of convergence. In our case, the convergence of T-rich aptamers at round 7 of both selection platforms suggests that this nucleotide plays an important role in the recognition and binding of LPS or lipid A.
Although there was a lack of identical or consensus sequence amongst the oligoaptamers derived from PolySorp-LPS and MaxiSorp-LPS selections, the highly homogeneous sequences of M7 as well as the majority of P7 (data not shown) displayed a tendency to form a T-rich ring structure where T residues in the variable region were clustered on one side of the ring and exposed (fig. 3d). This suggests that T-rich ring structures contribute to the recognition and binding of LPS. As a pyrimidine member that is hydrophobic due to the additional methyl group at the 5’ carbon of its ribose ring, we postulate that exposed T-rich region could recognize LPS specifically through hydrophobic interaction between its methyl group and the lipid A acyl chains. Studies have shown that various endotoxin-binding proteins and peptides recognize and bind the lipid A region of LPS to exert their LPS-activating or -neutralizing effects. LBP (LPS-binding protein), BPI (bactericidal permeability-increasing factor) and LALF (Limulus anti-LPS factor) are LPS-binding proteins [5, 38]. The LBP holoprotein is known to enhance the presentation of LPS to TLR4 [4, 5, 6]. On the other hand, synthetic peptides derived from these LPS-binding proteins bind LPS at the lipid A domain [37, 38] and block its bioactivity. Hence, it is conceivable that 93-mer T-rich oligoaptamers bind to the lipid A domain of LPS, and the binding affinity was remarkably high, at KD in the nanomolar range (fig. 6).
Notwithstanding the multiple rounds of SELEX on both the hydrophobic and hydrophilic surfaces of the immobilized LPS, the lack of distinctive sequence identity amongst the M7 oligonucleotides and the lack of LPS-binding ability by the short oligonucleotides (18–39 mer) prompted us to examine the relationship between the lengths of various ssDNAs and their LPS-binding efficacy. Interestingly, we noted that with LPS or lipid A as a target ligand, there was no apparent sequence specificity but only a length dependency of LPS antagonism by the ssDNAs. Furthermore, we demonstrate that oligonucleotides of at least ˜93 bases appeared optimal and essential for anti-LPS activity to achieve ˜95% endotoxin neutralization (fig. 4b). Compared to the parental 93-mer library with random bases in the 40-base variable region, the M7-2 and M7-5 showed improved LPS neutralization ability which suggests that sequence specificity does contribute somewhat to LPS inhibition, although to a lesser extent than the exertion from the length of the oligoaptamers. This deduction is also supported by the result from the 23-mer that has higher affinity and LPS antagonism activity than the slightly longer counterparts of 30- and 39-mers. Notably, the 23-mer oligoaptamer is A,T-rich and also fold into a large ring-like structure exposing all its T residues (fig. 5). Similar to the T-rich ring structure of M7-2 and M7-5, the T residues of the 23-mer probably recognize the lipid A domain and neutralize the endotoxicity, hence making it a better antagonist than the 30- and 39-mer oligoaptamers. However with its shorter length, it is still less efficient than the 93-mers in terms of affinity and lipid A neutralization (fig. 6). Nevertheless, it would be pertinent to examine the specificity of the bases in the 23-mer in future, perhaps through site-specific mutations. However, in cell cultures, the oligoaptamers seem to function indiscriminately of their lengths resulting in an overall suppression of up to ˜40% LPS-induced TNF-α production. Probably, some unknown factor(s) necessary for cellular anti-LPS activity are lacking under the culture condition. Further work is also needed to modify the aptamers in order to increase their differential efficiencies in vivo, and this can be readily performed on nucleotide aptamers. Consideration may also be given to the use of combinations of aptamers with a view to allowing the mixture of aptamers to confer greater molecular recognition and target at different moieties of the LPS to increase their anti-LPS efficiency.
Taken together, it can be concluded that the binding and inhibition of LPS/lipid A by the oligoaptamers is mostly influenced by the length and to a certain degree by the nucleotide composition which influences the folding structure of the oligoaptamers.
Acknowledgements
This work was supported by grants from the A*STAR-NUS-UCSD Joint Research Programme and the Ministry of Education (NUS FRC and Tier 2), Singapore.
References
- 1.Downey JS, Han J. Cellular activation mechanisms in septic shock. Front Biosci. 1998;3:d468–d476. doi: 10.2741/a293. [DOI] [PubMed] [Google Scholar]
- 2.Tall A. Plasma lipid transfer proteins. Annu Rev Biochem. 1995;64:235–257. doi: 10.1146/annurev.bi.64.070195.001315. [DOI] [PubMed] [Google Scholar]
- 3.Brun-Buisson C, Doyon F, Carlet J, Dellamonica P, Gouin F, Lepoutre A, Mercier JC, Offenstadt G, Regnier B. Incidence, risk factors, and outcome of severe sepsis and septic shock in adults. A multicenter prospective study in intensive care units. French ICU Group for Severe Sepsis. JAMA. 1995;274:968–974. [PubMed] [Google Scholar]
- 4.Lamping N, Hoess A, Yu B, Park TC, Kirschning CJ, Pfeil D, Reuter D, Wright SD, Herrmann F, Schumann RR. Effects of site-directed mutagenesis of basic residues (Arg 94, Lys 95, Lys 99) of lipopolysaccharide (LPS)-binding protein on binding and transfer of LPS and subsequent immune cell activation. J Immunol. 1996;157:4648–4656. [PubMed] [Google Scholar]
- 5.Schumann RR, Leong SR, Flaggs GW, Gray PW, Wright SD, Mathison JC, Tobias PS, Ulevitch RJ. Structure and function of lipopolysaccharide binding protein. Science. 1990;249:1429–1431. doi: 10.1126/science.2402637. [DOI] [PubMed] [Google Scholar]
- 6.Wright SD, Ramos RA, Tobias PS, Ulevitch RJ, Mathison JC. CD14, a receptor for complexes of lipopolysaccharide (LPS) and LPS binding protein. Science. 1990;249:1431–1433. doi: 10.1126/science.1698311. [DOI] [PubMed] [Google Scholar]
- 7.Rietschel ET, Brade H. Bacterial endotoxins. Sci Am. 1992;267:54–61. doi: 10.1038/scientificamerican0892-54. [DOI] [PubMed] [Google Scholar]
- 8.Watson RW, Redmond HP, Bouchier-Hayes D. Role of endotoxin in mononuclear phagocyte-mediated inflammatory responses. J Leukoc Biol. 1994;56:95–103. doi: 10.1002/jlb.56.1.95. [DOI] [PubMed] [Google Scholar]
- 9.Rietschel ET, Brade H, Holst O, Brade L, Muller-Loennies S, Mamat U, Zahringer U, Beckmann F, Seydel U, Brandenburg K, Ulmer AJ, Mattern T, Heine H, Schletter J, Loppnow H, Schonbeck U, Flad HD, Hauschildt S, Schade UF, Di Padova F, Kusumoto S, Schumann RR. Bacterial endotoxin: chemical constitution, biological recognition, host response, and immunological detoxification. Curr Top Microbiol Immunol. 1996;216:39–81. doi: 10.1007/978-3-642-80186-0_3. [DOI] [PubMed] [Google Scholar]
- 10.Bone RC, Balk RA, Fein AM, Perl TM, Wenzel RP, Reines HD, Quenzer RW, Iberti TJ, Macintyre N, Schein RM. A second large controlled clinical study of E5, a monoclonal antibody to endotoxin: results of a prospective, multicenter, randomized, controlled trial. The E5 Sepsis Study Group. Crit Care Med. 1995;23:994–1006. doi: 10.1097/00003246-199506000-00003. [DOI] [PubMed] [Google Scholar]
- 11.Greenman RL, Schein RM, Martin MA, Wenzel RP, MacIntyre NR, Emmanuel G, Chmel H, Kohler RB, McCarthy M, Plouffe J, et al. A controlled clinical trial of E5 murine monoclonal IgM antibody to endotoxin in the treatment of Gram-negative sepsis. The XOMA Sepsis Study Group. JAMA. 1991;266:1097–1102. [PubMed] [Google Scholar]
- 12.Abraham E, Anzueto A, Gutierrez G, Tessler S, San Pedro G, Wunderink R, Dal Nogare A, Nasraway S, Berman S, Cooney R, Levy H, Baughman R, Rumbak M, Light RB, Poole L, Allred R, Constant J, Pennington J, Porter S. Double-blind randomised controlled trial of monoclonal antibody to human tumour necrosis factor in treatment of septic shock. NORASEPT II Study Group. Lancet. 1998;351:929–933. [PubMed] [Google Scholar]
- 13.Cohen J, Carlet J. INTERSEPT: an international, multicenter, placebo-controlled trial of monoclonal antibody to human tumor necrosis factor-α in patients with sepsis. International Sepsis Trial Study Group. Crit Care Med. 1996;24:1431–1440. doi: 10.1097/00003246-199609000-00002. [DOI] [PubMed] [Google Scholar]
- 14.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 15.Robertson DL, Joyce GF. Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature. 1990;344:467–468. doi: 10.1038/344467a0. [DOI] [PubMed] [Google Scholar]
- 16.Ellington AD, Szostak JW. Selection in vitro of single-stranded DNA molecules that fold into specific ligand-binding structures. Nature. 1992;355:850–852. doi: 10.1038/355850a0. [DOI] [PubMed] [Google Scholar]
- 17.Jayasena SD. Aptamers: an emerging class of molecules that rival antibodies in diagnostics. Clin Chem. 1999;45:1628–1650. [PubMed] [Google Scholar]
- 18.Gebhardt K, Shokraei A, Babaie E, Lindqvist BH. RNA aptamers to S-adenosylhomocysteine: kinetic properties, divalent cation dependency, and comparison with anti-S-adenosylhomocysteine antibody. Biochemistry. 2000;39:7255–7265. doi: 10.1021/bi000295t. [DOI] [PubMed] [Google Scholar]
- 19.Dwarakanath S, Bruno JG, Shastry A, Phillips T, John AA, Kumar A, Stephenson LD. Quantum dot-antibody and aptamer conjugates shift fluorescence upon binding bacteria. Biochem Biophys Res Commun. 2004;325:739–743. doi: 10.1016/j.bbrc.2004.10.099. [DOI] [PubMed] [Google Scholar]
- 20.Li C, Ng ML, Zhu Y, Ho B, Ding JL. Tandem repeats of Sushi3 peptide with enhanced LPS-binding and -neutralizing activities. Protein Eng. 2003;16:629–635. doi: 10.1093/protein/gzg078. [DOI] [PubMed] [Google Scholar]
- 21.Ding JL, Ho B. A new era in pyrogen testing. Trends Biotechnol. 2001;19:277–281. doi: 10.1016/s0167-7799(01)01694-8. [DOI] [PubMed] [Google Scholar]
- 22.Glauser MP, Zanetti G, Baumgartner JD, Cohen J. Septic shock: pathogenesis. Lancet. 1991;338:732–736. doi: 10.1016/0140-6736(91)91452-z. [DOI] [PubMed] [Google Scholar]
- 23.Riedemann NC, Guo RF, Ward PA. Novel strategies for the treatment of sepsis. Nat Med. 2003;9:517–524. doi: 10.1038/nm0503-517. [DOI] [PubMed] [Google Scholar]
- 24.Blank M, Weinschenk T, Priemer M, Schluesener H. Systematic evolution of a DNA aptamer binding to rat brain tumor microvessels. Selective targeting of endothelial regulatory protein pigpen. J Biol Chem. 2001;276:16464–16468. doi: 10.1074/jbc.M100347200. [DOI] [PubMed] [Google Scholar]
- 25.Fukusaki E, Hasunuma T, Kajiyama S, Okazawa A, Itoh TJ, Kobayashi A. SELEX for tubulin affords specific T-rich DNA aptamers. Systematic evolution of ligands by exponential enrichment. Bioorg Med Chem Lett. 2001;11:2927–2930. doi: 10.1016/s0960-894x(01)00584-4. [DOI] [PubMed] [Google Scholar]
- 26.Wang C, Zhang M, Yang G, Zhang D, Ding H, Wang H, Fan M, Shen B, Shao N. Single-stranded DNA aptamers that bind differentiated but not parental cells: subtractive systematic evolution of ligands by exponential enrichment. J Biotechnol. 2003;102:15–22. doi: 10.1016/s0168-1656(02)00360-7. [DOI] [PubMed] [Google Scholar]
- 27.Aurell CA, Wistrom AO. Critical aggregation concentrations of Gram-negative bacterial lipopolysaccharides. Biochem Biophys Res Commun. 1998;253:119–123. doi: 10.1006/bbrc.1998.9773. [DOI] [PubMed] [Google Scholar]
- 28.Luderitz O, Galanos C, Lehmann V, Mayer H, Rietschel ET, Weckesser J. Chemical structure and biological activities of lipid A's from various bacterial families. Naturwissenschaften. 1978;65:578–585. doi: 10.1007/BF00364907. [DOI] [PubMed] [Google Scholar]
- 29.Ruckman J, Green LS, Beeson J, Waugh S, Gillette WL, Henninger DD, Claesson-Welsh L, Janjic N. 2′-Fluoropyrimidine RNA-based aptamers to the 165-amino acid form of vascular endothelial growth factor (VEGF165). Inhibition of receptor binding and VEGF-induced vascular permeability through interactions requiring the exon 7-encoded domain. J Biol Chem. 1998;273:20556–20567. doi: 10.1074/jbc.273.32.20556. [DOI] [PubMed] [Google Scholar]
- 30.Hicke BJ, Watson SR, Koenig A, Lynott CK, Bargatze RF, Chang YF, Ringquist S, Moon-McDermott L, Jennings S, Fitzwater T, Han HL, Varki N, Albinana I, Willis MC, Varki A, Parma D. DNA aptamers block L-selectin function in vivo. Inhibition of human lymphocyte trafficking in SCID mice. J Clin Invest. 1996;98:2688–2692. doi: 10.1172/JCI119092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wen JD, Gray CW, Gray DM. SELEX selection of high-affinity oligonucleotides for bacteriophage Ff gene 5 protein. Biochemistry. 2001;40:9300–9310. doi: 10.1021/bi010109z. [DOI] [PubMed] [Google Scholar]
- 32.Yang Q, Goldstein IJ, Mei HY, Engelke DR. DNA ligands that bind tightly and selectively to cellobiose. Proc Natl Acad Sci USA. 1998;95:5462–5467. doi: 10.1073/pnas.95.10.5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Padmanabhan K, Padmanabhan K, Ferrara JD, Sadler JE, Tulinsky A. The structure of α-thrombin inhibited by a 15-mer single-stranded DNA aptamer. J Biol Chem. 1993;268:17651–17654. doi: 10.2210/pdb1hut/pdb. [DOI] [PubMed] [Google Scholar]
- 34.Huizenga DE, Szostak JW. A DNA aptamer that binds adenosine and ATP. Biochemistry. 1995;34:656–665. doi: 10.1021/bi00002a033. [DOI] [PubMed] [Google Scholar]
- 35.Weiss S, Proske D, Neumann M, Groschup MH, Kretzschmar HA, Famulok M, Winnacker EL. RNA aptamers specifically interact with the prion protein PrP. J Virol. 1997;71:8790–8797. doi: 10.1128/jvi.71.11.8790-8797.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schneider D, Gold L, Platt T. Selective enrichment of RNA species for tight binding to Escherichia coli rho factor. FASEB J. 1993;7:201–207. doi: 10.1096/fasebj.7.1.7678562. [DOI] [PubMed] [Google Scholar]
- 37.Dankesreiter S, Hoess A, Schneider-Mergener J, Wagner H, Miethke T. Synthetic endotoxin-binding peptides block endotoxin-triggered TNF-α production by macrophages in vitro and in vivo and prevent endotoxin-mediated toxic shock. J Immunol. 2000;164:4804–4811. doi: 10.4049/jimmunol.164.9.4804. [DOI] [PubMed] [Google Scholar]
- 38.Marra MN, Wilde CG, Collins MS, Snable JL, Thornton MB, Scott RW. The role of bactericidal/permeability-increasing protein as a natural inhibitor of bacterial endotoxin. J Immunol. 1992;148:532–537. [PubMed] [Google Scholar]