Abstract
Macrophages express various classes of pattern recognition receptors involved in innate immune recognition of artificial, microbial and host-derived ligands. These include the scavenger receptors (SRs), which are important for phagocytosis, and the Toll-like receptors (TLRs) involved in microbe sensing. The class A macrophage scavenger receptor (SR-A) and macrophage receptor with a collagenous structure (MARCO) display similar domain structures and ligand-binding specificity, which has led to the assumption that these two receptors may be functionally redundant. In this study we show that SR-A and MARCO differentially recognise artificial polyanionic ligands as well as surface proteins from the pathogenic bacterium Neisseria meningitidis. We show that, while acetylated low-density lipoprotein (AcLDL) is a strong ligand for SR-A, it is not a ligand for MARCO. Of the neisserial proteins that were SR ligands, some were ligands for both receptors, while other proteins were only recognised by either SR-A or MARCO. We also analysed the potential of these ligands to act as TLR agonists and assessed the requirement for SR-A and MARCO in pro-inflammatory cytokine induction. SR ligation alone did not induce cytokine production; however, for proteins that were both SR and TLR ligands, the SRs were required for full activation of TLR pathways.
Key Words: Ligand, Receptor collaboration, Cytokine, Macrophage, Scavenger receptor, Toll-like receptor, <italic>Neisseria</italic>
Introduction
Macrophages (Mϕ) express different classes of innate pattern recognition receptors (PRRs), including phagocytic scavenger receptors (SRs) and the microbe sensing Toll-like receptors (TLRs). Members of both receptor families recognise a variety of microbial, host-derived and artificial ligands, including proteins, lipids, carbohydrates and nucleic acids [1, 2, 3].
The TLRs are type I transmembrane proteins with an extracellular ligand recognition domain containing leucine-rich repeats (LRRs) and a cytoplasmic TIR domain with homology to the cytoplasmic portion of interleukin-1 Annette Plüddemann and Subhankar Mukhopadhyay contributed equally to this study. receptor [4]. The cytoplasmic TIR domain interacts with other TIR domain-containing adaptor molecules, including MyD88, TRIF, TRAM and MAL/TIRAP, thereby inducing signal transduction pathways culminating in the activation of transcription factors including NF-κB. Only a restricted number of TLRs have been identified in mammalian systems. All of these express an extracellular LRR domain, yet TLRs can recognise a wide range of chemically varied ligands [1]. Similarly, the limited numbers of TLRs utilise an even smaller number of adaptor proteins to generate diverse biological responses. Differential and combinatorial use of adaptors and the capacity of some TLRs to form functional homo- or hetero-dimers only partially explain the great diversity of TLR-mediated ligand recognition and biological response. Our understanding of the fine specificities in TLR-mediated ligand recognition and responses is still limited.
The TLRs have not been shown to be phagocytic: this function is performed by other Mϕ surface receptors, including the SRs. SRs are broadly defined by their ability to bind modified low-density lipoproteins (mLDL) and other polyanions. Based on their multidomain structures, SRs have been further classified into 8 different classes (classes A–H) [5]. Two important members of the class A scavenger receptor family are Mϕ scavenger receptor A (SR-A) and Mϕ receptor with a collagenous structure (MARCO). These two receptors were primarily grouped together based on their highly similar domain structures and their ability to bind mLDL, common polyanions and Gram-positive and Gram-negative organisms. The shared structural features and common ligands between SR-A and MARCO have led to the assumption that these receptors may be functionally redundant. Subsequently, subtle but distinct differences in ligand-binding domains, tissue distribution and regulation of these two molecules have been reported, but careful comparison of their biological functions has not been carried out.
Both SR-A and MARCO are trimeric type II transmembrane glycoproteins consisting of a cytoplasmic tail, transmembrane domain, spacer region, collagenous domain and C-terminal SR cysteine-rich domain. Additionally, SR-A expresses an α-helical coiled coil domain between the spacer region and the collagenous domain; however, MARCO lacks this coiled coil domain and exhibits a longer collagenous domain [6]. The functional implication of this distinct domain organisation is still unclear. Three naturally occurring isoforms of SR-A have been reported (SR-AI, SR-AII and SR-AIII), which are alternative splice variants of the same gene and are collectively referred to as SR-A [7, 8, 9]. Among these isoforms, SR-AIII is not expressed on the cell surface and is nonfunctional. SR-AII is characterised by a truncated C-terminus, compared to the full-length SR cysteine-rich domain of SR-AI. However, no functional difference has been observed between these two isoforms. In contrast, no alternative splicing has been reported for the gene encoding MARCO. Despite similar domain organisation, SR-A and MARCO differ in their ligand-binding domains. Whereas for SR-A the collagenous region has been identified as the ligand-binding domain [7, 10], the ligand-binding region for MARCO lies within the cysteine-rich domain [6]. Furthermore, for SR-A ligand binding is divalent cation independent [11] whereas for MARCO ligand binding is dependent on the presence of divalent cations [12].
Both SR-A and MARCO are primarily myeloid-restricted molecules. However, SR-A is a pan-myeloid marker, constitutively expressed on most mature tissue Mϕ and on bone marrow-derived dendritic cells and splenic dendritic cells [13] but not on their immature precursor monocytes [14]. In addition, SR-A expression has also been shown on mast cells [15]. Apart from the myeloid lineage, selected endothelial and smooth muscle cells can also express SR-A [16]. In contrast, constitutive MARCO expression is strictly restricted to resident peritoneal, spleen marginal zone and lymph node medullary cord Mϕ. However, MARCO expression can be readily induced in most isolated and tissue-resident Mϕ by various infectious or inflammatory TLR stimuli (e.g. bacteria/lipopolysaccharide, LPS) but not by the major proinflammatory and anti-inflammatory cytokines [17]. TLR-mediated induction of MARCO requires MyD88-dependent activation of the p38 MAPK signalling pathway and transcription factor NF-κB [18]. In contrast, SR-A expression is not modulated by TLR stimulation but highly induced by an Mϕ-specific growth factor CSF-1 [19].
SR-A and MARCO share a wide range of polyanionic ligands of artificial, microbial and endogenous origin [20, 21, 22]. These include polyribonucleotides (polyinosinic and polyguanylic acid), polysaccharides (dextran sulphate, DxSO4), mLDL, i.e. acetylated LDL (AcLDL) and oxidised LDL, modified proteins (maleylated bovine serum albumin, malBSA) and intact Gram-positive and Gram-negative bacteria. LPS, lipoteichoic acid (LTA), peptidoglycan (PGN) and CpG DNA are some of the ligands proposed for SR-A and MARCO on bacteria [23, 24, 25, 26]. Many of these ligands are also recognised by members of the TLR family [27]. TLR3 has been shown to recognise the artificial polyribonucleotide, poly(I) [28] and TLR2 and TLR4 have been demonstrated to play a role in atherosclerotic lesion development [29]. With regard to bacterial recognition, TLR2 recognises Gram-positive peptidoglycan, LTA and lipoproteins [30, 31], TLR4 recognises LPS from Gram-negative bacteria [32, 33] and CpG DNA is a ligand for TLR9 [34]. However, the functional relationships and ligand specificities between these two receptor families are not clear. Recently we have shown that both SR-A and MARCO recognise and phagocytose the Gram-negative diplococcus Neisseria meningitidis, but LPS, previously shown to be a ligand for these receptors, was not required for recognition [35, 36]. However, LPS was essential for a cytokine response, which was independent of SR-A but mediated by TLR4. We subsequently identified N. meningitidis surface proteins to be ligands for SR-A [37].
Considering several distinct differences between SR-A and MARCO biology, we hypothesize that SR-A and MARCO are not merely redundant receptors, but may have differential functional properties. Furthermore, despite distinct structural differences, SR-A, MARCO and TLRs show remarkable ligand overlap, indicating a possible functional collaboration. Similar collaboration between other members of SRs and TLRs has been previously reported [38, 39]. In this study, we set out to compare the ligand repertoire and specificity between SR-A and MARCO by screening a range of known artificial and host-derived polyanions or protein ligands from the medically important Gram-negative organism, N. meningitidis. We further tested the potential of these SR ligands to induce TLR-mediated cytokine production and the requirement of SR-A and MARCO in such responses.
Materials and Methods
Chemicals and Reagents
Unless stated otherwise, all chemicals were from Sigma. AcLDL was obtained from Molecular Probes (Eugene, Oreg., USA). LPS-free BSA was from Calbiochem (La Jolla, Calif., USA); malBSA was made according to published protocols [40]. The rat anti-mouse SR-A monoclonal antibody (2F8) has been described previously [11] and is available from AbD Serotec (Oxford, UK). The monoclonal anti-MARCO antibody ED31 was a kind gift from G. Kraal [41] and the monoclonal anti-EMR1 antibody BC9 was kindly supplied by M. Stacey. The ELISA kits for murine tumour necrosis factor-α (TNF-α) and interleukin-6 (IL-6) were from BD Biosciences Pharmingen (San Diego, Calif., USA). Serogroup B N. meningitidis proteins were expressed in Escherichia coli as previously described and purified using standard methods for His- and GST-tagged proteins [42]. All proteins were tested for LPS contamination using the Limulus amebocyte lysate test for endotoxins (E-Toxate, Sigma, UK). The test is sensitive to 0.05–0.1 endotoxin units per millilitre and no detectable levels of endotoxin were measured.
Animals
The following knock-out mouse strains were used: MyD88–/–, SR-A–/–, MARCO–/– and an SR-A-MARCO double knock-out (SR-A-MARCO–/–), along with their wild-type (WT) control strain, C57BL/6J. SR-A–/– and MARCO–/– animals were developed and bred onto C57BL/6J background using standard molecular biology techniques and SR-A-MARCO–/– animals were generated by mating SR-A–/– and MARCO–/– animals [43, 44]. All animals were bred and housed under specific pathogen-free conditions and all procedures involving animals were conducted according to the requirements of the United Kingdom Home Office Animals (Scientific Procedures) Acts, 1986.
Cell Culture
Biogel-elicited peritoneal Mϕ (Bg-PMϕ) were prepared by intraperitoneal injection of 1 ml polyacrylamide gel P-100 beads (Bio-Rad) (2% w/v in endotoxin-free water). After 4 days, peritoneal cells were harvested by lavage with phosphate-buffered saline (PBS). Resident peritoneal Mϕ (RPMϕ) were isolated from uninjected animals by lavage with PBS. Both Bg-PMϕ and RPMϕ were plated on bacteriological plastic dishes in Opti-MEM®, supplemented with 50 IU/ml penicillin-streptomycin and 2 mM L-glutamine. After 3–4 h, adherent cells were washed three times to remove non-adherent cells and biogel beads. After washing, the purity of Mϕ in the adherent monolayer was >95% as characterised previously [45, 46]. Murine bone marrow-derived Mϕ (BMMϕ) were obtained and cultured by standard procedures. Mϕ were routinely cultured in RPMI (Gibco) supplemented with 50 IU/ml penicillin G, 50 µg/ml streptomycin and 2 mM L-glutamine (PSG), 10% fetal calf serum (FCS) and 15% L-cell-conditioned medium. For preparation of lysates, Mϕ were washed five times in ice-cold PBS and cells were lysed using 1 ml NP-40 protein lysis buffer [150 mM NaCl, 10 mM EDTA, 10 mM NaN3, 10 mM Tris (pH 8.0), 1 mM PMSF, 5 mM iodoacetamide and 1% Nonidet P-40] per 1 × 107 cells. Lysates were then centrifuged for 15 min at 12,000 g at 4°C to remove nuclei and cellular debris. Supernatant aliquots were stored at −80°C until required. The protein concentration of the lysates was measured using the Bicinchoninic Acid Protein Assay Reagent Kit (Pierce Chemical Company, Chester, UK).
ELISA Overlay Assays
SR-A ELISA overlay assays were performed as described previously [47]. Briefly, 96-well OPTI-EIA plates (Corning, NewYork, N.Y., USA) were coated overnight with candidate ligands (10 µg/ml unless stated otherwise) at 4°C in replicates of 10. The wells were blocked with 10% LPS-free BSA before incubation with post-nuclear supernatant prepared as above from WT or SR-A–/– BMMϕ for 2 h, in the presence of 5 mM EDTA. Five wells of each candidate were incubated with WT or SR-A–/– lysate. The wells were washed with PBS containing 0.1% Tween 20 and incubated with 10 µg/ml 2F8 for 2 h at room temperature. Binding of the SR-A antibody was detected by incubating the wells with a horseradish peroxidase (HRP)-conjugated anti-rat antibody and visualized using TMB reagent according to the manufacturer's instructions. The average of 5 replicates for each condition was plotted and results are representative of at least 3 independent experiments. The statistical significance of results was determined using the paired Student's t test and significance tested at the 95% confidence level (p ≤ 0.05). The MARCO overlay ELISA assay was optimized by adjusting the above protocol as follows: ELISA plates were coated with potential ligands as above and 5% milk was used as a blocking agent. Ligands were incubated with recombinant extracellular MARCO (sMARCO) at a final concentration of 3 µg/ml. sMARCO was produced in 293/EBNA cells and purified using a His-tag as described previously [48]. In parallel, we incubated the ligands with another recombinantly produced, His-tagged version of an unrelated Mϕ surface protein EMR1 (sEMR1) as a control for nonspecific binding. EMR1 is a Mt surface receptor with 6 extracellular EGF domains. A His-tagged version of EGF domain 4–5 was produced recombinantly following a similar methodology as sMARCO. We detected sMARCO and sEMR1 binding by using their specific monoclonal antibodies ED31 and BC9, respectively, together with an HRP-conjugated secondary antibody. During optimisation of this protocol, we also included appropriate isotype-matched controls for ED31 and BC9. Since no non-specific binding was observed for ED31 and BC9, we excluded these isotype controls for subsequent assays.
Measurement of IL-6 and TNF-α
105 Biogel-elicited peritoneal Mϕ were plated in 96-well bacteriological plastic dishes. After 3–4 h, monolayers were washed three times with PBS to remove any non-adherent cells and biogel beads, and incubated overnight with 200 (µl of fresh media containing a final concentration of 1 µg/ml of the various polyanions or N. meningitidis proteins. Cells were separately stimulated with E. coli LPS (100 ng/ml final concentration), which served as a positive control for cytokine production. TNF-α and IL-6 were measured using the commercially available murine TNF-α and IL-6 ELISA kits (BD Biosciences), following the manufacturer's protocol. All experiments were repeated at least three times and all data are from representative experiments.
Results
Receptor Recognition of Polyanionic Ligands
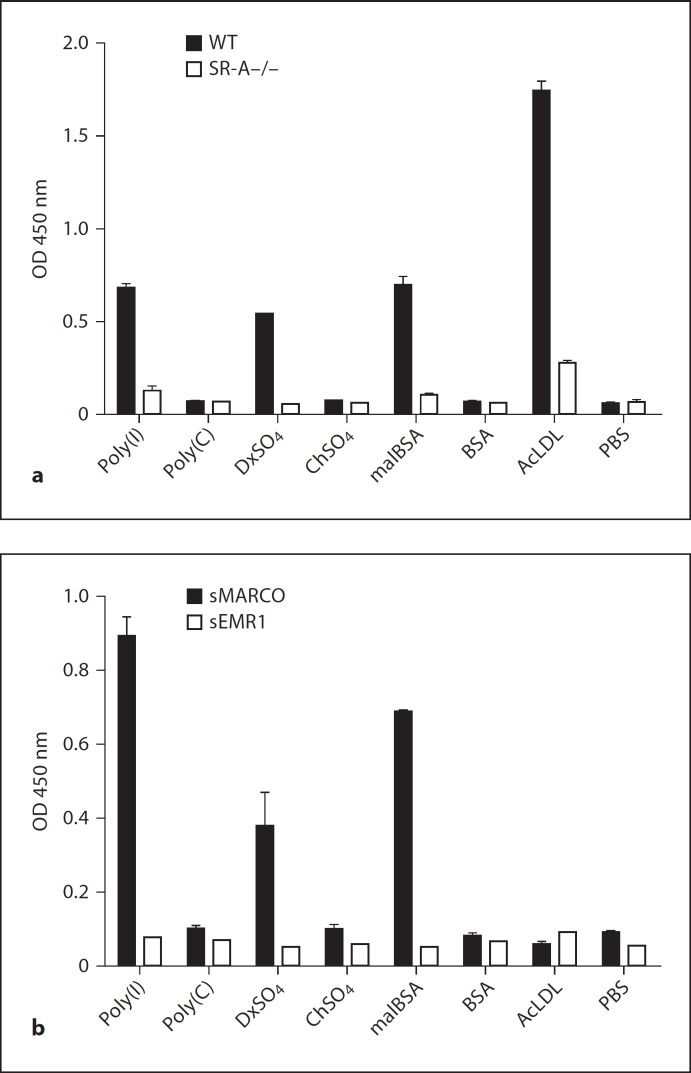
We compared differential ligand-binding activity of SR-A and MARCO by screening a range of non-microbial polyanionic ligands (and their closely related non-ligand controls) reported to interact with SRs, using an ELISA overlay assay (fig. 1a, b). SR-A-specific binding was detected by measuring ligand activity in lysate from wild-type BMMϕ, using a specific anti-SR-A monoclonal antibody (2F8) and comparing the ligand activity with any non-specific binding determined in lysate derived from SR-A–/– BMMϕ. MARCO is not expressed on BMMϕ and other readily available Mϕ populations [25]; therefore, recombinant His-tagged purified sMARCO was used to detect ligand binding to this receptor in combination with a specific anti-MARCO monoclonal antibody (ED31). To determine whether any His-tag-mediated binding of sMARCO to its potential ligands occurred, we used another unrelated recombinant His-tagged soluble Mϕ surface receptor, EMR1 (sEMR1) as a control in conjunction with a specific anti-EMR1 antibody BC9. Most commercially available anti-His antibodies showed high non-specific background binding in our assay; therefore, we avoided the use of anti-His antibodies to detect sMARCO and sEMR1. Our results confirmed that SR-A and MARCO both recognised the known ligands polyinosinic acid [poly(I)] and dextran sulphate (DxSO4) whereas the related non-binding control molecules polycytidylic acid [poly(C)] and chondroitin sulphate (ChSO4) showed no binding to either receptor. Similarly, malBSA showed binding to both receptors while native BSA was not recognised. AcLDL is a ligand for most SRs, including SR-A [49], and has been used to define the family of SRs. Previous reports suggested that AcLDL was also a ligand for MARCO [25]. Our data confirmed that AcLDL is an excellent ligand for SR-A, but AcLDL did not exhibit any ligand activity for MARCO in this assay. We have shown that SR-A and MARCO pre-dictively recognised most known ligands, which further validated the use of the ELISA method for high-throughput screening of receptor-ligand interactions. Our unexpected observation that AcLDL is not a ligand for MARCO again suggested that SR-A and MARCO are not redundant. This observation may have further implications in dissecting the differential role of these two receptors in atherosclerosis.
Fig. 1.
SR-A and MARCO bind to various polyanionic ligands. a Known ligands and non-ligands for SR-A were coated into wells of a 96-well plate and overlaid with post-nuclear cell lysate from WT and SR-A knock-out (SR-A−/−) BMMϕ. Binding to SR-A was detected with a rat anti-mouse SR-A antibody, 2F8, and visualized with a secondary anti-rat HRP antibody and TMB reagent. b In parallel, wells of a 96-well plate were coated with the same series of polyanionic molecules as in a and overlaid with recombinant sMARCO or recombinant sEMR1, an unrelated Mϕ surface receptor, as a control for non-specific binding. Binding of sMARCO and sEMR1 was detected by using their specific monoclonal antibodies ED31 and BC9, respectively, together with an HRP-conjugated secondary antibody.
Recognition of Gram-Negative N. meningitidis Ligands
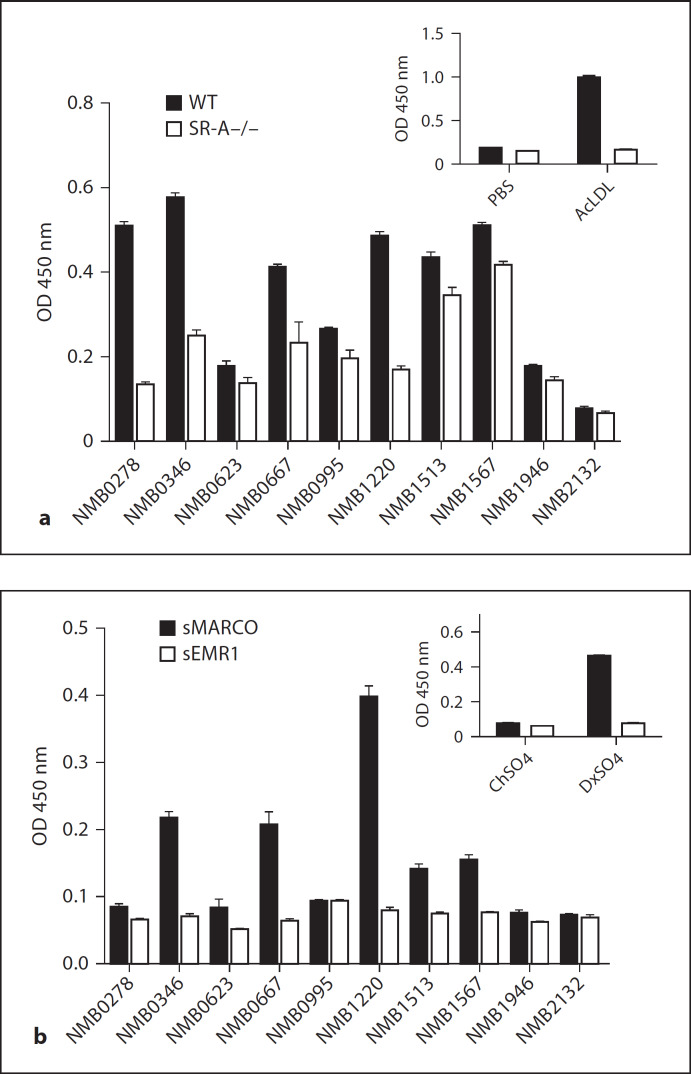
SR-A and MARCO bind and phagocytose Gram-negative N. meningitidis independent of LPS [35, 36]. We previously tested a series of 10 potential candidate surface proteins from N. meningitidis for their ability to bind SR-A, and identified several SR-A ligands [37]. To further test whether SR-A and MARCO recognise common ligands on the surface of N. meningitidis, we tested these surface proteins in parallel for their ability to bind to SR-A and MARCO (table 1). The proteins were coated onto ELISA plates overnight at a concentration of 10 µg/ml, and SR-A and MARCO binding was detected using WT BMMϕ lysate or sMARCO as described above (fig. 2a, b). Equal protein coating of the wells was determined by standard His-tag or GST-tag ELISA methods (data not shown). A range of SR-A- and MARCO-mediated binding was observed. NMB0346, NMB0667 and NMB1220 showed ligand activity for both receptors, while NMB0278 was a strong ligand for SR-A but showed no binding to MARCO. Similarly, NMB1513 and NMB1567 were intermediate ligands for MARCO; however, no specific binding to SR-A was detected. NMB0623, NMB1946 and NMB2132 were not recognised by either receptor. These data show that both SR-A and MARCO specifically bind to selected N. meningitidis surface proteins and that SR-A and MARCO display overlapping but differential recognition of these ligands.
Table 1.
N. meningitidis proteins
| Protein | MW | Annotation |
|---|---|---|
| NMB0278 | 24 | thiol:disulfide interchange protein DsbA |
| NMB0346 | 28 | hypothetical protein |
| NMB0623 | 40 | spermidine/putrescine ABC transporter |
| NMB0667 | 47 | hypothetical protein with some homology to E. coli cell division protein zipA |
| NMB0995 | 20 | Mϕ infectivity potentiator-related protein |
| NMB1220 | 33 | stomatin/Mec-2 family protein |
| NMB1513 | 23 | conserved hypothetical protein |
| NMB1567 | 28 | Mϕ infectivity potentiator |
| NMB1946 | 30 | lipoprotein of the NlpA family of lipoproteins, similar to HlpA of Haemophilus influenzae |
| NMB2132 | 42 | lipoprotein with low similarity to transferrin-binding proteins |
Fig. 2.
SR-A and MARCO differentially bind to N. meningitidis surface proteins. Wells of duplicate 96-well plates were coated with 10 µg/ml of various purified His- or GST-tagged N. meningitidis surface proteins. a SR-A binding was detected using Mt lysates from WT and SR-A–/– BMMϕ along with the SR-A-specific monoclonal antibody 2F8. PBS and AcLDL were used as a negative and a positive control for binding to SR-A, respectively (inset). b Binding to MARCO was detected using recombinant soluble MARCO and the anti-MARCO monoclonal antibody ED31. Recombinant soluble EMR1 acted as a control for non-specific binding. ChSO4 and DxSO4 were used as a negative and a positive control for binding to MARCO, respectively (inset).
TLR-Mediated Cytokine Induction by SR-A and MARCO Ligands
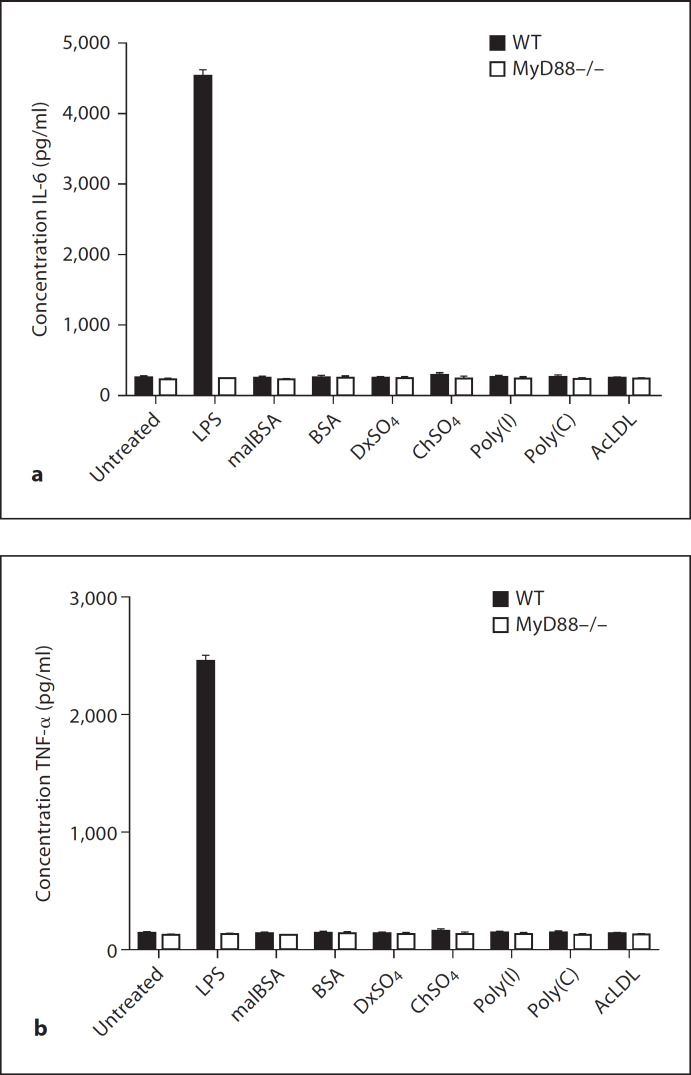
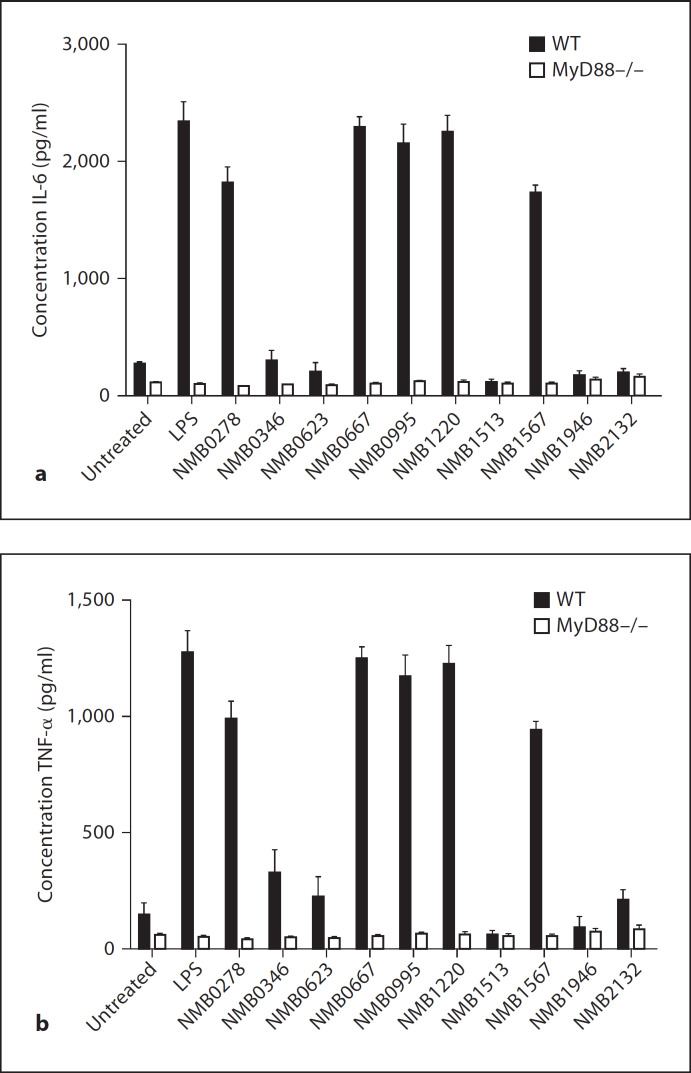
Since SR-A, MARCO and TLRs recognise many common ligands, we were interested in determining the ability of these ligands to induce cytokines and the relative contribution of these receptors. First, we stimulated biogel-elicited peritoneal Mϕ from WT and MyD88–/– mice with the series of polyanionic ligands and measured the production of IL-6 and TNF-α (fig. 3). No IL-6 or TNF-α production was detected for any of the molecules, regardless of their ability to bind to SR-A or MARCO. These data show that the known SR ligands AcLDL, DxSO4, malBSA and poly(I) do not induce TLR activation, further confirming that the samples do not contain impurities such as LPS, which may give false-positive ligand-binding data. LPS, a common ligand for SR-A, MARCO and TLR4, is known to be a potent inducer of cytokines [50] and induced high levels of IL-6 and TNF-α in WT Mϕ, but not in MyD88–/– Mϕ. Apart from serving as a positive control for the cytokine assay, the above data also confirmed that ligation of SRs by LPS alone is not sufficient to induce cytokines in the absence of TLR4 signalling, as proposed in previous studies [51]. We then tested whether the surface proteins from Gram-negative N. meningitidis could also act as TLR agonists and induce IL-6 and TNF-α production (fig. 4). Five proteins, namely NMB0278, NMB0667, NMB0995, NMB1220 and NMB1567, mediated strong induction of both cytokines in WT Mϕ and two proteins, namely NMB0346 and NMB0623, showed low levels of cytokine induction. Cytokine induction by these proteins was abrogated in Mϕ from MyD88–/– animals, confirming that these ligands were TLR agonists. Of the proteins that induced cytokine production, NMB0667 and NMB1220 were also ligands for both SR-A and MARCO. In some cases, the TLR agonist was only a ligand for either SR-A (NMB0278) or MARCO (NMB1567) while some proteins that were TLR agonists showed no binding to either SR (NMB0995) (table 1). Importantly, NMB1513, NMB1946 and NMB2132 showed no induction of IL-6 and TNF-α in either WT or MyD88–/– Mϕ. indicating the specificity of the other neisserial proteins as TLR agonists. The inability of NMB1513, NMB1946 and NMB2132 to induce TLR-mediated cytokine production served as an internal negative control in our assay and eliminated the possibility that these neisserial proteins are contaminated with other TLR agonists (e.g. LPS) as they were produced following identical procedures. Altogether, our results show that ligand recognition by SR-A and MARCO and cytokine induction seem to be independent and that TLRs are required for the latter. Furthermore, SR-A, MARCO and TLRs differentially recognise overlapping sets of neisserial proteins.
Fig. 3.
Polyanionic ligands do not stimulate TLR-mediated cytokine induction. Bg-PMϕ from WT and MyD88–/– mice were stimulated with 1 µg/ml of each polyanionic ligand and the induction of IL-6 (a) and TNF-α (b) was measured. Cells were separately stimulated with E. coli LPS (100 ng/ml), which served as a positive control for cytokine induction.
Fig. 4.
Selected N. meningitidis surface proteins stimulate TLR-mediated cytokine induction. Bg-PMϕ from WT and MyD88–/– mice were stimulated with 1 µg/ml of the purified recombinant N. meningitidis surface proteins and the induction of IL-6 (a) and TNF-α (b) was measured. Cells were separately stimulated with E. coli LPS (100 ng/ml), which served as a positive control for cytokine induction.
Cytokine Induction in SR-A and MARCO Knock-Out Strains
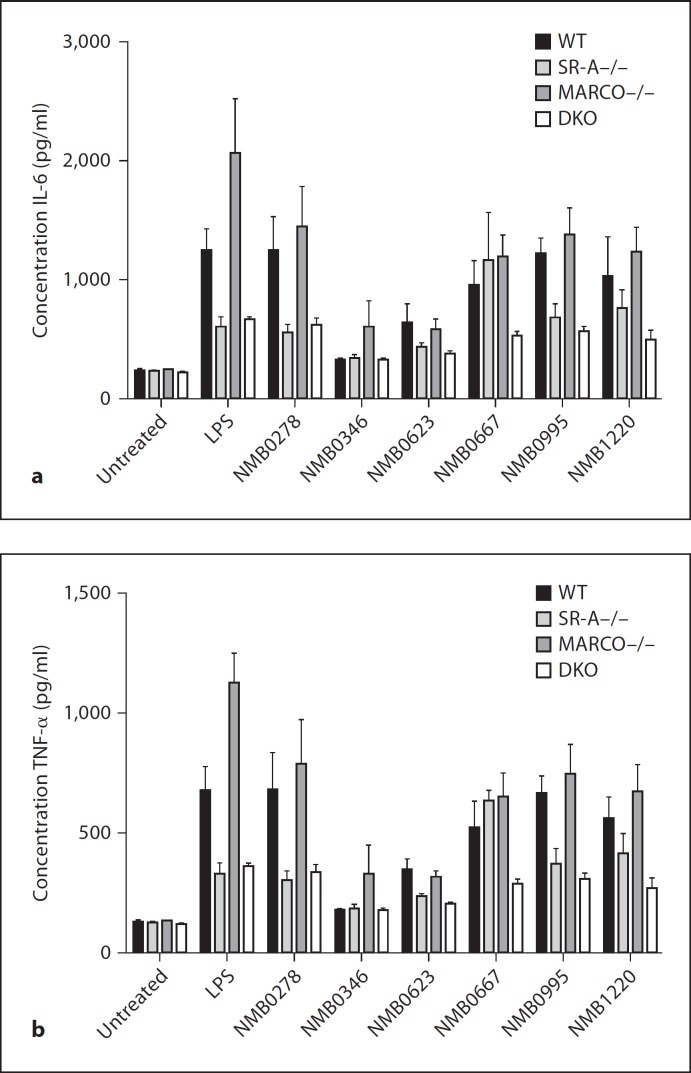
Finally, we tested whether either SR-A or MARCO contribute to the TLR-mediated cytokine secretion induced by neisserial proteins. We stimulated resident peritoneal Mϕ from WT, SR-A–/–, MARCO–/– and SR-A-MARCO–/– double knock-out animals with those N. meningitidis proteins that were shown to be TLR agonists in figure 4 and measured induction of IL-6 and TNF-α (fig. 5). We also separately stimulated each strain of Mϕ with purified LPS, which served as a positive control in our assay. Our results confirmed that, similar to figure 4, NMB0346 (which is a ligand for SR-A and MARCO) induced very little cytokine response compared to other proteins, indicating that this protein is not an agonist for TLRs. No difference in cytokine production was detected between the WT and three different knock-out strains. Similarly, NMB0623, which is negative for both SR-A and MARCO binding, was also a very weak agonist for TLRs. Consistent with this result, NMB0623 showed no difference in cytokine production between WT, SR-A–/–, MARCO–/– and SR-A-MARCO–/– animals. NMB0278 (which is a strong ligand for SR-A and TLRs, but not for MARCO) showed a ˜50% decrease in cytokine levels in SR-A–/– and SR-A-MARCO–/– Mϕ but possibly a slightly increased cytokine secretion in MARCO–/– Mϕ compared to WT cells. In contrast, NMB0667 and NMB1220 (which are strong ligands for both SR-A, MARCO and TLRs) showed no significant decrease in cytokine production by SR-A–/– and MARCO–/– Mϕ, but induced significantly less cytokines in SR-A-MARCO–/– Mϕ. These data indicate that selected neisserial proteins such as NMB0278, NMB0667 and NMB1220 require either SR-A, MARCO or both along with TLRs to induce a maximal cytokine response, indicating a possible functional collaboration between these two classes of receptors.
Fig. 5.
Cytokine induction in SR-A–/–, MARCO–/– and double knock-out Mϕ. RPMϕ from WT, SR-A–/–, MARCO–/– and SR-A-MARCO–/– double knock-out animals were stimulated with 1 µg/ml of the N. meningitidis proteins that were showntobeTLR agonists in figure 4 and the induction of IL-6 (a) and TNF-α (b) was measured. Each strain of Mϕ was also stimulated with E. coli LPS (100 ng/ml), which served as a positive control for cytokine induction.
Discussion
SR-A, MARCO and TLRs are major innate immune receptors for phagocytosis and the innate inflammatory response, respectively. However, the mechanism by which these two receptor systems interact to mount an efficient anti-microbial immunity or maintain homeostasis is not clear. The structural similarity of SR-A and MARCO has led to the assumption that they may be functionally redundant; however, the fine specificity of ligand interaction with these two receptors had previously not been studied in detail. In the present study, we compared the ligand specificities between SR-A and MARCO by screening a range of known artificial and host-derived polyanions or protein ligands from the Gram-negative pathogen, N. meningitidis. Furthermore, we also assessed the potential of the SR-A and MARCO ligands to stimulate TLR-mediated cytokine secretion and the requirement for SR-A and MARCO in that process, indicating a functional collaboration between members of the SR and TLR family. Our results confirmed that SR-A and MARCO recognise overlapping but distinct sets of artificial, endogenous and microbial ligands, confirming fine specificities in the ligand recognition repertoire of these two receptors. SR-A and MARCO showed significant overlap in the recognition of artificial or endogenous polyanions; however, ligand diversity is more marked in the recognition of microbial proteins. We also established that none of the artificial and endogenous polyanions tested in this study stimulated a TLR-mediated cytokine response. However, most of the microbial protein ligands induced a cytokine response although there were some exceptions. Furthermore, among these microbial TLR agonists, not all are recognised by SR-A and MARCO, indicating that TLRs are able to recognise these microbial proteins in the absence of SR-A and MARCO. Similarly, not all the microbial ligands for SR-A and MARCO stimulated TLRs. However, those TLR ligands which are also recognised by either SR-A, MARCO or both depend on these two molecules for full activation of TLR pathways.
Our data show that known polyanionic SR ligands poly(I), DxSO4 and malBSA are ligands for both MARCO and SR-A. However, contrary to current literature [25], AcLDL is not a ligand for MARCO. These data provided a hint that SR-A and MARCO, though structurally similar, show differential recognition of ligands. Various studies have reported the role of SR-A and MARCO in bacterial recognition and the bacterial ligands that were identified included LPS [26, 48], LTA [23, 52] and unmethylated bacterial CpG DNA [53, 54].
The Gram-negative diplococcus N. meningitidis is the causative agent of meningitis and meningococcal septicaemia. Studies in our group showed that both class A scavenger receptors mediate uptake of N. meningitidis in the absence of LPS [35, 36] and we identified several unmodified bacterial surface proteins as ligands for SR-A. Although LPS and LTA were reported to be ligands for MARCO based on biacore studies, careful analysis of the data suggests that these molecules are weak ligands for this receptor [23]. In this study we tested whether bacterial surface proteins from N. meningitidis were also ligands for MARCO and we identified three proteins that were ligands for both receptors (NMB0346, NMB0667 and NMB1220). The function of these proteins is unknown; however, the C-terminus of NMB0667 has 20% homology with the zipA protein from E. coli, which is involved in septum formation during cell division [55], and NMB1220 belongs to the stomatin/Mec-2 protein family, which are oligomeric lipid raft-associated integral proteins that regulate the function of ion channels and transporters [55]. Antibodies raised against NMB1220 also tested positive in a serum bactericidal assay [S. Savino, pers. commun.]. One protein, NMB0278, a possible thiol-disulphide interchange protein, was a strong ligand for SR-A, but was not recognised by MARCO. Furthermore, two proteins, NMB1513 (a conserved hypothetical protein) and NMB1567 (an Mϕ infectivity potentiator), which showed no specific interaction with SR-A, were intermediate ligands for MARCO. Most of these neisserial proteins are highly conserved over the different neisserial species, and closely related homologues are also present in other bacteria, thus making them ideal pathogen-associated molecular patterns and targets for PRRs. Our data clearly show that MARCO and SR-A differentially recognise neisserial proteins, suggesting that these two receptors may have evolved to increase the repertoire of innate immune recognition.
Some studies have linked SR-A ligation directly with cytokine induction [51, 56]. Here we show that SR ligation alone does not induce cytokine production, as stimulation of Mϕ with malBSA, DxSO4, poly(I) and AcLDL did not induce production of IL-6 and TNF-α. Cytokine induction is most likely mediated by the TLR family of receptors, as shown with LPS, a known ligand for TLR4 [50]. We wanted to determine whether the N. meningitidis proteins that were ligands for SR-A and MARCO were also ligands for TLR. Several proteins induced IL-6 and TNF-α production in WT Mϕ, which was abrogated in Mϕ from MyD88–/– mice, indicating a TLR-mediated effect. However, not all proteins that were SR-A and/or MARCO ligands were also TLR agonists and some proteins that induced cytokine production were not ligands for either receptor. These data suggested that binding to SR-A and/or MARCO was not required for TLR-mediated cytokine induction; therefore, we stimulated resident peritoneal Mϕ from WT, SR-A–/–, MARCO–/– and SR-A-MARCO–/– animals with the neisserial proteins that were ligands for either or both receptors and measured pro-inflammatory cytokine production. Surprisingly, cytokine induction was reduced by ˜50% in the absence of SR-A when LPS and two SR-A ligands (NMB0278 and NMB1220) were used as agonists. Deletion of MARCO alone did not affect cytokine production; however, when cells from a double knock-out mutant lacking both SR-A and MARCO were used, cytokine levels were ˜50% lower throughout when compared to WT cells. While ligation of SR-A and MARCO does not directly mediate IL-6 and TNF-α production, these data nevertheless indicate that SR ligation is required for efficient induction of pro-inflammatory cytokines. Several recent studies have reported collaboration between SRs and TLRs. Hoebe et al. [38] showed that CD36, a class B SR, is a non-redundant sensor of microbial diacylglycerides that signal via the TLR2/6 heterodimer. In a separate study, Jeannin et al. [39] showed that, while Klebsiella pneumoniae outer membrane protein A activates Mϕ in a TLR2-dependent manner, binding is mediated by the class E and F SRs, LOX-1 and SREC-I, respectively. They also showed that LOX-1 co-localizes and co-operates directly with TLR2 in triggering cellular responses. Our data suggest a similar co-operation between SR-A and MARCO and one or more members of the TLR family.
In vivo studies have already reported that SR-A and MARCO are important receptors for bacterial clearance during infection [44, 57, 58]. Two major challenges in eliciting protective immunity during successful vaccine development are efficient delivery of antigen to the antigen-presenting cells and activation of these cells to promote proper presentation of that antigen to the T-helper cells. Common adjuvants such as Freund's complete adjuvant and monophosphoryl lipid A adjuvant, frequently used in animal studies, activate the innate immune system via TLRs [59]. However, these are often associated with significant morbidity and are not useful for humans. On the other hand, adjuvants currently in use have also exhibited limitations. Therefore, antigens with inherent adjuvant activity would be a useful target for vaccine development. At present there is no vaccine available for serogroup B N. meningitidis. Similarly, other polysaccharide-based vaccines do not provide lasting protection. Therefore there is a need for a protein-based protective vaccine for serogroup B Neisseria and other human pathogens. Two N. meningitidis proteins (NMB0667 and NMB1220) were strong ligands for both SR-A and MARCO and were also TLR agonists, highlighting their potential as interesting vaccine candidates. The high-throughput system we described here to analyse SR-A and MARCO ligand activity could be extended to other antigen-capturing receptors to expedite the identification of vaccine candidates for a variety of medically important pathogens.
Table 2.
Summary of receptor interaction with N. meningitidis proteins
| N. meningitidis protein ligand | Receptor-ligand interaction |
TLR-mediated cytokine induction | |
|---|---|---|---|
| SR-A | MARCO | ||
| NMB0278 | + | – | + |
| NMB0346 | + | + | +/– |
| NMB0623 | – | – | +/– |
| NMB0667 | + | + | + |
| NMB0995 | – | – | + |
| NMB1220 | + | + | + |
| NMB1513 | – | +/– | – |
| NMB1567 | – | +/– | + |
| NMB1946 | – | – | – |
| NMB2132 | – | – | – |
Acknowledgements
The authors wish to thank T. Areschoug for helpful discussions and critical review of the manuscript. Work in the laboratory of SG was supported by the Medical Research Council, UK. S.M. was funded by the E.P. Abraham Trust, Oxford, UK.
References
- 1.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 2.Gordon S. Pattern recognition receptors: doubling up for the innate immune response. Cell. 2002;111:927–930. doi: 10.1016/s0092-8674(02)01201-1. [DOI] [PubMed] [Google Scholar]
- 3.Taylor PR, Martinez-Pomares L, Stacey M, Lin H-H, Brown GD, Gordon S. Macrophage receptors and immune recognition. Annu Rev Immunol. 2005;23:901–944. doi: 10.1146/annurev.immunol.23.021704.115816. [DOI] [PubMed] [Google Scholar]
- 4.O'Neill LA. How Toll-like receptors signal: what we know and what we don't know. Curr Opin Immunol. 2006;18:3–9. doi: 10.1016/j.coi.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 5.Krieger M. The other side of scavenger receptors: pattern recognition for host defense. Curr Opin Lipidol. 1997;8:275–280. doi: 10.1097/00041433-199710000-00006. [DOI] [PubMed] [Google Scholar]
- 6.Elomaa O, Sankala M, Pikkarainen T, Bergmann U, Tuuttila A, Raatikainen-Ahokas A, Sariola H, Tryggvason K. Structure of the human macrophage MARCO receptor and characterization of its bacteria-binding region. J Biol Chem. 1998;273:4530–4538. doi: 10.1074/jbc.273.8.4530. [DOI] [PubMed] [Google Scholar]
- 7.Gough PJ, Greaves DR, Gordon S. A naturally occurring isoform of the human macrophage scavenger receptor (SR-A) gene generated by alternative splicing blocks modified LDL uptake. J Lipid Res. 1998;39:531–543. [PubMed] [Google Scholar]
- 8.Matsumoto A, Naito M, Itakura H, Ikemoto S, Asaoka H, Hayakawa I, Kanamori H, Aburatani H, Takaku F, Suzuki H, et al. Human macrophage scavenger receptors: primary structure, expression, and localization in atherosclerotic lesions. Proc Natl Acad Sci USA. 1990;87:9133–9137. doi: 10.1073/pnas.87.23.9133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rohrer L, Freeman M, Kodama T, Penman M, Krieger M. Coiled-coil fibrous domains mediate ligand binding by macrophage scavenger receptor type II. Nature. 1990;343:570–572. doi: 10.1038/343570a0. [DOI] [PubMed] [Google Scholar]
- 10.Kodama T, Freeman M, Rohrer L, Zabrecky J, Matsudaira P, Krieger M. Type I macrophage scavenger receptor contains alpha-helical and collagen-like coiled coils. Nature. 1990;343:531–535. doi: 10.1038/343531a0. [DOI] [PubMed] [Google Scholar]
- 11.Fraser I, Hughes D, Gordon S. Divalent cation-independent macrophage adhesion inhibited by monoclonal antibody to murine scavenger receptor. Nature. 1993;364:343–346. doi: 10.1038/364343a0. [DOI] [PubMed] [Google Scholar]
- 12.Ojala JR, Pikkarainen T, Tuuttila A, Sandalova T, Tryggvason K. Crystal structure of the cysteine-rich domain of scavenger receptor MARCO reveals the presence of a basic and an acidic cluster that both contribute to ligand recognition. J Biol Chem. 2007;282:16654–16666. doi: 10.1074/jbc.M701750200. [DOI] [PubMed] [Google Scholar]
- 13.Becker M, Cotena A, Gordon S, Platt N. Expression of the class A macrophage scavenger receptor on specific subpopulations of murine dendritic cells limits their endotoxin response. Eur J Immunol. 2006;36:950–960. doi: 10.1002/eji.200535660. [DOI] [PubMed] [Google Scholar]
- 14.Hughes DA, Fraser IP, Gordon S. Murine macrophage scavenger receptor: in vivo expression and function as receptor for macrophage adhesion in lymphoid and non-lymphoid organs. Eur J Immunol. 1995;25:466–473. doi: 10.1002/eji.1830250224. [DOI] [PubMed] [Google Scholar]
- 15.Brown JM, Swindle EJ, Kushnir-Sukhov NM, Holian A, Metcalfe DD. Silica-directed mast cell activation is enhanced by scavenger receptors. Am J Respir Cell Mol Biol. 2007;36:43–52. doi: 10.1165/rcmb.2006-0197OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Naito M, Suzuki H, Mori T, Matsumoto A, Kodama T, Takahashi K. Coexpression of type I and type II human macrophage scavenger receptors in macrophages of various organs and foam cells in atherosclerotic lesions. Am J Pathol. 1992;141:591–599. [PMC free article] [PubMed] [Google Scholar]
- 17.van der Laan LJ, Kangas M, Dopp EA, Broug-Holub E, Elomaa O, Tryggvason K, Kraal G. Macrophage scavenger receptor MARCO: in vitro and in vivo regulation and involvement in the anti-bacterial host defense. Immunol Lett. 1997;57:203–208. doi: 10.1016/s0165-2478(97)00077-1. [DOI] [PubMed] [Google Scholar]
- 18.Doyle SE, O'Connell RM, Miranda GA, Vaidya SA, Chow EK, Liu PT, Suzuki S, Suzuki N, Modlin RL, Yeh WC, Lane TF, Cheng G. Toll-like receptors induce a phagocytic gene program through p38. J Exp Med. 2004;199:81–90. doi: 10.1084/jem.20031237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Villiers WJ, Fraser IP, Hughes DA, Doyle AG, Gordon S. Macrophage-colony-stimulating factor selectively enhances macrophage scavenger receptor expression and function. J Exp Med. 1994;180:705–709. doi: 10.1084/jem.180.2.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gough PJ, Gordon S. The role of scavenger receptors in the innate immune system. Microbes Infect. 2000;2:305–11. doi: 10.1016/s1286-4579(00)00297-5. [DOI] [PubMed] [Google Scholar]
- 21.Plüddemann A, Neyen C, Gordon S. Macrophage scavenger receptors and host-derived ligands. Methods. 2007;43:207–217. doi: 10.1016/j.ymeth.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 22.Plüddemann A, Mukhopadhyay S, Gordon S. The interaction of macrophage receptors with bacterial ligands. Expert Rev Mol Med. 2006;8:1–25. doi: 10.1017/S1462399406000159. [DOI] [PubMed] [Google Scholar]
- 23.Chen Y, Sankala M, Ojala JRM, Sun Y, Tuuttila A, Isenman DE, Tryggvason K, Pikkarainen T. A phage display screen and binding studies with acetylated LDL provide evidence for the importance of the scavenger receptor cysteine-rich (SRCR) domain in the ligand-binding function of MARCO. J Biol Chem. 2006;281:12767–12775. doi: 10.1074/jbc.M513628200. [DOI] [PubMed] [Google Scholar]
- 24.Dunne DW, Resnick D, Greenberg J, Krieger M, Joiner KA. The type I macrophage scavenger receptor binds to gram-positive bacteria and recognizes lipoteichoic acid. Proc Natl Acad Sci USA, 1994;91:1863–1867. doi: 10.1073/pnas.91.5.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elomaa O, Kangas M, Sahlberg C, Tuukkanen J, Sormunen R, Liakka A, Thesleff I, Kraal G, Tryggvason K. Cloning of a novel bacteria-binding receptor structurally related to scavenger receptors and expressed in a subset of macrophages. Cell. 1995;80:603–609. doi: 10.1016/0092-8674(95)90514-6. [DOI] [PubMed] [Google Scholar]
- 26.Hampton RY, Golenbock DT, Penman M, Krieger M, Raetz CR. Recognition and plasma clearance of endotoxin by scavenger receptors. Nature. 1991;352:342–344. doi: 10.1038/352342a0. [DOI] [PubMed] [Google Scholar]
- 27.Takeda K, Kaisho T, Akira S. Toll-like receptors. Annu Rev Immunol. 2003;21:335–376. doi: 10.1146/annurev.immunol.21.120601.141126. [DOI] [PubMed] [Google Scholar]
- 28.Marshall-Clarke S, Downes JE, Haga IR, Bowie AG, Borrow P, Pennock JL, Grencis RK, Rothwell P. Polyinosinic acid is a ligand for toll-like receptor 3. J Biol Chem. 2007;282:24759–24766. doi: 10.1074/jbc.M700188200. [DOI] [PubMed] [Google Scholar]
- 29.Edfeldt K, Swedenborg J, Hansson GK, Yan ZQ. Expression of toll-like receptors in human atherosclerotic lesions: a possible pathway for plaque activation. Circulation. 2002;105:1158–1161. [PubMed] [Google Scholar]
- 30.Hashimoto M, Tawaratsumida K, Kariya H, Kiyohara A, Suda Y, Krikae F, Kirikae T, Gotz F. Not lipoteichoic acid but lipoproteins appear to be the dominant immunobiologically active compounds in Staphylococcus aureus. J Immunol. 2006;177:3162–3169. doi: 10.4049/jimmunol.177.5.3162. [DOI] [PubMed] [Google Scholar]
- 31.Henneke P, Dramsi S, Mancuso G, Chraibi K, Pellegrini E, Theilacker C, Hubner J, Santos-Sierra S, Teti G, Golenbock DT, Poyart C, Trieu-Cuot P. Lipoproteins are critical TLR2 activating toxins in group B streptococcal sepsis. J Immunol. 2008;180:6149–6158. doi: 10.4049/jimmunol.180.9.6149. [DOI] [PubMed] [Google Scholar]
- 32.da Silva Correia J, Soldau K, Christen U, Tobias PS, Ulevitch RJ. Lipopolysaccharide is in close proximity to each of the proteins in its membrane receptor complex transfer from CD14 to TLR4 and MD-2. J Biol Chem. 2001;276:21129–21135. doi: 10.1074/jbc.M009164200. [DOI] [PubMed] [Google Scholar]
- 33.Dziarski R, Wang Q, Miyake K, Kirschning CJ, Gupta D. MD-2 enables Toll-like receptor 2 (TLR2)-mediated responses to lipopolysaccharide and enhances TLR2-mediated responses to Gram-positive and Gram-negative bacteria and their cell wall components. J Immunol. 2001;166:1938–1944. doi: 10.4049/jimmunol.166.3.1938. [DOI] [PubMed] [Google Scholar]
- 34.Dalpke A, Frank J, Peter M, Heeg K. Activation of toll-like receptor 9 by DNA from different bacterial species. Infect Immun. 2006;74:940–946. doi: 10.1128/IAI.74.2.940-946.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mukhopadhyay S, Chen Y, Sankala M, Peiser L, Pikkarainen T, Kraal G, Tryggvason K, Gordon S. MARCO, an innate activation marker of macrophages, is a class A scavenger receptor for Neisseria meningitidis. Eur J Immunol. 2006;36:940–949. doi: 10.1002/eji.200535389. [DOI] [PubMed] [Google Scholar]
- 36.Peiser L, de Winther MPJ, Makepeace K, Hollinshead M, Coull P, Plested J, Kodama T, Moxon ER, Gordon S. The class A macrophage scavenger receptor is a major pattern recognition receptor for Neisseria meningitidis which is independent of lipopolysaccharide and not required for secretory responses. Infect Immun. 2002;70:5346–5354. doi: 10.1128/IAI.70.10.5346-5354.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peiser L, Makepeace K, Plüddemann A, Savino S, Wright JC, Pizza M, Rappuoli R, Moxon ER, Gordon S. Identification of Neisseria meningitidis nonlipopolysaccharide ligands for class A macrophage scavenger receptor by using a novel assay. Infect Immun. 2006;74:5191–5199. doi: 10.1128/IAI.00124-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoebe K, Georgel P, Rutschmann S, Du X, Mudd S, Crozat K, Sovath S, Shamel L, Hartung T, Zahringer U, Beutler B. CD36 is a sensor of diacylglycerides. Nature. 2005;433:523–527. doi: 10.1038/nature03253. [DOI] [PubMed] [Google Scholar]
- 39.Jeannin P, Bottazzi B, Sironi M, Doni A, Rusnati M, Presta M, Maina V, Magistrelli G, Haeuw JF, Hoeffel G, Thieblemont N, Corvaia N, Garlanda C, Delneste Y, Mantovani A. Complexity and complementarity of outer membrane protein A recognition by cellular and humoral innate immunity receptors. Immunity. 2005;22:551–560. doi: 10.1016/j.immuni.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 40.Butler PJG, Hartley BS. Maleylation of amino groups. Methods Enzymol. 1972;25:191–199. doi: 10.1016/S0076-6879(72)25016-9. [DOI] [PubMed] [Google Scholar]
- 41.van der Laan LJ, Dopp EA, Haworth R, Pikkarainen T, Kangas M, Elomaa O, Dijkstra CD, Gordon S, Tryggvason K, Kraal G. Regulation and functional involvement of macrophage scavenger receptor MARCO in clearance of bacteria in vivo. J Immunol. 1999;162:939–947. [PubMed] [Google Scholar]
- 42.Pizza M, Scarlato V, Masignani V, Giuliani MM, Arico B, Comanducci M, Jennings GT, Baldi L, Bartolini E, Capecchi B, Galeotti CL, Luzzi E, Manetti R, Marchetti E, Mora M, Nuti S, Ratti G, Santini L, Savino S, Scarselli M, Storni E, Zuo P, Broeker M, Hundt E, Knapp B, Blair E, Mason T, Tettelin H, Hood DW, Jeffries AC, Saunders NJ, Granoff DM, Venter JC, Moxon ER, Grandi G, Rappuoli R. Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science. 2000;287:1816–1820. doi: 10.1126/science.287.5459.1816. [DOI] [PubMed] [Google Scholar]
- 43.Chen Y, Pikkarainen T, Elomaa O, Soininen R, Kodama T, Kraal G, Tryggvason K. Defective microarchitecture of the spleen marginal zone and impaired response to a thymus-independent type 2 antigen in mice lacking scavenger receptors MARCO and SR-A. J Immunol. 2005;175:8173–8180. doi: 10.4049/jimmunol.175.12.8173. [DOI] [PubMed] [Google Scholar]
- 44.Suzuki H, Kurihara Y, Takeya M, Kamada N, Kataoka M, Jishage K, Ueda O, Sakaguchi H, Higashi T, Suzuki T, Takashima Y, Kawabe Y, Cynshi O, Wada Y, Honda M, Kurihara H, Aburatani H, Doi T, Matsumoto A, Azuma S, Noda T, Toyoda Y, Itakura H, Yazaki Y, Kodama T, et al. A role for macrophage scavenger receptors in atherosclerosis and susceptibility to infection. Nature. 1997;386:292–296. doi: 10.1038/386292a0. [DOI] [PubMed] [Google Scholar]
- 45.Fauve RM, Jusforgues H, Hevin B. Maintenance of granuloma macrophages in serum-free medium. J Immunol Methods. 1983;64:345–351. doi: 10.1016/0022-1759(83)90442-8. [DOI] [PubMed] [Google Scholar]
- 46.Rabinowitz S, Horstmann H, Gordon S, Griffiths G. Immunocytochemical characterization of the endocytic and phagolysosomal compartments in peritoneal macrophages. J Cell Biol. 1992;116:95–112. doi: 10.1083/jcb.116.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Plüddemann A, Neyen C, Gordon S, Peiser L. A sensitive solid-phase assay for identification of class A macrophage scavenger receptor ligands using cell lysate. J Immunol Methods. 2008;329:167–175. doi: 10.1016/j.jim.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 48.Sankala M, Brannstrom A, Schulthess T, Bergmann U, Morgunova E, Engel J, Tryggvason K, Pikkarainen T. Characterization of recombinant soluble macrophage scavenger receptor MARCO. J Biol Chem. 2002;277:33378–33385. doi: 10.1074/jbc.M204494200. [DOI] [PubMed] [Google Scholar]
- 49.Goldstein JL, Ho YK, Basu SK, Brown MS. Binding site on macrophages that mediates uptake and degradation of acetylated low density lipoprotein, producing massive cholesterol deposition. Proc Natl Acad Sci USA. 1979;76:333–337. doi: 10.1073/pnas.76.1.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Raetz CR, Garrett TA, Reynolds CM, Shaw WA, Moore JD, Smith DC, Ribeiro AA, Murphy RC, Ulevitch RJ, Fearns C, Reichart D, Glass CK, Benner C, Subramaniam S, Harkewicz R, Bowers-Gentry RC, Buczynski MW, Cooper JA, Deems RA, Dennis EA. Kdo2-Lipid A of Escherichia coli, a defined endotoxin that activates macrophages via TLR-4. J Lipid Res. 2006;47:1097–1111. doi: 10.1194/jlr.M600027-JLR200. [DOI] [PubMed] [Google Scholar]
- 51.Coller SP, Paulnock DM. Signaling pathways initiated in macrophages after engagement of type A scavenger receptors. J Leukoc Biol. 2001;70:142–148. [PubMed] [Google Scholar]
- 52.Greenberg JW, Fischer W, Joiner KA. Influence of lipoteichoic acid structure on recognition by the macrophage scavenger receptor. Infect Immun. 1996;64:3318–3325. doi: 10.1128/iai.64.8.3318-3325.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu FG, Reich CF, Pisetsky DS. The role of the macrophage scavenger receptor in immune stimulation by bacterial DNA and synthetic oligonucleotides. Immunology. 2001;103:226–234. doi: 10.1046/j.1365-2567.2001.01222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jozefowski S, Sulahian TH, Arredouani M, Kobzik L. Role of scavenger receptor MARCO in macrophage responses to CpG oligodeoxynucleotides. J Leukoc Biol. 2006;80:870–879. doi: 10.1189/jlb.0705357. [DOI] [PubMed] [Google Scholar]
- 55.Hale CA, de Boer., PA Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell. 1997;88:175–185. doi: 10.1016/s0092-8674(00)81838-3. [DOI] [PubMed] [Google Scholar]
- 56.Hsu HY, Chiu SL, Wen MH, Chen KY, Hua KF. Ligands of macrophage scavenger receptor induce cytokine expression via differential modulation of protein kinase signaling pathways. J Biol Chem. 2001;276:28719–28730. doi: 10.1074/jbc.M011117200. [DOI] [PubMed] [Google Scholar]
- 57.Arredouani M, Yang Z, Ning Y, Qin G, Soininen R, Tryggvason K, Kobzik L. The scavenger receptor MARCO is required for lung defense against pneumococcal pneumonia and inhaled particles. J Exp Med. 2004;200:267–272. doi: 10.1084/jem.20040731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thomas CA, Li Y, Kodama T, Suzuki H, Silverstein SC, El Khoury., J Protection from lethal gram-positive infection by macrophage scavenger receptor-dependent phagocytosis. J Exp Med. 2000;191:147–156. doi: 10.1084/jem.191.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zaks K, Jordan M, Guth A, Sellins K, Kedl R, Izzo A, Bosio C, Dow S. Efficient immunization and cross-priming by vaccine adjuvants containing TLR3 or TLR9 agonists complexed to cationic liposomes. J Immunol. 2006;176:7335–7345. doi: 10.4049/jimmunol.176.12.7335. [DOI] [PubMed] [Google Scholar]