Abstract
Monoclonal antibodies, engineered antibodies, and antibody fragments have become important biological therapeutic platforms. The IgG format with bivalent binding sites has a modular structure with different biological roles, i.e., effector and binding functions, in different domains. We demonstrated the reconstruction of an IgG-like domain structure in vitro by protein ligation using protein trans-splicing. We produced various binding domains to replace the binding domain of IgG from Escherichia coli and the Fc domain of human IgG from Brevibacillus choshinensis as split-intein fusions. We showed that in vitro protein ligation could produce various Fc-fusions at the N-terminus in vitro from the independently produced domains from different organisms. We thus propose an off-the-shelf approach for the combinatorial production of Fc fusions in vitro with several distinct binding domains, particularly from naturally occurring binding domains. Antiviral lectins from algae are known to inhibit virus entry of HIV and SARS coronavirus. We demonstrated that a lectin could be fused with the Fc-domain in vitro by protein ligation, producing an IgG-like molecule as a “lectibody”. Such an Fc-fusion could be produced in vitro by this approach, which could be an attractive method for developing potential therapeutic agents against rapidly emerging infectious diseases like SARS coronavirus without any genetic fusion and expression optimization.
Keywords: protein trans-splicing, split intein, immunoglobulin G, lectin, protein ligation, Fc, scytovirin, segmental isotopic labeling, lectibody
1. Introduction
Immunoglobulin G (IgG) is a multidomain protein that consists of four polypeptide chains. Two identical heavy chains and two identical light chains form a Y-shaped molecule connected by disulfide bonds (Figure 1a). While the heavy chain is composed of four structural domains called immunoglobulin domains (VH, CH1, CH2, and CH3), the light chain has two immunoglobulin domains (VL and CL) (Figure 1a) [1]. Variable domains VH and VL constitute an antigen-binding region, bearing two binding sites per IgG. The enormous diversity of the variable domains plays an essential role in recognizing specific antigens, such as pathogens and foreign molecules in immunity [1]. Constant domains CH2 and CH3 of the two heavy chains construct the Fc domain, which mediates various effector functions such as antibody-dependent, cell-mediated cytotoxicity (ADCC) and the activation of complement pathways in immunity. Thus, domain structures of antibodies can be dissected based on their functional roles. Antibodies from camelids (like camel and llama) lack light chains, and are therefore termed as heavy-chain antibodies (Figure 1b) [2]. Antigen-binding regions of the heavy-chain antibodies contain only one immunoglobulin fold termed VHH. The VHH domain replaces the functional role of the variable domains VH and VL in conventional immunoglobulins [2]. The smaller size of VHH provides several advantages, such as simple production and engineering [2]. The VHH domain has been widely used as a standalone agent for biotechnological applications [3,4].
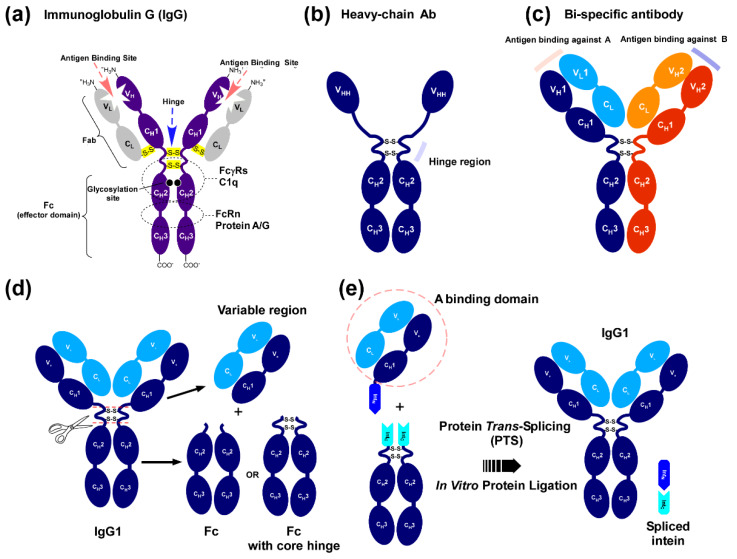
Figure 1.
Domain structures of immunoglobulin G (IgG) and IgG variants. (a) Domains in IgG with their functional roles. (b) Domain structure of a heavy-chain antibody. (c) Domain structure of an engineered bispecific antibody showing each unique chain highlighted in different colors. Dissection of immunoglobulin G (IgG). (d) IgG can be split into the Fab region, which contains an antigen-binding site, and the effector domain Fc at two positions in the hinge region. (e) Reconstitution of IgG-like molecules by protein ligation using protein trans-splicing (PTS) with a split intein. The figure was adapted from [5].
Monoclonal antibodies such as IgG have increasingly become indispensable biological therapeutic drugs for cancer and autoimmune diseases, with currently more than 70 antibody drugs on the market [6]. Because of their therapeutic success, antibodies represent the fastest growing sector in the pharmaceutical industry. Therapeutic antibodies typically comprise intact monoclonal IgG molecules and IgG derivatives [6]. Additionally, antibody fragments have been fused with various functional moieties, including enzymes, toxins, radionuclides, and viruses, for new therapeutic functions [3,4]. Since the therapeutic efficacy of ADCC by antibodies is mitigated by the fact that not all immune cells express antibody receptors, bispecific antibodies have been artificially engineered to simultaneously bind to two different types of antigens (Figure 1c) [7]. All IgGs, heavy-chain antibodies, and bispecific antibodies commonly contain an Fc domain with some sequence variation, while the binding sites are unique to individual formats and antigens. mAbs, Fc fusion proteins, and bispecific antibodies are usually produced from a single cell line for each BsAb, mAb, and Fc fusion by making stable cell lines to produce the genetically engineered proteins, despite the likely presence of the Fc domain among these proteins [8]. Whereas a binding domain such as VHH can be obtained by in vitro selection with directed evolution [9], the production of each new mAb from a mammalian cell line can be labor-intensive and time-consuming, especially for bispecific antibodies requiring four separate constructs (encoding each of the four polypeptide chains for the production of BsAbs) to be engineered [10].
We aimed to develop an efficient method to produce diverse N-terminal fusion proteins of the Fc domain in vitro from dissected domains by protein trans-splicing (Figure 1d,e). Protein splicing is an autocatalytic selfexcision process catalyzed by intein and concomitantly ligating the flanking protein sequences (exteins), catalyzed by an intein (Figure 2a) [11,12,13]. Canonical inteins contain a homing endonuclease (EN) domain. Inteins lacking EN domains are also found in nature and called mini-inteins (Figure 2b) [12]. Split inteins can catalyze bimolecular protein splicing, namely protein splicing in trans (Figure 2b) [12]. Since protein trans-splicing (PTS) was established as a protein-ligation tool of foreign protein sequences with a peptide bond, it has become an essential protein engineering tool (Figure 2d) [14,15]. The split fragments of an intein are termed N-intein (IntN) and C-intein (IntC) for N-terminal and C-terminal fragments, respectively [13,14]. Many artificially and naturally split inteins have been developed and characterized for in vitro protein ligation (Figure 2c) [14,16,17]. In vitro protein ligation of Fc domain with diverse binding domains could circumvent the fusion of different genes on the DNA level with an identical Fc gene, facilitating in vitro production of Fc fusions with various binding capabilities using an off-the-shelf approach (Figure 1e and Figure 3) [18,19].
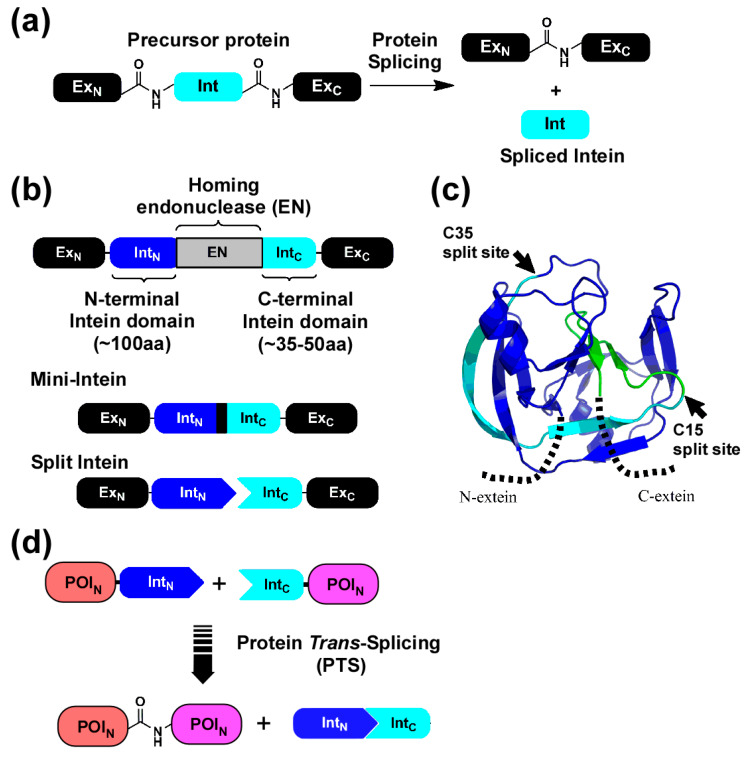
Figure 2.
Protein splicing in cis and trans. (a) Protein splicing in cis. An internal protein (Intein, Int) is spliced out from a precursor protein, concomitantly ligating the two flanking proteins (N-extein and C-extein). ExN and ExC indicate N-extein and C-extein, respectively. (b) Canonical intein with homing endonuclease domain (EN), Mini-intein without EN, and split intein. (c) A cartoon model of NpuDnaE intein [20,21]. The natural split-site (C35) and engineered split-site (C15) are marked with arrows [22]. The N-terminal split fragment of the natural split intein is colored in blue. IntN used in this study is colored in blue or cyan. The region corresponding to the IntC fragment used in this study is highlighted in green. (d) Protein ligation by PTS using a split intein. PTS ligates the N-terminal fragment of a protein of interest (POIN) fused with the N-terminal split intein (IntN) and the C-terminal fragment of POI (POIC) fused with the C-terminal split intein (IntC). PTS results in ligated POIN-POIC and excised IntN and IntC. The figures were adapted from [5].
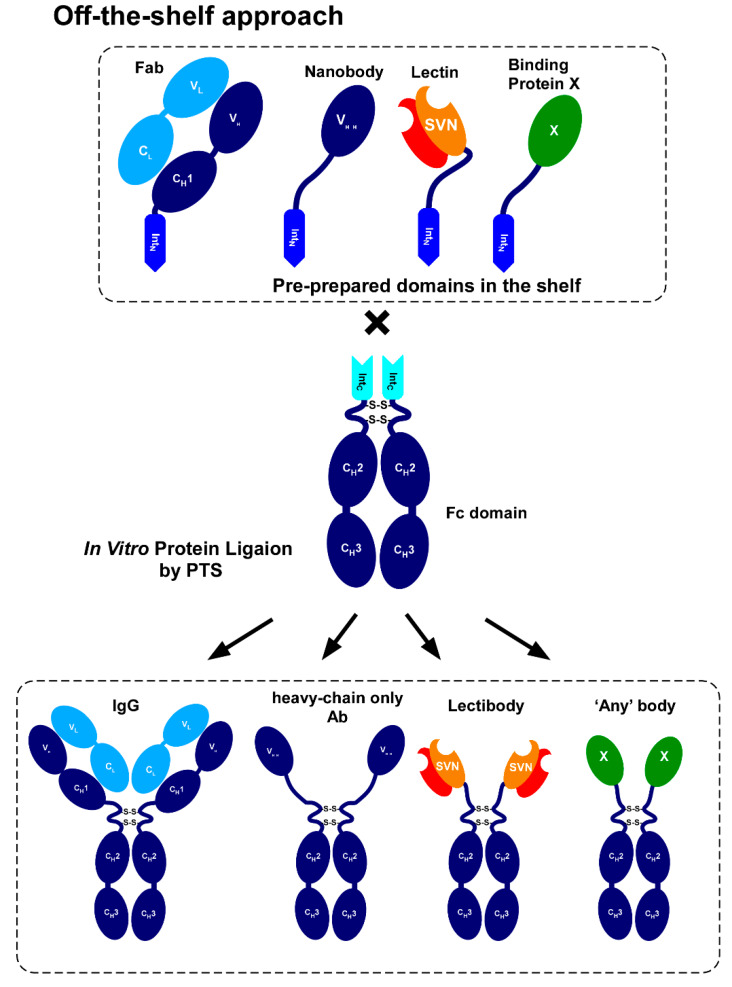
Figure 3.
Off-the-shelf approach for in vitro production of Fc fusion by protein ligation. Various binding domains (protein of interest, POI), such as lectins and VHH, fused with IntN are separately prepared. The Fc domain fused with IntC is also individually made. The N-terminal POI and C-terminal Fc domain are ligated in vitro by PTS, concomitantly splicing out the two intein fragments IntN and IntC. The ligated Fc fusion is a bivalent Y-shape molecule like an IgG molecule.
In this study, we prepared split-intein fusions with the human Fc domain produced in Brevibacillus choshinensis, a gram-positive bacterium. Brevibacillus choshinensis has an excellent ability to produce many exogenous proteins extracellularly [23,24]. We tested in vitro protein ligation between the Fc domain and different model proteins, including the cyanobacterial lectin scytovirin tin (SVN). SVN has antiviral activity against a variety of human pathogens, including the HIV-1 and SARS viruses, by binding to glycosylated viral surface proteins [25,26,27,28]. The lectin-Fc fusion might thus function as a carbohydrate-targeting antibody, namely ‘lectibody’ (Figure 3) [29].
2. Results
2.1. Strategy to Produce Fc Fusions In Vitro
We dissected the IgG format at two different sites within the hinge region (Figure 1d and Figure 4a). The hinge region can be divided into the upper, core, and lower hinges [1,30]. The Fc domain with and without the core region was fused with the C-terminal fragment (IntC) of the naturally split DnaE intein from Synechocystis species, strain PCC6803 (SspDnaEC16) (Figure 4b). We chose Npu/SspDnaE chimeric intein for protein ligation because it has been used successfully in the ligation of many protein domains in vitro, and has a higher tolerance of amino acids at the +2 position [21]. Naturally split SspDnaE and NpuDnaE inteins have a 36-residue IntC fragment (Figure 2c) [21]. Therefore, we termed the split-site of these inteins as C35 (excluding the start codon Met residue) [12]. However, we selected C15 as the intein split site, and used the engineered pair of NpuDnaEΔC16/SspDnaEC16, because this pair has worked better for some model proteins than the naturally split C35 pair of split inteins (Figure 2c) [22]. We hypothesized that the shorter 15-residue IntC would disturb less the folding of the Fc domain. Three different IntC-Fc fusion proteins (IntC-Fc, IntC+1S-Fc, and IntC-CPPC-Fc) were constructed in this study (Figure 4b).
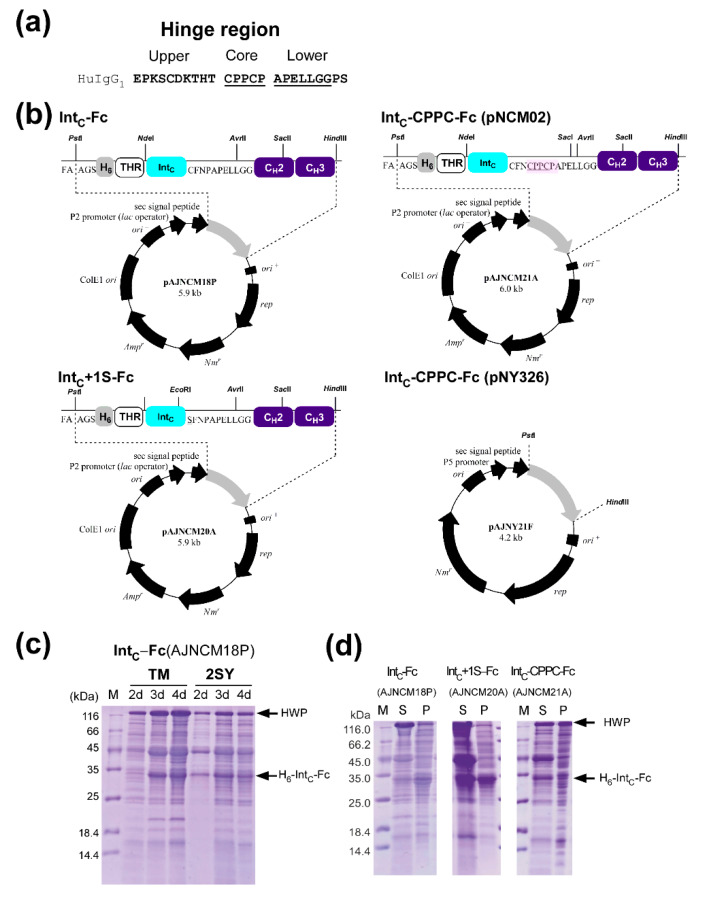
Figure 4.
Vector designs and protein production of IntC-Fc. (a) Primary structure of the hinge region of human IgG. (b) Vector maps of three constructs (IntC-Fc, IntC+1S-Fc, and IntC-CPPC-Fc) in the backbone of pNCM02 and pNY326 vector. (c) Optimization of the growth media. Production of IntC-Fc was compared in TM and SY media. Lanes 2d, 3d, 4d indicate SDS-PAGE analysis of the culture supernatant after 2 days, 3 days, and 4 days of protein expression, respectively. (d) Comparison of protein production between IntC-Fc, IntC+1S-Fc, and IntC-CPPC-Fc by SDS-PAGE analysis. Lanes S and P indicate supernatant and pellet fractions from the culture medium after four days of protein expression, respectively. The control cell wall protein (HWP) and IntC-Fc are marked with arrows.
2.2. Cloning and Protein Expression of IntC-Fc in Brevibacillus choshinensis
The Fc domain without the hinge-region has been produced in Escherichia coli [31,32,33]. We decided to use the commercially available Brevibacillus expression system which is suitable for the secretory production of heterologous proteins up to 3.7 g/L [34,35]. Following the protocol of the commercial Brevibacillus expression system (TakaraBio), we cloned the gene of IntC-Fc fusion protein with or without the core hinge into pNY326 and pNCM02 with the signal peptide of HWP from the kit (Figure 4b). Brevibacillus choshinensis has a distinctive cell surface structure containing one surface protein layer formed by HPD31 cell wall protein (HWP) under the regulation of five tandem promoters, named P1–P5. While pNY326 use P5 promoter, pNCM02 is a high-copy-number plasmid harboring a strong promoter, P2, and a modified Sec signal sequence for efficient secretory expression in B. choshinensis [35].
We then compared the protein expression of the Fc fusions in TM and 2SY media (Figure 4c and Supplemental Figure S1). We found that the cells harboring the pNCM02-backbone vector secreted IntC-Fc fusion with the core hinge more quickly in 2SY than the cells bearing pNY326 backbone vector in TM and SY media (Supplemental Figure S1).
2.3. Production of IntC-Fc for Protein Trans-Splicing
For the production of IntC-Fc fusions for protein ligation, we chose three different constructs IntC-Fc (AJNCM18P), IntC+1S-Fc (AJNCM20A), and IntC-CPPC-Fc (AJNCM21A) (Figure 4). We constructed IntC-Fc with or without the core hinge. Additionally, we created a variant replacing Cys+1 with Ser+1 for the active site of the intein at the so-called +1 position [21]. We first optimized the culture media and analyzed the time-course of the expression using AJNCM18P (IntC-Fc without the core hinge) as a model protein. The presence of the IntC-Fc was detected in both TM and 2SY media after three days, and no significant increase was observed after four days (Figure 4c). Based on these results, three different IntC-Fc fusion proteins were expressed and secreted under the P2 promoter in a 4-mL scale using TM medium and compared after four days (Figure 4d). All three fusion proteins were successfully secreted into the culture medium. Whereas the control protein HWP with the molecular weight of 118 kDa accumulated mostly in the supernatant fraction, IntC-Fc and IntC+1S-Fc were also detected in the pellet fraction, suggesting incomplete secretion of the fusion proteins or incorrect folding of the protein. In contrast, IntC-CPPC-Fc was mainly present in the supernatant fraction.
2.4. Purification of IntC-Fc Fusions
We purified IntC-Fc fusions using an N-terminal hexahistidine (His-tag) incorporated in IntC by Immobilized Metal Chelate Affinity Chromatography (IMAC) (Figure 4b and Figure 5). The yields were 27 mg and 15 mg per liter of the culture medium for IntC+1S-Fc and IntC-CPPC-Fc, respectively. These yields were calculated from pooled 4-mL cultures after three days of protein expression. It is noteworthy that we were unable to scale up to over 50 mL, abolishing the protein secretion of interest at a larger scale. After the first IMAC purification, we tested the binding of IntC-Fc fusions to the Staphylococcal protein A column by affinity chromatography. All three IntC-Fc fusions bound to a protein A sepharose column and could be eluted by acid elution as used for IgG purification, indicating that the overexpressed Fc domain is capable of binding to the protein A column, despite the N-terminal fusion of IntC; 35–49% of the total protein could be recovered after the protein A affinity chromatography and subsequent dialysis. The molecular mass of IntC-CPPC-Fc produced from B. choshinensis was confirmed (Supplemental Figure S2).
Figure 5.
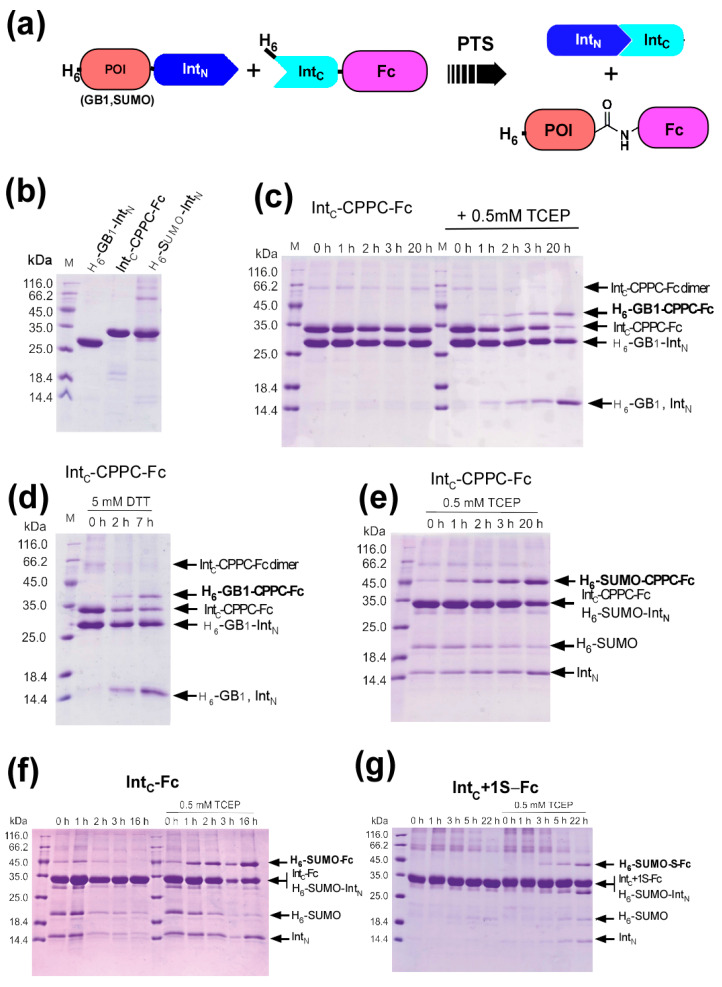
In vitro Fc fusion by PTS (a) Schematic drawing of the ligation of model proteins (SUMO and GB1) with the C-terminal Fc domain by PTS. (b) SDS-PAGE analysis of the N- and C-terminal precursor proteins (H6-GB1-IntN, IntC-CPPC-Fc, and H6-SUMO-IntN) independently purified by IMAC. (c) Time course of the ligation reaction between H6-GB1-IntN and IntC-CPPC-Fc in the absence (left panel) or presence (right panel) of 0.5 mM TCEP. Samples were taken after 0, 1, 2, 3, and 20 h of incubation, and analyzed by SDS-PAGE. (d) The effect of 5 mM DTT instead of 0.5 mM TCEP was tested for the reaction mixture of H6-GB1-IntN and IntC-CPPC-Fc. Samples were analyzed by SDS-PAGE after 0, 2, and 7 h of incubation. (e–g) Comparison between the three Fc constructs (IntC-CPPC-Fc (e), IntC-Fc (f), and IntC+1S-Fc (g)). Protein ligation with H6-SUMO-IntN was analyzed with and without 0.5 mM TCEP by SDS-PAGEs. Incubation times in hours (h) are indicated above the lanes of SDS-PAGEs.
2.5. Model Proteins for Protein Ligation with IntC-Fc
To test the protein ligation of the purified IntC-Fc fusions, we prepared the B1 domain of IgG binding protein G (GB1) and yeast SMT3 protein (SUMO domain) as the N-terminal target proteins. Both GB1 and SUMO domains were fused with an N-terminal His-tag and with the N-terminal split fragment (IntN) of NpuDnaEΔC16 intein (NpuDnaEN123) for PTS. Both model proteins were previously prepared and tested for protein ligation with other model proteins [22]. H6-GB1-IntN (NpuDnaEΔC16) and H6-SUMO-IntN (NpuDnaEΔC16) were independently produced in E.coli and purified by IMAC using the N-terminal His-tag (Figure 5b).
2.6. Protein Ligation of IntC-CPPC-Fc by Protein Trans-Splicing
The individually purified N-terminal precursor proteins (H6-GB1-IntN and H6-SUMO-IntN) and three Fc fusion proteins (IntC-Fc, IntC+1S-Fc, and IntC-CPPC-Fc) were tested for in vitro protein ligation (Figure 5). The incubation of a reaction mixture of IntC-CPPC-Fc and H6-GB1-IntN did not result in any ligated product (H6-GB1-CPPC-Fc) (Figure 5c). However, the same reaction mixture in the presence of a reducing agent, 0.5 mM TCEP (tris(2-carboxyethyl)phosphine), produced the ligated product H6-GB1-CPPC-Fc, together with the spliced-out intein fragment. Despite the slow reaction, i.e., requiring over 20 h, the in vitro ligation of IntC-CPPC-Fc with GB1 was possible under reducing conditions (Figure 5c). We also tested in vitro ligation using 5 mM DTT instead of TCEP, yielding a slightly faster reaction (Figure 5d). The requirement of a reducing agent suggests that disulfide bonds in the core hinge region might hinder the PTS reaction. Indeed, we observed the dimer formation of the Fc domain without a reducing agent, which could be prevented by removing the core hinge and Cys at the +1 position of IntC (Supplemental Figure S3). Next, we tested the SUMO domain as another model protein for in vitro protein ligation. Similar to GB1, we could produce H6-SUMO-CPPC-Fc in the presence of 0.5 mM TCEP (Figure 5e–g). In the case of H6-SUMO, we observed some premature cleavage of H6-SUMO-IntN, as reported previously [22]. Since two precursor proteins, i.e., H6-SUMO-IntN and IntC-CPPC-Fc, have the same migration in SDS-PAGE, we could not estimate the reaction yield precisely (Figure 5e). However, the ligation with the SUMO domain accumulated the ligated product more than the ligation with GB1. We believe that this was because GB1 has an affinity to the Fc domain, thereby limiting its access to IntC-Fc for PTS.
2.7. The Influence of the Core Hinge on Protein Trans-Splicing
The core hinge in IntC-CPPC-Fc is more rigid due to two disulfide bonds, and could therefore complicate the folding of the Fc domain during protein production [30]. To evaluate the effect of these disulfide bridges on PTS, we tested in vitro protein ligation of IntC-Fc and IntC+1S-Fc, which lack the core hinge (Figure 5f,g). IntC+1S-Fc is expected to produce Fc fusion without any cysteine in the hinge region, while both IntC-CPPC-Fc and IntC-Fc result in an additional “CFN” sequence in the hinge region, which could form an interchain disulfide bond (Figure 4). The reaction mixture of H6-SUMO-IntN and IntC-Fc produced the ligated product H6-SUMO-Fc only in the presence of 0.5 mM TCEP, similar to IntC-CPPC-Fc (Figure 5e,f). Premature cleavage of H6-SUMO-IntN was observed both with IntC-Fc and IntC-CPPC-Fc, whereas the reaction of H6-SUMO-IntN and IntC+1S-Fc did not show any premature cleavage of H6-SUMO-IntN. This observation is presumably because the hydroxyl group of serine is less nucleophilic than the thiol group of cysteine. Despite the slow reaction, the reaction mixture of H6-SUMO-IntN and IntC+1S-Fc produced the ligated product H6-SUMO-S-Fc fusion. We expected that H6-SUMO-IntN and IntC+1S-Fc would produce the ligated product without any reducing agent, because the mutation of Cys to Ser was introduced at the active site of the +1 position (Supplemental Figure S3). However, IntC+1S-Fc still required a reducing agent, suggesting that other cysteine residues in the intein, such as the first residue of NpuDnaE intein, need reducing conditions.
2.8. In Vitro Production of the Lectin Fusion Protein by Protein Trans-Splicing
Encouraged by the results from the model proteins, we attempted to create ‘lectibody’ by replacing the antigen-binding domains of IgG with a small lectin by PTS (Figure 3 and Figure 6). Lectins are a group of proteins that recognize and bind carbohydrates with high specificity. Since there are characteristic carbohydrates, for example, on the surfaces of pathogens and cancer cells, lectins are promising candidates as neutralizing and drug-targeting agents [26,35]. For example, cyanobacterial lectin scytovirin (SVN) is an antiviral lectin from Scytonema varium that binds to high mannose oligosaccharides on the surface of the human immunodeficiency virus (HIV) [27]. Bivalent structures of the IgG format could increase the binding of lectin when it is fused with Fc. Additionally, the Fc domain could improve the in vivo stability of a small lectin protein.
Figure 6.
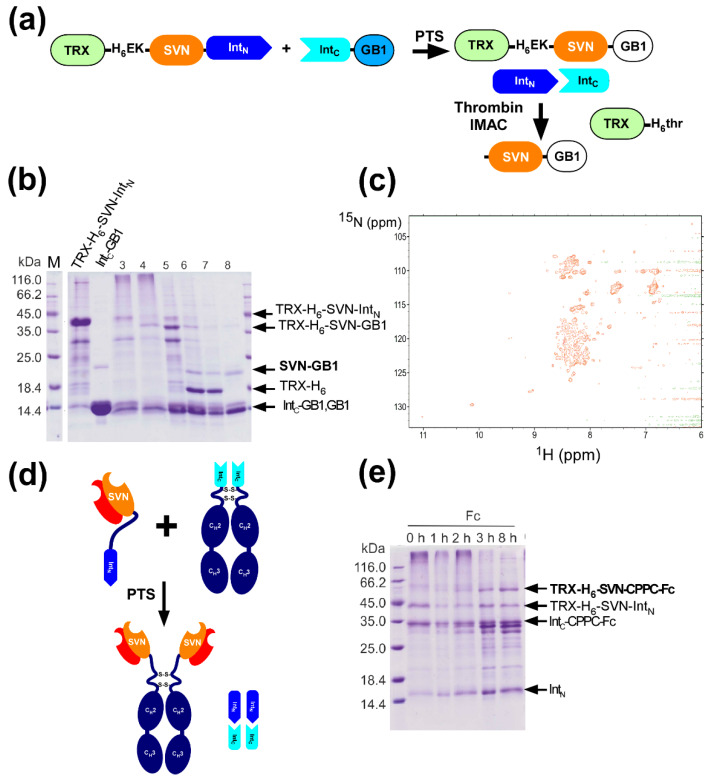
A lectin fusion protein, ‘lectibody’ (a) Schematic drawing of the ligation of a lectin (SVN) with a model protein (GB1) by PTS. (b) SDS-PAGE analysis of the protein ligation of SVN and GB1. The first two lanes show separately prepared N- and C-precursor proteins (TRX-H6-SVN-IntN and IntC-GB1). Lanes 3 and 4 show the ligation reaction immediately after the mixture and after 8 h of incubation, respectively. Lane 5 and 6 indicate samples before and after the proteolytic digestion by enterokinase (EK), respectively. Lanes 7 and 8 show samples before loading on a protein A sepharose column and the elution fraction from the column, respectively. (c) [1H,15N]-HSQC spectrum of the segmentally isotopic labeled [15N]-SVN-[14N]-GB. (d) Schematic drawing of the production of SVN-Fc fusion by PTS. (e) SDS-PAGE analysis of the time-course of the ligation between TRX-H6-SVN-IntN and IntC-CPPC-Fc by PTS.
We produced SVN as a fusion protein with thioredoxin (TRX) as previously reported, but additionally fused it with IntN (NpuDnaEΔC16) [27]. The construct TRX-H6-SVN-IntN was expressed in E. coli under T7 promoter and purified with a His-tag by IMAC (Figure 6a,b). We expressed and purified the model protein GB1 with IntC, as previously reported [22,36]. The reaction mixture of TRX-H6-SVN-IntN and IntC-GB1 in the presence of 0.5 mM TCEP produced the ligated product TRX-H6-SVN-GB1 after 8 h (Figure 6b). The ligated product was immediately subjected to proteolytic digestion by enterokinase (EK) using an EK site engineered between the His-tag and SVN. The proteolysis of TRX-H6-SVN-GB1 resulted in two proteins, TRX-H6 and SVN-GB1 (Figure 6a,b). After digestion with EK, we removed the His-tagged TRX-H6 by affinity chromatography using IgG sepharose (Figure 6b, lane 8). SVN contains ten cysteines which need to form five disulfide bonds correctly, despite the presence of free cysteines of IntN. These disulfide bonds facilitate SVN folding to the native conformation [27,28]. Since we had to use reducing conditions for PTS, we prepared segmentally isotope-labeled SVN-GB1 for the structural assessment of SVN. We used 15N-labeled TRX-H6-SVN-IntN produced in 15N-labeled medium and unlabeled IntC-GB1, resulting in segmentally labeled [15N]-SVN-[14N]-GB1 (Figure 6c) [36,37]. The molecular weight of the ligated [15N]-SVN-[14N]-GB1 was also confirmed by mass-spectrometry (Supplemental Figure S4). The [1H, 15N]-HSQC spectra of SVN with invisible GB1 tag did not show well-dispersed peaks typical for the well-folded globular domain, indicating that SVN was incorrectly folded or unfolded. The fusion with IntN or the ligation in reducing conditions likely affected the correct folding of SVN. We assume that the ligated SVN-GB1 prepared in this study was biologically inactive.
2.9. In vitro Production of a Lectibody by Protein Trans-Splicing
Despite the analysis of SVN-GB1, we tested IntC-CPPC-Fc and TRX-H6-SVN-IntN for protein ligation in the presence of 0.5 mM TCEP (Figure 6d,e). As observed with SUMO-IntN, cleavage reactions were apparent. The ligated product TRX-H6- SVN-CPPC-Fc accumulated after 3 h of reaction (Figure 6e). This observation confirmed that it was feasible to produce ’lectibody’ in vitro by ligating the Fc domain and a lectin using PTS, even though SVN bearing five disulfide bonds is not ideal for PTS, which requires reducing conditions. Several algal lectins contain no or few disulfide bridges such as griffithsin, which could be better candidates for ‘lectibody’ production by PTS [38].
3. Discussion
Inspired by heavy-chain antibodies from camelids composed of only two heavy chains, we decided to create Fc fusion proteins in vitro at the N-terminus of the Fc region. In heavy-chain antibodies, 50-kDa antigen-binding domains of IgG, which consist of VH, VL, CL, and CH1 domains, are replaced with a smaller 12-kDa variable domain (VHH). We hypothesized that carbohydrate-binding proteins like lectins could be able to replace the role of the variable domain of VHH as a binding domain in IgG format when fused at the N-terminus of Fc domain (Figure 3 and Figure 6d). Such Fc fusion proteins might function like an antibody, i.e., ‘lectibody’, with a bivalent binding capability [29]. Previously, a lectibody was engineered as a genetic fusion of a lectin and Fc domain, and produced in plants [29]. The production of full-length antibodies or antibody fragments in large quantities can be challenging in bacterial overexpression systems because the disulfide bonds need to form correctly [39]. Mammalian expression systems have been established for the large-scale production of monoclonal antibodies, but establishing a cell-line for each new monoclonal antibody or a fusion protein requires time-consuming optimizations [40]. Additionally, protein production in mammalian cells can be more expensive and slower compared with bacterial expression systems [39]. Heterogenous expression using mammalian cells could also be more problematic than bacterial expression for engineered foreign proteins from different organisms, such as from plants and bacteria, even in small quantities. Therefore, we developed an in vitro protein ligation approach, which provides an alternative solution for the production of Fc fusions. The in vitro protein ligation approach enabled us to produce various fusion proteins using off-the-shelf domains that were preproduced in different suitable host organisms for each domain. This off-the-shelf protein ligation approach avoids time-consuming steps to optimize the expression and purification for each fusion protein. Once individual domains have been prepared and stored, only the protein ligation step needs optimization.
In this study, we demonstrated the feasibility of producing N-terminal fusion proteins of the Fc domain in vitro by PTS. PTS successfully ligated the Fc domain made in Brevibacillus and other domains produced in E. coli. The method worked for the Fc domain, both with and without the core hinge region, although the reaction required reducing conditions. C-terminal fusions of a full-length IgG and Fc domains by PTS have been demonstrated previously, resulting in functional IgGs with a high ligation efficiency (80%) under reducing reaction conditions [41,42]. Reducing conditions had to be employed in previous reports, as well as in this study. The reducing conditions affected neither antibody affinity nor integrity by the protein ligation using PTS [41,42]. This observation indicated that Fc-fusion proteins generated by PTS could be further exploited for various in vitro products of Fc fusions, including bispecific antibodies, if two orthogonal split inteins could be used [42,43]. We successfully demonstrated the production of an Fc fusion of SVN, which is a model lectin bearing five disulfide bridges, and the N-terminal split intein in E. coli, as previously reported [27,28]. However, the SVN domain in the ligated product by PTS appeared to be unfolded or misfolded, as revealed by NMR analysis using segmental isotopic labeling of SVN [36,37]. We believe that the folding problem of SVN was due to the reducing condition used in the protein ligation reaction by PTS, and that the ligated product was functionally inactive. We chose SVN from the cyanobacterium Scytonema varium as a model system. SVN binds to various viruses, including HIV glycoproteins, SARS coronavirus, and Ebola virus [28,38,44]. It might be possible to use the fusion protein of antiviral lectins as a therapeutic anti-HIV or anti-SARS drugs as a lectibody. In our study, however, SVN bearing five disulfide bonds turned out to be a nonideal candidate for in vitro protein ligation by PTS, at least with split DnaE inteins that require reducing conditions for PTS. Many other lectins contain no disulfide bridges, such as griffithsin from red algae, which also binds to gp120 and gp41 of HIV with high affinity, as well as to SARS coronavirus [38,45,46]. These lectins bearing no disulfide bonds could be more easily developed as lectibodies with therapeutic potential when the intein-Fc fusions are prepared from mammalian cells for the effector functions of IgG, such as ADCC. As the Fc domain could improve the pharmacokinetic properties, the Fc domain may be a good fusion partner for other binding domains [29].
We used the chimeric Npu/SspDnaE split intein with a cysteine residue at the +1 position, which is required for protein splicing reactions and remains in the ligated product. We replaced the cysteine with a serine residue at the +1 position in one of the IntC-Fc precursors (IntC+1S-Fc). This construct, however, still requires reducing conditions for PTS reaction, despite the lack of cysteine in both the intein and hinge regions. This result suggests that other cysteines, such as the first residue of the intein, which is responsible for the first step of the splicing reaction, still require reducing conditions. However, the produced Fc domain retained the capability of protein A binding, indicating the proper three-dimensional structure of the Fc domain by preserving the protein A binding surface. The structure was preserved presumably because two buried intramolecular disulfide bridges in Ig folds (CH2 and CH3) were not influenced by a reducing agent such as 0.5 mM TCEP (Supplemental Figure S3). In the case of the Fc domain with core hinge, the two disulfide-bridges (CPPC) in the core hinge probably have to be reduced to avoid steric hindrance with the IntN fragment. Several inteins in nature contain no cysteine residue within their sequence, including the +1 position [47]. These cysteine-free inteins might be alternatively exploited for PTS.
4. Materials and Methods
4.1. Bacterial Strains and Growth Conditions
B. choshinensis strain HPD31-SP3 (Takara Bio, Shiga, Japan), and E. coli Rosetta-gami 2 (DE3) pLacI (Merck, Kenilworth, NJ, US) and T7 Express (New England Biolabs, Ipswich, MA, US) were used for protein expression. MT medium, MT supplemented with 1.6 mM MgCl2, and 2SY media were used for culturing B. choshinensis [35]. E. coli was cultured in Luria-Bertani (LB) medium. B. choshinensis was transformed by electroporation with a protocol adjusted from the previous report [48].
4.2. Construction of Plasmids
For the N-terminal target proteins, an E.coli expression vector for H6-SUMO-IntN was constructed by transferring the gene of IntN from pHYDuet36 [22] into pHYRSF53 vector [49] (addgene #64696) using BamHI and HindIII restriction sites, resulting in pHYRSF53-36. The gene encoding TRX-H6-SVN was amplified from pET(SVN) by PCR with two oligonucleotides, HK901: 5′-TACACCATGGGAAGCGATAAAATTATTCAC and HK900: 5′-TAGGATCCAC TACCACCTCCCGCAGCCGC GTGACCCGC, followed by cloning into pHYRSF53-36 between NcoI and BamHI sites, resulting in pAJRSF25A bearing the gene of TRX-H6-SVN-IntN (NpuDnaEΔC15).
Expression vectors of IntC-Fc fusions in B. choshinensis were cloned as follows. The gene encoding the DnaE C-intein (IntC) from Synechocystis sp. (SspDnaEC16) was constructed as described previously and transferred into the pBAD vector, resulting in pSABAD7PG [25]. The gene of Fc domain bearing CH2 and CH3from human immunoglobulin G1 was amplified from the L1-Fc vector (addgene #15124) and cloned into pSABAD7PG, resulting in pGVBAD049. The N-terminal hexahistidine tag followed by a thrombin cleavage site introduced by cloning the gene into pRSF vector with PstI site introduced by PCR using two oligonucleotides of GV065: 5′- CAGGTAGCCATCATCATCATCATCACAGCAGC and GV066: 5′-CAGTATATCTCCTTATTAAAGTTAAACAAAATTATTTCTACAGG, resulting in pGVRSF55-049#2. The lower hinge sequence of ‘PAPELLGG’ was introduced by PCR assembly using the four oligonucleotides GV065, AJ001: 5′-CCAGCACCTGAACTCCTAGGGGGACCGTCAGTCTTCCTC, J002: 5′-CCCTAGGAGTTCAGGTGCTGGGTTAAAACAGTTGGCGGCGATAG, and HK548: 5′-TACAAGCTTATTTACCCGGAGACAGGG, and cloned into pGVRSF55-049#2 and pNCM02 vector using PstI-HindIII restriction sites, resulting in pAJRSF18A and pAJNCM18P, respectively for the expression of IntC-Fc. For the production of IntC+1S-Fc in B. choshinensis, the vector pAJNCM18P was mutated by inverse PCR using a pair of the two primers HK866: 5′- GGTGCTATCGCCGCGAATTCTTTTAACCCAGCACCTG and HK867: 5′-CAGGTGCTGGGTTAAAAGAATTCGCGGCGATAGCACC, resulting in pAJNCM20A. For IntC-CPPC-Fc, the core hinge ‘CPPC’ was introduced into pAJRSF18P by inverse PCR using the two primers HK881: 5′-TTGCCCACCGTGCCCAGCACCTGAGCTCCTAGGGGGAC and HK882: 5′-GGTGCTGGGCACGGTGGGSAATTAAAACAGTTGGCGGCG, resulting in pAJRSF21B. The gene corresponding to IntC-CPPC-Fc was transferred from pAJRSF21B to pNCM02 or pNY326, resulting in pAJNCM21A and pAJNY21F vectors with two different promoters. The pAJNCM18P, pAJNCM20A, pAJNCM21A, pAJNY21F, pHYRSF53-36, and pAJRSF25A will be available from Addgene (www.addgene.org) with addgene #153166, #153167, #153168, #153169, #153170, and #153171, respectively.
4.3. Expression and Purification of Fc Fusions from B. choshinensis
The plasmids pAJNCM18P, pAJNCM20A, and pAJNCM21A were transformed into B. choshinensis for protein expression and incubated in 4 mL of TM media at 30 °C, 200 min−1 overnight. The precultures were then inoculated by a 100-fold dilution and incubated in 4 mL 2SY medium at 30 °C. After 3–4 days, the culture supernatants were collected and subjected to SDS-PAGE analysis of the protein incubation. The culture supernatants were collected, centrifuged (42,740× g, 50 min, 4 °C), and loaded onto a 5-mL HisTrap FF column (GE Healthcare, Chicago, IL, USA). Unbound contaminants were washed off with 35 mM imidazole, and bound proteins were eluted with a 15- or 20-mL linear gradient of imidazole (35 mM–250 mM) in 50 mM sodium phosphate, 300 mM NaCl, pH 8.0. Fractions were collected and monitored by measuring absorbance at 280 nm (A280), and their purity was confirmed by SDS-PAGE. The fractions containing target proteins were pooled and dialyzed overnight at 4 °C against 20 mM sodium phosphate, pH 6.5 (protein A binding buffer) using 6–8 kDa molecular weight cut-off (MWCO) membrane. The dialyzed samples were concentrated using a centrifugal filter device (Vivaspin 20, 3 kDa MWCO, GE Healthcare) and clarified by centrifugation (20,817× g, 15 min, 4 °C) prior to loading on a 1-mL HiTrap Protein A HP column. After washing, the bound proteins were eluted with a 2-mL linear gradient of 100 mM sodium citrate, pH 3.5 (0–100%). Fractions were collected and monitored by measuring A280 and by SDS-PAGE examination. The fractions containing the target proteins were immediately neutralized with 20% (v/v) 1 M Tris, pH 9.5, pooled, and dialyzed overnight at 4 °C against 10 mM Tris, 500 mM NaCl, 1 mM EDTA, pH 7 (ligation buffer). When necessary, the dialyzed samples were concentrated using a centrifugal filter device (Microcon YM-10, 10kDa MWCO, Millipore, Temecula, CA, US).
4.4. Expression and Purification of the N-Terminal Model Domains
Expression vectors pHYDuet36 [22], pHYRSF53-36, pAJRSF25A, and pSZRS7PG [22] were transformed into E. coli Rosetta-gami 2(DE3)pLacI. A single transformant was grown overnight in 50 mL LB medium, at 30–37 °C, 220 min−1. The overnight culture was diluted into 2 L of LB medium in three shake flasks and cultured at 37 °C, 220 min−1. When OD600 reached 0.5–0.6, protein expression was induced with IPTG in the final concentration of 40 μg/mL. After 4-h protein expression, cells were harvested by centrifugation (8,980× g, 10 min, 4 °C) and resuspended into 50 mM sodium phosphate, 300 mM NaCl, pH 8.0. The cell suspension was flash-frozen with liquid nitrogen and stored at −70 °C. After thawing, cells were lysed by sonication. Cell debris was removed by centrifugation (42,740× g, 50 min, 4 °C), and the expressed proteins were further purified by polyhistidine tag IMAC with a linear gradient of 50–250 mM imidazole. Elution from the HisTrap FF column was dialyzed overnight at 4 °C against 10–20 mM Tris, 500 mM NaCl, 0–1 mM EDTA, pH 7–8 using 6–8 kDa MWCO membrane.
4.5. Protein Trans-Splicing Assays
For ligation reactions, equal amounts of the two precursor proteins were mixed at a final concentration of 10–18 µM. The reactions were conducted using a bench-top shaker (Thermomixer Comfort R, Eppendorf) at 25 °C with mild shaking (350 rpm) in either reducing or nonreducing conditions. For reducing conditions, 0.5 mM TCEP and 5 mM DTT were tested. The reactions were monitored periodically after mixing by reducing SDS-PAGE. Each sample for SDS-PAGE was mixed 1:1 with 2× SDS sample buffer containing 200 mM DTT and heated at 95 °C for 5 min to stop the reaction. The samples were either loaded on a gel immediately or stored overnight at −20 °C.
SDS-PAGE and protein A affinity chromatography were used to analyze the effect of protein trans-splicing reaction conditions on intramolecular disulfide bonds. Proteins AJNCM18P, AJNCM20A, and AJNCM21A at a concentration of 15 µM were incubated in the presence of 0.5 mM TCEP or 5 mM DTT at 25 °C, 350 rpm (Thermomixer Comfort R, Eppendorf, Hamburg, Germany). Control reactions were incubated without a reducing agent. After 3 h, samples were mixed with the SDS sample buffer and analyzed on a gel. AJNCM21A at a concentration of 14 µM was incubated in the presence of 0.5 mM TCEP for 1.5 h and stored overnight at 4 °C. The buffer was exchanged for a protein A binding buffer in a centrifugal filter device (Amicon Ultra-4, 3-kDa MWCO, Millipore, Temecula, USA), and the sample was loaded onto the protein A column.
5. Conclusions
We demonstrated an off-the-shelf approach to producing Fc fusion proteins at their N-terminus by in vitro protein ligation using PTS. Independently prepared N-terminal domains made in E. coli were ligated in vitro with the C-terminal Fc domain produced in Brevibacillus choshinensis. The in vitro product could constitute an IgG-like Y-shape structure with bivalent binding sites. In nature, there are many natural bioactive proteins, peptides, and domains like lectins that bind to various receptors and ligands but which have not been widely used as a binding domain in IgG-like format. The production of fusion proteins with such bioactive domains in vitro using an off-the-shelf approach paves a new way for the rapid production of IgG-like molecules with known therapeutic potential without any time-consuming genetic engineering and optimization. The off-the-shelf approach using premade domains could be advantageous for drug development against rapidly emerging infectious diseases.
Acknowledgments
We appreciate A. Wlodawer and B.R. O’Keefe for providing the plasmid containing SVN. We acknowledge the Finnish Biological NMR center for the NMR measurements and the proteomic unit at the Institute of Biotechnology for mass-spectrometry.
Abbreviations
| VH | variable heavy chain domain |
| VL | variable light chain domain |
| VHH | the variable domain of heavy chain-only Ab |
| mAb | monoclonal antibody |
| BsAb | bispecific antibody |
| scFv | single-chain variable fragment |
| BiTE | bispecific T-cell engager |
| TRX | thioredoxin |
| NMR | nuclear magnetic resonance |
| HSQC | heteronuclear single quantum coherence |
| SVN | scytovirin |
| Fc | the Fc domain of Immunoglobulin G |
| GB1 | the B1 domain of IgG binding protein G |
| PTS | Protein trans-splicing |
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/21/11/4011/s1.
Author Contributions
Conceptualization, H.I.; methodology, A.J. G.V., and H.I.; validation, H.I., and A.J.; formal analysis, H.I. and A.J.; investigation, A.J.; resources, H.I.; data curation, H.I. and A.J.; writing—original draft preparation, H.I. and A.J.; writing—review and editing, H.I., G.V. and A.J.; visualization, A.J. and H.I.; supervision, H.I.; project administration, H.I.; funding acquisition, H.I. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Academy of Finland, grant number 1118385.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Schroeder H.W., Jr., Cavacini L. Structure and function of immunoglobulins. J. Allergy Clin. Immunol. 2010;125:S41–S52. doi: 10.1016/j.jaci.2009.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hamers-Casterman C., Atarhouch T., Muyldermans S., Robinson G., Hamers C., Songa E.B., Bendahman N., Hamers R. Naturally occurring antibodies devoid of light chains. Nature. 1993;363:446–448. doi: 10.1038/363446a0. [DOI] [PubMed] [Google Scholar]
- 3.Kolkman J.A., Law D.A. Nanobodies—From llamas to therapeutic proteins. Drug Discov. Today Technol. 2010;7:e139–e146. doi: 10.1016/j.ddtec.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 4.Beghein E., Gettemans J. Nanobody technology: A versatile toolkit for microscopic imaging, Protein–Protein interaction analysis, and protein function exploration. Front. Immunol. 2017;8:771. doi: 10.3389/fimmu.2017.00771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jaakkonen A. Master’s Thesis. Aalto University; Espoo, Finland: 2012. [(accessed on 8 April 2020)]. Production of Antibody Fc Fusion Proteins by Intein-Mediated Splicing. INSSI record number: 44596. Available online: http://web.lib.aalto.fi/en/oa/db/INSSI/ [Google Scholar]
- 6.Strohl W.R. Current progress in innovative engineered antibodies. Protein Cell. 2018;9:86–120. doi: 10.1007/s13238-017-0457-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ridgway J.B., Presta L.G., Carter P. ‘Knobs-into-holes’ engineering of antibody CH3 domains for heavy chain heterodimerization. Protein Eng. 1996;9:617–621. doi: 10.1093/protein/9.7.617. [DOI] [PubMed] [Google Scholar]
- 8.Li F., Vijayasankaran N., Shen A.Y., Kiss R., Amanullah A. Cell culture processes for monoclonal antibody production. MAbs. 2010;2:466–479. doi: 10.4161/mabs.2.5.12720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nemoto N., Kumachi S., Arai H. In vitro selection of single-domain antibody (VHH) using cDNA Display. In: Nevoltris D., Chames P., editors. Methods in Molecular Biology. Humana Press; New York, NY, USA: 2018. p. 1827. Antibody Engineering. [DOI] [PubMed] [Google Scholar]
- 10.Sedykh S.E., Prinz V.V., Buneva V.N., Nevinsky G.A. Bispecific antibodies: Design, therapy, perspectives. Drug Des. Devel Ther. 2018;12:195–208. doi: 10.2147/DDDT.S151282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirata R., Ohsumk Y., Nakano A., Kawasaki H., Suzuki K., Anraku Y. Molecular structure of a gene, VMA1, encoding the catalytic subunit of H(+)-translocating adenosine triphosphatase from vacuolar membranes of Saccharomyces cerevisiae. J. Biol. Chem. 1990;265:6726–6733. [PubMed] [Google Scholar]
- 12.Gogarten J.P., Senejani A.G., Zhaxybayeva O., Olendzenski L., Hilario E. Inteins: Structure, function, and evolution. Annu. Rev. Microbiol. 2002;56:263–287. doi: 10.1146/annurev.micro.56.012302.160741. [DOI] [PubMed] [Google Scholar]
- 13.Perler F.B., Davis E.O., Dean G.E., Gimble F.S., Jack W.E., Neff N., Noren C.J., Thorner J., Belfort M. Protein splicing elements: Inteins and exteins--a definition of terms and recommended nomenclature. Nucleic Acids Res. 1994;22:1125–1127. doi: 10.1093/nar/22.7.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aranko A.S., Wlodawer A., Iwaï H. Nature’s recipe for splitting inteins. Protein Eng. Des. Sel. 2014;27:263–271. doi: 10.1093/protein/gzu028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wood D.W., Camarero J.A. Intein applications: From protein purification and labeling to metabolic control methods. J. Biol. Chem. 2014;289:14512–14519. doi: 10.1074/jbc.R114.552653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ciragan A., Aranko A.S., Tascon I., Iwaï H. Salt-inducible protein splicing in cis and trans by inteins from extremely halophilic archaea as a novel protein-engineering tool. J. Mol. Biol. 2016;428:4573–4588. doi: 10.1016/j.jmb.2016.10.006. [DOI] [PubMed] [Google Scholar]
- 17.Aranko A.S., Oeemig J.S., Zhou D., Kajander T., Wlodawer A., Iwaï H. Structure-based engineering and comparison of novel split inteins for protein ligation. Mol. Biosyst. 2014;10:1023–1034. doi: 10.1039/C4MB00021H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han L., Chen J., Ding K., Zong H., Xie Y., Jiang H., Zhang B., Lu H., Yin W., Gilly J., et al. Efficient generation of bispecific IgG antibodies by split intein mediated protein trans-splicing system. Sci. Rep. 2017;7:8360. doi: 10.1038/s41598-017-08641-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haga N., Asano R., Nakazawa H., Hattori T., Takeda D., Sugiyama A., Kurotani R., Kumagai I., Umetsu M., Makabe K. Generation of camelid VHH bispecific constructs via in-cell intein-mediated protein trans-splicing. Protein Eng. Des. Sel. 2017;30:15–21. doi: 10.1093/protein/gzw057. [DOI] [PubMed] [Google Scholar]
- 20.Oeemig J.S., Aranko A.S., Djupsjöbacka J., Heinämäki K., Iwaï H. Solution structure of DnaE intein from Nostoc punctiforme: Structural basis for the design of a new split intein suitable for site-specific chemical modification. FEBS Lett. 2009;583:1451–1456. doi: 10.1016/j.febslet.2009.03.058. [DOI] [PubMed] [Google Scholar]
- 21.Iwai H., Züger S., Jin J., Tam P.H. Highly efficient protein trans-splicing by a naturally split DnaE intein from Nostoc punctiforme. Febs Lett. 2006;580:1853–1858. doi: 10.1016/j.febslet.2006.02.045. [DOI] [PubMed] [Google Scholar]
- 22.Aranko A.S., Züger S., Buchinger E., Iwaï H. In vivo and in vitro protein ligation by naturally occurring and engineered split DnaE inteins. PLoS ONE. 2009;4:e5185. doi: 10.1371/journal.pone.0005185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Udaka S., Yamagata H. High-level secretion of heterologous proteins by Bacillus brevis. Methods Enzym. 1993;217:23–33. doi: 10.1016/0076-6879(93)17053-8. [DOI] [PubMed] [Google Scholar]
- 24.Takagi H., Kadowaki K., Udaka S. Screening and characterization of protein-hyperproducing bacteria without detectable exoprotease activity. Agric. Biol. Chem. 1989;53:691–699. [Google Scholar]
- 25.Bies C., Lehr C.M., Woodley J.F. Lectin-mediated drug targeting: History and applications. Adv. Drug Deliv. Rev. 2004;56:425–435. doi: 10.1016/j.addr.2003.10.030. [DOI] [PubMed] [Google Scholar]
- 26.Akkouh O., Ng T.B., Singh S.S., Yin C., Dan X., Chan Y.S., Pan W., Cheung R.C. Lectins with anti-HIV activity: A review. Molecules. 2015;20:648–668. doi: 10.3390/molecules20010648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McFeeters R.L., Xiong C., O’Keefe B.R., Bokesch H.R., McMahon J.B., Ratner D.M., Castelli R., Seeberger P.H., Byrd R.A. The novel fold of scytovirin reveals a new twist for antiviral entry inhibitors. J. Mol. Biol. 2007;369:451–461. doi: 10.1016/j.jmb.2007.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiong C., O’Keefe B.R., Botos I., Wlodawer A., McMahon J.B. Overexpression and purification of scytovirin, a potent, novel anti-HIV protein from the cultured cyanobacterium Scytonema varium. Protein. Expr. Purif. 2006;46:233–239. doi: 10.1016/j.pep.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 29.Hamorsky K.T., Kouokam J.C., Dent M.W., Grooms T.N., Husk A.S., Hume S.D., Rogers K.A., Villinger F., Morris M.K., Hanson C.V., et al. Engineering of a lectibody targeting high-mannose-type glycans of the HIV envelope. Mol. Ther. 2019;2:2038–2052. doi: 10.1016/j.ymthe.2019.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim H., Matsunaga C., Yoshino A., Kato K., Arata Y. Dynamical structure of the hinge region of immunoglobulin G as studied by 13C nuclear magnetic resonance spectroscopy. J. Mol. Biol. 1994;236:300–309. doi: 10.1006/jmbi.1994.1136. [DOI] [PubMed] [Google Scholar]
- 31.Verma R., Boleti E., George A.J. Antibody engineering: Comparison of bacterial, yeast, insect and mammalian expression systems. J. Immunol. Methods. 1998;216:165–181. doi: 10.1016/S0022-1759(98)00077-5. [DOI] [PubMed] [Google Scholar]
- 32.Gong R., Vu B.K., Feng Y., Prieto D.A., Dyba M.A., Walsh J.D., Prabakaran P., Veenstra T.D., Tarasov S.G., Ishima R., et al. Engineered human antibody constant domains with increased stability. J. Biol. Chem. 2009;284:14203–14210. doi: 10.1074/jbc.M900769200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu D., Cocco M.J., Rosenfied R., Lewis J.K., Ren D., Li L., Remmele R.L., Jr., Brems D.N. Assignment of backbone 1H, 13C and 15N resonances of human IgG1 Fc (51.4 kDa) Biomol. Nmr. Assign. 2007;1:233–235. doi: 10.1007/s12104-007-9065-5. [DOI] [PubMed] [Google Scholar]
- 34.Yashiro K., Lowenthal J.W., O’Neil T.E., Ebisu S., Takagi H. High-level production of recombinant chicken interferon-g by Brevibacillus choshinensis. Protein. Expr. Purif. 2001;23:113–120. doi: 10.1006/prep.2001.1481. [DOI] [PubMed] [Google Scholar]
- 35.Takara Brevibacillus Expression System. Takara Bio. Inc.; Shiga, Japan: 2015. Product Manual. [Google Scholar]
- 36.Züger S., Iwai H. Intein-based biosynthetic incorporation of unlabeld protein tags into isotopically labeled proteins for NMR studies. Nat. Biotech. 2005;23:736–740. doi: 10.1038/nbt1097. [DOI] [PubMed] [Google Scholar]
- 37.Muona M., Aranko A.S., Raulinaitis V., Iwaï H. Segmental isotopic labelling of multi-domain and fusion proteins by protein trans-splicing in vivo and in vitro. Nat. Prot. 2010;5:574–587. doi: 10.1038/nprot.2009.240. [DOI] [PubMed] [Google Scholar]
- 38.Mitchell C.A., Ramessar K., O’Keefe B.R. Antiviral lectins: Selective inhibitors of viral entry. Antivir. Res. 2017;142:37–54. doi: 10.1016/j.antiviral.2017.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frenzel A., Hust M., Schirrmann T. Expression of recombinant antibodies. Front. Immunol. 2013;4:217. doi: 10.3389/fimmu.2013.00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kunert R., Reinhart D. Advances in recombinant antibody manufacturing. Appl. Microbiol. Biotechnol. 2016;100:3451–3461. doi: 10.1007/s00253-016-7388-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mohlmann S., Bringmann P., Greven S., Harrenga A. Site-specific modification of ED-B-targeting antibody using intein-fusion technology. BMC Biotechnol. 2011;11:76. doi: 10.1186/1472-6750-11-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hofmann T., Schmidt J., Ciesielski E., Becker S., Rysiok T., Schütte M., Toleikis L., Kolmar H., Doerner A. Intein mediated high throughput screening for bispecific antibodies. MAbs. 2020;12:e1731938. doi: 10.1080/19420862.2020.1731938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Busche A.E.L., Aranko A.S., Talebzadeh-Farooji M., Bernhard F., Dötsch V., Iwaï H. Segmental isotopic labelling of a central domain in a multi-domain protein by protein trans-splicing using only one robust DnaE intein. Angew. Chem. Int. Ed. 2009;48:6128–6131. doi: 10.1002/anie.200901488. [DOI] [PubMed] [Google Scholar]
- 44.Garrison A.R., Giomarelli B.G., Lear-Rooney C.M., Saucedo C.J., Yellayi S., Krumpe L.R., Rose M., Paragas J., Bray M., Olinger G.G., et al. The cyanobacterial lectin scytovirin displays potent in vitro and in vivo activity against Zaire Ebola virus. Antivir. Res. 2014;112:1–7. doi: 10.1016/j.antiviral.2014.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li G., De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV) Nat. Rev. Drug Discov. 2020;19:149–150. doi: 10.1038/d41573-020-00016-0. [DOI] [PubMed] [Google Scholar]
- 46.Moulaei T., Alexandre K.B., Shenoy S.R., Meyerson J.R., Krumpe L.R., Constantine B., Wilson J., Buckheit R.W., Jr., McMahon J.B., Subramaniam S., et al. Griffithsin tandemers: Flexible and potent lectin inhibitors of the human immunodeficiency virus. Retrovirology. 2015;12:6. doi: 10.1186/s12977-014-0127-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perler F.B. InBase: The Intein Database. Nucleic Acids Res. 2002;30:383–384. doi: 10.1093/nar/30.1.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Okamoto A., Kosugi A., Koizumi Y., Yanagida F., Udaka S. High-efficiency transformation of Bacillus brevis by electroporation. Biosci. Biotechnol. Biochem. 1997;61:202–203. doi: 10.1271/bbb.61.202. [DOI] [PubMed] [Google Scholar]
- 49.Guerrero F., Ciragan A., Iwaï H. Tandem SUMO fusion vectors for improving soluble protein expression and purification. Protein Expr. Purif. 2015;116:42–49. doi: 10.1016/j.pep.2015.08.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.