Abstract
Eucalyptus leaf polyphenols extract (EPE) has been proved to have various bioactivities, but few reports focus on its antioxidant mechanism in vivo. The purpose of this study was to elucidate the effect and mechanism of EPE dietary supplements on antioxidant capacity in chicken. A total of 216 chickens were randomly selected for a 40-day experiment. Four treatment groups received diets including the control diet only, the control diet + low EPE (0.6 g/kg), the control diet + moderate EPE (0.9 g/kg), and the control diet + high EPE (1.2 g/kg). Compared with control group, the glutathione peroxidase (GSH-Px) activity and glutathione (GSH) content in the breast muscle of the moderate EPE treatment group was significantly higher (p < 0.05), while the malonaldehyde (MDA) content in the moderate EPE group was reduced (p < 0.05). Moreover, proteomic and transcriptomic analyses of the breast muscle revealed that glutathione metabolism and the peroxisome were the two crucial metabolic pathways responsible for increased antioxidant capacity of the muscle. Accordingly, nine candidate genes and two candidate proteins were identified related to improved antioxidant status induced by EPE supplements. This research provides new insights into the molecular mechanism of antioxidant capacity in chickens treated with EPE dietary supplements.
1. Introduction
The genus Eucalyptus comprises over 900 species of economically valuable plants endemic to Australia [1, 2]. In China, the eucalyptus plantation area reached 4.5 million hm2 by the end of 2016 [3]. There is a growing need to develop alternative uses for eucalyptus products, and eucalyptus plants have attracted broad interest among researchers to investigate their functionality and potential applications [4–6]. Eucalyptus leaves are known to contain numerous bioactive substances including, but not limited to, terpenoids, tannins, flavonoids, and phloroglucinol derivatives, and eucalyptus has also been shown to function as antioxidants [7, 8]. Accordingly, bioactive components derived from eucalyptus leaves and their functions for use in the food industry are widely researched, with many studies focused on their antioxidant capacity [9–11].
Recent technological advances in proteomics and transcriptomics have substantially improved the understanding of how variation in proteins and genes impact living organisms [12–15]. In particular, these methods may be used to elucidate the molecular mechanism of antioxidant capacity. Using proteomic techniques, Wang et al. [16] discovered that broiler chickens given albusin B supplements could upregulate the expression of GST, Prdx6, PPIA, alfatoxin aldehyde reductase, and superoxide dismutase to improve antioxidant defense.
Our previous study revealed that EPE exhibited a strong antioxidant activity in chemical-based and cellular-based assay [7]. Moreover, EPE treatment could protect acute-induced oxidative damage by improving antioxidant enzymes (GSH-Px, T-SOD) in chicken [7]. Several researches also reported that diet with eucalyptus leaves extract significantly improves antioxidant status [17, 18]. Eucalyptus leaves extracts with immense biomass and great antioxidant potential benefited to improve growth performance and health status of broilers, which could be useful for the poultry industry [19, 20]. However, few studies have investigated the antioxidant mechanism of EPE in chicken. Therefore, this study was performed to reveal EPE dietary supplements on the antioxidant mechanism of chicken using proteomic and transcriptomic analysis.
2. Materials and Methods
2.1. Experimental Material
All the Huiyang beard chickens used in this study were provided by Xingtai Modern Agricultural Limited Company of Huizhou in Guangdong, China.
EPE: Eucalyptus leaves (Eucalyptus grandis × Eucalyptus urophylla GL9) were picked in October in Zhanjiang and air-dried naturally (7~9 d, 28 ± 2°C, moisture content 15.53 ± 2.11%). The eucalyptus leaves were extracted by 70% ethanol solvent (v/v) at 75°C. And then the extraction solution was vacuum-concentrated and spray-dried (the yield of EPE was 25.78 ± 2.03%), the EPE was stored at 4°C for the subsequent experiment.
The total polyphenol content in EPE was determined using the Folin-phenol method [21]. Additionally, the EPE was analyzed using high-performance liquid chromatography (HPLC) with a Diamonsil C18 column (250 × 4.6 mm, 5 μm, Diamonsil, China). EPE was reconstituted in buffer A (H2O) and loaded onto the column. It was then eluted using gradient buffer B (Methanol) in the following order: 10% at 0 min and 10~90% at 60 min. The flow rate was 1 mL/min and the injection volume was 10 μL. The column temperature was 37°C and a detection wavelength of 270 nm.
2.2. Animal Experimental Design
A total of 216 chickens (90 days old) were randomly assigned to one of four treatments (6 pens/treatment and 6 chicken/pen), and the duration of the trial was 40 days (from 90 to 130 days). Four treatment groups received diets including control diet only (Table 1), low EPE (control diet+0.6 g/kg EPE), moderate EPE (control diet+0.9 g/kg EPE), and high EPE (control diet+1.2 g/kg EPE). The experimental design and procedures were approved by following the requirements of the Regulations for the Administration of Affairs Concerning Experimental Animals of China.
Table 1.
Ingredients and nutrient composition of the experimental diets.
| Ingredients (%) | Content | Nutrient level | Content |
|---|---|---|---|
| Corn | 64 | Metabolic energy (mcal∕kg) | 2.96 |
| Wheat shorts | 2 | Crude protein (%) | 15.5 |
| Rice bran | 3 | Lysine (%) | 0.65 |
| Bean pulp | 9.7 | Calcium (%) | 1.19 |
| Peanut bran | 4 | Phosphorus (%) | 0.57 |
| Corn gluten meal | 2 | Sodium (%) | 0.206 |
| Refined three worm powder CP45% | 6 | Chlorine (%) | 0.196 |
| Rock flour | 1.5 | Potassium (%) | 0.47 |
| Calcium hydrophosphate | 1.3 | Methionine (%) | 0.25 |
| Soybean oil | 2.5 | Arginine (%) | 0.83 |
| 338 gunk | 4 | ||
| Total | 100 |
Note: the vitamin/mineral premix includes (per kg feed): vitamin A, 15750 IU; vitamin D, 3500 IU; vitamin E 35 mg; Menadione, 4.4 mg; Thiamine, 3.5 mg; Riboflavin, 10.5 mg; vitamin B6, 7 mg; vitamin B12, 35 mg; Nicotinic acid, 70 mg; Pantothenic acid, 21 mg; Folic acid, 1.75 mg; Biotin, 0.175 mg.
2.3. Experimental Basic Fodder and Management
Ingredients and nutrient compositions of the diets are shown in Table 1, and the diets were used throughout the whole experimental period. The chickens were raised in cages in a controlled environment and had free access to food and drinking water. The temperature in the house was 24~29°C, and the relative humidity was 60~78%. Pens were routinely disinfected to maintain appropriate standards of cleanliness. All animal procedures were conducted under the protocol (SCAU-AEC-2010-0416) approved by the Animal Ethics Committee of South China Agricultural University.
2.4. Sample Collection
After 40 days, 12 chickens were randomly selected from each treatment group (2 broiler chickens from each replication), and blood samples were drawn from the neck vein with a sterile syringe. The blood was centrifuged at 3000 × g for 15 min to obtain serum samples, and they were stored at -80°C until analysis. Chickens were then euthanized, and the whole breast muscle tissues were preserved in liquid nitrogen.
2.5. Antioxidant Activity in Serum and Muscle Tissue
About 0.25 g muscle tissue was mixed with 9 times (m/v) 0.9% physiological saline, and then, homogenized with a tissue grinder (Lawson, Japan) at 4°C. After being centrifuged for 10 min at 4000 r/min (Eppendorf, Germany), the supernatant was used for subsequent analysis. The activities of total superoxide dismutase (T-SOD, A001-1-2), GSH-Px (A005-1-2), total antioxidant capacity (T-AOC, A015-1-2), the contents of malondialdehyde (MDA, A003-1-2), and GSH (A061-1-1) in serum and breast muscle were determined spectrophotometrically using the commercial kits obtained from Nanjing Jiancheng Institute of Bioengineering (Nanjing, Jiangsu, China) [22]. The quantities of T-SOD, GSH-Px, and T-AOC are expressed as units (U) per milligram of protein. The MDA and GSH content are expressed as nanomoles per milligram of protein and milligram per gram of protein, respectively.
2.6. Screening and Annotation of Differentially Expressed Genes (DEGs) in Muscle Induced by EPE Dietary Supplements in Chicken Based on RNA-Seq Techniques
Based on the evaluation of antioxidant activity in this study and a previous study [23], the moderate EPE treatment group tended to exhibit higher antioxidant effects compared with the other group. Therefore, breast muscles were obtained from the control group and the moderate EPE treatment group (0.9 g/kg) for transcriptome sequencing, with two replicates per group [24].
2.6.1. Total RNA Isolation, Labeling, and Sequencing
Total RNA was extracted from muscle samples. RNA concentration and purity were evaluated by Nanodrop 2000 (Thermo, USA), and RNA integrity was determined by 1% agarose gel electrophoresis. Then, mRNA was isolated from the total RNA using Oligo (dT) beads. Fragmentation buffer was used to randomly shear mRNA into 200 bp fragments. Using reverse transcriptase, random hexamers were added to synthesize a strand of cDNA from the mRNA template, followed by two-strand synthesis to form a stable double-stranded structure. The viscous end of the double-stranded cDNA structure was repaired using End Repair Mix (Enzymatics, USA), followed by the addition of an A base at the 3′ end to form the Y-form linker. The final library was sequenced on an Illumina Hiseq (Hokkaido System Science, Sapporo, Japan) [25, 26].
2.6.2. Data Analysis and Bioinformatics of Genes
Chicken transcriptome sequencing was carried out using an Illumina sequencing platform (2 × 150 bp, 375 bp insert size). Quality control of the sequenced data was completed, and the transcriptome data were analyzed using an established bioinformatics method [27]. DEGs were calculated based on gene read count data by the edgeR software, and the screening criteria for DEGs were False discovery rate (FDR) < 0.05, p value < 0.05 and fold change (FC) > 2. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) were used to annotate and enrich the DEGs.
2.7. Screening and Annotation of Differentially Expressed Proteins (DEPs) in Muscle Induced by EPE Dietary Supplements in Chicken Based on iTRAQ Techniques
The breast muscle was obtained from the control group and the moderate (0.9 g/kg) EPE group for proteomic analysis, with two replicates per group [28].
2.7.1. Two-Dimensional Fluorescence Difference Gel Electrophoresis
About 15 mg of breast muscle was manually ground with liquid nitrogen, and then, 8 M urea containing 1% sodium dodecyl sulfate (Sinopharm Chemical Reagent Co., Ltd, China) and protease inhibitor were added in a 1 : 5 ratio. The solution was placed on ice in an ultrasound wave for 2 min and then centrifuged at 4°C for 20 min. The supernatant was taken to determine the protein concentration via gel electrophoresis. A 100 microgram aliquot of protein sample was dissolved with 100 μl 8 M urea containing 1% sodium dodecyl sulfate, and then, 10 mM TCEP solution (Thermo, USA) was added to the sample and incubated at 37°C for 60 min. 40 mM iodoacetamide (Sigma, USA) was then added, and the solution was incubated at room temperature for 40 min. Precooled acetone (Sinopharm Chemical Reagent Co., Ltd, China) was added in a 6 : 1 ratio (acetone : sample). Samples were precipitated at -20°C for 4 h, followed by centrifugation at 10000 g for 20 min. Then, samples were digested overnight at 37°C with 100 μL 100 mM triethylammonium bicarbonate buffer (TEAB, Sigma, USA), and trypsin was added according to a mass ratio of 1 : 25 (enzyme : protein) [29].
2.7.2. Identification of DEPs
Muscle was digested with trypsin, and the peptide was dried with a vacuum pump and redissolved with a 0.4 M TEAB solution. The iTRAQ reagent (AB Sciex, USA) was added per 100 μg of peptide, and samples were incubated for 2 h, followed by adding 50 μL ultrapure water and incubating for 30 min. Each group of labeled product was mixed and dried. The peptide samples were reconstituted with UPLC loading buffer and separated by C18 column during the pH liquid phase.
The first-dimensional separation was performed using Waters' ACQUITY UPLC BEH C18 column (3 mm × 150 mm, 1.7 μm, Waters, USA) with a flow rate of 400 μL/min. The detection wavelength was 214 nm. The labeled peptides were reconstituted in buffer A (2% acetonitrile, pH 10.0) and loaded onto the column. The peptides were then eluted at 37°C using gradient buffer B (80% acetonitrile, pH 10.0) in the following order: 0% in 2 min, 0~3.8% in 15 min, 3.8~24% in 18 min, 24~30% in 3 min, 30~43% in 1 min, 43~100% in 1 min, 100~0% in 6 min, and 0% keeping for 20 min.
The second-dimensional separation was performed using Liquid-mass spectrometry. The chromatographic instrument was EASY-nLC 1200 (75 μm × 25 cm, Thermo, USA), the mass spectrometer was Q-Exactive (Thermo, USA), and the data acquisition software was Thermo Xcalibur 4.0 (Thermo, USA). The samples were reconstituted in buffer A (2% acetonitrile with 0.1% formic acid) and loaded onto the column. The peptides were then eluted at 37°C using gradient buffer B (80% acetonitrile with 0.1% formic acid) in the following order: 0~5% in 1 min, 5~23% in 62 min, 23~48% in 25 min, 48~100% in 1 min, 100% keeping for 6 min, 100~0% in 5 min, and 0% keeping for 20 min. Mass spectrometry conditions were as follows: MS scan range (m/z) 350-1300, acquisition mode DDA; Top 20 (select the strongest signal in the parent ion 20 for secondary fragmentation); first-order mass spectrometry resolution of 70000, fragmentation HCD; Resolution 17500, dynamic exclusion time 18 s.
2.7.3. Bioinformatic Analysis of Proteins
The GO analysis and the KEGG pathways analysis were implemented in KOBAS [30], and significance was evaluated using Fisher's exact test.
2.8. Statistical Analysis
Data were analyzed by one-way ANOVA (SPSS, 2010). Differences among treatment groups were evaluated using Duncan's multiple range tests, with a significance threshold of p < 0.05.
3. Results
3.1. Quality of EPE
According to the Folin-reagent method described by Wang et al. (Gallic acid as the equivalent polyphenol) [21], the total polyphenols content in EPE was 317.08 ± 16.49 mg/g. Further, HPLC of EPE (Figure 1) indicated that the main active substance was of comparable quality to the EPE reported in our previous research, based on the standard substance retention time [23].
Figure 1.
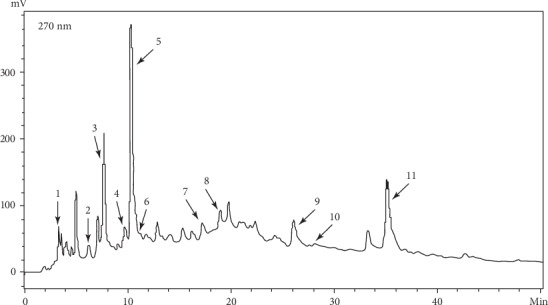
High-performance liquid chromatography of EPE. Note: 1 Gemin D; 2, 4 Pedunculagin; 3 Gallic acid; 5 Oenothein B; 6,7 TellimagrandinI; 8 Chlorongenic acid; 9 Ethyl gallate; 10 1,2,3,4,6-O-pentagalloylglucose; 11 Hyperoside.
3.2. Variation of Antioxidant Capacity in Serum
As shown in Table 2, the concentrations of GSH-Px, T-SOD, T-AOC, and MDA in serum were not significantly different among the treatment groups (p > 0.05). Compared with the control group, the T-SOD activity and T-AOC increased in the EPE treatment groups, and the GSH-Px activity in the moderate EPE group increased (0.9 g/kg) by 13.8% (p > 0.05).
Table 2.
Effect of EPE dietary supplements on antioxidant capacity of serum (n = 12).
| Item | Control group | 0.6 g/kg EPE group | 0.9 g/kg EPE group | 1.2 g/kg EPE group |
|---|---|---|---|---|
| T-SOD (U/mgprot) | 69.03 ± 1.43 | 69.23 ± 1.63 | 70.99 ± 1.96 | 69.48 ± 1.70 |
| T-AOC (U/mgprot) | 11.43 ± 0.66 | 11.44 ± 0.57 | 11.56 ± 0.46 | 11.81 ± 0.63 |
| GSH-Px (U/mgprot) | 775.61 ± 45.28 | 803.12 ± 38.08 | 815.64 ± 25.76 | 882.27 ± 57.32 |
| MDA (nmol/mL) | 2.45 ± 0.20 | 2.13 ± 0.28 | 1.96 ± 0.25 | 2.12 ± 0.21 |
3.3. Variation of Antioxidant Capacity in Muscle Tissue
As shown in Table 3, the T-SOD activity and T-AOC of the breast muscle in the moderate EPE (0.9 g/kg) and high EPE (1.2 g/kg) groups showed an increasing trend compared to the control group (p > 0.05). The GSH-Px activity of the breast muscle tissue in the moderate (0.9 g/kg) EPE group was 44.5%, higher than that of the control group (p < 0.05), and there was an upward trend in the low (0.6 g/kg) and high (1.2 g/kg) EPE groups (p > 0.05). The GSH content of breast muscle in the moderate and high EPE groups increased by 23.3% and 20.7% compared with the control group (p < 0.05). Additionally, MDA content in the breast muscle of the moderate (0.9 g/kg) EPE group was reduced by 25.4% (p < 0.05), though the other two groups did not show a significant change (p > 0.05).
Table 3.
Effect of EPE dietary supplements on antioxidant capacity of breast muscle (n = 12).
| Item | Control group | 0.6 g/kg EPE group | 0.9 g/kg EPE group | 1.2 g/kg EPE group |
|---|---|---|---|---|
| T-SOD (U/mgprot) | 28.31 ± 5.60 | 27.48 ± 5.63 | 33.39 ± 7.82 | 30.12 ± 4.41 |
| T-AOC (U/mgprot) | 7.43 ± 0.81 | 7.42 ± 1.45 | 8.39 ± 0.98 | 8.96 ± 1.75 |
| GSH-Px (U/mgprot) | 127.77 ± 24.61b | 133.64 ± 30.06ab | 184.63 ± 38.69a | 159.26 ± 33.58ab |
| GSH (mg/gprot) | 3.47 ± 0.25b | 3.73 ± 0.21ab | 4.28 ± 0.31a | 4.19 ± 0.13a |
| MDA (nmol/mgprot) | 13.74 ± 1.79b | 11.26 ± 0.93ab | 10.25 ± 1.81a | 11.65 ± 1.50ab |
a-bMeans within a row with different superscripts differ significantly (p < 0.05).
3.4. Screening and Annotation of DEGs in Muscle Induced by EPE Dietary Supplements in Chicken Based on RNA-Seq Techniques
To ensure high quality data, raw sequencing reads were filtered based on coverage, sequence saturation, the redundant distribution frequency map, and the distribution of sequencing reads across the chromosomes. A total of 14621 genes were identified as differentially regulated, of which 289 were significantly different between the EPE treatment group and the control group. The overall distribution of gene expression differences was visualized by a volcanic map (Figure 2).
Figure 2.
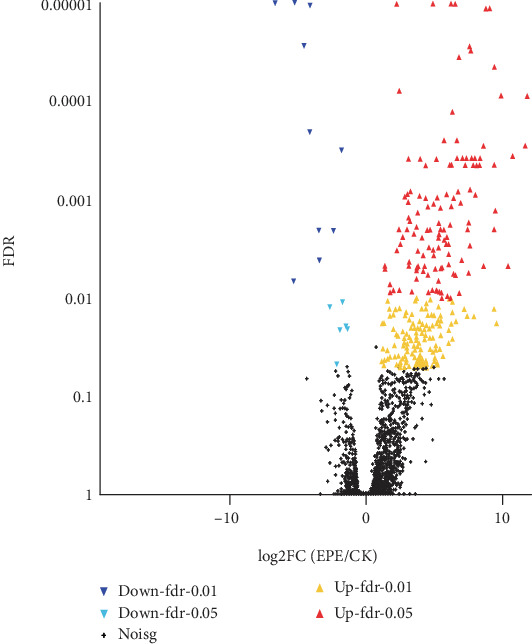
Volcano plots of DEGs. Note: differentially expressed genes (DEGs). The abscissa is the fold change of the gene's expression difference. The ordinate is the statistical test value of the difference in the amount of gene arrival, and the higher the p value, the more significant the difference in expression. Red dots indicate significantly upregulated genes, blue dots indicate significantly down-regulated genes, and black dots are nonsignificant differentially expressed genes.
3.4.1. Gene Ontology Enrichment Analysis of the DEGs
Gene Ontology (GO) enrichment analysis was used to annotate these DEGs by biological process, cellular component, and molecular function (Figure 3). Overall, 47 different biological processes were enriched, including oxygen transport, oxidation-reduction, aminophospholipid transport, phospholipid translocation, and lipid translocation. Cellular component analysis revealed that the apical plasma membrane, membrane region, membrane part, cellular component, integral component of membrane, plasma membrane region, brush border, hemoglobin complex, and substrate-specific transporter activity were significantly enriched. Additionally, oxygen binding, oxygen transporter activity, phospholipid-translocating ATPase activity, and other 22 molecular function items were identified by the molecular function analysis.
Figure 3.
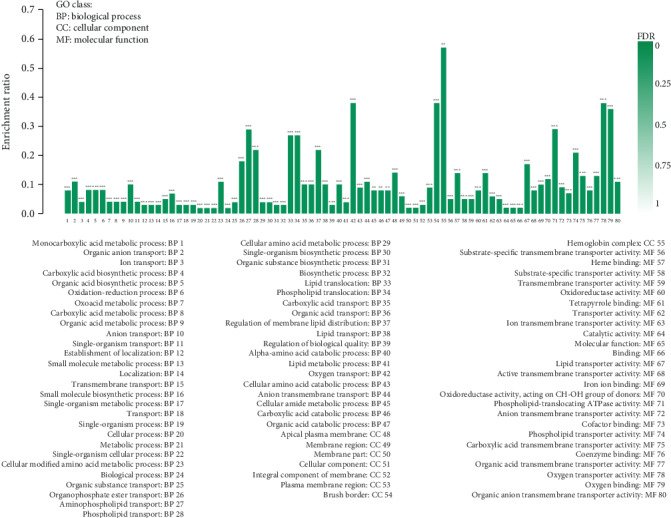
GO enrichment histogram for DEGs. Note: differentially expressed genes (DEGs). Each column in the figure is a GO term, and the abscissa text indicates the name and classification of the GO term. The height of the column, that is, the ordinate, indicates the enrichment rate. The color indicates the significance of enrichment, that is, FDR. The darker the color, the more significant the enrichment of the GO term, wherein the mark with FDR < 0.001 is ∗∗∗, the mark with FDR < 0.01 is ∗∗, and the mark with FDR < 0.05 is ∗, the right color gradient indicates the FDR size.
Based on the above GO enrichment analysis, glutathione metabolic process (biological process), glutathione transferase activity (molecular function), and peroxisome (cellular component) were the enriched GO terms most clearly associated with antioxidant capacity (Table 4). Overall, 4 antioxidant genes (gamma-glutamyltransferase 1, GGT1; microsomal glutathione S-transferase 1, MGST1; glutathione S-transferase alpha 4, GSTA4L; and glutathione S-transferase class-alpha, GSTAL1) in the EPE group were substantially enhanced in glutathione metabolic process relative to the control group. Similarly, three of these genes (MGST1, GSTA4L, and GSTAL1) were also associated with glutathione metabolic process and glutathione transferase activity, which play an important role in muscle antioxidant activity. Additionally, hydroxyacid oxidase 1 (HAO1), hydroxyacid oxidase 2 (HAO2), and bile acid-CoA: amino acid N-acyltransferase (BAAT) are peroxisome genes known to be involved in the oxidation of fatty acids, regulation of oxygen concentration, and decomposition of hydrogen peroxide to improve the antioxidant status in breast muscle.
Table 4.
GO enrichment analysis for DEGs related antioxidant capacity.
| GO id | Description of GO enrichment | Seq id | p value | Regulate | Description | Symbol name |
|---|---|---|---|---|---|---|
| GO:0006749 | Glutathione metabolic process | ENSGALG00000006565 | <0.01 | Up | Gamma-glutamyltransferase 1 | GGT1 |
| GO:0006749/GO:0016705 | Glutathione metabolic process/glutathione transferase activity | ENSGALG00000013098 | <0.01 | Up | Microsomal glutathione S-transferase 1 | MGST1 |
| ENSGALG00000016324 | <0.01 | Up | Glutathione S-transferase alpha 4-like | GSTA4L | ||
| ENSGALG00000038652 | <0.01 | Up | Glutathione S-transferase class-alpha-like 1 | GSTAL1 | ||
| GO:0005777 | Peroxisome | ENSGALG00000008845 | <0.01 | Up | Hydroxyacid oxidase 1 | HAO1 |
| ENSGALG00000014766 | <0.01 | Up | Hydroxyacid oxidase 2 | HAO2 | ||
| ENSGALG00000040619 | <0.01 | Up | Bile acid-CoA: amino acid N-acyltransferase | BAAT |
Note: differentially expressed genes (DEGs). The screening criteria for significantly GO enrichment analysis were p < 0.05.
3.4.2. Kyoto Encyclopedia of Genes and Genomes (KEGG) Analysis of the DEGs
DEGs were enriched in 148 pathways in the KEGG enrichment analysis, and 38 KEGG pathways were identified to have significant changes. Based on the previous analysis of physiological indicators, the peroxisomes and the glutathione metabolism pathway were selected for further evaluation of the antioxidant capacity of the muscle by KEGG enrichment metabolic pathway analysis. Significant DEGs that were enriched in the peroxisomes and glutathione metabolism pathway showed some overlap with the genes related to antioxidant status identified by the GO enrichment analysis, including HAO1, HAO2, and GGT1 (Table 5).
Table 5.
KEGG enrichment pathways for DEGs associated with antioxidant capacity.
| Name of KEGG pathway | KEGG ID | Number of different gene | p value | Description | Symbol name |
|---|---|---|---|---|---|
| Peroxisome | ko04146 | 6 | <0.01 | Solute carrier family 27 member 5, alanine-glyoxylate and serine--pyruvate aminotransferase, | SLC27A5, AGXT, PIPOX, HAO1, HAO2, BAAT |
| Peroxisomal sarcosine oxidase, hydroxyacid oxidase 1, hydroxyacid oxidase 2, bile acid-CoA: amino acid N-acyltransferase | |||||
| Glutathione metabolism | ko00480 | 4 | <0.05 | Gamma-glutamyltranspeptidase 1, microsomal glutathione S-transferase 1, glutathione S-transferase class-alpha, glutathione S-transferase alpha 4 | GGT1, MGST1, GSTAL1, GSTA4L |
Note: differentially expressed genes (DEGs). The screening criteria for significantly KEGG enrichment pathways were p < 0.05. Hydroxyacid oxidase 1 (HAO1); hydroxyacid oxidase 2 (HAO2); bile acid-CoA: amino acid N-acyltransferase (BAAT); microsomal glutathione S-transferase 1 (MGST1); glutathione S-transferase class-alpha (GSTAL1); glutathione S-transferase alpha 4 (GSTA4L).
3.5. Screening and Annotation of DEPs in Muscle Induced by EPE Dietary Supplements in Chicken Based on iTRAQ Techniques
3.5.1. Identification and GO Analysis of DEPs Induced by EPE
The total number of tested proteins was 1430, and there were 149 protein differences between the EPE group and the control group. According to the standard of differential protein screening, there were 14 significant differences in protein concentration (>1.2-fold change, p < 0.05), 10 of which were upregulated and 4 were downregulated (Table 6). These proteins were annotated (cellular components, molecular functions, and biological processes) by GO enrichment analysis (Figure 4). Biological process analysis revealed that most proteins were associated with 31 different GO terms, including p53 class mediator, threonyl-tRNA aminoacylation, and regulation of DNA damage checkpoint. When DEPs were annotated by the cell components, significantly enriched terms included chylomicron, plasma lipoprotein particle, very-lower-density lipoprotein particle, triglyceride-rich lipoprotein particle, protein-lipid complex, and five other cellular components terms. A total of five DEPS were annotated by molecular function, and there was significant enrichment of nineteen GO terms, including threonine-tRNA ligase activity, nutrient reservoir activity, p53 binding, and glutathione transferase activity.
Table 6.
Significantly DEPs in the proteomics analysis.
| Proteins | Description | Fold change | p value | Regulate |
|---|---|---|---|---|
| Q7LZS1 | 12K serum protein, beta-2-m cross-reactive (fragment) | 0.71 | 0.04 | Down |
| A0A1D5NXR4 | Uncharacterized protein | 0.34 | 0.003 | Down |
| F1P372 | Uncharacterized protein | 1.24 | 0.03 | Up |
| F1NG89 | Ubiquitin carboxyl-terminal hydrolase 10 | 1.20 | 0.02 | Up |
| A0A1D5PFH3 | Uncharacterized protein | 1.28 | 0.02 | Up |
| F1NHM9 | Phosphoglycerate mutase | 0.82 | 0.02 | Down |
| E1C4V1 | ATP synthase-coupling factor 6, mitochondrial | 1.20 | 0.02 | Up |
| A0A0A0MQ61 | Uncharacterized protein | 1.48 | 0.03 | Up |
| A7UEB0 | Alpha-1-acid glycoprotein | 0.69 | 0.01 | Down |
| Q5ZLR5 | Cytochrome b-c1 complex subunit Rieske, mitochondrial | 1.25 | 0.01 | Up |
| P02659 | Apovitellenin-1 | 1.41 | 0.002 | Up |
| Q5ZHZ0 | Spliceosome RNA helicase DDX39B | 1.21 | 0.05 | Up |
| Q91968 | Alpha-tropomyosin | 1.49 | 0.04 | Up |
| A0A1D5NZ55 | Uncharacterized protein | 1.30 | 0.05 | Up |
Note: differentially expressed proteins (DEPs). The screening criteria for significantly differentially expressed proteins were p < 0.05 and (FC < 0.83 or FC > 1.20).
Figure 4.
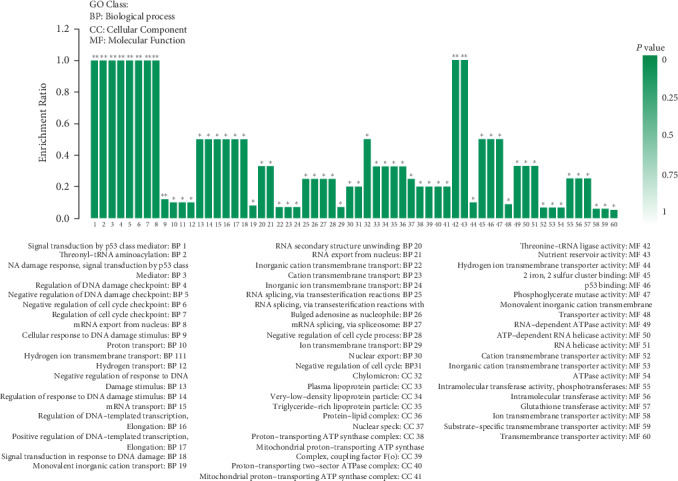
GO enrichment histogram for DEPs. Note: differentially expressed proteins (DEPs). Each column in the figure is a GO term, and the abscissa text indicates the name and classification of the GO. The height of the column, that is, the ordinate, indicates the enrichment rate. The color indicates the significance of enrichment, that is, FDR. The darker the color, the more significant the enrichment of the GO term, wherein the mark with FDR < 0.001 is ∗∗∗, the mark with FDR < 0.01 is ∗∗, and the mark with FDR < 0.05 is ∗, the right color gradient indicates the FDR size.
3.5.2. KEGG Pathway Analysis of the DEPs
DEPs were analyses by KEGG pathway analysis and the results indicated that cytochrome b-c1 complex subunit Rieske (Q5ZLR5), A0A0A0MQ61, and F1P372 were main proteins associated with oxidative phosphorylation, glutathione metabolism, aminoacyl-tRNA biosynthesis, metabolism of xenobiotics by cytochrome P450, etc. (Table 7).
Table 7.
KEGG enrichment pathways for DEPs.
| Pathway | Pathway definition | Number of proteins | Proteins |
|---|---|---|---|
| ko00190 | Oxidative phosphorylation | 1 | Q5ZLR5 (connectin/fragment) |
| ko01100 | Metabolic pathways | 1 | |
| ko04260 | Cardiac muscle contraction | 1 | |
| ko04932 | Nonalcoholic fatty liver disease (NAFLD) | 1 | |
| ko05010 | Alzheimer's disease | 1 | |
| ko05012 | Parkinson's disease | 1 | |
| ko05016 | Huntington's disease | 1 | |
| ko00480 | Glutathione metabolism | 1 | A0A0A0MQ61 (uncharacterized protein) |
| ko05204 | Chemical carcinogenesis | 1 | |
| ko00980 | Metabolism of xenobiotics by cytochrome P450 | 1 | |
| ko00982 | Drug metabolism-cytochrome P450 | 1 | |
| ko00970 | Aminoacyl-tRNA biosynthesis | 1 | F1P372 (uncharacterized protein) |
Note: differentially expressed proteins (DEPs). Q5ZLR5 (cytochrome b-c1 complex subunit Rieske, mitochondrial), A0A0A0MQ61 (uncharacterized protein), F1P372 (uncharacterized protein).
3.6. Combined Analysis of Transcriptomics and Proteomics
All proteins and their associated transcripts in both the transcriptomic and proteomic analyses were classified into nine categories (Figure 5). Of the 61 proteins and genes that were upregulated in the EPE group, the A0A0A0MQ61 protein and the glutathione S-transferase alpha-like 2 gene (GSTAL2) played a crucial role in enhancing antioxidant status. The glutathione metabolism in the transcriptomic analysis indicated that GGT1, MGST1, and GSTA4L were downstream genes regulated by glutathione, which correspond to the observed increase in GSH-Px activity in muscle tissue induced by EPE supplements. Accordingly, A0A0A0MQ61, the protein product of the glutathione S-transferase gene, was substantially upregulated in glutathione metabolism, which was in line with the expression of GGT1, MGST1, and GSTA4L (Figure 6). Therefore, EPE may enhance the antioxidant status of chicken by improving the activity of specific antioxidant proteins.
Figure 5.
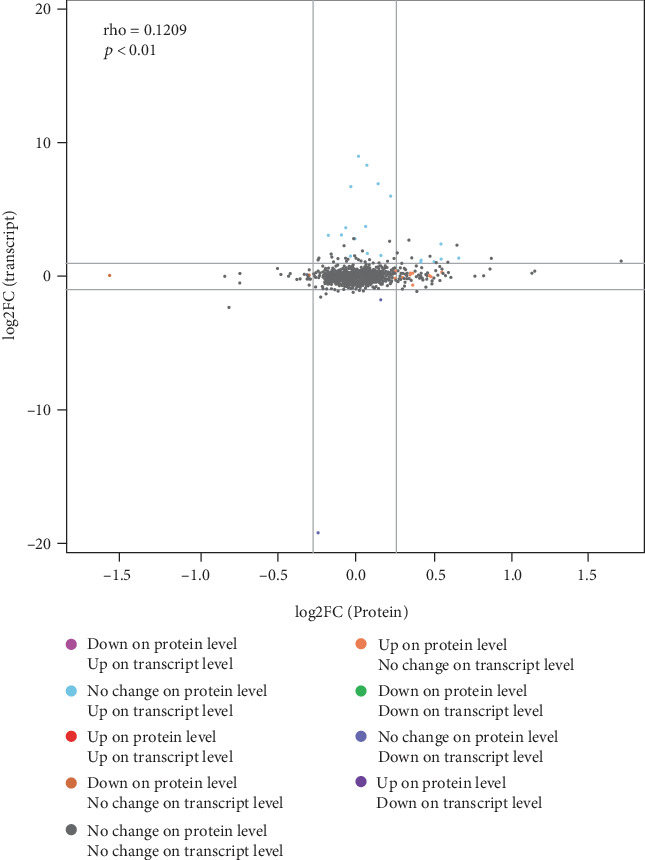
A scatter plots of the expression levels of all proteins and their associated transcripts in both groups. Note: the abscissa in the figure indicates the difference in the expression of the protein in the EPE treatment group and the control group, and the ordinate indicates the difference in the FPKM value of the corresponding transcript in the experimental group and the control group. Each point represents a protein and its associated transcript; in the upper left corner, rho represents the Pearson's correlation coefficient between the two omics, p represents the correlation test p value; when rho > 0, it is called the negative correlation; When rho < 0, it is called positive correlation; when rho = 0, it is called zero correlation, that is, there is no correlation; the larger the∣rho∣, the greater the correlation between the two omics.
Figure 6.
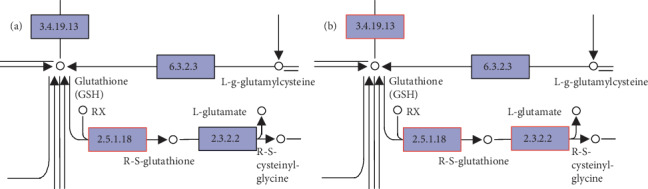
Upregulated protein in the proteome (a) and genes in the transcriptome (b) in glutathione metabolism. Note: the genes with red borders in the figure belong to the DEGs or DEPs detected by this sequencing, in which red represents the upregulated gene.
4. Discussion
Phytogenic feed additives have been gaining attention in improving the health status of the flocks [31, 32]. Our previous study revealed that diet with polyphenols obtained from eucalyptus leaves exhibited a positive effect on growth performance in laying hens [23]. In the present study, different concentrations of EPE in chicken diet did not exhibit significant positive or negative effect on chicken performance, including average daily gain, average daily feed take, and feed conversion rate (data not shown), which is in agreement with the report of Sedaghat and Torshizi [33], and might be due to that the chickens were in the adulthood.
Recently, natural polyphenols used in animal diets have been reported to build an integrated antioxidant system to prevent from damage led by free radicals [34–37]. This antioxidant capacity is assumed to result from the free radical-scavenging properties of phenolic compounds [38–40]. Our previous study reported that EPE effectively scavenged DPPH• and ABTS• free radicals in vitro [21]. In the present study, the antioxidant effects in serum were not obviously affected by the EPE supplement. However, an increasing trend in GSH-Px was observed with a higher concentration of the EPE diet, whereas the MDA content showed the opposite effects. This result was supported by the previous report that diet with 0.8 g/kg polyphenols from eucalyptus leaves increased the GSH-Px activity in serum of laying hen and an insignificant decrease in MDA was observed in quails supplement with eucalyptus leaves [23, 31]. Interestingly, our study revealed that EPE dietary supplements significantly improved GSH-Px activity and GSH content and decreased MDA content in breast muscle tissues. Higher concentration of the EPE diet led to higher GSH-Px activity, which is consistent with the report by Fathi et al. [31]. Similarly, MDA content can reflect the degree of lipid peroxidation; thereby, indirectly reflect cell damage and freshness of meat [41]. In agreement with our results, a diet with green tea extract led to a remarkably decreasing MDA content in meat tissue [42]. Polyphenols might be accumulated to exhibit antioxidant effects in meat tissues [35]. To explain the antioxidant capacity of polyphenols, it is crucial to understand how they are absorbed, metabolized, and eliminated from the body. It is stated that the intestinal utilization of polyphenols depended on their degree of polymerization and galloylation [43]. Monomeric and some oligomeric polyphenols tended to be easily absorbed at small intestine compared with the polymeric forms of polyphenols [44]. This suggested that monomeric and some oligomeric polyphenols in EPE, such as gallic acid, pedunculagin, hyperoside, and other compounds, could be absorbed and functioned as an antioxidant in chickens. This hypothesis was supported by Chamorro et al. that monomeric (catechin, epicatechin, gallic acid, and epicatechin-O-gallate) and dimeric (procyanidin B1 and procyanidin B2) catechins in grape pomace were easily digested and absorbed in chickens [45]. Overall, EPE dietary supplements are able to improve the antioxidant capacity of chicken.
The results of the transcriptomic analysis indicated that glutathione transferase activity, glutathione metabolic process, and the peroxisome were the three GO enrichment terms related to antioxidant activity. That is, several significantly upregulated antioxidant genes, such as GGT1, MGST1, GSTA4L, HAO1, and HAO2, are all believed to improve antioxidant status. Moreover, previous research indicated that GGT1 and PGDS play an important role in the synthesis of glutathione, and its upregulation exerts a prooxidant action [46]. Shinno et al. [47] reported that microsomal glutathione S-transferase 1 (MGST1) could be activated by gallic acid to protect the membrane against damage caused by oxidative stress. These results suggested that these antioxidant genes were vital to improve antioxidant status in chicken. Likewise, the KEGG pathway enrichment analysis also indicated that the peroxisomes and the glutathione metabolism pathway were the two crucial antioxidant pathways. Peroxisomes are rich in enzymes, mainly including oxidases, catalase, and peroxidase. Catalase, in particular, is known to protect cells by hydrolyzing the hydrogen peroxide generated in redox reactions [48, 49]. Additionally, several differentially expressed antioxidant-related genes were significantly enriched in the glutathione metabolism pathway, and the content of glutathione may be altered in cells as a result of EPE supplements. Glutathione is known to effectively scavenge free radicals and other reactive oxygen species, and it can be oxidized to form GSSG [50, 51]. In this study, significantly improved GSH-Px converted GSSG into GSH to enhance antioxidant status. In addition, SOD and APOA4 genes were notably identified to be upregulated in this study, though the protein products remained unchanged. They may also contribute to the antioxidation of muscle, given the known functions of SOD and the known APOA4 [52]. In the present study, the gene expression variation of superoxide dismutase was consistent with the increasing trend of SOD enzyme activity in the breast muscle in the moderate (0.9 g/kg) EPE treatment groups compared to the control group. This result suggests that EPE improved the activity of SOD, but supplementation with EPE over a long period of time or an increased dose of EPE in chicken feed also substantially impacted the animal's antioxidant capacity. Previous research indicated that the expressions of GST, Prdx6, PPIA, alfatoxin aldehyde reductase, and SOD regulated by albusin B were enhanced to activate the systemic antioxidant defense [16], and SOD, catalase (CAT), GST, and GSH-Px were considered as AOE to protect against oxidative stress [53]. Overall, the functions of these upregulated genes were highly correlated with antioxidant status and could be linked to the improvement of antioxidant capacity induced by EPE supplements.
Many previous studies reported that glutathione is associated with glutathione metabolism pathways and that it likely functions as a reducing milieu to improve antioxidant function in cells [54]. As such, the results of the proteomic analysis indicated that antioxidant-related glutathione transferase activity was significantly increased in response to EPE supplements. The corresponding protein was a kind of glutathione S-transferase (A0A0A0MQ61, EC2.5.1.18) that is known to function in glutathione metabolism in Gallus gallus. Additionally, A0A0A0MQ61, an antioxidant protein, was significantly increased as a result of EPE dietary supplements. Further, the upregulated peroxiredoxin-1 protein in the peroxisome metabolic pathway is a known redox-regulating protein and is considered to be an antioxidant enzyme to eliminate various ROS [55]. Based on the above analyses, the antioxidant mechanism of muscle tissue in chicken treated with EPE dietary supplements is inferred and given in Figure 7. As shown, EPE treatments significantly improved GSH-Px activity and decreased MDA content of the breast muscle. Transcriptomic and proteomic analyses revealed that nine candidate genes and two candidate proteins were identified, which are responsible for improving antioxidant status induced by EPE supplements. Overall, this study promotes the understanding of the antioxidant mechanism in chicken regulated by EPE treatment.
Figure 7.
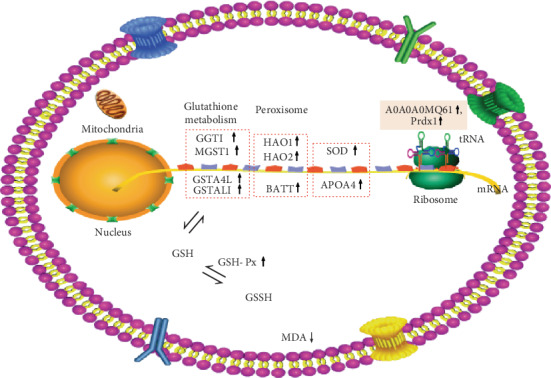
Potential antioxidant mechanism in muscle regulated by EPE.
5. Conclusion
Overall, EPE dietary supplements appeared to improve the antioxidant status of chicken by enhancing GSH-Px activity and reducing MDA content in muscle tissues. Glutathione metabolism and the peroxisome were considered the key metabolic pathways underlying the upregulation of AOE. Furthermore, all of the changes induced by EPE supplements may contribute to the systemic antioxidant defense of chicken, and these altered proteins and genes should be further explored in the future.
Acknowledgments
This work was supported by the National Key R&D Program of China (2016YFD0600806), the Guangdong Modern Agricultural Industry Technology System Innovation Team Project (2019KJ117), and the National Natural Science Foundation of China (31700501) for their financial support.
Abbreviations
- EPE:
Eucalyptus leaf polyphenols extract
- GSH-Px:
Glutathione peroxidase
- MDA:
Malonaldehyde
- T-SOD:
Total superoxide dismutase
- T-AOC:
Total antioxidant capacity
- DEGs:
Differentially expressed genes
- DEPs:
Differentially expressed proteins
- GO:
Gene Ontology
- KEGG:
Kyoto Encyclopedia of Genes and Genomes
- GGT1:
Gamma-glutamyltransferase 1
- MGST1:
Microsomal glutathione S-transferase 1
- GSTA4L:
Glutathione S-transferase alpha 4
- GSTAL1:
Glutathione S-transferase class-alpha
- HAO1:
Hydroxyacid oxidase 1
- HAO2:
Hydroxyacid oxidase 2
- BAAT:
Bile acid-CoA: amino acid N-acyltransferase
- SLC27A5:
Solute carrier family 27 member 5
- AGXT:
Alanine-glyoxylate and serine--pyruvate aminotransferase
- PIPOX:
Peroxisomal sarcosine oxidase
- GGT1:
Gamma-glutamyltranspeptidase 1
- ROS:
Reactive oxygen species
- GSH:
Glutathione
- CAT:
Catalase
- GST:
Glutathione-S-transferase
- GPX:
Glutathione peroxidase
- AOE:
Antioxidant enzymes.
Data Availability
The data used to support the findings of this study are included within the article and the supplementary information file.
Conflicts of Interest
The authors declare no conflicts of interest.
Authors' Contributions
Wei Li conceptualized, did the methodology, and wrote the original draft. Ze-Qi He did the methodology and resources. Xiao-Ying Zhang conceptualized and did the methodology. Yun-Jiao Chen did the resources, wrote the review, and edited. Jian-Jun Zuo wrote the review and edited. Yong Cao supervised, wrote the review, and edited.
References
- 1.Harkat-Madouri L., Asma B., Madani K., et al. Chemical composition, antibacterial and antioxidant activities of essential oil of Eucalyptus globulus from Algeria. Industrial Crops and Products. 2015;78:148–153. doi: 10.1016/j.indcrop.2015.10.015. [DOI] [Google Scholar]
- 2.Poersch N. L., Filho L. R. T. F., Miguel E. P., Cruz G. H. M. d., Francisquette K. L., Cavalheiro S. B. Influence of climate variables in the initial growth of Corymbia citriodora and different species of Eucalyptus. Bioscience Journal. 2017;33(6):1452–1464. doi: 10.14393/bj-v33n6a2017-36735. [DOI] [Google Scholar]
- 3.Engler B., Becker G., Hoffmann S. Process mechanization models for improved Eucalyptus plantation management in Southern China based on the analysis of currently applied semi-mechanized harvesting operations. Biomass and Bioenergy. 2016;87:96–106. doi: 10.1016/j.biombioe.2016.02.021. [DOI] [Google Scholar]
- 4.Tolba H., Moghrani H., Benelmouffok A., Kellou D., Maachi R. Essential oil of Algerian Eucalyptus citriodora: chemical composition, antifungal activity. Journal de Mycologie Médicale/Journal of Medical Mycology. 2015;25(4):e128–e133. doi: 10.1016/j.mycmed.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 5.Zhu L. W., Zhao P., Wang Q., et al. Stomatal and hydraulic conductance and water use in a eucalypt plantation in Guangxi, Southern China. Agricultural and Forest Meteorology. 2015;202:61–68. doi: 10.1016/j.agrformet.2014.12.003. [DOI] [Google Scholar]
- 6.Anigboro A. A., Avwioroko O. J., Cholu C. O. Phytochemical constituents, antimalarial efficacy, and protective effect of Eucalyptus camaldulensis aqueous leaf extract in plasmodium berghei-infected mice. Preventive Nutrition and Food Science. 2020;25(1):58–64. doi: 10.3746/pnf.2020.25.1.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Y., Wang J., Ou Y., et al. Cellular antioxidant activities of polyphenols isolated from Eucalyptus leaves (Eucalyptus grandis × Eucalyptus urophylla GL9) Journal of Functional Foods. 2014;7:737–745. doi: 10.1016/j.jff.2013.12.003. [DOI] [Google Scholar]
- 8.Sebei K., Sakouhi F., Herchi W., Khouja M. L., Boukhchina S. Chemical composition and antibacterial activities of seven Eucalyptus species essential oils leaves. Biological Research. 2015;48(1):p. 7. doi: 10.1186/0717-6287-48-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gullón B., Muñiz-Mouro A., Lú-Chau T. A., Moreira M. T., Lema J. M., Eibes G. Green approaches for the extraction of antioxidants from eucalyptus leaves. Industrial Crops and Products. 2019;138:p. 111473. doi: 10.1016/j.indcrop.2019.111473. [DOI] [Google Scholar]
- 10.González-Burgos E., Liaudanskas M., Viškelis J., Žvikas V., Janulis V., Gómez-Serranillos M. P. Antioxidant activity, neuroprotective properties and bioactive constituents analysis of varying polarity extracts from Eucalyptus globulus leaves. Journal of Food and Drug Analysis. 2018;26(4):1293–1302. doi: 10.1016/j.jfda.2018.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vitta Y., Figueroa M., Calderon M., Ciangherotti C. Synthesis of iron nanoparticles from aqueous extract of Eucalyptus robusta Sm and evaluation of antioxidant and antimicrobial activity. Materials Science for Energy Technologies. 2020;3:97–103. doi: 10.1016/j.mset.2019.10.014. [DOI] [Google Scholar]
- 12.Pan Z., Zeng Y., An J., Ye J., Xu Q., Deng X. An integrative analysis of transcriptome and proteome provides new insights into carotenoid biosynthesis and regulation in sweet orange fruits. Journal of Proteomics. 2012;75(9):2670–2684. doi: 10.1016/j.jprot.2012.03.016. [DOI] [PubMed] [Google Scholar]
- 13.Zhang C., Wei Y., Xiao D., et al. Transcriptomic and proteomic analyses provide new insights into the regulation mechanism of low-temperature-induced leafy head formation in Chinese cabbage. Journal of proteomics. 2016;144:1–10. doi: 10.1016/j.jprot.2016.05.022. [DOI] [PubMed] [Google Scholar]
- 14.Zhang J., Wang S., Song S., Xu F., Pan Y., Wang H. Transcriptomic and proteomic analyses reveal new insight into chlorophyll synthesis and chloroplast structure of maize leaves under zinc deficiency stress. Journal of Proteomics. 2019;199:123–134. doi: 10.1016/j.jprot.2019.03.001. [DOI] [PubMed] [Google Scholar]
- 15.Soewarto J., Hamelin C., Bocs S., et al. Transcriptome data from three endemic Myrtaceae species from New Caledonia displaying contrasting responses to myrtle rust (Austropuccinia psidii) Data in Brief. 2019;22:794–811. doi: 10.1016/j.dib.2018.12.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang H.-T., Li Y.-H., Chou I.-P., Hsieh Y.-H., Chen B.-J., Chen C.-Y. Albusin B modulates lipid metabolism and increases antioxidant defense in broiler chickens by a proteomic approach. Journal of the Science of Food and Agriculture. 2013;93(2):284–292. doi: 10.1002/jsfa.5754. [DOI] [PubMed] [Google Scholar]
- 17.Li W., Zhang X., He Z., et al. In vitro and in vivo antioxidant activity of eucalyptus leaf polyphenols extract and its effect on chicken meat quality and cecum microbiota. Food Research International. 2020;136:p. 109302. doi: 10.1016/j.foodres.2020.109302. [DOI] [PubMed] [Google Scholar]
- 18.Fathi M., Abdelsalam M., Al-Homidan I., et al. Supplemental effects of eucalyptus (Eucalyptus camaldulensis) leaves on growth performance, carcass characteristics, blood biochemistry and immune response of growing rabbits. Annals of Animal Science. 2019;19(3):779–791. doi: 10.2478/aoas-2019-0023. [DOI] [Google Scholar]
- 19.Mashayekhi H., Mazhari M., Esmaeilipour O. Eucalyptus leaves powder, antibiotic and probiotic addition to broiler diets: effect on growth performance, immune response, blood components and carcass traits. Animal. 2018;12(10):2049–2055. doi: 10.1017/S1751731117003731. [DOI] [PubMed] [Google Scholar]
- 20.Salman N. T. S. A., Al-Gharawi J. K. M. Effect of eucalyptus of leaves water extract on some productive trait of broilers. Plant archives. 2019;19:920–923. [Google Scholar]
- 21.Wang J. L., Xiao S. Y., Chen Y. J., Chen X. X., Tang J., Cao Y. Antioxidant activity of polyphenol extracts from leaves of E. grandis× E. urophylla Guanglin No. 9. Food Science. 2012;33(1):20–24. [Google Scholar]
- 22.Cao F. L., Zhang X. H., Yu W. W., Zhao L. G., Wang T. Effect of feeding fermented Ginkgo biloba leaves on growth performance, meat quality, and lipid metabolism in broilers. Poultry Science. 2012;91(5):1210–1221. doi: 10.3382/ps.2011-01886. [DOI] [PubMed] [Google Scholar]
- 23.Chen Y., Chen H., Li W., et al. Polyphenols in eucalyptus leaves improved the egg and meat qualities and protected against ethanol-induced oxidative damage in laying hens. Journal of Animal Physiology and Animal Nutrition. 2018;102(1):214–223. doi: 10.1111/jpn.12680. [DOI] [PubMed] [Google Scholar]
- 24.Gleason L. U., Burton R. S. RNA-seq reveals regional differences in transcriptome response to heat stress in the marine snail Chlorostoma funebralis. Molecular ecology. 2016;24(3):610–627. doi: 10.1111/mec.13047. [DOI] [PubMed] [Google Scholar]
- 25.Watson H., Videvall E., Andersson M. N., Isaksson C. Transcriptome analysis of a wild bird reveals physiological responses to the urban environment. Scientific Reports. 2017;7(1):p. 44180. doi: 10.1038/srep44180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun H., Jiang R., Xu S., et al. Transcriptome responses to heat stress in hypothalamus of a meat-type chicken. Journal of Animal Science and Biotechnology. 2015;6(1):p. 6. doi: 10.1186/s40104-015-0003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sevane N., Bialade F., Velasco S., et al. Dietary inulin supplementation modifies significantly the liver transcriptomic profile of broiler chickens. Plos One. 2014;9(6, article e989426) doi: 10.1371/journal.pone.0098942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan L.-B., Yu Y.-J., Zhang Q.-B., et al. Identification of p 90 ribosomal S6 kinase 2 as a novel host protein in HBx augmenting HBV replication by iTRAQ-based quantitative comparative proteomics. Proteomics-Clinical Applications. 2018;12(3):p. e1700090. doi: 10.1002/prca.201700090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu X., Yang J., Ning Z., Zhang X. Proteomic analysis of intestinal tissues from mice fed with Lentinula edodes-derived polysaccharides. Food & Function. 2016;7(1):250–261. doi: 10.1039/C5FO00904A. [DOI] [PubMed] [Google Scholar]
- 30.Xie C., Mao X., Huang J., et al. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic acids research. 2011;39(suppl_2):W316–W322. doi: 10.1093/nar/gkr483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fathi M. M., Al-Homidan I., Ebeid T. A., Abou-Emera O. K., Mostafa M. M. Dietary supplementation of Eucalyptus leaves enhances eggshell quality and immune response in two varieties of Japanese quails under tropical condition. Poultry Science. 2019;99(1):879–885. doi: 10.1016/j.psj.2019.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hashemi S. R., Davoodi H. Phytogenics as new class of feed additive in poultry industry. Journal of Animal and veterinary Advances. 2010;9(17):2295–2304. [Google Scholar]
- 33.Sedaghat A., Karimi Torshizi M. A. Immune responses, intestinal microbiota, performance and blood characteristics of Japanese quail fed on diets containing camphor. Animal. 2017;11(12):2139–2146. doi: 10.1017/S1751731117001148. [DOI] [PubMed] [Google Scholar]
- 34.Ramay M. S., Yalçın S. Effects of supplemental pine needles powder (Pinus brutia) on growth performance, breast meat composition, and antioxidant status in broilers fed linseed oil-based diets. Poultry Science. 2020;99(1):479–486. doi: 10.3382/ps/pez542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Surai P. F. Polyphenol compounds in the chicken/animal diet: from the past to the future. Journal of Animal Physiology and Animal Nutrition. 2014;98(1):19–31. doi: 10.1111/jpn.12070. [DOI] [PubMed] [Google Scholar]
- 36.Alagawany M., El-Hack M. E. A., Farag M. R., et al. Dietary supplementation of Yucca schidigera extract enhances productive and reproductive performances, blood profile, immune function, and antioxidant status in laying Japanese quails exposed to lead in the diet. Poultry science. 2018;97(9):3126–3137. doi: 10.3382/ps/pey186. [DOI] [PubMed] [Google Scholar]
- 37.Xiang J., Apea-Bah F. B., Ndolo V. U., Katundu M. C., Beta T. Profile of phenolic compounds and antioxidant activity of finger millet varieties. Food Chemistry. 2019;275:361–368. doi: 10.1016/j.foodchem.2018.09.120. [DOI] [PubMed] [Google Scholar]
- 38.Kishawy A. T., Amer S. A., El-Hack M. E. A., Saadeldin I. M., Swelum A. A. The impact of dietary linseed oil and pomegranate peel extract on broiler growth, carcass traits, serum lipid profile, and meat fatty acid, phenol, and flavonoid contents. Asian-Australasian journal of animal sciences. 2019;32(8):1161–1171. doi: 10.5713/ajas.18.0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Acero N., Gradillas A., Beltran M., García A., Muñoz Mingarro D. Comparison of phenolic compounds profile and antioxidant properties of different sweet cherry (Prunus avium L.) varieties. Food Chemistry. 2019;279:260–271. doi: 10.1016/j.foodchem.2018.12.008. [DOI] [PubMed] [Google Scholar]
- 40.Wang J., Jia R., Celi P., et al. Green tea polyphenol epigallocatechin-3-gallate improves the antioxidant capacity of eggs. Food & Function. 2020;11(1):534–543. doi: 10.1039/C9FO02157D. [DOI] [PubMed] [Google Scholar]
- 41.Tsikas D. Assessment of lipid peroxidation by measuring malondialdehyde (MDA) and relatives in biological samples: analytical and biological challenges. Analytical Biochemistry. 2017;524:13–30. doi: 10.1016/j.ab.2016.10.021. [DOI] [PubMed] [Google Scholar]
- 42.Chi X., Zhang Y., Ma X., et al. Antioxidative stress of oral administration of tea extract granule in chickens. Poultry Science. 2020;99(4):1956–1966. doi: 10.1016/j.psj.2019.11.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Henning S. M., Choo J. J., Heber D. Nongallated compared with gallated flavan-3-ols in green and black tea are more bioavailable. Journal of Nutrition. 2008;138(8):1529S–1534S. doi: 10.1093/jn/138.8.1529S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ou K., Gu L. Absorption and metabolism of proanthocyanidins. Journal of Functional Foods. 2014;7:43–53. doi: 10.1016/j.jff.2013.08.004. [DOI] [Google Scholar]
- 45.Chamorro S., Viveros A., Rebolé A., et al. Addition of exogenous enzymes to diets containing grape pomace: effects on intestinal utilization of catechins and antioxidant status of chickens. Food Research International. 2017;96:226–234. doi: 10.1016/j.foodres.2017.02.010. [DOI] [PubMed] [Google Scholar]
- 46.Laskaj R., Slavica D., Cepelak I., Kuzman I. Gamma-glutamyltransferase activity and total antioxidant status in serum and platelets of patients with community-acquired pneumonia. Archives of Medical Research. 2007;38(4):424–431. doi: 10.1016/j.arcmed.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 47.Shinno E., Shimoji M., Imaizumi N., Kinoshita S., Sunakawa H., Aniya Y. Activation of rat liver microsomal glutathione S-transferase by gallic acid. Life Science. 2005;78(1):99–106. doi: 10.1016/j.lfs.2005.04.034. [DOI] [PubMed] [Google Scholar]
- 48.Sahay S., Gupta M. An update on nitric oxide and its benign role in plant responses under metal stress. Nitric Oxide. 2017;67:39–52. doi: 10.1016/j.niox.2017.04.011. [DOI] [PubMed] [Google Scholar]
- 49.Del Río L. A., López-Huertas E. ROS generation in peroxisomes and its role in cell signaling. Plant and Cell Physiology. 2016;57(7):1364–1376. doi: 10.1093/pcp/pcw076. [DOI] [PubMed] [Google Scholar]
- 50.Wu G. Y., Fang Y. Z., Yang S., Lupton J. R., Turner N. D. Glutathione metabolism and its implications for health. Journal of Nutrition. 2004;134(3):489–492. doi: 10.1093/jn/134.3.489. [DOI] [PubMed] [Google Scholar]
- 51.Liu S. M., Eady S. J. Glutathione: its implications for animal health, meat quality, and health benefits of consumers. Australian of Agricultural Research. 2005;56(8):775–780. doi: 10.1071/AR05053. [DOI] [Google Scholar]
- 52.Wang F., Kohan A. B., Lo C.-M., Liu M., Howles P., Tso P. Apolipoprotein A-IV: a protein intimately involved in metabolism. Journal of Lipid Research. 2015;56(8):1403–1418. doi: 10.1194/jlr.R052753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hu R., He Y., Arowolo M. A., Wu S., He J. Polyphenols as potential attenuators of heat stress in poultry production. Antioxidants. 2019;8(3):p. 67. doi: 10.3390/antiox8030067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aquilano K., Baldelli S., Ciriolo M. R. Glutathione: new roles in redox signaling for an old antioxidant. Frontiers in pharmacology. 2014;5(196) doi: 10.3389/fphar.2014.00196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ding C., Fan X., Wu G. Peroxiredoxin 1- an antioxidant enzyme in cancer. Journal of Cellular and Molecular Medicine. 2017;21(1):193–202. doi: 10.1111/jcmm.12955. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the findings of this study are included within the article and the supplementary information file.


