Abstract
Emerging evidence has revealed significant roles for small nucleolar RNAs (snoRNAs) in tumorigenesis. However, the genetic and pharmacogenomic landscape of snoRNAs has not been characterized. Using the genotype and snoRNA expression data from The Cancer Genome Atlas, we characterized the effects of genetic variants on snoRNAs across 29 cancer types and further linked related alleles with patient survival as well as genome-wide association study risk loci. Furthermore, we characterized the impact of snoRNA expression on drug response in patients to facilitate the clinical utility of snoRNAs in cancer. We also developed a user-friendly data resource, GPSno (http://hanlab.uth.edu/GPSno), with multiple modules for researchers to visualize, browse, and download multi-dimensional data. Our study provides a comprehensive genetic and pharmacogenomic landscape of snoRNAs, which will shed light on future clinical considerations for the development of snoRNA-based targeted therapies.
Keywords: Small nucleolar RNA, Genetic variants, Pharmacogenomics, Cancer
Main text
Small nucleolar RNAs (snoRNAs) are a group of regulatory RNAs that mainly reside in the nucleolus and post-transcriptionally modify ribosomal RNA (rRNA) and small nuclear RNA (snRNA). Emerging evidence has demonstrated significant roles of snoRNAs in cancer [1]. For example, deletion of SNORD50A/B may cooperate with oncogenic KRAS mutations in cancer to drive Ras-MAPK hyperactivation [2]. Overexpression of SNORD46 has been shown to promote cancer cell growth, migration, and invasion [3]. Recent studies characterized the expression landscape and copy number variations of snoRNAs in large numbers of cancer patients [2, 3]. It remains challenging to further understand the functional roles of snoRNAs in cancer. For example, it is unclear whether genetic variants will affect the expression level of snoRNAs, and whether the alterations of snoRNAs are associated with drug response in patients.
In this study, we investigated the effects of genetic variants on snoRNA expression and characterized the effects of snoRNA expression on drug response. To facilitate broad access of these data for the biomedical research community, we developed a user-friendly database, GPSno. We expect this study to have a significant clinical impact on the future development of snoRNA-based targeted therapies.
Results and discussions
Impact of genetic variants on snoRNA expression
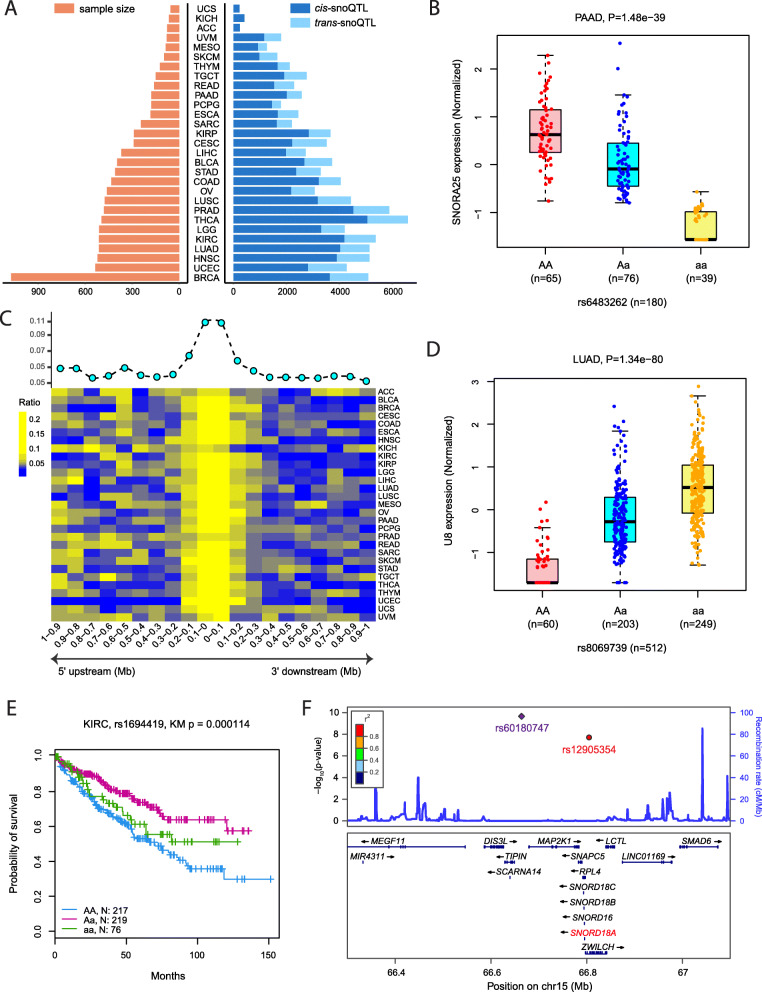
To comprehensively characterize the impact of genetic variants on snoRNA expression across different cancer types, we performed snoRNA expression quantitative trait loci (QTL) analysis [4] across 29 cancer types with at least 50 patients having both genotype data and snoRNA expression data in The Cancer Genome Atlas (TCGA) (additional file: Fig. S1A and Table S1). A total of 9449 tumor samples were included, with the sample size of each cancer type ranging from 56 in uterine carcinosarcoma (UCS) to 1073 in breast invasive carcinoma (BRCA) (Fig.1a). After imputation and quality control for the genotype data, we obtained a median of 4,358,817 SNPs for each cancer type. There were on average 435 snoRNAs (reads per kilobase per million reads [RPKM] ≥1) for each cancer type (additional file: Table S2). For analysis of proximal genetic regulation of snoRNAs (SNP within 1 Mb from the snoRNA location), a total of 69,557 significant SNP–snoRNA pairs in 29 cancer types were identified at a per-tissue false discovery rate (FDR) < 0.05 (Fig. 1a), which corresponded to a median P-value < 8.34 × 10− 9. The number of cis-snoQTLs ranged from 227 in UCS to 5067 in thyroid carcinoma (THCA), with a median of 2184 cis-snoQTLs per cancer type (Fig. 1a; additional file: Table S2). For example, in pancreatic adenocarcinoma (PAAD), rs6483262 alleles demonstrated significant effects on regulating the expression of SNORA25 (P-value = 1.48 × 10− 39) (Fig. 1b). To be noted, SNORA25 was reported as a promising biomarker for the early detection of pancreatic cancer [5]. We also examined the relative location distribution of cis-snoQTLs in regard to paired snoRNAs, and found that cis-snoQTLs were preferentially located in proximity to paired snoRNAs (Fig. 1c). For analysis of the remote genetic regulation of snoRNAs (SNP beyond 1 Mb from the snoRNA location), a total of 34,151 SNP–snoRNA pairs in 29 cancer types were identified at a per-tissue FDR < 0.05 (Fig. 1a), which corresponded to a median P-value < 7.40 × 10− 15. The number of trans-snoQTLs ranged from 0 in adrenocortical carcinoma (ACC) to 1525 in THCA, with a median of 821 trans-snoQTLs per cancer type (Fig. 1a; additional file: Table S2). For example, rs8069739 alleles showed remote regulation of the expression of U8 in lung adenocarcinoma (LUAD) (Fig. 1d). It was reported that U8 depletion triggers a remarkably potent p53-dependent anti-tumor stress response in lung adenocarcinoma [6]. The numbers of cis-snoQTLs and trans-snoQTLs were both significantly correlated with the number of samples (Spearman’s correlation, cis-snoQTL: Rs = 0.92, P-value = 3.48 × 10− 12; trans-snoQTL: Rs = 0.87, P-value = 6.52 × 10− 10) (additional file: Fig. S1B). This suggests that the effects of some SNPs may be underestimated due to insufficient sample size in certain cancer types, and this could be improved with additional patient samples in the future.
Fig. 1.
Summary of pan-cancer snoQTL analysis. a Numbers of samples included and numbers of snoQTLs identified in different cancer types. b Association between cis-snoQTL rs6483262 alleles and SNORA25 levels in PAAD. c Relative location distribution of cis-snoQTLs in regard to their paired snoRNAs. d Association between trans-snoQTL rs8069739 alleles and U8 levels in LUAD. e Kaplan–Meier plot displaying the association between rs1694419 genotypes and overall survival times of KIRC patients. f snoQTL rs12905354 located in TCGT GWAS locus
To investigate the clinical relevance of these genetic variants associated with snoRNA expression, we identified 475 SNP alleles that correlated with patients’ overall survival times in different cancer types with FDR < 0.05. For instance, kidney renal clear cell carcinoma (KIRC) patients carrying the homozygous rs1694419 AA had worse overall survival than those carrying the heterozygous Aa and the homozygote aa (log-rank test, P-value = 1.14 × 10− 4) (Fig. 1e). Patients carrying the homozygous rs1694419 AA had significantly higher expression level of SNORD45B than patients carrying the heterozygous Aa and the homozygous aa in 26 out of 29 (89.7%) cancer types, including KIRC (P-value = 3.93 × 10− 34, additional file: Fig. S2A). SNORD45B was significantly upregulated in KIRC tumor tissues compared to adjacent normal tissues (Student’s t-test, P-value = 0.011; additional file: Fig. S2B), and KIRC patients with higher expression of SNORD45B also had worse overall survival than those with lower expression of SNORD45B (log-rank test, P-value = 0.0041; additional file: Fig. S2C). Taken together, our results demonstrated the effects of snoQTLs on patients’ survival through regulating the expression of snoRNAs.
To identify genome-wide association study (GWAS)-related snoQTLs, 28,345 trait/disease-related SNPs were extracted from the GWAS catalog, and 1,167,961 SNPs were obtained that were located in the GWAS linkage disequilibrium (LD) regions. We identified 29,795 snoQTL–snoRNA pairs in which the snoQTLs overlapped with known disease/trait-associated loci in different cancer types. For example, the Testicular Cancer Consortium found that rs60180747 is significantly associated with testicular germ cell tumor (TGCT) risk (odds ratio [OR] = 1.23; P = 1.10 × 10− 10) [7]. The SNP marker rs60180747 marks a 261 kb haploblock on 15q22.31 that contains several genes, including TIPIN, MAP 2 K1, DIS3L, SNAPC5, RPL4 and ZWILCH (Fig. 1f), and rs60180747 is located in the intron of gene TIPIN. However, this risk allele is not correlated with any protein-coding genes in 15q22.31 in TGCT patients [8]. We further examined nearby SNPs, and observed that rs12905354, which is in LD with rs60180747 (LD r2 = 1.0), significantly correlated with the expression of SNORD18A (additional file: Fig. S3) rather than protein-coding genes located in this risk locus. These findings suggest that SNORD18A may be a causal target in this TGCT GWAS loci.
Pharmacogenomic landscape of snoRNAs across different cancer types
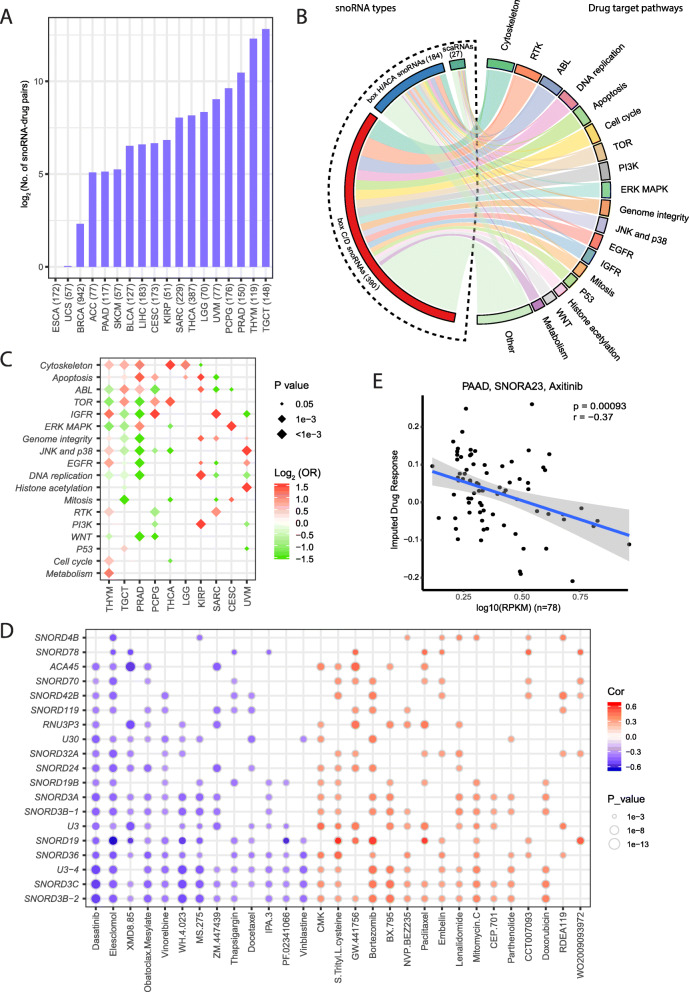
To understand the effects of snoRNA expression on drug response, we performed an integrative analysis to assess the associations between the variance of snoRNA expression and the response to anticancer drugs in TCGA patients. We acquired imputed drug response data of TCGA patients from a previous study [9]. Eighteen cancer types in TCGA with at least 50 patients having both imputed drug response data and snoRNA expression data were included for drug response analysis. We identified 16,393 significantly correlated snoRNA–drug response pairs at FDR < 0.05 from 18 cancer types, ranging from 0 in esophageal carcinoma (ESCA) to 7226 in TGCT, with a median of 113 snoRNA–drug pairs per cancer type (Fig. 2a). The number of snoRNA–drug pairs did not correlate with the number of samples (Spearman’s correlation, P-value = 0.75). The snoRNAs (390 box C/D snoRNAs, 184 box H/ACA snoRNAs, and 27 scaRNAs) had extensive impact on patients’ responses to drugs within various drug target pathways across different cancer types. Among these drug target pathways, drugs related to the cytoskeleton pathway showed the largest number of drug–snoRNA pairs (Fig. 2b). We further used Fisher’s exact test to evaluate the enrichment of each drug target pathway in 10 cancer types with at least 100 significantly correlated snoRNA–drug response pairs identified, and found that the cytoskeleton pathway was significantly enriched in 5 cancer types (P-value < 0.05; Fig. 2c).
Fig. 2.
Pharmacogenomic landscape of snoRNAs. a Sample size included and significant snoRNA–drug pairs identified. b Association between snoRNA expression and imputed drug response. c Enrichment of various drug target pathways of significant snoRNA–drug response pairs. d Significantly correlated snoRNA–drug response pairs identified in PRAD. Drugs significantly associated with at least 5 snoRNAs are shown in the plot. e Association between SNORA23 expression and response to the drug axitinib in PAAD patients
Taking prostate adenocarcinoma (PRAD) as an example, the response to 29 anti-cancer drugs significantly correlated with the levels of at least five snoRNAs (Fig. 2d). Intriguingly, various snoRNAs showed great consistency in having either a positive or negative effect on the response to one certain drug, which was likely due to the high correlation among the expression of these snoRNAs in each cancer type (additional file: Fig. S4). In patients with PAAD, the expression level of SNORA23 was highly associated with the response to the drug axitinib (R = − 0.37, P-value = 0.93 × 10− 3; Fig. 2e). SNORA23 has been shown to promote tumor growth and metastasis in pancreatic cancer [10]. Therefore, the expression level of SNORA23 may need to be considered in future clinical trials for axitinib. These results suggest that appreciable levels of snoRNAs could contribute to response to drug therapy. Future experiments are necessary to validate the effects of drug response for these snoRNAs.
A comprehensive data resource to explore the genetic impacts and pharmacogenomic landscape of snoRNAs in cancer
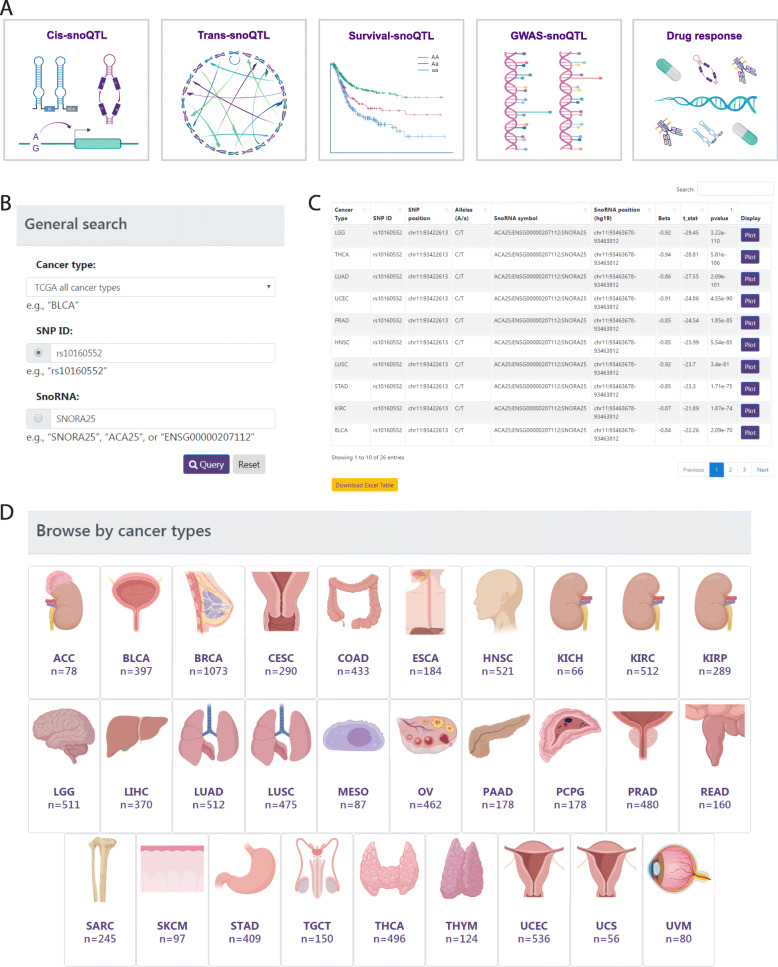
We developed a user-friendly data portal, GPSno (https://hanlab.uth.edu/GPSno/), to facilitate visualizing, searching and browsing of data by the biomedical research community. GPSno contains five main modules: cis-snoQTLs, trans-snoQTLs, survival-snoQTLs, GWAS-snoQTLs, and drug response (Fig. 3a). Several entryways are provided for querying, and each supports user-defined filters such as cancer type, SNP ID, and snoRNA ID. Users can enter different pages to search SNPs or snoRNAs of interest. We also provide a search section for users to query the data based on cancer type, SNP ID, or snoRNA (Fig. 3b). Querying on the cis/trans-snoQTL page, a table with SNP ID, SNP genomic position, SNP alleles, snoRNA ID, snoRNA position, beta value (effect size of SNP on gene expression), and P-value of snoQTL will be returned (Fig. 3c). For each record, a vector diagram of a boxplot is provided to display the association between SNP genotypes and snoRNA levels. Querying on the survival-snoQTL page, details with SNP ID, SNP genomic position, SNP alleles, log-rank test P-value and median survival times of different genotypes will be displayed. A vector diagram of the Kaplan–Meier plot is embedded in each record to display the association between snoQTL and overall survival times. Querying on the GWAS-snoQTLs page will return the SNP information, snoRNA information and related GWAS traits. Querying on the drug response page will return the snoRNA and related drug information, and a diagram is also provided to display the association between snoRNA levels and drug response. To facilitate researchers studying different cancer types, we also designed a cancer-type-specific module for querying results (Fig. 3d). Tables of querying results can be downloaded in XLSX format, and figures of results can be downloaded as a PDF. The document page includes an introduction, construction pipeline, and interpretation guidance for the database. This database is a valuable resource and will be of great interest to the research community, which will provide a unique resource to select candidate snoRNAs for future experiments.
Fig. 3.
Web design and querying of GPSno. a Five main modules in GPSno. b General search section for querying. c Example of resulting list after querying on the cis/trans-snoQTL page. d Specific modules for querying results by cancer type
Conclusions
In light of the significance of snoRNAs in oncogenesis, we systematically investigated the impact of genetic variants and the pharmacogenomic landscape of snoRNAs in multiple cancer types from TCGA. We also developed GPSno as the first comprehensive data resource for the genetic and pharmacogenomic landscape of snoRNAs. Our study will shed light on future clinical considerations for the development of snoRNA-based targeted therapies.
Supplementary information
Additional file 1 Supplementary Figure 1. (A) Workflow of snoQTL analysis. Blue boxes are software or packages utilized; green boxes are detailed modules or analysis used. Other boxes are input or output data in QTL analysis. (B) Relationship between snoQTLs identified and sample size included. Supplementary Figure 2. (A) Association between snoQTL rs1694419 alleles and SNORD45B levels in KIRC patients. (B) SNORD45B significantly upregulated in KIRC tumor tissues compared to adjacent normal tissues. (C) KIRC patients with higher expression of SNORD45B have worse overall survival than those with lower expression of SNORD45B. Supplementary Figure 3. Association between snoQTL rs12905354 alleles and SNORD18A levels in TGCT patients. Supplementary Figure 4. Co-expression of snoRNAs related to drug response found in PRAD.
Additional file 2 Supplementary Table 1. Abbreviations of cancer types. Supplementary Table 2. Analysis Summary
Acknowledgments
This work was supported by the Cancer Prevention & Research Institute of Texas (RR150085 and RP190570) to CPRIT Scholar in Cancer Research (L.H.). This work was also supported by UTHealth Innovation for Cancer Prevention Research Training Program Post-doctoral Fellowship (Cancer Prevention and Ressearch Institute of Texas grant #RP160015). We gratefully acknowledge contributions from TCGA Research Network. We thank LeeAnn Chastain for editorial assistance.
Abbreviations
- ACC
Adrenocortical carcinoma
- BRCA
Breast invasive carcinoma
- ESCA
Esophageal carcinoma
- GWAS
Genome-wide association study
- KIRC
Kidney renal clear cell carcinoma
- LD
Linkage disequilibrium
- LUAD
Lung adenocarcinoma
- PAAD
Pancreatic adenocarcinoma
- PRAD
Prostate adenocarcinoma
- QTL
Quantitative trait loci
- RPKM
Reads per kilobase per million reads
- rRNA
Ribosomal RNA
- snoQTL
snoRNA expression quantitative trait loci
- snoRNA
Small nucleolar RNA
- snRNA
Small nuclear RNA
- TCGA
The Cancer Genome Atlas
- THCA
Thyroid carcinoma
- UCS
Uterine carcinosarcoma
Authors’ contributions
L.H., L.D., and X.Z. conceived and supervised the project. Y.L. and L.H. designed and performed the research. Y.L. performed the data analyses. H.R. and Y.L. constructed the data portal. Y.L., H.R., S.L., Y.Y., W.H., J.G., Z.Z., Y.J., X.Z., L.D. and L.H. interpreted the results. Y.L. and L.H. wrote the paper with input from all the other authors. The authors read and approved the final manuscript.
Funding
This work was supported by the Cancer Prevention & Research Institute of Texas (RR150085 and RP190570) to CPRIT Scholar in Cancer Research (L.H.). This work was also supported by UTHealth Innovation for Cancer Prevention Research Training Program Post-doctoral Fellowship (Cancer Prevention and Research Institute of Texas grant # RP160015).
Availability of data and materials
The genotype and miRNAseq data of tumor patients can be found at the GDC portal (https://portal.gdc.cancer.gov/). The software and resources used in our analyses are described in each method section in the supplementary files. All results generated in this study can be found in our database, GPSno (http://hanlab.uth.edu/GPSno).
Ethics approval and consent to participate
The patient data we used were acquired as publicly available datasets that were collected with patients’ informed consent.
Consent for publication
All authors give consent for the publication of this manuscript in Molecular Cancer.
Competing interests
The authors declare no potential conflicts of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yaoming Liu and Hang Ruan contributed equally to this work.
Contributor Information
Xiulan Zhang, Email: zhangxl2@mail.sysu.edu.cn.
Lixia Diao, Email: ldiao@mdanderson.org.
Leng Han, Email: leng.han@uth.tmc.edu.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12943-020-01228-z.
References
- 1.Williams GT, Farzaneh F. Are snoRNAs and snoRNA host genes new players in cancer? Nat Rev Cancer. 2012;12:84–88. doi: 10.1038/nrc3195. [DOI] [PubMed] [Google Scholar]
- 2.Siprashvili Z, Webster DE, Johnston D, Shenoy RM, Ungewickell AJ, Bhaduri A, et al. The noncoding RNAs SNORD50A and SNORD50B bind K-Ras and are recurrently deleted in human cancer. Nat Genet. 2015;48:53–58. doi: 10.1038/ng.3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gong J, Li Y, jie LC, Xiang Y, Li C, Ye Y, et al. A pan-cancer analysis of the expression and clinical relevance of small Nucleolar RNAs in human Cancer. Cell Rep. 2017;21:1968–1981. doi: 10.1016/j.celrep.2017.10.070. [DOI] [PubMed] [Google Scholar]
- 4.Ye Y, Zhang Z, Liu Y, Diao L, Han L. A multi-Omics perspective of quantitative trait loci in precision medicine. Trends Genet Elsevier. 2020:1–19. [DOI] [PubMed]
- 5.Kitagawa T, Taniuchi K, Tsuboi M, Sakaguchi M, Kohsaki T, Okabayashi T, et al. Circulating pancreatic cancer exosomal RNAs for detection of pancreatic cancer. Mol Oncol. 2019;13:212–227. doi: 10.1002/1878-0261.12398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Langhendries JL, Nicolas E, Doumont G, Goldman S, Lafontaine DLJ. The human box C/D snoRNAs U3 and U8 are required for prerRNA processing and tumorigenesis. Oncotarget. 2016;7:59519–59534. doi: 10.18632/oncotarget.11148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Z, McGlynn KA, Rajpert-De Meyts E, Bishop DT, Chung CC, Dalgaard MD, et al. Meta- a nalysis of five genome-wide association studies identifies multiple new loci associated with testicular germ cell tumor. Nat Genet. 2017;49:1141–1146. doi: 10.1038/ng.3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gong J, Mei S, Liu C, Xiang Y, Ye Y, Zhang Z, et al. PancanQTL: systematic identification of cis -eQTLs and trans -eQTLs in 33 cancer types. Nucleic Acids Res. 2018;46:D971–D976. doi: 10.1093/nar/gkx861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geeleher P, Zhang Z, Wang F, Gruener RF, Nath A, Morrison G, et al. Discovering novel pharmacogenomic biomarkers by imputing drug response in cancer patients from large genomics studies. Genome Res. 2017;27:1743–1751. doi: 10.1101/gr.221077.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cui L, Nakano K, Obchoei S, Setoguchi K, Matsumoto M, Yamamoto T, et al. Small Nucleolar Noncoding RNA SNORA23, Up-Regulated in Human Pancreatic Ductal Adenocarcinoma, Regulates Expression of Spectrin Repeat-Containing Nuclear Envelope 2 to Promote Growth and Metastasis of Xenograft Tumors in Mice. Gastroenterology. 2017;153:292–306. doi: 10.1053/j.gastro.2017.03.050. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1 Supplementary Figure 1. (A) Workflow of snoQTL analysis. Blue boxes are software or packages utilized; green boxes are detailed modules or analysis used. Other boxes are input or output data in QTL analysis. (B) Relationship between snoQTLs identified and sample size included. Supplementary Figure 2. (A) Association between snoQTL rs1694419 alleles and SNORD45B levels in KIRC patients. (B) SNORD45B significantly upregulated in KIRC tumor tissues compared to adjacent normal tissues. (C) KIRC patients with higher expression of SNORD45B have worse overall survival than those with lower expression of SNORD45B. Supplementary Figure 3. Association between snoQTL rs12905354 alleles and SNORD18A levels in TGCT patients. Supplementary Figure 4. Co-expression of snoRNAs related to drug response found in PRAD.
Additional file 2 Supplementary Table 1. Abbreviations of cancer types. Supplementary Table 2. Analysis Summary
Data Availability Statement
The genotype and miRNAseq data of tumor patients can be found at the GDC portal (https://portal.gdc.cancer.gov/). The software and resources used in our analyses are described in each method section in the supplementary files. All results generated in this study can be found in our database, GPSno (http://hanlab.uth.edu/GPSno).