Abstract
Circular RNA (circRNA) is a novel class of regulatory noncoding RNA (ncRNA) molecules with a unique covalently closed loop structure. Next-generation sequencing shows that thousands of circRNAs are widely and stably expressed in multiple eukaryotes. As novel regulatory ncRNAs, circRNAs possess several specific molecular functions, including regulating gene transcription and translation, acting as miRNA sponges, and interacting with functional proteins. Ovarian cancer (OvCa) is one of the most aggressive malignant diseases affecting the lives of thousands of women worldwide, and the majority of OvCa cases are diagnosed at advanced stages. Accumulating evidence has revealed the significant roles of circRNAs in the occurrence and progression of OvCa, indicating the function of circRNAs as promising biomarkers and their therapeutic relevance in this disease. This review aims to summarize the mechanisms by which circRNAs mediate OvCa progression as well as their diagnostic and prognostic values in OvCa.
Keywords: Circular RNA, Ovarian cancer, Biomarker
Background
Ovarian cancer (OvCa) is one of the most aggressive female malignancies and is estimated to account for approximately 21,750 new cases and 13,940 deaths worldwide in 2020 [1]. OvCa tends to be diagnosed at advanced stages (stage III or IV) with poor prognosis because the ovary is located deep in the abdominal cavity and the early symptoms of OvCa are not obvious [2]. With the development of medical technology, the prognosis of OvCa has significantly improved, but the 5-year overall survival (OS) rate remains low, between 35 and 40% [3]. Hence, discovery of the molecular biological mechanism of OvCa tumorigenesis and development has captured extensive attention from researchers, and novel OvCa-associated biomarkers may be essential for enhancing the diagnosis and prognosis of these patients.
Noncoding RNAs (ncRNAs), which constitute more than 98% of the human genome, play essential roles in gene regulation and translation. Importantly, a growing number of studies have proven the role of ncRNAs in tumorigenesis, including cell proliferation, apoptosis, migration, and invasion [4, 5]. CircRNA is a class of recently re-recognized ncRNA with a distinct structure, which has not been well studied and was originally regarded as the result of mis-splicing or as a byproduct of pre-mRNA processing with low abundance over the past 30 years [6]. Currently, the rapid expansion of next-generation sequencing, extranuclear enrichment tools, and bioinformatics analyses have confirmed the critical roles of circRNAs in the pathogenesis of a large number of human diseases, including heart diseases [7], diabetes [8], nervous system diseases [9], immune system diseases [10] and cancers [11]. In addition, increasing numbers of studies have shown that circRNAs may be involved in cancer proliferation, invasion, metastasis, and apoptosis, indicating the use of circRNAs as promising biomarkers and their therapeutic relevance [12–14]. In particular, circRNAs may serve as essential regulatory factors in OvCa occurrence and progression according to recent publications [11, 15, 16]. Thus, the current review discusses the dysregulated expression of circRNAs and circRNA-mediated OvCa progression as well as the diagnostic and prognostic values of circRNAs in OvCa.
Characteristics of circRNAs
CircRNAs, differing from linear RNAs, are a class of re-recognized ncRNAs with covalently closed structures that were first reported in the viroid genome by Sanger et al. in 1976 [17]. In recent years, scholars have found that circRNAs are present in a stable structure in various types of eukaryotic cells, including in protists, fungi, plants, fish, worms, insects, and mammals [18]. CircRNAs often share the following characteristics: (1) circRNAs are characterized by their covalently closed loop in which the 5′ and 3′ ends are connected to each other; (2) circRNAs are prone to be highly resistant to endonuclease cleavage in that they have no exposed 5′ or 3′ end, and circRNAs are more stable due to their closed structure in comparison with their linear counterparts [19]; (3) circRNAs are generally expressed in eukaryotes, and their expression levels vary extremely in a tissue-specific and developmental-stage-specific manner. For example, Ahmed et al. [20] examined the differential expression of circRNAs in primary and metastatic OvCa tissues; (4) circRNAs are mainly derived from protein-coding genes, and one gene may generate various circRNAs through selective circularization [21]; (5) most circRNAs are evolutionarily and sequentially conserved in various species [22]; and (6) circRNAs could be divided into three subtypes, namely, exonic circRNAs, composed of only exons, are primarily found in the cytoplasm; intronic circRNAs, originating from introns, are principally present in the nucleus; and exon–intron circRNAs, which contain both introns and exons and are mainly present in the nucleus [23]. Overall, conservation, specificity, and stability are the three major characteristics of circRNAs. These features indicate that circRNAs potentially play functional roles in gene expression at the transcriptional and posttranscriptional levels and may therefore influence diagnoses, treatment, and prognoses of numerous diseases.
Biogenesis of circRNAs
In eukaryotic genes, pre-mRNA undergoes alternative splicing, thus removing the noncoding introns and reattaching the exons [24]. After that, the noncoding introns that are removed by the splicing process can be shaped into linear or lariat molecules. Accumulating evidence has demonstrated that circRNAs are generated with the aid of RNA polymerase II (Pol II) in the splicing process [23]. On the basis of RNA composition and biogenesis mechanisms, circRNAs can be grouped into three subclasses: exonic circRNAs (ecircRNAs), circular intronic RNAs (ciRNAs), and exon–intron circRNAs (EIciRNAs) [23]. EcircRNAs, which contain only exons, are generated in a process called back-splicing circularization or exon skipping event [25]. Back-splicing circularization, also called circle splicing or head-to-tail splicing, is a process where the downstream 5′ end of an exon binds to the upstream 3′ end of another exon, giving rise to a circular RNA. A single exon may also generate a circular RNA; in this case, the 3′ end of one exon is linked to the 5′ end of the same exon [18]. Exon skipping is the other process of generating ecircRNAs, where pre-mRNAs first undergo exon skipping and then form an exon-containing lariat with the aid of looped introns during internal splicing [26].
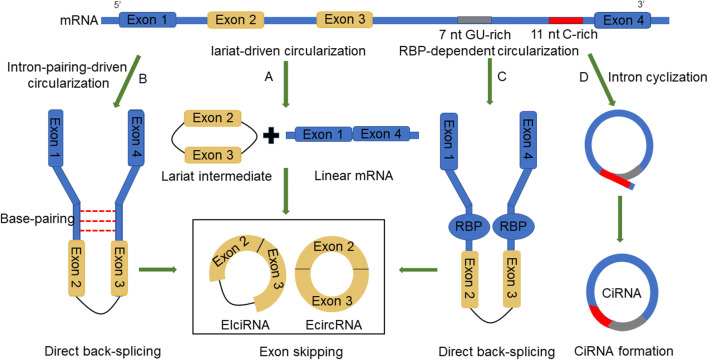
Lariat-driven circularization, intron pairing-driven circularization, and RNA binding protein (RBP)-dependent circularization are the major models to explain the formation of ecircRNA or EIciRNA. Lariat-driven circularization, an exon skipping process, first generates a lariat intermediate in which the 5ʹ end donor of an exon binds to the 3ʹ end acceptor of another exon and subsequently forms an ecircRNA by removing the introns between the exons [22] (Fig. 1a). Intron pairing-driven circularization, a direct back-splicing mechanism, is a process that induces cyclization with the aid of reverse complementary sequences and thus generated an EIciRNA or ecircRNA via intron removal [21] (Fig. 1b). RBP-driven circularization, the other model of the back-splicing mechanism, starts with back-splicing, and then the flanking introns bind to each other closely with the aid of RBPs, resulting in the formation of a circRNA [27] (Fig. 1c). However, unlike ecircRNA, ciRNA formation is dependent on a conserved sequence near both sides of the spliceosome [28]. When a 3′ splice acceptor binds to a 5′ splice donor, thus generating a lariat intron, the conserved sequence composed of a 7 nt GU-rich element close to the 5′ splice site and an 11 nt C-rich element near the branchpoint site helps the intronic lariat evade degradation by debranching enzymes [29] (Fig. 1d).
Fig. 1.
Biogenesis of circRNAs: four models explain circRNA formation. a Lariat-driven circularization first generates a lariat intermediate in which the 5′ end donor of an exon binds to the 3′ end acceptor of another exon and subsequently forms an ecircRNA by removing the introns between the exons. b Intron pairing-driven circularization begins with cyclization with the aid of reverse complementary sequences and thus generates an EIciRNA or ecircRNA via intron removal. c RBP-driven circularization starts with back-splicing, and then the flanking introns bind to each other closely with the aid of RBPs, resulting in the formation of a circRNA. d CiRNA formation is dependent on the conserved sequence near both sides of the spliceosome. The conserved sequence assists the intronic lariat to evade degradation by debranching enzymes
In conclusion, the biogenesis of circRNAs has not been completely elucidated, and the mechanism described above represents only a limited point of view. In future years, by the use of advanced computing tools, it is likely that the mechanism of circRNA biogenesis will be further better understood.
Functions of circRNAs
CircRNAs act as ceRNAs or miRNA sponges
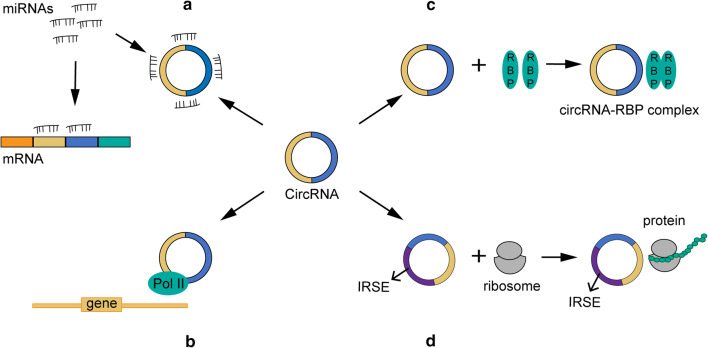
Multiple studies have uncovered the function of circRNAs as competing endogenous RNAs (ceRNAs) or miRNA sponges, whereby the indirectly regulate gene expression by competing for miRNA binding (Fig. 2a). CeRNAs are transcripts that possess miRNA response elements (MREs) and include long noncoding RNAs (lncRNAs), mRNAs, pseudogenic RNAs, and circRNAs, among which circRNAs have the highest binding capacity to miRNAs. It is no exaggeration to say that circRNA has become a novel research hotspot in the ceRNA field due to its unique characteristics and has attracted tremendous attention from scholars [30, 31]. The ceRNA hypothesis indicates that circRNA shares a similar seed sequence with the target mRNA, thus sponging miRNAs that regulate the function and expression levels of the target gene [32]. EcircRNAs, which contain MREs with a stable circular structure, are reported to act as functional ceRNAs that possess the capability to bind to miRNAs and prevent their binding to target mRNAs [32]. It has been reported that several types of malignant diseases are associated with an aberrant circRNA–miRNA–mRNA axis. For instance, CiRS-7, which possesses 73 miR-7 binding sites, can sponge miR-7, thus inhibiting the biological function of miR-7 target genes. It has been confirmed that CiRS-7 mediates apoptosis by sponging miR-7 to regulate the PTEN/PI3K/AKT pathway in gastric cancer [33]. In addition, circ-ITCH is reported to increase the cancer-suppressing ability of its parental gene by sponging miR-7 and miR-214 and consequently attenuating lung cancer cell proliferation [34]. Hsa_circ_0012673 promotes cell proliferation in lung cancer tissues and cells through regulation of LIM domain kinase 1 (LIMK1) levels by sponging miR-320a [15]. In conclusion, circRNAs may commonly exert their regulatory functions in cancers by acting as miRNA sponges. However, to date, scholars have characterized only a small fraction of these circRNAs as well as their sponge functions, and extensive research is needed to explore the rest of the circRNA family.
Fig. 2.
Functions of circRNAs. a miRNA sponges; b Regulation of gene transcription; c Interaction with functional proteins; d Translation into peptides and/or proteins
CircRNAs regulate gene transcription
In addition to acting as sponges for miRNAs, a portion of circRNAs can cis- or trans-regulate gene transcription, thus impacting gene expression [35] (Fig. 2b). CiRNAs, primarily located in the nucleus, are reported to modulate the transcription and expression of their parental genes by binding to RNA Pol II [28]. Typical examples are ci-ankrd52 and ci-sirt7. It has been confirmed that both circRNAs can positively regulate the transcription of their parental genes by interacting with Pol II [28]. Additionally, EIciRNAs are also thought to regulate gene expression. For instance, Li et al. found that both EIciRNAs, circ-EIF3J, and circ-PAIP2, can increase the expression of their parental genes in HEK293 and HeLa cells via their involvement in the interaction between U1 small nuclear ribonucleic proteins (snRNPs) and RNA Pol II in the promoter region of the host gene [36]. In summary, circRNAs can function as regulator factors to impact gene expression by binding with the RNA pol II complex and transcription-related proteins.
CircRNAs interact with functional proteins
Although most studies have focused on circRNA sponge activity, several studies have demonstrated that circRNAs harbor numerous protein-binding sites for RBPs and exert biological functions by binding to RBPs (Fig. 2c). For example, CircFoxo3, which is mainly expressed in the cytoplasm, was proven to interact with the anti-senescence proteins ID1 and E2F1 and the anti-stress proteins FAK and HIF1a, arresting them in the cytoplasm and preventing them from moving to the nucleus, thus inhibiting their antisenescence and anti-stress functions [37]. In addition, circFoxo3 was reported to bind to the cell cycle proteins cyclin-dependent kinase 2 (CDK2) and cyclin-dependent kinase inhibitor 1 (p21), giving rise to a circ-Foxo3-p21-CDK2 ternary complex, thus arresting the cell cycle and blocking the transition from G1 to S phase [38]. Yang et al. [39] detected a circRNA, derived from angiomotin-like 1, which was colocalized with an oncogenic protein called c-myc in the nucleus, thus preventing the degradation of this protein and promoting the tumorigenesis of breast cancer. Overall, circRNAs could exert biological functions by interacting with cancer-associated proteins, suggesting their potential as novel promising targets in cancer therapy.
CircRNAs can be translated into proteins
CircRNAs are regarded as a type of distinct noncoding RNAs that cannot be translated into peptides due to their closed loop structure and lack of polyadenylated tail and internal ribosome entry site (IRES). In contrast, the majority of circRNAs are derived from protein-coding genes and contain complete exons, indicating the potential of circRNAs to be translated into polypeptides (Fig. 2d). To date, a portion of circRNAs has been proven to share this function. For example, Chen et al. [40] found that after inserting an IRES into a synthetic circRNA, circRNAs were able to bind to the eukaryotic ribosome and initiate the translation process both in vitro and in vivo. Additionally, it has been confirmed that viral circRNAs can be translated into proteins in eukaryotes. A typical example is circRNAs in hepatitis D virus (HDV), which possess an IRES and can be translated into the hepatitis D virus antigen (HDAg) after infection of human cells [41]. CircZNF609 is another circRNA that is proven to translate protein, providing a novel example of protein-coding circRNA in eukaryotic cells [42]. This circRNA possesses an open reading frame (ORF) with a start codon and a stop codon on both sides and can generate a protein due to the detected structure [42]. Besides, inserting an artificial circRNA into the green fluorescent protein (GFP) ORF could result in the production of extremely long protein chains in Escherichia coli [43]. Additionally, studies by Pamudurti et al. discovered that circMbl3 could be translated into polypeptides in fly heads [44].
As mounting evidence has determined the function of circRNAs as protein-coding RNAs, an increasing number of studies are focused on the specific mechanism of circRNA being translated into peptides. For example, Naoko et al. indicated that circRNAs possessing an infinite ORF can produce long protein chains via a rolling cycle amplification mechanism [45, 46]. In addition, studies of Yang et al. demonstrated a common mechanism of circRNAs being translated in a m6A-driven manner. N6-methyladenosine (m6A), one of the most abundant modifications in ncRNAs, was proved to facilitate translation initiation of circRNAs via acting as IRESs and researchers investigated that such a single modification is sufficient to initiate translation [47].
CircRNAs in OvCa
Currently, circRNAs have gradually become a novel research hotspot among ncRNAs and have subsequently captured the attention of the cancer research field. Numerous studies have demonstrated that circRNAs are differentially expressed in a variety of cancerous tissues. Moreover, an increasing number of studies have proven the presence of aberrantly expressed circRNAs in OvCa tissues, suggesting the relevance of circRNAs in the tumorigenesis and progression of OvCa. We discuss the potential diagnostic and prognostic values of circRNAs in OvCa and summarize the biological and cellular functions of several representative circRNAs in OvCa (Table 1).
Table 1.
Summarization of the cellular functions of circRNAs in tumorigenesis of OvCa
| CircRNAs | Circbase ID | Role in OvCa | Cancer phenotypes | Sponge miRNAs | Target genes | References |
|---|---|---|---|---|---|---|
| circMUC16 | hsa_circ_0049116 | Oncogene | Promoted proliferation and invasion | miR-199a-5p | Beclin1/RUNX1 | [55] |
| circCSPP1 | hsa_circ_0001806 | Oncogene | Promoted proliferation, migration, invasion and EMT | miR-1236-3p | ZEB1 | [58] |
| circEPSTI1 | hsa_circ_0000479 | Oncogene | Promoted proliferation, invasion and induced apoptosis | miR-942 | EPSTI1 | [61] |
| / | hsa_circ_0051240 | Oncogene | Promoted proliferation, migration, and invasion | miR-637 | KLK4 | [62] |
| circWHSC1 | hsa_circ_0001387 | Oncogene | Promoted proliferation, migration, invasion, EMT and induced apoptosis | miR-145/miR-1182 | MUC1/hTERT | [64] |
| / | hsa_circ_0061140 | Oncogene | Promoted proliferation, invasion and EMT | miR-370 | FOXM1 | [16] |
| circHIPK3 | hsa_circ_0000284 | Tumor suppressor | Inhibited proliferation, migration and invasion and promoted apoptosis | miR-10a-5p/miR-148b | / | [77] |
| circLARP4 | has_circ_101057 | Tumor suppressor | Inhibited proliferation and migration | miR-513b-5p | LARP4 | [81] |
| circPLEKHM3 | hsa_circ_0001095 | Tumor suppressor | Inhibited proliferation and migration | miR-9 | BRCA1/NAJB6/KLF4 | [82] |
| CircITCH | / | Tumor suppressor | Inhibited proliferation, migration, invasion and promoted apoptosis | miR-10a-α/miR-145 | RASA1 | [83, 84] |
CircRNAs function as diagnostic and prognostic biomarkers
Most functional circRNAs tend to exhibit differential expression patterns between OvCa patients and healthy individuals. Furthermore, circRNAs can reflect cancer progression and can be used to monitor the prognosis of OvCa patients, and thus have the potential to act as biomarkers in clinical practice.
Circulating cell-free circRNAs extracted from blood is a hotspot in the field of diagnostic biomarkers. According to published literature, circBNC2 could be an OvCa-specific diagnostic biomarker distinguishing early-stage OvCa patients from benign and healthy cohorts [48]. Circulating circABCB10 and circMAN1A2 also acted as diagnostic biomarkers for OvCa [49, 50]. Furthermore, Sheng et al. identified the circRNA profiles in OvCa specimens and demonstrated that circBNC2, circEXOC6B, circFAM13B, circN4BP2L2, circRHOBTB3, and circCELSR1 were notably dysregulated in OvCa tissues and may be promising diagnostic biomarkers for OvCa [51].
Since the prognosis of OvCa patients is quite poor, prognostic evaluation is essential for both clinicians and patients. Positive circABCB10 expression was significantly associated with poor OS in OvCa patients [50]. Low expression of circEXOC6B and circN4BP2L2 exhibited a tight association with OS and PFS in OvCa patients [51]. In addition, circRNA1656 was reported to be downregulated in both high-grade ovarian serous carcinoma (HGSOC) tissue and OvCa cell lines and correlated with the International Federation of Gynecology and Obstetrics (FIGO) stage of HGSOC, suggesting its potential to serve as a novel tumor marker for HGSOC [52].
On the other hand, several circRNAs have been shown to function as tumor suppressors in OvCa and serve as prognostic indicators. CircHIPK3 expression was higher in OvCa tissues than in matched adjacent noncancerous tissues, and higher circHIPK3 expression was positively associated with lymph node invasion, advanced FIGO stage, and worse DFS and OS in OvCa patients [53]. In addition, circLARP4 was proposed to decrease in OvCa tissues compared to adjacent normal tissues, and its lower expression was positively associated with advanced tumor FIGO stage, lymph node metastases, and worse disease-free survival (DFS) and OS [54].
CircRNAs act as tumor promoters
Hsa_circ_0049116
Hsa_circ_0049116, also named circMUC16, is spliced from the MUC16 gene. The expression of this circRNA was significantly elevated in OvCa tissues and serum of patients with OvCa. Moreover, highly expressed circMUC16 was reported to be positively associated with tumor stage and tumor grade. Gan et al. revealed that circMUC16 mediated autophagic flux in OvCa by regulating the circMUC16-miR-199a-5p-Beclin1/RUNX1 axis, thereby accelerating the proliferation and invasion of OvCa cells [55]. In addition, several studies have revealed that transcription factors can modulate the expression of circRNAs, and circMUC16 is a typical example. The expression of this circRNA was reported to be impacted by the downstream target runt-related transcription factor 1 (RUNX1) via transcriptional promotion. Gan et al. discovered that overexpression of RUNX1 facilitated MUC16 promoter activity and increased the expression of circMUC16, while silencing RUNX1 inhibited MUC16 promoter activity and decreased circMUC16 expression [55].
Hsa_circ_0001806
Hsa_circ_0001806, also called circCSPP1, is generated from the centrosome/spindle pole-associated protein 1 (CSPP1) gene. CSPP1, an oncogene located in the 8q13.2 region of the human genome, has been thoroughly investigated in luminal breast cancer and B-cell lymphoma [56, 57]. Additionally, circCSPP1 was found to be preferentially upregulated in borderline and tumor compared to benign and cancer specimens, and the expression of this circRNA was positively correlated with stage II–V [58]. Subsequently, Li et al. demonstrated that knockdown of circCSPP1 suppressed cell proliferation, migration, and invasion, while circCSPP1 overexpression promoted cancer behavior. In addition, miR-1236-3p, a tumor suppressor in OvCa, was negatively correlated with circCSPP1 in OvCa cell lines [58]. MiR-1236-3p was able to suppress cancer properties by targeting zinc finger E-box binding homeobox1 (ZEB1) in OvCa, while circCSPP1 was reported to sponge miR-1236-3p and suppress the silencing effect on ZEB1, thereby stimulating epithelial-to-mesenchymal transition (EMT) and cancer progression in OvCa [58, 59]. Moreover, Li et al. revealed that circCSPP1 exerted a positive impact on the expression of oncogenic proteins in OvCa, including vascular endothelial growth factor A (VEGFA) and matrix metalloproteinase-2 (MMP2) [58].
Hsa_circ_0000479
Hsa_circ_0000479, also named circEPSTI1, was reported to be upregulated in triple-negative breast cancer (TNBC) specimens, and the high expression of circEPSTI1 was related to decreased survival in these cases. It was demonstrated that circEPSTI1 was able to facilitate cell growth and suppress apoptosis by binding to miR-4753 and miR-6809 to upregulate BCLI1A expression in TNBC [60]. Recently, an increasing number of studies have confirmed the critical role of circEPSTI1 in OvCa development. CircEPSTI1 was remarkably upregulated in OvCa specimens compared to adjacent nontumor counterparts. In addition, circEPSTI1 was correlated with tumor size and lung metastasis in mouse xenografts, and this circRNA may potentially enhance OvCa cell proliferation and invasion and inhibit cell apoptosis in vivo [61]. Moreover, miR-942, a tumor suppressor in OvCa, was found to directly target the linear form of circEPSTI1, while circEPSTI1 was proven to sponge miR-942, suggesting a potential effect of circEPSTI1 on EPSTI1, which was downregulated in OvCa tissues [61].
Hsa_circ_0051240
Hsa_circ_0051240, derived from the gene CEACAM5, is another circRNA that is upregulated in OvCa. This highly expressed circRNA was reported to enhance cell proliferation, invasion and migration by targeting miR-637 to regulate the expression of Kallikrein-related peptidases 4 (KLK4), and KLK4 was reported to be involved in many steps of OvCa tumorigenesis and progression and to exert modulatory effects on a subset of cancer-related genes and proteins in several cancers [62, 63].
Hsa_circ_0001387
Hsa_circ_0001387, also called circWHSC1, is derived from the WHSC1 gene through the exonic back-splicing process. This circRNA was confirmed to function as an oncogenic circRNA and may represent a novel promising biomarker and therapeutic strategy for OvCa. Zong et al. found that circWHSC1 was highly expressed in OvCa and was able to adsorb miR-145 and miR-1182 to upregulate the expression of downstream targets Mucin 1 (MUC1) and human telomerase reverse transcriptase (hTERT) [64]. MUC1 is a highly expressed gene in a variety of cancers, including OvCa, and is known to participate in tumorigenicity, initiate the EMT process, and coordinate cancer metastasis [65–67]. HTERT was found to be essential in cancer cell behavior, and ectopic expression of this oncogene could induce EMT by upregulating Slug [68, 69]. Importantly, circWHSC1 can be secreted into exosomes. Zong et al. claimed that this circRNA can be absorbed by peritoneal mesothelial cells in the form of exosomes and facilitates EMT changes, thus triggering tumor distribution in the peritoneal cavity and promoting the development of OvCa [64].
Hsa_circ_0061140
Chen et al. demonstrated that hsa_circ_0061140 was significantly upregulated in OvCa cell lines using real-time qPCR. Subsequent experimental data suggested that hsa_circ_0061140 silencing caused an arrest in cell proliferation and invasion as well as EMT in SKOV3 and A2780 cells by sponging miR-370 to inhibit the expression of forkhead box protein M1 (FOXM1) [16]. MiR-370 is known to be decreased in OvCa and acts as an anticancer gene. Overexpression of miR-370 was reported to inhibit cell growth and invasion in a variety of cancers, including breast cancer [70], bladder cancer [71], and hepatocellular carcinoma [72], while silencing of hsa_circ_0061140 was reported to increase miR-370 expression. Bioinformatics analyses revealed that FOXM1, a transcription factor that is involved in the cell cycle and cell proliferation, may potentially be the downstream target of miR-370 [16]. Therefore, overexpression of FOXM1 was detected to rescue miR-370-mediated inhibition of migration by wound healing and transwell analyses and rescue miR-370-induced EMT inhibition by western blot [16]. Overall, hsa_circ_0061140 upregulated FOXM1 expression by binding to miR-370, providing a promising therapeutic target for OvCa.
CircRNAs act as tumor suppressors
Hsa_circ_0000284
Hsa_circ_0000284, also called circHIPK3, is localized primarily in the cytoplasm of OvCa cells and was found to exhibit different functions in different cells or tissues. Zheng et al. and Shan et al. discovered that knockdown of circHIPK3 could inhibit the proliferation of retinal endothelial cells and HEK-293 T cells [73, 74]. Zeng et al. claimed that silencing circHIPK3 inhibited cell invasion and migration and promoted apoptosis in colorectal cancer [75]. In contrast, Li et al. found that overexpression of this circRNA inhibited cell migration and invasion in bladder cancer [76].
Interestingly, the concrete function of circHIPK3 in OvCa varies extremely. In the study by Teng et al., qRT-PCR and the expression profile of circHIPK3 both confirmed that this circRNA was decreased in epithelial OvCa cell lines compared with normal ovarian tissue cells, while Liu et al. discovered that the expression of this circRNA was significantly increased in OvCa tissues compared to their matched adjacent noncancerous counterparts [53]. The same study by Teng et al. revealed that in both normal ovarian epithelial cells (IOSE80) and OvCa cells (A2780 and SKOV3) silencing of circHIPK3 could facilitate OvCa processes, including cell proliferation, migration, and invasion, and inhibited cell apoptosis, providing a novel example of cell-type-specific expression and function of circRNA [77].
Additionally, a growing number of studies have concentrated on the function of circHIPK3 as a miRNA sponge. For instance, circHIPK3 has been shown to sponge miR-30a-3p in diabetes mellitus, miR-558 in bladder cancer cells, miR-7 in colorectal cancer, and miR-124 in HeLa cells [73–76]. It was reported that this circRNA was capable of sponging miR-193-3p to restore MCL1 expression, which was remarkably expressed in numerous cancers [78]. In the study by Teng et al., circHIPK3 was found to possess 1–3 binding sites each of 12 miRNAs in OvCa, namely, miR-224-3p, miR-579-3p, miR-10a-5p, miR-148b-3p, miR-876-5p, miR-193a-3p, miR-30d-3p, miR-499a-5p, miR-522-3p, miR-1278, miR-106a-3p and miR-30b-3p [77]. Moreover, a miR-106a inhibitor was demonstrated to restore OvCa cell proliferation in xenograft mice [79]. MiR-148b was overexpressed in 92.21% of OvCa tissues and might exert a significant function in the tumorigenesis and progression of OvCa [80]. Therefore, circHIPK3 might exhibit its biological function in the tumorigenesis of OvCa by sponging one or several of the 12 miRNAs.
Hsa_circ_101057
Hsa_circ_101057, also termed circLARP4, is derived from exons 9 and 10 within the La ribonucleoprotein domain family member 4 (LARP4) locus, which is located on chromosome 12q13.12. This circRNA may serve as a tumor suppressor in OvCa. Additionally, Lin et al. discovered that circRNA expression was significantly decreased in OvCa cell lines compared with the normal group. Additionally, circLARP4 could upregulate its host gene LARP4 posttranscriptionally by sponging miR-513b-5p, thereby inhibiting cell proliferation and migration [81]. Overall, circLARP4 suppressed the development of OvCa via the miR-513b-5p/LARP4 axis, providing a promising therapeutic target for OvCa treatment. However, further clinical investigations with larger samples are needed to confirm the suppressive function of this circRNA.
Hsa_circ_0001095
Hsa_circ_0001095, also called circPLEKHM3, is spliced from exons of PLEKHM3 on the reverse strand of chromosome 2. CircPLEKHM3 was remarkably downregulated in OvCa tissues and predicted poor prognosis. Mechanistically, circPLEKHM3 inactivated the AKT1 and canonical Wnt/β-catenin signaling pathways by sponging miR-9 to modulate the expression of KLF4, DNAJB6, and BRCA1, thus suppressing OvCa cell proliferation and migration. Interestingly, Zhang et al. suggested that the combination of MK-2206 and Taxol exhibits a synergetic therapeutic effect on OvCa patients, and this synergetic treatment was elevated in OvCa cells with a loss of circPLEKHM3 expression, providing a novel example of circRNA involved in drug treatment cases [82].
CircITCH
Several studies have shown that miR-10a, a noncoding miRNA located on chromosome 17, is capable of promoting the progression of several cancers, and emerging studies have focused on the function of this miRNA in OvCa. Luo et al. detected a decrease in circITCH expression in human OvCa cell lines compared to normal human ovarian epithelial cell lines and determined the effect of circITCH on miR-10a levels. Overexpression of circITCH significantly suppressed cell growth and promoted cell apoptosis via direct interaction with miR-10a [83]. Additionally, Hu and colleagues claimed that circITCH acted as a ceRNA to interact with miR-145, upregulated the level of RASA1, and inhibited the cancerous properties of OvCa cells by regulating the circITCH-miR-145-RASA1 axis in vitro and in vivo [84]. Hence, circITCH may inhibit the malignant progression of OvCa by targeting different miRNAs. However, further investigations are needed to identify the specific downstream gene affected by circITCH acting as a miRNA sponge.
Future perspective
Accumulating studies have confirmed the critical function of circRNAs in the pathogenesis of numerous diseases. More recently, functional peptides produced by circRNAs have emerged into scientific attention. For example, SHPRH-146aa, encoded by circ-SHPRH, inhibits progression of central nervous system cancer via regulating protein ubiquitination pathways to protect the quality of SNF2 histone linker PHD RING helicase (SHPRH) gene [85, 86]. AKT3-174aa, another tumor-related functional peptide encoded by circ-AKT3, was less expressed in glioblastoma in comparison with normal brain tissues and was proved to suppress cancer properties through negatively regulating the PI3K/Akt signaling pathway in glioblastoma [87]. Evidence have certificated the existence and function of circRNA encoded peptides, however, the systematic detection of such peptides remains difficult due to the technical challenges as well as the similarity between circRNA-encoded peptides and the counterpart of mRNA. Therefore, it is essential to conduct in-depth researches on circRNA-encoded peptides including the specific mechanism of such peptides involving in the pathogenesis of diseases, whether such peptides undergo modification post-translationally as well as the associated condition and factors regulating the translation of circRNAs.
Also, the function of circRNAs in the tumor microenvironment (TME) have been studied, including regulating tumor immune surveillance via serving as immune system antigens [88], mediating immune escape via circRNA-miRNA-PD-1/PD-L1 axis [89], regulating the cytotoxicity of natural killer cells by acting as miRNA sponges and modulating associated proteins [90], promoting or inhibiting angiogenesis through multiple mechanisms [91] and inhibiting the permeability of endothelial cell [92]. Initial results have demonstrated the impact of circRNAs on TME, however, researches in correlation with this topic are in its infancy and further researches need to identify the clinical relevance on this topic.
As well known, hypoxia plays an essential role in cancer aggressiveness and recent researches have focused on the correlation between hypoxia and circRNAs [93]. Several circRNAs have been reported to be impacted by hypoxia including circDENND4C [94], circDENND2A [95], circ-0000977 [96], and circ-0010729 [97]. Detecting the mechanism of circRNAs in hypoxia conditions may provide a new insight into the potential utilities of circRNAs.
Conclusion
It is no exaggeration to say that circRNAs have become a novel frontier in cancer research and have captured extensive attention from researchers. OvCa-associated circRNAs exert their biological functions in OvCa principally by regulating cell proliferation, invasion, migration, apoptosis, EMT, and cell cycle. Currently, an increasing number of circRNA transcriptomes in OvCa have been revealed, and a few circRNAs have been identified as functional ncRNAs associated with clinical performance. Notably, the characterized cancer-related circRNAs have been shown to play significant roles in the tumorigenesis and progression of OvCa and display superiority in clinical application due to their advantages in diagnosis, prognosis, and therapy. However, the number of experimental samples used to investigate the expression profile and specific function of circRNAs has been relatively small. Therefore, it is urgent to conduct comprehensive studies in a larger number of specimens to confirm the transcriptome sequencing data and functions of circRNAs in OvCa. In particular, the current research mainly revealed the function of circRNAs as miRNA sponges, suggesting that further investigations are needed to explore other potential mechanisms of circRNAs in the pathogenesis of OvCa.
Acknowledgements
Not applicable.
Abbreviations
- circRNA
Circular RNA
- ncRNAs
Noncoding RNAs
- OvCa
Ovarian cancer
- OS
Overall survival
- Pol II
Polymerase II
- ecircRNA
Exonic circRNA
- ciRNA
Circular intronic RNA
- EIciRNAs
Exon–intron circRNA
- RBP
RNA binding protein
- ceRNA
Competing endogenous RNA
- MREs
miRNA response elements
- lncRNA
Long noncoding RNA
- LIMK1
LIM domain kinase 1
- snRNPs
Small nuclear ribonucleic proteins
- CDK2
Cyclin-dependent kinase 2
- p21
Cyclin-dependent kinase inhibitor 1
- IRES
Internal ribosome entry site
- HDV
Hepatitis D virus
- HDAg
Hepatitis D virus antigen
- ORF
Open reading frame
- GFP
Green fluorescent protein
- m6A
N6-methyladenosine
- HGSOC
High-grade ovarian serous carcinoma
- FIGO
International Federation of Gynecology and Obstetrics
- DFS
Disease-free survival
- RUNX1
Runt-related transcription factor 1
- EMT
Epithelial-to-mesenchymal transition
- VEGFA
Vascular endothelial growth factor A
- MMP2
Matrix metalloproteinase-2
- TNBC
Triple-negative breast cancer
- KLK4
Kallikrein-related peptidase 4
- MUC1
Mucin 1
- hTERT
Human telomerase reverse transcriptase
- FOXM1
Forkhead box protein M1
- LARP4
La ribonucleoprotein domain family member 4
- SHPRH
SNF2 histone linker PHD RING helicase
- TME
Tumor microenvironment
Authors’ contributions
CL and JD conceived the study and participated in the study design, performance, coordination and manuscript writing. XY, JM, HW and DG carried out the literature review and graphics production. CL and JD revised the manuscript. All authors reviewed the final manuscript. All authors read and approved the final manuscript.
Funding
This work was founded by the Natural Science Foundation of Jiangsu Province (BE2017626), the Six talent peaks project in Jiangsu Province (WSN-186) and the Foundation of Wuxi Health Commission (QNRC003).
Availability of data and materials
All data are included in the article.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no conflicts of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xuejing Yang and Jie Mei contributed equally to this work
Contributor Information
Xuejing Yang, Email: yxjdoctor@163.com.
Jie Mei, Email: meijie1996@njmu.edu.cn.
Huiyu Wang, Email: wanghuiyu2011@126.com.
Dingyi Gu, Email: 961228667@qq.com.
Junli Ding, Email: dingjunliletters@163.com.
Chaoying Liu, Email: liuchaoying666@163.com.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 2.Coleman RL, Monk BJ, Sood AK, Herzog TJ. Latest research and treatment of advanced-stage epithelial ovarian cancer. Nat Rev Clin Oncol. 2013;10(4):211–224. doi: 10.1038/nrclinonc.2013.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Waldmann A, Eisemann N, Katalinic A. Epidemiology of malignant cervical, corpus uteri and ovarian tumours—current data and epidemiological trends. Geburtshilfe Frauenheilkd. 2013;73(2):123–129. doi: 10.1055/s-0032-1328266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davalos V, Esteller M. Disruption of long noncoding RNAs targets cancer hallmark pathways in lung tumorigenesis. Cancer Res. 2019;79(12):3028–3030. doi: 10.1158/0008-5472.CAN-19-0910. [DOI] [PubMed] [Google Scholar]
- 5.Chan JJ, Tay Y. Noncoding RNA: RNA regulatory networks in cancer. Int J Mol Sci. 2018;19(5):1310. doi: 10.3390/ijms19051310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shen T, Han M, Wei G, Ni T. An intriguing RNA species-perspectives of circularized RNA. Protein Cell. 2015;6(12):871–880. doi: 10.1007/s13238-015-0202-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aufiero S, Reckman YJ, Pinto YM, Creemers EE. Circular RNAs open a new chapter in cardiovascular biology. Nat Rev Cardiol. 2019;16(8):503–514. doi: 10.1038/s41569-019-0185-2. [DOI] [PubMed] [Google Scholar]
- 8.Fang Y, Wang X, Li W, Han J, Jin J, Su F, et al. Screening of circular RNAs and validation of circANKRD36 associated with inflammation in patients with type 2 diabetes mellitus. Int J Mol Med. 2018;42(4):1865–1874. doi: 10.3892/ijmm.2018.3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Q, Qu L, Chen X, Zhao Y-H, Luo Q. Progress in understanding the relationship between circular RNAs and neurological disorders. J Mol Neurosci. 2018;65(4):546–556. doi: 10.1007/s12031-018-1125-z. [DOI] [PubMed] [Google Scholar]
- 10.Ouyang Q, Wu J, Jiang Z, Zhao J, Wang R, Lou A, et al. Microarray expression profile of circular RNAs in peripheral blood mononuclear cells from rheumatoid arthritis patients. Cell Physiol Biochem. 2017;42(2):651–659. doi: 10.1159/000477883. [DOI] [PubMed] [Google Scholar]
- 11.Lei M, Zheng G, Ning Q, Zheng J, Dong D. Translation and functional roles of circular RNAs in human cancer. Mol Cancer. 2020;19(1):30. doi: 10.1186/s12943-020-1135-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Verduci L, Strano S, Yarden Y, Blandino G. The circRNA-microRNA code: emerging implications for cancer diagnosis and treatment. Mol Oncol. 2019;13(4):669–680. doi: 10.1002/1878-0261.12468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kristensen LS, Hansen TB, Venø MT, Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37(5):555–565. doi: 10.1038/onc.2017.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meng S, Zhou H, Feng Z, Xu Z, Tang Y, Li P, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16(1):94. doi: 10.1186/s12943-017-0663-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qin H, Liu J, Du ZH, Hu R, Yu YK, Wang QA. Circular RNA hsa_circ_0012673 facilitates lung cancer cell proliferation and invasion via miR-320a/LIMK18521 axis. Eur Rev Med Pharmacol Sci. 2020;24(4):1841–1852. doi: 10.26355/eurrev_202002_20362. [DOI] [PubMed] [Google Scholar]
- 16.Chen Q, Zhang J, He Y, Wang Y. hsa_circ_0061140 knockdown reverses FOXM1-mediated cell growth and metastasis in ovarian cancer through miR-370 sponge activity. Mol Ther Nucleic Acids. 2018;13:55–63. doi: 10.1016/j.omtn.2018.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci USA. 1976;73(11):3852–3856. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–691. doi: 10.1038/s41576-019-0158-7. [DOI] [PubMed] [Google Scholar]
- 19.Harland R, Misher L. Stability of RNA in developing Xenopus embryos and identification of a destabilizing sequence in TFIIIA messenger RNA. Development. 1988;102(4):837–852. doi: 10.1242/dev.102.4.837. [DOI] [PubMed] [Google Scholar]
- 20.Ahmed I, Karedath T, Andrews SS, Al-Azwani IK, Mohamoud YA, Querleu D, et al. Altered expression pattern of circular RNAs in primary and metastatic sites of epithelial ovarian carcinoma. Oncotarget. 2016;7(24):36366–36381. doi: 10.18632/oncotarget.8917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang X-O, Wang H-B, Zhang Y, Lu X, Chen L-L, Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159(1):134–147. doi: 10.1016/j.cell.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 22.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17(4):205–211. doi: 10.1038/nrm.2015.32. [DOI] [PubMed] [Google Scholar]
- 24.He J, Xie Q, Xu H, Li J, Li Y. Circular RNAs and cancer. Cancer Lett. 2017;396:138–144. doi: 10.1016/j.canlet.2017.03.027. [DOI] [PubMed] [Google Scholar]
- 25.Ebbesen KK, Kjems J, Hansen TB. Circular RNAs: identification, biogenesis and function. Biochim Biophys Acta. 2016;1859(1):163–168. doi: 10.1016/j.bbagrm.2015.07.007. [DOI] [PubMed] [Google Scholar]
- 26.Barrett SP, Wang PL, Salzman J. Circular RNA biogenesis can proceed through an exon-containing lariat precursor. Elife. 2015;4:e07540. doi: 10.7554/eLife.07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160(6):1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, et al. Circular intronic long noncoding RNAs. Mol Cell. 2013;51(6):792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 29.Petkovic S, Müller S. RNA circularization strategies in vivo and in vitro. Nucleic Acids Res. 2015;43(4):2454–2465. doi: 10.1093/nar/gkv045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao R, Li FQ, Tian LL, Shang DS, Guo Y, Zhang JR, et al. Comprehensive analysis of the whole coding and non-coding RNA transcriptome expression profiles and construction of the circRNA-lncRNA co-regulated ceRNA network in laryngeal squamous cell carcinoma. Funct Integr Genomics. 2019;19(1):109–121. doi: 10.1007/s10142-018-0631-y. [DOI] [PubMed] [Google Scholar]
- 31.Xiong DD, Dang YW, Lin P, Wen DY, He RQ, Luo DZ, et al. A circRNA–miRNA–mRNA network identification for exploring underlying pathogenesis and therapy strategy of hepatocellular carcinoma. J Transl Med. 2018;16(1):220. doi: 10.1186/s12967-018-1593-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pan H, Li T, Jiang Y, Pan C, Ding Y, Huang Z, et al. Overexpression of circular RNA ciRS-7 abrogates the tumor suppressive effect of miR-7 on gastric cancer via PTEN/PI3K/AKT signaling pathway. J Cell Biochem. 2018;119(1):440–446. doi: 10.1002/jcb.26201. [DOI] [PubMed] [Google Scholar]
- 34.Wan L, Zhang L, Fan K, Cheng Z-X, Sun Q-C, Wang J-J. Circular RNA-ITCH suppresses lung cancer proliferation via inhibiting the Wnt/β-catenin pathway. Biomed Res Int. 2016;2016:1579490. doi: 10.1155/2016/1579490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, et al. Circular RNA: a new star of noncoding RNAs. Cancer Lett. 2015;365(2):141–148. doi: 10.1016/j.canlet.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 36.Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22(3):256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 37.Du WW, Yang W, Chen Y, Wu Z-K, Foster FS, Yang Z, et al. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur Heart J. 2017;38(18):1402–1412. doi: 10.1093/eurheartj/ehw001. [DOI] [PubMed] [Google Scholar]
- 38.Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44(6):2846–2858. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang Q, Du WW, Wu N, Yang W, Awan FM, Fang L, et al. A circular RNA promotes tumorigenesis by inducing c-myc nuclear translocation. Cell Death Differ. 2017;24(9):1609–1620. doi: 10.1038/cdd.2017.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen CY, Sarnow P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science. 1995;268(5209):415–417. doi: 10.1126/science.7536344. [DOI] [PubMed] [Google Scholar]
- 41.Kos A, Dijkema R, Arnberg AC, van der Meide PH, Schellekens H. The hepatitis delta (delta) virus possesses a circular RNA. Nature. 1986;323(6088):558–560. doi: 10.1038/323558a0. [DOI] [PubMed] [Google Scholar]
- 42.Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell. 2017;66(1):22–37.e9. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perriman R, Ares M., Jr Circular mRNA can direct translation of extremely long repeating-sequence proteins in vivo. RNA. 1998;4(9):1047–1054. doi: 10.1017/s135583829898061x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, et al. Translation of CircRNAs. Mol Cell. 2017;66(1):9–21.e7. doi: 10.1016/j.molcel.2017.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abe N, Matsumoto K, Nishihara M, Nakano Y, Shibata A, Maruyama H, et al. Rolling circle translation of circular RNA in living human cells. Sci Rep. 2015;5:16435. doi: 10.1038/srep16435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abe N, Hiroshima M, Maruyama H, Nakashima Y, Nakano Y, Matsuda A, et al. Rolling circle amplification in a prokaryotic translation system using small circular RNA. Angew Chem Int Ed Engl. 2013;52(27):7004–7008. doi: 10.1002/anie.201302044. [DOI] [PubMed] [Google Scholar]
- 47.Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. 2017;27(5):626–641. doi: 10.1038/cr.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu Y, Zhu Y, Zhang W, Lang J, Ning L. Utility of plasma circBNC2 as a diagnostic biomarker in epithelial ovarian cancer. Onco Targets Ther. 2019;12:9715–9723. doi: 10.2147/OTT.S211413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fan CM, Wang JP, Tang YY, Zhao J, He SY, Xiong F, et al. circMAN1A2 could serve as a novel serum biomarker for malignant tumors. Cancer Sci. 2019;110(7):2180–2188. doi: 10.1111/cas.14034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen Y, Ye X, Xia X, Lin X. Circular RNA ABCB10 correlates with advanced clinicopathological features and unfavorable survival, and promotes cell proliferation while reduces cell apoptosis in epithelial ovarian cancer. Cancer Biomark. 2019;26(2):151–161. doi: 10.3233/CBM-190064. [DOI] [PubMed] [Google Scholar]
- 51.Ning L, Long B, Zhang W, Yu M, Wang S, Cao D, et al. Circular RNA profiling reveals circEXOC6B and circN4BP2L2 as novel prognostic biomarkers in epithelial ovarian cancer. Int J Oncol. 2018;53(6):2637–2646. doi: 10.3892/ijo.2018.4566. [DOI] [PubMed] [Google Scholar]
- 52.Gao Y, Zhang C, Liu Y, Wang M. Circular RNA profiling reveals circRNA1656 as a novel biomarker in high grade serous ovarian cancer. Biosci Trends. 2019;13(2):204–211. doi: 10.5582/bst.2019.01021. [DOI] [PubMed] [Google Scholar]
- 53.Liu N, Zhang J, Zhang LY, Wang L. CircHIPK3 is upregulated and predicts a poor prognosis in epithelial ovarian cancer. Eur Rev Med Pharmacol Sci. 2018;22(12):3713–3718. doi: 10.26355/eurrev_201806_15250. [DOI] [PubMed] [Google Scholar]
- 54.Zou T, Wang PL, Gao Y, Liang WT. Circular RNA_LARP4 is lower expressed and serves as a potential biomarker of ovarian cancer prognosis. Eur Rev Med Pharmacol Sci. 2018;22(21):7178–7182. doi: 10.26355/eurrev_201811_16250. [DOI] [PubMed] [Google Scholar]
- 55.Gan X, Zhu H, Jiang X, Obiegbusi SC, Yong M, Long X, et al. CircMUC16 promotes autophagy of epithelial ovarian cancer via interaction with ATG13 and miR-199a. Mol Cancer. 2020;19(1):45. doi: 10.1186/s12943-020-01163-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Adélaïde J, Finetti P, Bekhouche I, Repellini L, Geneix J, Sircoulomb F, et al. Integrated profiling of basal and luminal breast cancers. Cancer Res. 2007;67(24):11565–11575. doi: 10.1158/0008-5472.CAN-07-2536. [DOI] [PubMed] [Google Scholar]
- 57.Patzke S, Hauge H, Sioud M, Finne EF, Sivertsen EA, Delabie J, et al. Identification of a novel centrosome/microtubule-associated coiled-coil protein involved in cell-cycle progression and spindle organization. Oncogene. 2005;24(7):1159–1173. doi: 10.1038/sj.onc.1208267. [DOI] [PubMed] [Google Scholar]
- 58.Li Q-H, Liu Y, Chen S, Zong Z-H, Du Y-P, Sheng X-J, et al. Circ-CSPP1 promotes proliferation, invasion and migration of ovarian cancer cells by acting as a miR-1236-3p sponge. Biomed Pharmacother. 2019;114:108832. doi: 10.1016/j.biopha.2019.108832. [DOI] [PubMed] [Google Scholar]
- 59.Wang Y, Yan S, Liu X, Zhang W, Li Y, Dong R, et al. miR-1236-3p represses the cell migration and invasion abilities by targeting ZEB1 in high-grade serous ovarian carcinoma. Oncol Rep. 2014;31(4):1905–1910. doi: 10.3892/or.2014.3046. [DOI] [PubMed] [Google Scholar]
- 60.Chen B, Wei W, Huang X, Xie X, Kong Y, Dai D, et al. CircEPSTI1 as a prognostic marker and mediator of triple-negative breast cancer progression. Theranostics. 2018;8(14):4003–4015. doi: 10.7150/thno.24106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xie J, Wang S, Li G, Zhao X, Jiang F, Liu J, et al. circEPSTI1 regulates ovarian cancer progression via decoying miR-942. J Cell Mol Med. 2019;23(5):3597–3602. doi: 10.1111/jcmm.14260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang M, Xia B, Xu Y, Zhang Y, Xu J, Lou G. Circular RNA (hsa_circ_0051240) promotes cell proliferation, migration and invasion in ovarian cancer through miR-637/KLK4 axis. Artif Cells Nanomed Biotechnol. 2019;47(1):1224–1233. doi: 10.1080/21691401.2019.1593999. [DOI] [PubMed] [Google Scholar]
- 63.Wang P, Magdolen V, Seidl C, Dorn J, Drecoll E, Kotzsch M, et al. Kallikrein-related peptidases 4, 5, 6 and 7 regulate tumour-associated factors in serous ovarian cancer. Br J Cancer. 2018;119(7):1–9. doi: 10.1038/s41416-018-0260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zong Z-H, Du Y-P, Guan X, Chen S, Zhao Y. CircWHSC1 promotes ovarian cancer progression by regulating MUC1 and hTERT through sponging miR-145 and miR-1182. J Exp Clin Cancer Res. 2019;38(1):437. doi: 10.1186/s13046-019-1437-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tagde A, Rajabi H, Bouillez A, Alam M, Gali R, Bailey S, et al. MUC1-C drives MYC in multiple myeloma. Blood. 2016;127(21):2587–2597. doi: 10.1182/blood-2015-07-659151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Deng J, Wang L, Chen H, Li L, Ma Y, Ni J, et al. The role of tumour-associated MUC1 in epithelial ovarian cancer metastasis and progression. Cancer Metastasis Rev. 2013;32(3–4):535–551. doi: 10.1007/s10555-013-9423-y. [DOI] [PubMed] [Google Scholar]
- 67.Roy LD, Sahraei M, Subramani DB, Besmer D, Nath S, Tinder TL, et al. MUC1 enhances invasiveness of pancreatic cancer cells by inducing epithelial to mesenchymal transition. Oncogene. 2011;30(12):1449–1459. doi: 10.1038/onc.2010.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Choi MJ, Cho KH, Lee S, Bae YJ, Jeong KJ, Rha SY, et al. hTERT mediates norepinephrine-induced Slug expression and ovarian cancer aggressiveness. Oncogene. 2015;34(26):3402–3412. doi: 10.1038/onc.2014.270. [DOI] [PubMed] [Google Scholar]
- 69.Takakura M, Kyo S, Kanaya T, Hirano H, Takeda J, Yutsudo M, et al. Cloning of human telomerase catalytic subunit (hTERT) gene promoter and identification of proximal core promoter sequences essential for transcriptional activation in immortalized and cancer cells. Cancer Res. 1999;59(3):551–557. [PubMed] [Google Scholar]
- 70.Hagstrom AD, Denham J. microRNAs in high and low responders to resistance training in breast cancer survivors. Int J Sports Med. 2018;39(6):482–489. doi: 10.1055/a-0592-7691. [DOI] [PubMed] [Google Scholar]
- 71.Huang X, Zhu H, Gao Z, Li J, Zhuang J, Dong Y, et al. Wnt7a activates canonical Wnt signaling, promotes bladder cancer cell invasion, and is suppressed by miR-370-3p. J Biol Chem. 2018;293(18):6693–6706. doi: 10.1074/jbc.RA118.001689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pan XP, Huang LH, Wang X. MiR-370 functions as prognostic marker in patients with hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 2017;21(16):3581–3585. [PubMed] [Google Scholar]
- 73.Shan K, Liu C, Liu B-H, Chen X, Dong R, Liu X, et al. Circular noncoding RNA HIPK3 mediates retinal vascular dysfunction in diabetes mellitus. Circulation. 2017;136(17):1629–1642. doi: 10.1161/CIRCULATIONAHA.117.029004. [DOI] [PubMed] [Google Scholar]
- 74.Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zeng K, Chen X, Xu M, Liu X, Hu X, Xu T, et al. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. 2018;9(4):417. doi: 10.1038/s41419-018-0454-8. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 76.Li Y, Zheng F, Xiao X, Xie F, Tao D, Huang C, et al. CircHIPK3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. EMBO Rep. 2017;18(9):1646–1659. doi: 10.15252/embr.201643581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Teng F, Xu J, Zhang M, Liu S, Gu Y, Zhang M, et al. Comprehensive circular RNA expression profiles and the tumor-suppressive function of circHIPK3 in ovarian cancer. Int J Biochem Cell Biol. 2019;112:8–17. doi: 10.1016/j.biocel.2019.04.011. [DOI] [PubMed] [Google Scholar]
- 78.Chen D, Lu X, Yang F, Xing N. Circular RNA circHIPK3 promotes cell proliferation and invasion of prostate cancer by sponging miR-193a-3p and regulating MCL1 expression. Cancer Manage Res. 2019;11:1415–1423. doi: 10.2147/CMAR.S190669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cai Z-H, Chen L-M, Liang Y-J, Shi J-R, You-Ju MA, Wang W-M, et al. Experimental study on the inhibition effect of miR-106a inhibitor on tumor growth of ovarian cancer xenografts mice. Asian Pac J Trop Med. 2016;9(7):698–701. doi: 10.1016/j.apjtm.2016.05.008. [DOI] [PubMed] [Google Scholar]
- 80.Chang H, Zhou X, Wang Z-N, Song Y-X, Zhao F, Gao P, et al. Increased expression of miR-148b in ovarian carcinoma and its clinical significance. Mol Med Rep. 2012;5(5):1277–1280. doi: 10.3892/mmr.2012.794. [DOI] [PubMed] [Google Scholar]
- 81.Lin W, Ye H, You K, Chen L. Up-regulation of circ_LARP4 suppresses cell proliferation and migration in ovarian cancer by regulating miR-513b-5p/LARP4 axis. Cancer Cell Int. 2020;20:5. doi: 10.1186/s12935-019-1071-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang L, Zhou Q, Qiu Q, Hou L, Wu M, Li J, et al. CircPLEKHM3 acts as a tumor suppressor through regulation of the miR-9/BRCA1/DNAJB6/KLF4/AKT1 axis in ovarian cancer. Mol Cancer. 2019;18(1):144. doi: 10.1186/s12943-019-1080-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Luo L, Gao YQ, Sun XF. Circular RNA ITCH suppresses proliferation and promotes apoptosis in human epithelial ovarian cancer cells by sponging miR-10a-α. Eur Rev Med Pharmacol Sci. 2018;22(23):8119–8126. doi: 10.26355/eurrev_201812_16503. [DOI] [PubMed] [Google Scholar]
- 84.Hu J, Wang L, Chen J, Gao H, Zhao W, Huang Y, et al. The circular RNA circ-ITCH suppresses ovarian carcinoma progression through targeting miR-145/RASA1 signaling. Biochem Biophys Res Commun. 2018;505(1):222–228. doi: 10.1016/j.bbrc.2018.09.060. [DOI] [PubMed] [Google Scholar]
- 85.Zhang M, Huang N, Yang X, Luo J, Yan S, Xiao F, et al. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene. 2018;37(13):1805–1814. doi: 10.1038/s41388-017-0019-9. [DOI] [PubMed] [Google Scholar]
- 86.Begum S, Yiu A, Stebbing J, Castellano L. Novel tumour suppressive protein encoded by circular RNA, circ-SHPRH, in glioblastomas. Oncogene. 2018;37(30):4055–4057. doi: 10.1038/s41388-018-0230-3. [DOI] [PubMed] [Google Scholar]
- 87.Xia X, Li X, Li F, Wu X, Zhang M, Zhou H, et al. A novel tumor suppressor protein encoded by circular AKT3 RNA inhibits glioblastoma tumorigenicity by competing with active phosphoinositide-dependent Kinase-1. Mol Cancer. 2019;18(1):131. doi: 10.1186/s12943-019-1056-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen YG, Kim MV, Chen X, Batista PJ, Aoyama S, Wilusz JE, et al. Sensing self and foreign circular RNAs by intron identity. Mol Cell. 2017;67(2):228–238.e5. doi: 10.1016/j.molcel.2017.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang X-L, Xu L-L, Wang F. Hsa_circ_0020397 regulates colorectal cancer cell viability, apoptosis and invasion by promoting the expression of the miR-138 targets TERT and PD-L1. Cell Biol Int. 2017;41(9):1056–1064. doi: 10.1002/cbin.10826. [DOI] [PubMed] [Google Scholar]
- 90.Ma Y, Zhang C, Zhang B, Yu H, Yu Q. circRNA of AR-suppressed PABPC1 91 bp enhances the cytotoxicity of natural killer cells against hepatocellular carcinoma via upregulating UL16 binding protein 1. Oncol Lett. 2019;17(1):388–397. doi: 10.3892/ol.2018.9606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang Q, Wang W, Zhou Q, Chen C, Yuan W, Liu J, et al. Roles of circRNAs in the tumour microenvironment. Mol Cancer. 2020;19(1):14. doi: 10.1186/s12943-019-1125-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li J, Li Z, Jiang P, Peng M, Zhang X, Chen K, et al. Circular RNA IARS (circ-IARS) secreted by pancreatic cancer cells and located within exosomes regulates endothelial monolayer permeability to promote tumor metastasis. J Exp Clin Cancer Res. 2018;37(1):177. doi: 10.1186/s13046-018-0822-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Peng X, Gao H, Xu R, Wang H, Mei J, Liu C. The interplay between HIF-1alpha and noncoding RNAs in cancer. J Exp Clin Cancer Res. 2020;39(1):27. doi: 10.1186/s13046-020-1535-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liang G, Liu Z, Tan L, Su AN, Jiang WG, Gong C. HIF1α-associated circDENND4C promotes proliferation of breast cancer cells in hypoxic environment. Anticancer Res. 2017;37(8):4337–4343. doi: 10.21873/anticanres.11827. [DOI] [PubMed] [Google Scholar]
- 95.Su H, Zou D, Sun Y, Dai Y. Hypoxia-associated circDENND2A promotes glioma aggressiveness by sponging miR-625-5p. Cell Mol Biol Lett. 2019;24:24. doi: 10.1186/s11658-019-0149-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ou Z-L, Luo Z, Wei W, Liang S, Gao T-L, Lu Y-B. Hypoxia-induced shedding of MICA and HIF1A-mediated immune escape of pancreatic cancer cells from NK cells: role of circ_0000977/miR-153 axis. RNA Biol. 2019;16(11):1592–1603. doi: 10.1080/15476286.2019.1649585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dang R-Y, Liu F-L, Li Y. Circular RNA hsa_circ_0010729 regulates vascular endothelial cell proliferation and apoptosis by targeting the miR-186/HIF-1α axis. Biochem Biophys Res Commun. 2017;490(2):104–110. doi: 10.1016/j.bbrc.2017.05.164. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are included in the article.