Abstract
Campylobacter fetus is commonly associated with venereal disease and abortions in cattle and sheep, and can also cause intestinal or systemic infections in humans that are immunocompromised, elderly, or exposed to infected livestock. It is also believed that C. fetus infection can result from the consumption or handling of contaminated food products, but C. fetus is rarely detected in food since isolation methods are not suited for its detection and the physiology of the organism makes culturing difficult. In the related species, Campylobacter jejuni, the ability to colonize the host has been linked to N-linked protein glycosylation with quantitative proteomics demonstrating that glycosylation is interconnected with cell physiology. Using label-free quantitative (LFQ) proteomics, we found more than 100 proteins significantly altered in expression in two C. fetus subsp. fetus protein glycosylation (pgl) mutants (pglX and pglJ) compared to the wild-type. Significant increases in the expression of the (NiFe)-hydrogenase HynABC, catalyzing H2-oxidation for energy harvesting, correlated with significantly increased levels of cellular nickel, improved growth in H2 and increased hydrogenase activity, suggesting that N-glycosylation in C. fetus is involved in regulating the HynABC hydrogenase and nickel homeostasis. To further elucidate the function of the C. fetus pgl pathway and its enzymes, heterologous expression in Escherichia coli followed by mutational and functional analyses revealed that PglX and PglY are novel glycosyltransferases involved in extending the C. fetus hexasaccharide beyond the conserved core, while PglJ and PglA have similar activities to their homologs in C. jejuni. In addition, the pgl mutants displayed decreased motility and ethidium bromide efflux and showed an increased sensitivity to antibiotics. This work not only provides insight into the unique protein N-glycosylation pathway of C. fetus, but also expands our knowledge on the influence of protein N-glycosylation on Campylobacter cell physiology.
Keywords: Campylobacter fetus, N-linked protein glycosylation, glycosyltransferase, proteomics, metal regulation, hydrogenase
Introduction
Asparagine-linked protein glycosylation is a post-translational modification present in species from all three domains of life. The prototypical bacterial protein N-glycosylation system (referred to as pgl) was first identified in Campylobacter jejuni over two decades ago (Szymanski et al., 1999). This system utilizes five glycosyltransferases (pglA, pglC, pglH, pglI, pglJ) to produce the heptasaccharide GalNAc-α1,4-GalNAc-α1,4-(Glc-β1,3-)GalNAc-α1,4-GalNAc-α1,4-GalNAc-α1,3-diNAcBac-β1,N-Asn (diNAcBac is 2,4-diacetamido-2,4,6-trideoxyglucopyranose) which is attached to protein (Figure 1; Glover et al., 2005, 2006; Linton et al., 2005). The assembly of the full-length glycan occurs on the cytoplasmic side of the inner membrane through the sequential transfer of nucleotide-activated sugars onto the lipid carrier undecaprenyl-phosphate. The lipid-linked heptasaccharide is then flipped into the periplasm by the flippase PglK (Alaimo et al., 2006; Kelly et al., 2006) and transferred to the asparagine residue within the consensus sequon D/E-X1-N-X2-S/T (where X1, X2 can be any amino acid except proline) by the oligosaccharyltransferase PglB (Kowarik et al., 2006; Chen et al., 2007; Scott et al., 2011), or is released as free oligosaccharide (Nothaft et al., 2009), a process that is conserved among Campylobacter species (Nothaft et al., 2012). In C. jejuni, the conserved heptasaccharide has been found on more than 80 periplasmic and membrane-bound proteins (Scott et al., 2011; Cain et al., 2019). Mutagenesis of the pgl genes indicates that this glycosylation system impacts multiple cell functions including: (i) colonization of chickens and mice; (ii) adherence and invasion of epithelial cells; (iii) functionality of the multidrug efflux complex CmeABC; (iv) stability of the type IV secretion system; and (v) interactions with the immune system (Nothaft and Szymanski, 2013; Dubb et al., 2020). More specifically, two recent proteomics studies of C. jejuni pglB mutants have revealed multiple physiological functions associated with N-glycosylation (Abouelhadid et al., 2019; Cain et al., 2019). These include increased expression of stress response proteins, decreased survival in high temperature and osmolarity, altered metabolic activities, decreased chemotaxis, impaired efflux, and decreased nitrate reductase activity (Abouelhadid et al., 2019; Cain et al., 2019).
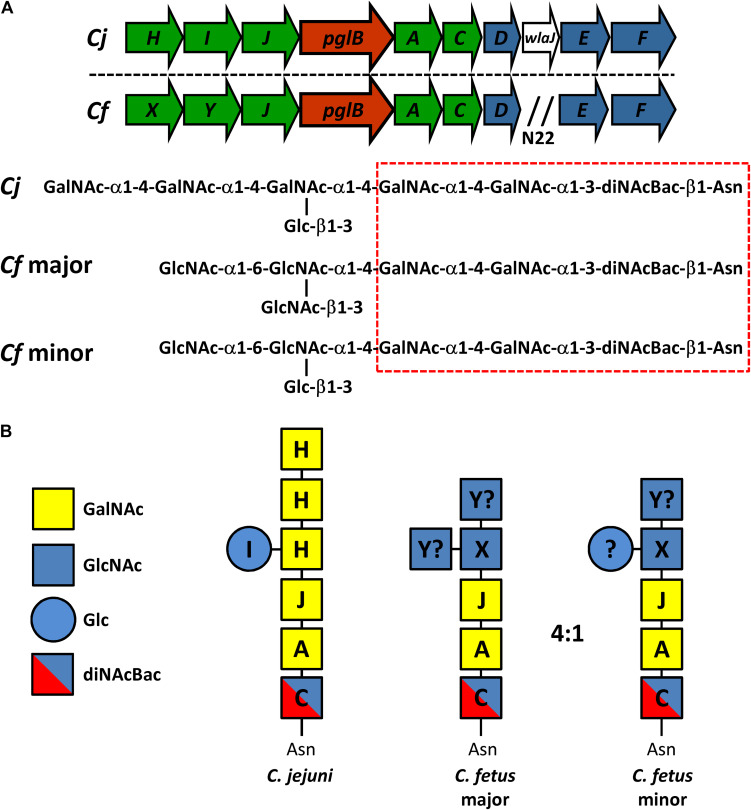
FIGURE 1.
Comparison of the N-linked protein glycosylation (pgl) pathway in C. jejuni and C. fetus. (A) The N-linked glycan structures and the genetic organization of the pgl locus for C. jejuni 11168 (Cj) and C. fetus fetus ATCC 27374 (Cf) (according to Nothaft et al., 2012) are shown. Genes encoding glycosyltransferases are in green, the oligosaccharyltransferase gene is in red, and genes for the biosynthesis of diNAcBac are in blue. N22 indicates an insertion of 22 genes (between pglD and pglE) in the Cff-pgl operon. The hatched red box indicates the conserved group of sugars at the reducing end. (B) Similar to Cj, we propose that Cf-PglC transfers diNAcBAc which is synthesized by Cj/Cf PglDEF (not shown) to undecaprenyl-phosphate (Nothaft and Szymanski, 2010). Subsequently, we show that Cf-PglA transfers the first α1–3 linked GalNAc followed by the second α1–4 linked GalNAc residue added by Cf-PglJ, comparable to the Cj homologs. To this trisaccharide, Cj-PglH transfers three α1–4 linked GalNAc residues and Cj-PglI subsequently transfers the β1–3 linked Glc branch (Kelly et al., 2006). For Cf, PglX most likely transfers the first HexNAc (α1–4 linked GlcNAc), but it remains to be determined if PglY can transfer the β1–3 linked Glc or the remaining two GlcNAc residues in the major and the minor glycan forms. The question mark indicates that another enzyme outside the gene cluster could be responsible for the addition of those sugar residues.
Orthologs of the pgl pathway have been found in all Campylobacter spp., select Helicobacter spp., Desulfovibrio desulfuricans, Wolinella succinogenes, Deferribacter desulfuricans, Sulfurovum sp., Nitratiruptor sp., and some less characterized δ- and ε-Proteobacteria (Nakagawa et al., 2007; Jervis et al., 2010; Ielmini and Feldman, 2011; Nothaft et al., 2012; Mills et al., 2016). Despite the conservation of the pgl pathway per se, different Campylobacter species produce N-glycans that vary in structure and composition (Jervis et al., 2012; Nothaft et al., 2012). This is particularly evident among the non-thermotolerant Campylobacter species which produce multiple N-linked glycoforms (Jervis et al., 2012; Nothaft et al., 2012). For instance, Campylobacter fetus synthesizes two distinct N-linked hexasaccharides: the major GlcNAc-α1-6-(GlcNAc-β1-3)-GlcNAc-α1-4-GalNAc-α1-4-GalNAc-α1-3-diNAcBac-β1,N-Asn and the minor GlcNAc-α1-6-(Glc-β1-3)-GlcNAc-α1-4-GalNAc-α1-4-GalNAc-α1-3-diNAcBac-β1,N-Asn at a 4:1 ratio, respectively (Nothaft et al., 2012).
Campylobacter fetus grows best between 25 and 37°C and consists of three subspecies: C. fetus subsp. fetus (Cff), C. fetus subsp. venerealis (Cfv), and the more recently described subspecies C. fetus subsp. testudinum (Cft) thought to originate from reptiles, but also associated with human infections (Patrick et al., 2013; Fitzgerald et al., 2014). Cff has the broadest host range and is found in cattle, sheep, reptiles, and humans (Tu et al., 2004; Wagenaar et al., 2014). In livestock, both Cfv and Cff are known to cause reproductive failure and infertility (Duncan et al., 2014), and although Cfv has been isolated from humans, it only causes disease in cattle (Holst et al., 1987). The majorities of human C. fetus infections are attributed to Cff and are associated with meningitis, acute diarrhea, and most commonly bacteremia (Wagenaar et al., 2014). Human infections are generally sporadic, with only a few reported outbreaks (Klein et al., 1986; Marchand-Senecal et al., 2017). Recent metagenomic analysis found C. fetus in 8% of feces from healthy humans, suggesting it is a possible pathobiont (Iraola et al., 2017).
In this study, we examined the role of several C. fetus pgl-encoded glycosyltransferases through mutagenesis and functional transfer into Escherichia coli. We demonstrate that the Cff-PglA and Cff-PglJ homologs have the same function as their counterparts in C. jejuni building the conserved GalNAc-α1,4-GalNAc-α1,3-diNAcBac reducing-end core. PglX (previously annotated as PglH1) and PglY (previously annotated as PglH2) are associated with the biosynthesis of the structurally variable region at the non-reducing end of the Cff-hexasaccharides. To assess the potential impact of the N-glycan truncations on other cellular functions, a label-free quantitative proteomics approach was used to examine the Cff-pglJ and pglX mutants. Proteomics demonstrated widespread changes in protein abundance with a notable impact on metal transport proteins, several (NiFe) hydrogenase subunits, and oxidative response proteins compared to the wild-type (WT). The results presented in this study provide new insights into the assembly and roles of N-linked glycoproteins in C. fetus.
Results
Characterization of Cff pgl Cluster
The C. fetus (Cf) pgl cluster is syntenic with the C. jejuni (Cj) pgl gene cluster (Jervis et al., 2012; Nothaft et al., 2012) apart from lacking pglI and possessing two homologs of pglH (Figure 1). The similarities between the two loci are reflected in their N-glycan structures, with both sharing the same three reducing end sugars. In C. jejuni, pglC, pglA, and pglJ are responsible for the formation of this initial diNAcBac-GalNAc2 trisaccharide that is conserved across nearly all Campylobacter species (Jervis et al., 2012; Nothaft et al., 2012). Previously, Cf was annotated to possess two pglH homologs; however, compositional and structural analyses of the Cf-pgl pathway products showed that it does not contain the three GalNAc residues added by the Cj-pglH gene product (Nothaft et al., 2012). Since it is the non-reducing end of the C. jejuni and C. fetus N-glycans that varies in structure, the pgl genes in the “variable” glycosyltransferase (GTase) region upstream of pglB most likely differ in function. We therefore named the two pglH homologs, pglX and pglY (Figure 1). Interestingly, both proteins contain the catalytic EX7E motif previously annotated in PglH (Cid et al., 2000; Troutman and Imperiali, 2009; Figure 2). In addition to this catalytic EX7E, PglY and PglX contain one and two additional EX7E motifs, respectively. K68 of Cj-PglH, which is believed to be involved in lipid-linked oligosaccharide (LLO) association, is altered to N67 and T70 in PglX and PglY, respectively. In addition, the binding site of the Cj-PglH catalytic EX7E motif that involves L269 and P270 was found to be altered in PglX and PglY. Both enzymes possess a G instead of a P at position P270; however, only PglY possesses an F at position 267 that corresponds to L269 in C. jejuni. These minor changes in the amino acid residues may explain the differences in enzyme specificity and the formation of the shorter glycans when compared to C. jejuni.
FIGURE 2.
Sequence alignment of C. fetus (Cf) PglX, PglY, and C. jejuni (Cj) PglH. Black boxes indicate specific amino acids associated with activity in PglH (Troutman and Imperiali, 2009; Ramirez et al., 2018). Black and dark gray amino acids represent functional residues that show non-conserved substitutions in PglX and PglY. Light gray highlighted sequences indicate EX7E motifs commonly found in glycosyltransferases (Cid et al., 2000; Coutinho et al., 2003; Troutman and Imperiali, 2009). The catalytic EX7E motif of Cj-PglH is located at residues E266 to E274. Cf-PglX CFF8240_1386, Cf-PglY CFF8240_1385, and Cj DDV78_00080 sequences were analyzed by Jalview (Waterhouse et al., 2009).
N-Glycan Analysis of Cff-pgl Mutants
To assess the functions of the “variable” GTases, we constructed mutants by insertion of a kanamycin resistance cassette (referred to as “kan”) into the respective gene loci. Both pglX (pglX:kan, further referred to as pglX-) and pglJ (pglJ:kan, further referred to as pglJ-) were constructed in the Cff strain ATCC 27374 (Supplementary Figure S1), however multiple attempts at generating mutants in pglY were unsuccessful.
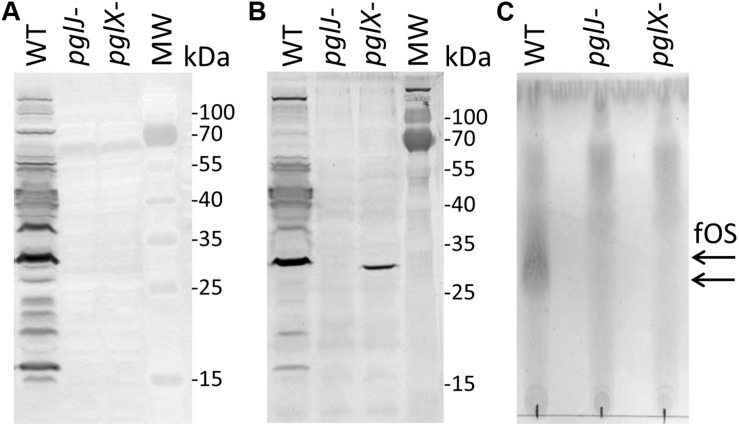
Insertion of the kan cassette in the pglJ and pglX genes was verified by PCR with oligonucleotides hybridizing outside of the recombination event (Supplementary Figure S1). When compared to the PCR product size obtained with chromosomal DNA isolated from Cff-WT, an increase in size by approximately 1.8 kb was observed when the kan cassette was present on the respective PCR product, clearly indicating insertion at the correct position within the Cff chromosome. To further investigate the effect of the mutations on N-glycan biosynthesis, western blot analysis of whole cell lysates probed with Cff-N-glycan specific serum was performed. Complete loss of serum reactivity in pglX- and pglJ- was observed when compared to the WT (Figure 3A and Supplementary Figure S6A). Lectin blotting with WGA confirmed those results, i.e., loss of reactivity in whole cell lysates of the pglJ mutant and strongly reduced reactivity (with only one signal present) in lysates of the pglX mutant (Figure 3B and Supplementary Figure S6B). Similarly, no free oligosaccharides (fOS) could be detected in the two pgl mutants when analyzed by thin layer chromatography (TLC) (Figure 3C). Here, two spots for the Cff-fOS variants could be seen when a fOS preparation of the WT was applied, confirming previous observations (Dwivedi et al., 2013), and these spots were absent in similar preparations from the pglX- and pglJ- strains.
FIGURE 3.
Product analysis of Cf WT, pglX- and pglJ- strains. (A) Western blot of whole-cell lysates with Cff-N-glycan-specific antiserum, and (B) wheat germ agglutinin reactivity of whole cell lysates of the WT, pglJ-, and pglX- strains. (C) Thin-layer chromatography (TLC)-free oligosaccharide (fOS) analysis of WT, pglJ-, and pglX- strains. Molecular weight (MW) markers for the western blots (in kDa) are indicated on the right; arrows indicate the migration of WT fOS on the TLC plate.
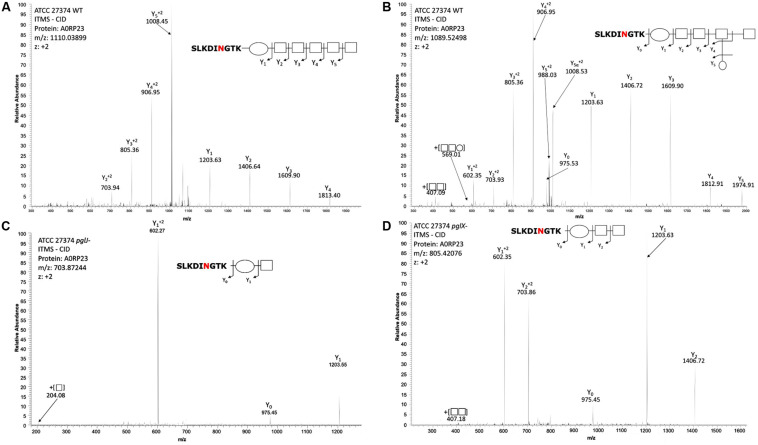
To investigate the N-glycan in the two Cff-pgl mutants in more detail, proteomics analysis of Cff-WT and the pgl mutants was performed. As the disruption of Cff-pgl was predicted to truncate the N-linked glycan, we examined whole cell lysates to avoid potential biases in the detection of glycoforms that can result from glycopeptide enrichment (Alagesan et al., 2019). Consistent with our previous work (Nothaft et al., 2012), we observed both GlcNAc-α1-6-(GlcNAc-β1-3)-GlcNAc-α1-4-GalNAc-α1-4-GalNAc-α1-3-diNAcBac and GlcNAc-α1-6-(Glc-β1-3)-GlcNAc-α1-4-GalNAc-α1-4-GalNAc-α1-3-diNAcBac glycans on multiple protein substrates within the WT (Figures 4A,B), which were absent within pglX- and pglJ- (Supplementary MS Data 1). Consistent with our western and lectin blotting assays, multiple truncated N-linked glycans were observed within pglX- and pglJ- including diNAcBac-HexNAc2 glycans, diNAcBac-HexNAc glycans (Figures 4C,D) as well as diNAcBac alone. Within pglX-, the diNAcBac-HexNAc2 glycan was the predominant glycoform (Supplementary MS Data 1) and is consistent with the Cff N-glycan core structure, diNAcBac-GalNAc2 (Nothaft et al., 2012). In contrast, multiple glycoforms were identified in pglJ- including diNAcBac-HexNAc2-, diNAcBac-HexNAc, and diNAcBac glycans (Supplementary MS Data 1). Taken together these results confirm the involvement of PglJ in the formation of the conserved reducing end trisaccharide and that PglX functions in extension of the non-conserved N-linked glycan structure.
FIGURE 4.
Mass-spectrometric analysis of Cff WT, pglJ-, and pglX- glycopeptides. Fragmentation of characteristic ions obtained by precursor ion scanning of digested Cff lysate samples using liquid chromatography-mass spectrometry. Red lettering indicates possible glycosylation site. Spectra of WT Cff have two glycans: (A) HexNAc5-diNAcBac and (B) HexNAc-[Hex]-HexNAc3-diNAcBac. (C) The peptide from the pglJ- mutant only shows the presence of a mass consistent with HexNAc-diNAcBac. (D) The peptide from the pglX- mutant indicates that it is modified with HexNAc-HexNac-diNAcBac.
Mutations in pglX and pglJ Have No Effect on Growth and the Expression of Downstream Genes but Reduces Motility
Growth curves were performed to investigate a potential influence of the pgl mutations. Although the pgl mutants reached a slightly higher optical density in the late logarithmic phase when compared to the WT, the final optical densities, as well as the growth rates in the early and mid-exponential phases, were similar among the three strains (Supplementary Figure S2). In addition, we did not observe a significant difference in pgl gene transcript levels after the insertion of the kan cassette in either pglX or pglJ (Supplementary Figure S3). This indicates that expression of the antibiotic cassette has no effect or that other transcriptional start sites in the Cff-pgl operon are compensating, as observed in the C. jejuni pgl operon (Szymanski et al., 1999; Dwivedi et al., personal communication). A downstream effect would influence expression of pglB but we see similar abundance of the PglB protein in WT when compared to either mutant (Supplementary Table S1). In addition, we still observe different forms of glycans on each mutant whereas in the absence of PglB we would not expect any glycans at all. However, we observed a significantly reduced swimming behavior in pglX- and pglJ- when compared to Cff-WT (Supplementary Figure S4) indicating that N-glycosylation either directly or indirectly affects motility.
Characterization of PglJ and PglA in E. coli
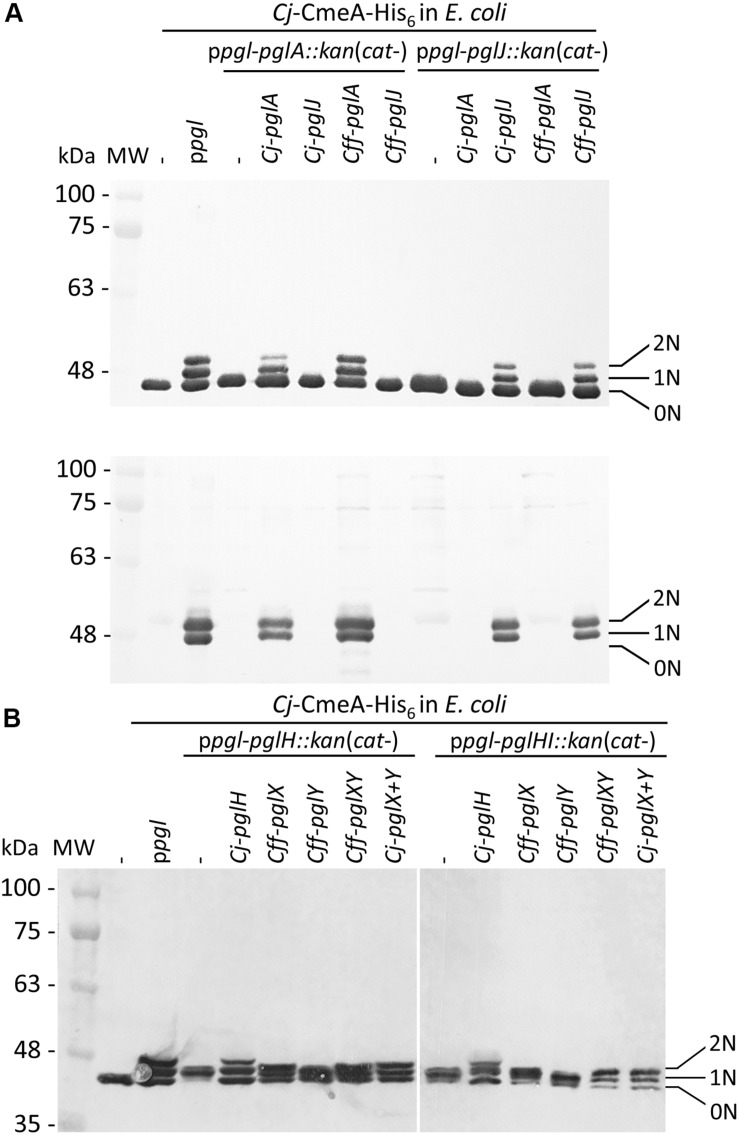
Since the N-glycan phenotype observed in the pglJ mutant was somewhat unexpected, the function of Cff-PglJ and Cff-PglA was further investigated by using a modified heterologous E. coli Cj/Cff hybrid glycosylation system (Wacker et al., 2002). Within this system, Cff-Pgl proteins are expressed in the presence of a mutant Cj-pgl operon (lacking select Cj-pgl genes). The glycans produced are then transferred to Cj-CmeA-His6 (N-glycosylation acceptor protein) via Cj-PglB. Western blotting of whole cell lysates of E. coli CLM24 prepared after co-expression of Cj-CmeA-His6 and Cj-pglA or Cj-pglJ in the presence of ppgl operon derivatives lacking either pglA or pglJ were probed with anti-His6 and anti-Cj-N-glycan antibodies (Figure 5A and Supplementary Figure S7A). The three Cj-Cme-His6-specific signals with anti-His (Figure 5A and Supplementary Figure S7A upper panel) and two N-glycans specific signals with the Cj-N-glycan specific R1 antiserum (Figure 5A and Supplementary Figure S7A lower panel) clearly identified the bands as non-(0N), mono-(1N), and di-(2N) glycosylated CmeA-His6. A similar Cj-CmeA-His6 pattern was produced in cells harboring the native Cj-pgl operon (from ppgl) and upon expression of Cff-pglA or Cff-pglJ (although with lower glycosylation efficiency) only when the Cj-homologous gene was knocked-out. In addition, no cross-complementation could be observed when Cj or Cff-pglA or pglJ were expressed in the presence of the ppgl plasmid lacking pglJ or pglA, respectively. These results confirm that Cff-PglA and Cff-PglJ fulfill the same functions as the homologous Cj-Pgl proteins, i.e., the addition of the second and third monosaccharide building blocks, respectively, to Und-diNAcBac, to form the diNAcBac-GalNAc2- trisaccharide. As expected, no Cj-CmeA-His6 glycosylation was observed in the absence of ppgl resulting in only non-glycosylated (0N) acceptor protein represented by a single band in the anti-His6 western blot and further confirmed by the absence of the N-glycan-specific signals in the anti-N-glycan (R1) blot (Figure 5A and Supplementary Figure S7A lower panel). Mass spectrometric analysis of isolated Cj-CmeA confirmed the modification of CmeA glycopeptides with the expected glycoforms supporting these western blot results (Supplementary Material and Supplementary MS Data 2). Here, the full length Cj-heptasaccharide was produced only when the Cj-pgl operon plasmids with mutations in pglA or pglJ were co-expressed with plasmids containing the corresponding pglA or pglJ from Cj or Cff.
FIGURE 5.
Functional analysis of Cff-Pgl pathway glycosyltransferases (GTases) in the heterologous E. coli glycosylation system. The GTase-activities of (A) PglA and PglJ were analyzed in western blots of Cj-CmeA-His6 with His6-tag antibodies (upper panel) and Cj-N-glycan specific (R1) antibodies (lower panel). (B) PglX and PglY activities were analyzed with His6-tag specific antibodies in western blots of CmeA-His6 used as the glycan acceptor to determine N-glycosylation activities. Whole cell extracts (5 μg) of E. coli CLM24 expressing the indicated gene/plasmid combinations are indicated above each lane. Non-, mono-, and di-glycosylated CmeA-His6 proteins are labeled as 0N, 1N, and 2N, respectively. Molecular weight markers (MW) in kDa are indicated on the left.
Characterization of Cff-pglX and pglY Using the E. coli Heterologous Glycosylation System
Since we could not obtain a mutant in Cff-pglY and therefore could not assign the functions of the two remaining GTases in the “variable” pgl region, we decided to analyze PglX and PglY using the heterologous E. coli glycosylation system (Wacker et al., 2002). In this case we employed the Cj-pgl operon lacking pglH that produces a trisaccharide (diNABacGalNAc2) identical to that found in Cff, potentially providing a substrate for PglX or PglY activity. In addition, we constructed and analyzed the complementation of a ppgl-pglHI:kan mutant plasmid (lacking Cj-pglH and Cj-pglI) to rule out the possibility of the Cj-PglI GTase adding or competing with the potential addition of a glucose residue to the N-glycan chain by either Cff-PglX or Cff-PglY. To do so, plasmid pCE111/28 derivatives expressing Cj-pglH (positive control), Cff-pglX, Cff-pglY, or Cff-pglXY served as complementation vectors. Western blots of whole cell lysates probed with anti-His6 antibodies were performed to investigate the Cj-CmeA-His6 glycosylation pattern in the underlying strains (Figure 5B and Supplementary Figure S7B). Expression of ppgl in combination with CmeA-His6 and CmeA-His6 alone served as positive and negative glycosylation controls, respectively. First, we demonstrated that expression of Cj-pglH in combination with the pgl operon lacking pglH resulted in a glycosylation pattern similar to the strain co-expressing CmeA-His6 and the Cj-WT pgl operon (on ppgl), i.e., production of non-(0N), mono-(1N), and di-(2N) glycosylated CmeA-His6, whereas in the absence the complementation plasmid, glycobands were migrating slightly faster due to the addition of only the trisaccharide N-glycan (missing the GalNAc3-Glc that is added by PglH and PglI in the full length Cj-heptasaccharide). Expression of Cff-pglY with ppgl-pglH:kan did not alter the migration behavior of the glycobands when compared to ppgl-pglH:kan alone, whereas transformation of Cff-pglX resulted in a slight mass increase compared to ppgl-pglH:kan/Cff-pglY, indicating that PglX, but not PglY, might be responsible for the addition of a sugar residue to the ppgl-pglH:kan glycan (Figure 5B). A slight increase in mass of the Cj-CmeA-His6 glycoprotein was also observed upon expression of Cff-pglXY (pglXY cloned as one PCR product), however a difference in the running behavior compared to ppgl-pglH:kan/Cff-pglY could not be resolved by SDS-PAGE and western blotting analysis alone (Figure 5B and Supplementary Figure S7B).
Similar results were obtained upon introduction of Cj-pglH and Cff pglX, Cff-pglY and Cff-pglXY into CLM24 expressing ppgl-pglHI:kan and Cj-CmeA-His6. Here, the glycobands in the Cj-pglH complements were expected to display a slightly faster running behavior when compared to the full length heptasaccharide due to the loss of the Glc residue; however, similar to the complementation analysis of the ppglH:kan strains, an obvious difference in the running behavior of the CmeA-His6 glycobands upon introduction of Cff-pglX, and Cff-pglXY could not be resolved (Figure 5B).
To further investigate the N-glycans produced upon expression of the different Cj-pgl operon mutants in combination with the Cff-pglX and pglY complementation plasmids, mass-spectrometric analyses of trypsinized CmeA was undertaken. While N-glycan structures observed upon complementation with the Cj-control (ppgl-pglH mutant expressing Cj-pglH) resulted in the formation of the expected full length Cj-N-glycan, only one plasmid combination, the expression of Cff-pglXY in the ppgl-pglH mutant background resulted in the formation of a structure that was similar in composition and sequence to the minor form of the native Cff-N-glycan, diNAcBac-HexNAc4-Hex (Supplementary Material and Supplementary MS Data 2).
Mutations in pglX and pglJ Have an Impact on Multiple Cellular Functions in Cff
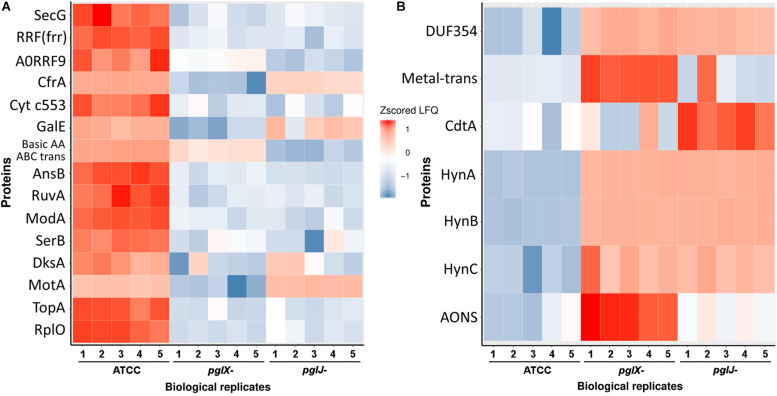
To further understand the role of N-glycosylation in Cff, label-free quantitative (LFQ) proteomics analysis of whole cell lysates of Cff-WT, and the pglX- and pglJ- was done. Across five biological replicates of each sample type (Supplementary Figure S5), 914 proteins were identified representing ∼77% of the Cff ATCC 27374 predicted proteome of 1,190 proteins (Supplementary Table S1). Quantitative proteome analyses revealed more than 100 proteins with significantly different abundance across various biological groups as shown in heat maps of the most prominent differences in abundance comparing WT to pglX- and pglJ- strains (Figures 6A,B). These results indicate that mutating glycosyltransferases involved in assembly of the N-linked glycan has a significant effect on abundance of numerous cellular proteins.
FIGURE 6.
Label-free quantification of proteins in Cff-pglJ- and pglX- strains compared to WT. (A,B) Heat maps of specific proteins with statistically significant decreases (A) or increases (B) in pglJ- and pglX- mutants compared to WT (labeled ATCC). Values are gray where MS did not identify fragments. This data represents samples from five biological replicates (B1–B5). The complete dataset is included in Supplementary Table S1.
Expression of the H2-Uptake Hydrogenase Complex HynABC Is Significantly Induced in Both pglJ and pglX N-Glycosylation Mutants
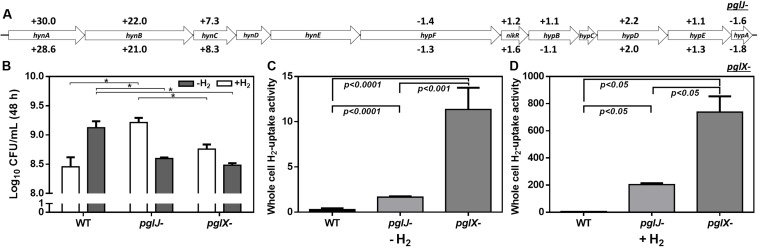
Among the proteins with increased abundance in both the pglJ and pglX mutants (compared to WT) were the three subunits (HynABC) of a putative nickel-iron (NiFe) H2-uptake hydrogenase complex (Benoit et al., 2020). In both pgl mutants, the expression levels of all three hydrogenase subunits, HynA, HynB, and HynC, were significantly higher compared to the WT (means of 29.3-fold, 21.5-fold, and 7.8-fold, respectively) (Figure 7A and Supplementary Table S1). This complex, found in a number of bacterial pathogens, enables the microbes to use the electron donor H2 as an energy source, thus providing an alternative respiratory pathway that is important for in vivo survival (Olson and Maier, 2002; Benoit and Maier, 2018). HynABC-associated proteins, such as hydrogenase accessory/maturation proteins (e.g., HypABCDEF) or the nickel specific transcriptional regulator (NikR) also showed moderate increases in protein levels in both mutants compared to WT (Figure 7A).
FIGURE 7.
Elevated hydrogenase levels in Cff-pgl mutants correlate with increased H2 uptake. (A) Gene arrangements of hyn and hyp clusters in Cff ATCC 27374 with LFQ proteomic protein abundance in pgl mutants vs WT are shown. Protein fold changes of pglJ- and pglX- with respect to WT are indicated above and below each gene respectively. Genes with no values associated had no coverage in our proteomics data analyses. Only HynABC showed significant protein level increases when compared to WT. (B) Effect of H2 on microaerobic growth of Cff WT, pglJ-, and pglX- strains. The data set is derived from three biological replicates (with two technical replicates each) of cells grown under the indicated condition. Number of cells obtained after 48 h of growth is represented by Log10 colony forming units per mL (CFU/mL). The error bars represent the standard error within each group. (C,D) Whole cell H2-uptake of Cff WT, pglX-, and pglJ- strains. Cff was grown on BHI agar in microaerophilic conditions for 24 h at 37°C in 10% H2 (+H2) or absence of hydrogen (–H2). Whole cell H2-uptake activity is expressed as nanomoles of H2 used per min per 109 cells. Results in (C) represent the mean ± SD of four independent assays; results in (D) represent the mean ± SD of two independent assays. p-values as analyzed by a two-tailed t-test are either indicated by an asterisk (*p-value ≤ 0.05 in B) or are directly included in the figure (for C,D).
Since H2 increases growth of various ε-Proteobacteria species, including Helicobacter pylori and Campylobacter concisus (Kuhns et al., 2016; Benoit and Maier, 2018), we determined whether higher hydrogenase expression in the C. fetus N-glycosylation mutants correlates with elevated H2-supported microaerobic growth. To do so, the cell yield (CFU/mL) of the WT and the pglJ and pglX mutants was assessed after 48 h of growth under microaerobic conditions in the presence or absence of 20% H2 (Figure 7B). We only determined the end point of growth due to the extended lag phase of Cff cultures grown under these conditions. With no added H2, Cff WT had a significantly higher growth yield compared to both mutants. However, in H2-enriched conditions, WT cells showed growth levels comparable to both pgl mutants. Although the addition of H2 was originally predicted to be beneficial for WT growth, we observed decreased growth in H2 for other Cf strains, Cft 03-427 and Cff 82-40 (data not shown). In contrast, we observed a significant increase in pglJ- growth compared to the other strains in the presence of H2. A slight increase in pglX- growth was also observed in the presence of H2, but it was not significant compared to WT. These results indicate that the pgl mutants have increased growth yield in H2 opposed to WT where H2 is deleterious.
H2-uptake in whole cells was examined to determine whether increased HynABC levels in the mutants correlate with increased H2-uptake activity. Cells were grown in microaerobic conditions in the presence or absence of supplemental H2 and hydrogenase activity was determined using a previously described amperometric method (Maier et al., 1996). The hydrogenase activity (expressed in nmoles of H2 oxidized per min per 109 cells) was 0.3 ± 0.07, 1.7 ± 0.04, and 11.4 ± 1.2 for WT pglJ-, and pglX, respectively, when cells where grown under microaerobic conditions in the absence of supplemental H2 (Figure 7C). This represented almost a 6-fold (for pglJ-) to 39-fold (for pglX-) increase in activity compared to WT. When cells were grown in the presence of 10% H2, we observed a 122- and 65-fold increase in hydrogenase activity in pglJ- and pglX-, respectively, and a 20-fold increase in WT (Figure 7D). The remarkable H2-uptake levels measured for pglJ- (204 ± 7 nmoles H2/min/109 cells) and pglX- (738 ± 82 nmoles H2/min/109 cells) mutants grown with H2 were the highest recorded values to date for a bacterial pathogen. Taken together, these results indicate an inverse correlation between N-glycosylation and H2 usage (i.e., hydrogenase synthesis and activity) in Cff.
N-Glycosylation Influences Transition Metal Profiles
Proteomics data indicate that multiple proteins associated with transition metals were significantly altered in both pgl mutants. These include ModA, involved in molybdenum transport (−49.9-fold in pglX- and −71.8-fold in pglJ-); the ZinT/AdcA family protein involved in zinc binding (−12.2-fold in pglX- and −6.5-fold in pglJ-); CfrA, a ferric receptor (−118.4-fold and −3.3-fold); and an iron ABC transporter (−5.1-fold and −7.6-fold) (Supplementary Table S1). Also, a copper/cadmium-translocating P-type ATPase protein was found to be significantly increased (51.9-fold) in pglX-; however, the increase was not significant (1.4-fold) in pglJ-.
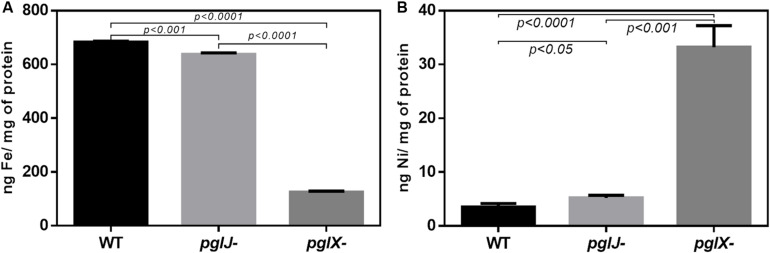
The increased levels, especially of the HynABC (Ni-Fe) hydrogenase observed in both pgl mutants, led us to further investigate nickel and iron levels in these strains. Using atomic absorption spectrometry (AAS) of lysed cells, we found that iron levels were dramatically decreased in pglX- (125.1 ng/mg protein), that is almost sixfold lower when compared to WT (683.2 ng/mg protein) whereas iron levels in pglJ- were modestly, but statistically significantly, decreased (Figure 8A). In addition, the pgl mutants had significantly higher levels of cellular nickel content compared to WT (Figure 8B); pglX- had a nickel content of 33.2 ng/mg protein that was almost 10-times higher than in the WT (3.5 ng/mg protein). Although still significantly higher when compared to the WT, pglJ- (5.2 ng/mg protein) had almost sixfold less nickel than pglX-. These results indicate that N-glycosylation might be vital in regulation of nickel homeostasis, iron, or both.
FIGURE 8.
Cellular iron and nickel content of Cff WT, pglX-, and pglJ- strains. Atomic absorption spectroscopy (AAS) was employed for the detection of (A) iron (Fe) and (B) nickel (Ni) in lysed cells of the indicated strain. Data are presented as the mean of at least three replicates, error bars depict the standard deviations. Statistically significant differences determined by a two-tailed t-test are indicated.
Antibiotic Sensitivity and Increased Membrane Efflux
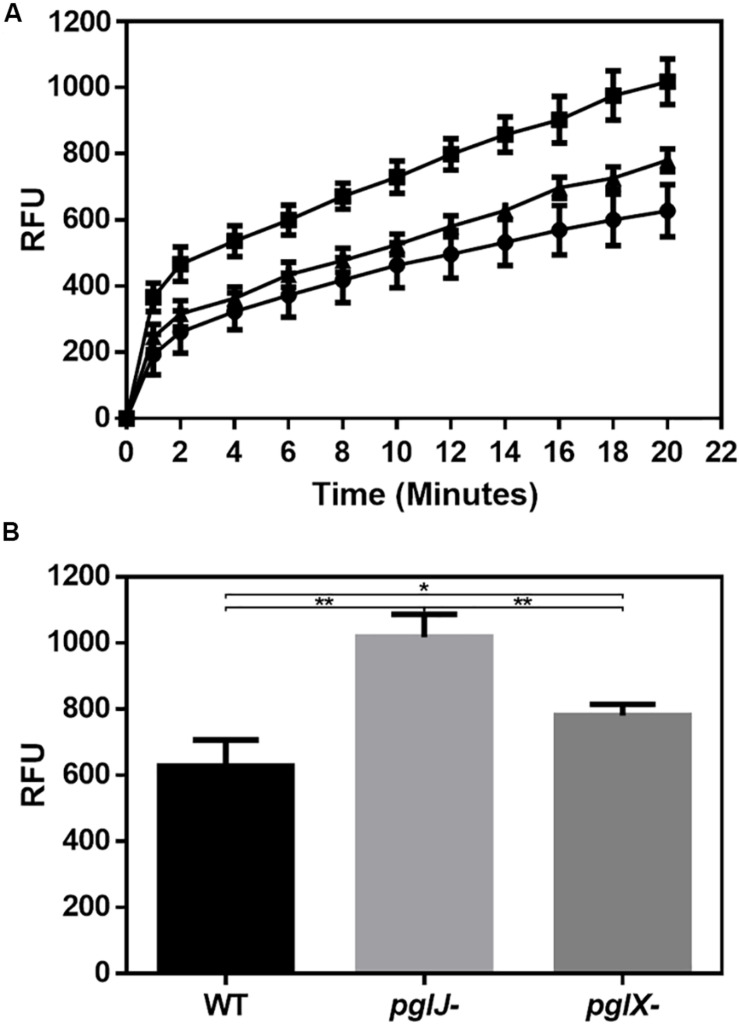
Our previous study showed that C. jejuni N-glycosylation was required for optimal activity of the CmeABC multidrug efflux pump necessary for antibiotic resistance (Dubb et al., 2020). In Cff, albeit not statistically significant, we found increased levels of CmeA, CmeB, and CmeC in pglX- and pglJ- (mean of both mutants: 2.0-fold CmeA, 1.9-fold CmeB, and 2.0-fold CmeC). Therefore, we examined the antibiotic sensitivity profiles of both Cff N-glycosylation mutants. As shown in Supplementary Table S2, the pglX- and pglJ- strains showed twofold increase in sensitivities to chloramphenicol, gentamicin, azithromycin, and sulfisoxazole, and a fourfold increase in sensitivity to ampicillin suggesting a correlation between N-glycosylation and antibiotic resistance in C. fetus, similar to previously observed in C. jejuni (Abouelhadid et al., 2019; Dubb et al., 2020). To explore this further, we used ethidium bromide (EtBr), a DNA intercalating agent, to quantitatively assess efflux pump activity over time. Both pglJ and pglX mutant strains showed significantly higher levels of EtBr accumulation compared to WT (Figure 9); however, accumulation was less pronounced in the pglX mutant. Taken together, these results suggest that N-glycosylation in C. fetus may be important for efflux pump activity and antibiotic sensitivity.
FIGURE 9.
Mutations in N-linked protein glycosylation decreased efflux in pglX- and pglJ- strains. (A) Accumulation of ethidium bromide in cultures of Cff WT (circle), pglJ- (square), and pglX- (triangle) over a time frame of 20 min. Relative fluorescent units (RFU) are indicated on the y-axis. (B) Bar graph depicting the relative fluorescence at t = 20 min. The error bars represent the standard error for each data set consisting of four biological replicates with three technical replicates each. ** p-value ≤ 0.001, * p-value ≤ 0.05 as determined by a two-tailed t-test.
Discussion
N-glycosylation is a conserved mechanism in all domains of life. The prototypical pgl N-glycosylation system, originally characterized in C. jejuni (Cj), has orthologs in many δ- and ε-Proteobacteria (Nakagawa et al., 2007; Jervis et al., 2010; Ielmini and Feldman, 2011; Nothaft et al., 2012; Mills et al., 2016). Non-thermotolerant Campylobacter species, like C. fetus (Cf), including C. fetus fetus (Cff) and C. fetus venerealis (Cfv), have been found to produce more than one N-glycan, unlike Cj which expresses one distinct heptasaccharide (Scott et al., 2011; Nothaft et al., 2012; Cain et al., 2019).
In our study, we generated mutants in PglX and PglJ in Cff strain ATCC 27374. Glycopeptides from the pglX- mutant showed fragmentation patterns consistent with the conserved diNAcBac-GalNAc2 (Nothaft et al., 2012), suggesting that PglX is responsible for the addition of the first GlcNAc residue to the Cf N-glycan structure (Figure 1). The loss of Cf N-glycan-specific serum reactivity and WGA lectin binding to lysates from the pglX- strain support this claim. Proteomics of the pglJ- strain resulted in a mixture of glycopeptides, primarily consisting of diNAcBac and a few fragments of diNAcBac-HexNAc, typically more characteristic of a Cj PglA mutant. To investigate this further, we used an E. coli expression system followed by MS-analyses and were able to show that the Cff-PglJ and Cff-PglA had similar transferase activities onto recombinantly expressed Cj-CmeA as the Cj homologs. However, we did not see reactivity with WGA or the Cff-N-glycan specific antiserum in the Cff-pgl mutants, except for a single band for pglX- in the WGA blot. These results suggest possible WGA interaction with another glycan, such as LPS, however the reason behind the absence of specific binding in the pglJ mutant strain has yet to be explained. Nevertheless our results suggest that the formation of the diNAcBac-GalNAc2 trisaccharide is conserved between Cj and Cff and that the observed differences in antigenicity (Jervis et al., 2010; Nothaft et al., 2012) stem from the non-reducing end.
Expression of Cff-PglX in E. coli showed a CmeA mass shift and with transfer of an additional sugar, consistent with transfer of an additional sugar and with our MS-analysis of glycopeptides from the native host (i.e., diNAcBac-GalNAc2-GlcNAc). Since we were unable to generate a pglY mutant in Cff, we also used the E. coli system to investigate Cff-PglY activity and detected a major glycoform with an additional sugar only when both PglX and PglY were co-expressed, suggesting that PglY’s activity is dependent on the initial modification by PglX. Based on our MS results, an N-linked glycan with a composition resembling the minor Cff N-glycan [i.e., GlcNAc-α1-6-(Glc-β1-3)-GlcNAc-α1-4-GalNAc-α1-4-GalNAc-α1-3-diNAcBac] was also observed in E. coli ppgl-pglH:kan expressing pglX in combination with pglY, although the addition of the Glc residue by Cj-PglI could not be ruled out since we observed less peptides containing the minor Cff-N-glycan in the ppgl-pglHI background, and also observed Glc addition in a ppgl-pglHI:kan mutant demonstrating that an E. coli enzyme could be contributing this residue. Thus, the GlcTF reaction requires further investigation either in Cff or in vitro. No N-glycan that resembles the major form of the Cff N-glycan could be detected with any Cj-pgl mutant/Cff-pgl gene combinations. Nevertheless, these data suggest that pglX and pglY can mediate the construction of a partial C. fetus N-linked glycan using the C. jejuni diNAcBac-GalNAc2 trisaccharide as a substrate. C. jejuni PglB does not have strict substrate specificity and can transfer full-length and truncated N-glycans and diverse O-antigen structures in E. coli and to a lesser extent, in the native host (Feldman et al., 2005; Linton et al., 2005). Therefore, we did not expect preferential transfer of certain Cj-Cff hybrid N-glycans to CmeA. However, since we only generated one potential variant of the Cff-N-glycan, this suggests that the GTase involved in the formation of the second Cff-N-glycan structure is either not fully functional in E. coli or is not part of the pgl locus, similar to the lack of pgl gene clustering in Helicobacter species and D. desulfuricans (Jervis et al., 2010; Nothaft and Szymanski, 2010).
To better understand the role of N-glycosylation in Cff, we utilized LFQ proteomics comparing Cff ATCC 27374 with two isogenic pgl mutants, pglX- and pglJ-. Through this approach, we were able to detect almost 77% of the (genome-inferred) total proteins. Analysis of proteins that were significantly up- or down-regulated indicated that more than 100 proteins were altered in the Cff pgl mutants in comparison to WT. It is worth noting that differences between pgl mutants may be due to differences in glycan length (diNAcBac-GalNAc in pglJ- and diNAcBac-GalNAc2 in pglX-) or differential feedback regulation in these two backgrounds. Although N-glycosylation was not completely eliminated, we observed a decrease in NapB (−6.7-fold in pglX- and −8.4-fold in pglJ-, Supplementary Table S1), similar to that previously seen in a Cj-pglB mutant (Cain et al., 2019). In C. jejuni, the nitrate reductase NapAB has been shown to be a two-subunit enzyme, with both subunits being N-glycosylated (Scott et al., 2011; Mintmier et al., 2018; Abouelhadid et al., 2019; Cain et al., 2019). In contrast, in Cff ATCC 27374, NapA lacks an N-glycosylation sequon, while at the same time NapB has two potential sequons. This may explain why we only observed a decrease in NapB (see above), while the difference in NapA protein levels was not significant (1.1-fold in both pgl mutants).
No effect on growth or on the expression of downstream genes was observed, but the pgl mutants were impaired in motility. Similarly, loss of pglB (and therefore complete loss of N-glycosylation) in C. jejuni JHH1 and C. jejuni 11168 also resulted in decreased motility when compared to WT cells (Scott et al., 2012; Cain et al., 2019). In addition, Cain et al. (2019) demonstrated that levels of specific proteins required for motility were expressed at significantly lower levels in the C. jejuni 11168 pglB mutant; among them MotA, MotB, and FlgP. We also observed lower levels of MotA and MotB (significantly lower in pglX-; 94- and 8.6-fold, respectively), but not in pglJ- (Supplementary Table S1); FliG [significantly lower in pglX- (5.7-fold) and pglJ- (4.7-fold) (Supplementary Table S1)], as well as the Cj-FlaA homolog flagellin protein [significantly lower in pglX- (9.7-fold) and pglJ- (7.1-fold) (Supplementary Table S1)]. This could imply that motility may be correlated with N-glycosylation changes in some Campylobacters. However, pglB, pglE, pglF, and pglH mutants in C. jejuni 81–178 were described to display WT levels of motility (Szymanski et al., 1999; Hendrixson and DiRita, 2004), therefore it seems that this regulatory network varies even among strains.
We did not observe a reduction in CmeABC in either the Cff pglX or pglY mutant. In contrast, we observed a slight, but not statistically significant, increase in these efflux proteins in both mutants. Despite that discrepancy, our pgl mutants still displayed decreased EtBr efflux activity compared to WT when cells were grown under the same conditions that were used to prepare whole cell lysates for proteomic analysis. This suggests that Cf N-glycosylation directly influences the activity of the efflux pump, an effect that has previously been described for C. jejuni (Abouelhadid et al., 2019; Dubb et al., 2020). However, the increased sensitivity to various classes of antibiotics observed in both Cff-pgl mutants is most likely indirect since not all of those antibiotics are substrates for the efflux pump in other Campylobacter species; however, variations in CmeABC substrate specificities have been observed even between strains (Lin et al., 2002; Akiba et al., 2006; Guo et al., 2010). One might speculate that membrane permeability increases due to lower abundance of certain periplasmic and/or membrane proteins or that loss of periplasmic fOS could result in a higher influx of those antibiotics and therefore lead to the observed decrease in MICs. It is worth noting that the observed effects were less pronounced in pglX- compared to pglJ-. This could be due to the fact that glycoproteins contain a longer N-glycan chain in pglX- compared to pglJ. Together these results indicate that N-glycosylation in Cf plays a role in efflux, although the mechanism is currently unknown.
Our proteomics data indicate that all three components (HynABC) of the (NiFe)-containing H2-uptake hydrogenase were significantly upregulated in both pgl mutants, suggesting that protein glycosylation plays a role in H2 utilization. Based on homology with hydrogenase complexes found in related ε-Proteobacteria, such as H. pylori, C. jejuni, and C. concisus (Olson and Maier, 2002; Weerakoon et al., 2009; Benoit and Maier, 2018), the Cff HynABC complex is likely to be involved in H2 oxidation. Consistent with the proteomics data, higher H2-mediated growth rates were observed in both pgl mutants compared to WT, with the highest growth rate seen in the pglJ- strain grown under H2 rich conditions. Surprisingly, H2-enriched conditions seemed to have a deleterious effect on WT growth. Nevertheless, we infer from these results that the improved growth observed in the mutants could be due to enhanced utilization of H2 from the drastically increased expression levels of the HynABC complex. In correlation with higher HynABC protein levels, H2-uptake activities were higher in both pglJ- and pglX mutants compared to WT in the absence and in the presence of H2. The increased (NiFe) hydrogenase synthesis (and activity) observed in the mutants might be linked to changes in metal homeostasis, particularly that pertaining to Fe and Ni. Studies conducted in the related organism H. pylori can provide insight into the respective roles of Fe and Ni with respect to transcriptional regulation of hydrogenase genes, through Fur and NikR regulators, respectively. For instance, H. pylori apo-Fur has been shown to repress hynABC (Ernst et al., 2005). Furthermore, addition of Ni to the medium leads to decreased hynABC expression; however, this repression was not observed in a nikR mutant background (Ernst et al., 2005) suggesting that either Ni-bound NikR represses or apo-NikR activates hydrogenase expression in H. pylori; in addition Ni-NikR has been shown to repress fur (Dosanjh et al., 2009). Taken together, these sets of results suggest the possible following mechanism in Cff: if Ni-bound NikR represses fur and (apo-) Fur represses hynABC, then elevated Ni levels (as observed in both pgl mutants) would be expected to de-repress Fur-controlled hynABC. The final outcome would be increased HynABC levels and increased hydrogenase activity, and indeed protein activities correlated well in cells and whole cell lysates grown under the same conditions. Obviously, the mechanism at play in Cff has yet to be elucidated. Nevertheless, taken together, our results indicate a clear link between N-glycosylation (or the lack thereof) and (NiFe) HynABC hydrogenase expression and/or enzymatic activity.
It is worth noting that Cff contains two additional hydrogenase complexes: a (FeFe) hydrogenase (HydA), hypothesized to be a H2-uptake type, and a (NiFe) H2-evolving complex (HycBCDEFG) predicted to be part of a formate hydrogen lyase (FHL) complex that links formate oxidation to hydrogen production (Benoit et al., 2020). Based on our proteomic study, neither HydA nor HycBCDEFG hydrogenase subunits were found to be expressed at different levels between WT and the N-glycosylation mutants.
The increase in (NiFe) HynABC and decrease in certain metal-related proteins prompted us to quantify Ni and Fe levels. In both pgl mutants we saw a significant decrease in iron; however, the decrease in iron for pglX- was fivefold lower than pglJ- and sixfold lower than WT. This may be because pglX- has a 118.4-fold decrease, and only 3.3-fold decrease in pglJ-, in the CfrA ferric enterobactin receptor present in Cj, which is responsible for high-affinity iron acquisition (Miller et al., 2009).
Although nickel is essential for both nickel containing hydrogenases in Cf, it is also toxic in excessive amounts, potentially causing oxidative stress and perturbing enzyme activities (Macomber and Hausinger, 2011). One mechanism of modulating nickel levels that was previously identified in E. coli is the nickel defense system (RcnA), which utilizes a proton gradient to translocate nickel to the periplasm where it can either be bound by sequestering proteins or effluxed from the cell (Macomber and Hausinger, 2011). We observed a 50-fold increase in a metal P-type ATPase in pglX- (A0RQS6), annotated as copper/cadmium-translocating P-type ATPase with similarly predicted activities. These metal P-type ATPase translocators are involved in detoxification of metals by transporting metals across the inner membrane (Ma et al., 2009). It is possible that this P-type metal translocator may be deficient at translocating; however, there is no clear link to N-glycosylation. These data are consistent with the cellular nickel levels of the pglX- strain, which were 6-times higher than the pglJ- strain. These increased nickel levels may be responsible for the higher hydrogenase activity levels measured in pglX- compared to the other two strains, while nickel toxicity could explain the decreased growth in H2 growth assays. Our data indicate that N-glycosylation regulates (NiFe)-hydrogenases HynABC, correlating with cellular nickel levels. Taken together, this suggests a possible link between our findings; however, their specific interaction with N-glycosylation is still unknown.
Our research connects N-glycosylation to HynABC hydrogenase regulation and nickel/iron homeostasis, two cellular processes which have been associated with pathogenicity in other bacteria (Palyada et al., 2004; Maier and Benoit, 2019; Benoit et al., 2020). The presented results deepen our understanding of the role of N-glycosylation in C. fetus cell physiology. In addition, the Cf-N-glycosylation system provides glycan diversity through PglX and PglY, which may further impact the biology of the microbe and warrants further investigation.
Materials and Methods
Bacterial Strains, Plasmids, Oligonucleotides and Growth Conditions
Oligonucleotides used in this study are listed in Supplementary Table S3. Bacterial strains and plasmids are listed in Supplementary Table S4. C. fetus was grown using Brain-Heart Infusion (BHI) medium (BHI-Hardy Diagnostics) and Columbia agar (CBA-Hardy Diagnostics) with 5% defibrinated horse blood (Hemostat, Dixon, CA, United States) under microaerobic conditions (10% CO2, 5% O2, 85% N2) at 37°C. E. coli was grown on 2xYT at 37°C or as indicated. If required, antibiotics were added to the following working concentrations: 100 μg/mL ampicillin, 25 μg/mL chloramphenicol, 50 μg/mL kanamycin, and 100 μg/mL spectinomycin.
Preparation of Whole Cell Lysates and Western Blotting
Whole cell lysates of bacterial cells were prepared as described previously (Liu et al., 2006). Protein concentrations were determined using either the NanoVue Plus Spectrophotometer (GE) at A280 or by the BioRad DC Bradford assay kit with bovine serum albumin as a protein standard. Samples were either analyzed immediately or were frozen at −20°C until further use. Western blot analyzes was carried out as described (Nothaft et al., 2010) with anti-His (1:2000) (Rockland), anti-Cff-N-glycan (1:5000) (Nothaft et al., 2012), anti-Cj-N-glycan (R1, 1:7500) (Nothaft et al., 2012) or anti-CmeA (1:5000) (Wacker et al., 2002) as the primary, and anti-rabbit IgG (1:2000) (Santa Cruz Biotechnology) as the secondary antibody or with alkaline phosphatase labeled wheat germ agglutinin (WGA, 1:500) (EY Labs). Antibody and WGA-lectin reactive bands were visualized directly on the membrane with nitro-blue tetrazolium chloride (NBT) and 5-bromo-4-chloro-3′-indolyphosphate p-toluidine salt (BCIP) alkaline phosphatase substrate solution (Roche) according to the protocol of the manufacturer.
Free Oligosaccharides Preparation and Analysis
Free oligosaccharides were obtained by ethanol extraction as described previously (Dwivedi et al., 2013) from 1 g of wet cell pellets. Free oligosaccharides (fOS) preparations were further purified using porous graphite carbon (PGC) columns as described (Liu et al., 2006). After elution and lyophilization fOS were dissolved in 100 μl of milliQ water and either stored at −20°C or directly analyzed by TLC as described (Dwivedi et al., 2013).
Generation of Cff pgl Gene Mutant Constructs
First, a PCR product containing Cff-pglKXYJ (4868 nt) was generated with oligo CS469 and CS470 using chromosomal DNA from Cff as a template and inserted into the EcoRV site of plasmid pPCR-Script Amp SK(+). After transforming E. coli DH5α, plasmid-containing cells were isolated on plates supplemented with Amp and X-gal (40 μg/ml) and plasmids isolated from white colonies were analyzed by restriction digestion. One positive candidate (pPCR-Script-Cffpgl) that had the PCR product with the Cff pgl genes inserted in opposite direction to the lacZ gene was processed further. Next, plasmid pPCR-Script-Cffpgl was digested with either EcoRV (1 site within pglX), AccI (1 site within pglY), or SpeI (1 site within pglJ). The linearized plasmid backbones were isolated and in the case of the AccI and SpeI digests, T4 DNA polymerase was used to generate blunt ends before the DNA fragments were purified by agarose gel extraction. The kanamycin (kan) resistance cassette obtained and isolated after SmaI digestion of plasmid pMW2 was ligated with each vector backbone preparation. Amp and Kan resistant colonies obtained after ligation and transformation were screened and verified by restriction analyzes. One positive clone in which the kan cassette is transcribed in the same orientation as the corresponding reading frame (pgl gene) was used to generate the gene-specific insertions by double homologous recombination into the chromosome of Cff.
Transformation and Insertion Mutagenesis of Cff
Natural transformation (on a BHI agar surface) (Wang and Taylor, 1990) and electroporation (Baillon et al., 1999) protocols were employed to introduce Cff-pgl gene:kan plasmid DNA for double homologous integration of the kan cassette into Cff. To do so, the corresponding suicide plasmids (pPCR-Script-CffpglX:kan, pPCR-Script-CffpglY:kan, and pPCR-Script-CffpglJ:kan) were isolated from either E. coli DH5α or E. coli JM110. The latter strain was used to generate non-methylated DNA to circumvent the Campylobacter restriction modification system. Transformants were selected on BHI plates for kanamycin resistance and individual colonies were isolated, streaked on fresh agar plates, and used to isolate chromosomal DNA. Candidate colonies were analyzed and verified by PCR with oligonucleotides hybridizing outside of the recombination event (Supplementary Figure S1) to confirm integration of the kan cassette at the correct position on the chromosome. One positive candidate (for pglX- and pglJ-) was used for further phenotypical analyzes, whereas (even after multiple attempts) no positive candidate could be obtained for the integration of the kan cassette into the Cff-pglY gene locus.
Growth Curves and Motility Assays
Growth comparison was performed in BHI broth and growth curves were recorded as described (Dubb et al., 2020). Motility assays were carried out as outlined previously (Golden and Acheson, 2002) with slight modifications. Briefly, Cff-WT and pgl mutant strains were grown for 18 h on BHI agar. Cells were harvested from the plates with 2 ml of BHI broth and cell suspensions were diluted to an OD600 of 0.05. Then, 1 μl of each cell suspension was spotted onto a BHI 0.3% agar plate and after 24 h of incubation, images were taken and the diameter of the motility zone was measured horizontally and vertically.
Reverse Transcriptase PCR
Reverse transcriptase (RT) PCR was performed according to Muraoka and Zhang (2011) with RNA extracted from cells grown on BHI agar for 18 h using the RNeasy Kit following the instructions of the manufacturer (Qiagen). PCR conditions after the RT-step were identical for each primer pair and were carried out as follows: 35 cycles with 30 s, 95°C; 30 s, 52°C and 20 s, 72°C followed by a 72°C finalizing step for 3 min. Samples were stored at 4°C before 15 μl of each 50 μl reaction were analyzed by 0.8% agarose gel electrophoresis.
Pgl Gene Expressing Plasmids
Gene-specific oligonucleotides (Supplementary Table S3) were used to amplify Cj-pglH, Cj-pglA, Cj-pglJ, Cff-pglA, Cff-pglJ, Cff-pglX, and Cff-pglY as well as Cff-pglXY for expression in E. coli. To do so, PCR products obtained with specific template DNA (plasmid ppgl for the C. jejuni pgl genes or chromosomal DNA from Cff) were purified, treated with restriction enzymes (see Supplementary Table S3), and inserted into plasmid pCE111/28 digested with the same enzymes. To generate the Cff-pglXY expression plasmid, a PCR product encompassing both open reading frames was generated; in addition, a second plasmid was generated by inserting the Cff-pglY PCR product into the pCE111/28 (Cff-pglX) product via PstI (introduced by PCR during the cloning of pglX) and XhoI simultaneously introducing an optimized RBS site upstream of the Cff-pglY start codon, as was done for all the other pgl genes. After ligation, transformation, and screening on selective (Cm) plates, plasmids isolated from candidate colonies were analyzed by restriction analyzes and verified by DNA sequencing. One positive candidate for each construct was used for further analysis.
Pgl Operon Expression Plasmids
To generate ppgl operon mutant plasmids that are compatible with the generated Cj and Cff-pgl gene expression plasmids (pCE111/28-derivatives, CmR), the cat cassette from all pgl operon plasmids with a kan cassette insertion in the various pgl genes (Supplementary Table S4; Linton et al., 2005) was deleted. To do so, plasmids ppgl-pglH:kan, ppgl-pglI:kan, ppgl-pglJ:kan, and ppgl-pglA:kan were treated with BsaAI excising the cat gene but leaving the rest of the plasmid intact. The complete DNA digest reactions were purified and directly re-ligated. To generate the pgl operon plasmid lacking pglH and pglI (ppgl-pglHI:kan), two PCR products were generated: the first reaction was performed with plasmid ppgl-pglH:kan as a template and with oligonucleotides pglHI-kan-R and pglHI-pACYC-F amplifying the 5-prime half of the kan cassette in pglH and the upstream part of the Cj-pgl operon. The second reaction was performed with plasmid ppgl-pglI:kan as template and with oligonucleotides pglHI-aph-FR and pglHI-pACYC-R amplifying the 3-prime half of the kan cassette in pglI, the pglI downstream region of the pgl operon, as well as the origin of replication. The obtained PCR products were purified and ligated without further treatment.
After transformation of DH5α candidate colonies for each ligation reaction were pre-screened on LB agar for KanR and CmS. The loss of the cat cassette and the correct gene organization on plasmids isolated form those colonies were further verified by restriction digest analyzes and DNA sequencing. One positive candidate for each construct (ppglop pgl-gene:kan, cat- derivative) was used for further analyzes.
Expression of CmeA-His6 in Glycosylation Competent E. coli Cells
Functional analysis of certain Cff-pgl proteins was performed in the heterologous E. coli glycosylation system. E. coli CLM24 was sequentially transformed with the individual pgl gene expression plasmids (pCE11/28 derivatives), the CmeA-His6 expression plasmid (pIH18, pEXT21-derivative), and either the plasmid carrying the WT Cj-pgl operon on ppgl or the compatible pgl operon mutant plasmids (cat- derivatives) with a kan cassette inserted into pglA, pglJ, pglH, and pglHI (double mutant). Cells stably maintaining the plasmid combinations were grown as 4 ml cultures overnight before inoculating 100 ml of fresh medium to a starting OD600 of 0.1. Cells were further grown until an OD600 of 0.5–0.7 was reached and CmeA-His6 expression (constitutively low expressed from the tetracycline promoter on pIH18) was further induced by the addition of IPTG to a final concentration of 0.5 mM. After growth for an additional 4 h, cells were cooled on ice for 10–15 min, pelleted by centrifugation (10 min, 12,000 rpm, 4°C), and washed twice with ice-cold 1 × PBS buffer. Then, 1/10 of the pellet (corresponding to 10 ml of culture volume) was used to produce whole cell lysates using Bacterial Protein Extraction Reagent B-PER (Thermo Fisher Scientific) according to the instructions of the manufacturer and the remainder of cells was used to generate whole cell lysates (as described in Liu et al., 2006) for the purification of the corresponding CmeA-His6 proteins by Ni-NTA affinity chromatography. To do so, whole cell extracts were filtered (0.22 μm) and loaded onto a 10 ml gravity-flow cartridge (Amersham Pharmacia Biosciences) pre-loaded with 0.5 ml Ni-NTA agarose and pre-equilibrated with 1 column volume 1 × PBS. The column was subsequently washed with at least 5 column volumes of 1 × PBS containing 20 mM imidazole and bound CmeA-His6 protein was eluted with 0.5–1.5 ml of PBS containing 0.5 M imidazole. Purified proteins were stored at 4°C until further use or immediately analyzed by 12.5% PAGE/mass spectrometry and/or western blotting.
Hydrogen Growth Conditions
Growth of Cff under hydrogen was performed as previously described (Benoit and Maier, 2018) with the following changes. Cff cells were grown for 48 h then streaked on BHI plates and further incubated for 12 h at 37°C under microaerobic conditions. Cells were resuspended in BHI broth and standardized to the same optical density at 600 nm (OD600), 3.0–4.0. Sealed 165 mL bottles containing 10 mL BHI were flushed with N2 gas for 10 min, then CO2 (10% headspace partial pressure, h.p.p.) and O2 (5% h.p.p.) were injected in every bottle. H2 (20% h.p.p.) was added as indicated. Cells were inoculated (1:100) and grown at 37°C while shaking at 200 rpm. Growth yields from three biological replicates (each performed in duplicate) were determined after 48 h by serially diluting in BHI and plating on CBA. Plates were incubated at 37°C in microaerobic conditions for three days before being counted.
Whole-Cell H2-Uptake Hydrogenase Assays
H2-uptake was performed as previously described (Maier et al., 1996). Cff cells were grown at 37°C for 24 h on BHI plates under microaerobic conditions either with 10% H2 or without. Cells were harvested and resuspended in phosphate buffered saline (PBS) to an optical density (OD600) of 1 which corresponds to ∼2.3 × 109 cells/mL. A 2 mL chamber was filled with cells followed by an injection with PBS saturated with H2. H2-uptake was monitored as previously described (Maier et al., 1996). Values are reported as nanomoles of H2 used per min per 109 cells and represent four independent measurements for cells grown in microaerobic conditions (and no H2) and two measurements for cells grown in microaerobic conditions with the addition of 10% H2.
Determination of Iron and Nickel Content
Campylobacter fetus subsp. fetus cells were grown at 37°C for 24 h on two BHI plates under microaerobic conditions and harvested with a loop in 1 mL metal-free double distilled water. Samples were centrifuged at 10,000 g for 5 min, washed once with water, resuspended and lyzed by sonication. A portion of lyzed sample was used to determine the protein concentration using the bicinchoninic acid (BCA, Thermo Scientific Pierce) assay. Samples were centrifuged at 15,000 × g for 5 min and the supernatant was analyzed for iron and nickel. The remaining sample portion was used for metal (Fe or Ni) content analysis. Briefly, Fe and Ni concentrations were measured by atomic absorption, using a Shimadzu AA-6701F spectrophotometer. All samples were diluted (in 1% HNO3) to be in the range of the standard curve (0 to 0.4 μM of either Fe or Ni) generated using atomic absorption-grade standard Fe or Ni solutions (Sigma). Results shown are means and standard deviations for 3–5 measurements.
Ethidium Bromide Accumulation Assay
Accumulation of EtBr was performed as previously described (Lin et al., 2002) with the following changes. Cff strains were grown overnight on BHI agar at 37°C in microaerobic conditions and harvested with MEM (Gibco). Cultures were adjusted to OD600 of 0.2 and then incubated at 37°C for 30 min in microaerobic conditions. EtBr was added to a final concentration of 2 μg/mL. Fluorescence was measured, with an excitation of 530 nm and emission of 600 nm, every 2 min over a 20 min time using a Bio Tek Synergy H1 plate reader. This was performed in three biological replicates, which included three technical replicates. Background fluorescence of MEM with EtBr was subtracted from these values.
Antibiotic MIC Assay
Campylobacter fetus subsp. fetus cells were grown for 24 h at 37°C in microaerobic conditions on CBA plates. Antibiotic MIC was assessed using the Sensititre (Trek Diagnostic Systems) platform. Sensititre plate EQUIN1F was used, following manufacturer’s instructions.
Preparation of Bacterial Whole Cell Proteome Samples
Campylobacter fetus subsp. fetus cells were grown for 24 h at 37°C under microaerobic conditions on BHI agar. Cells were harvested with ice-cold PBS and inactivated with PBS, 10% sodium azide for 30 min at 4°C. Cell pellets obtained after centrifugation (4000 × g for 15 min) were lyophilized and stored at −20°C until further use. Cell lysates for proteomic analyses were prepared as follows: cells were solubilized in 4% SDS, 100 mM Tris pH 8.0, and 20 mM DTT and boiled at 95°C with shaking at 2000 rpm for 10 min. Insoluble material was removed by centrifugation at 17,000 × g for 10 min at room temperature and the supernatant was collected. Protein concentrations were determined using the bicinchoninic acid assay (Thermo Scientific Pierce) and 200 μg of protein from each sample was acetone-precipitated overnight at −20°C by mixing volumes of ice-cold acetone with one volume of sample. Samples were then spun down at 16,000 × g for 10 min at 4°C. The precipitated protein pellets were resuspended with 80% ice-cold acetone and precipitated for an additional 4 h at −20°C. Samples were spun down at 17,000 × g for 10 min at 4°C to collect the precipitated protein.
Digestion of Complex Protein Lysates
Dried protein pellets were resuspended in 6 M urea, 2 M thiourea, 40 mM NH4HCO3 and reduced/alkylated prior to digestion with Lys-C (1/200 w/w) and then trypsin (1/50 w/w) overnight as previously described (Scott et al., 2011). Digested samples were acidified to a final concentration of 0.5% formic acid and desalted with home-made high-capacity StageTips composed on 5 mg EmporeTM C18 material (3M, Maplewood, Minnesota) and 5 mg of OLIGO R3 reverse phase resin (Thermo Fisher Scientific) according to the protocol of Ishihama and Rappsilber (Ishihama et al., 2006; Rappsilber et al., 2007). Bound peptides were eluted with buffer B, dried and stored at −20°C.
Reversed Phase Liquid Chromatography-Mass Spectrometry
Purified peptides were resuspended in Buffer A∗ and separated using a two-column chromatography set up comprising a PepMap100 C18 20 mm × 75 μm trap and a PepMap C18 500 mm × 75 μm analytical column (Thermo Fisher Scientific). Samples were concentrated onto the trap column at 5 μl/min for 5 min and infused into an Orbitrap EliteTM Mass Spectrometer (Thermo Fisher Scientific) at 300 nl/min via the analytical column using a Dionex Ultimate 3000 UPLC (Thermo Fisher Scientific). Then, 180 min gradients were run altering the buffer composition from 3% buffer B to 28% B over 150 min, then from 28% B to 40% B over 10 min, then from 40% B to 100% B over 2 min, followed by the composition held at 100% B for 3 min, and then dropped to 3% B over 5 min and held at 3% B for another 10 min. The Orbitrap Mass Spectrometer was operated in a data-dependent mode automatically switching between the acquisition of a single Orbitrap MS scan (60,000 resolution) followed by one data-dependent HCD (resolution 15 k AGC target of 4 × 105 with a maximum injection time of 250 ms, NCE 40) and CID (ion trap, AGC target of 5 × 104 with a maximum injection time of 100 ms, NCE 35) event for each precursor (total of five precursors per cycle with 45 s dynamic exclusion enabled).
Proteome Data Analyses
Proteome analysis to assess the expression of proteins within Cff strains was undertaken with MaxQuant [v1.5.3.30 (Cox and Mann, 2008)]. Database searching was carried out against the C. fetus subsp. fetus strain ATCC 27374 proteome (generated from a Maxquant generated six frame translation of the in-house sequenced strain). Searches were undertaken with the following search parameters: carbamidomethylation of cysteine as a fixed modification; oxidation of methionine, acetylation of protein N-terminal trypsin/P cleavage with a maximum of two missed cleavages. To enhance the identification of peptides between samples, the Match between Runs option was enabled with a precursor match window set to 2 min and an alignment window of 10 min. For label free quantitation, the MaxLFQ option within Maxquant was enabled in addition to the re-quantification module (Cox et al., 2014). The resulting outputs were processed within the Perseus (v1.5.0.9) analysis environment to remove reverse matches and common proteins contaminations prior to further analysis (Tyanova et al., 2016). Statistical analysis was undertaken in Perseus by grouping biological replicates, imputing missing values based on observed values (downshifted by 2.5 standard deviations with a width of 0.3 standard deviations) and then comparing groups using a student t-test. To define an appropriate p-value threshold, multiple hypothesis correction was undertaken using a Benjamini-Hochberg correction with a FDR of 0.05. All statistical outputs are provided within Supplementary Table S1. All mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (Vizcaino et al., 2016) with the dataset identifier PXD014538 [LFQ experiments of C. fetus fetus mutants (Supplementary MS Data 1) accessible using the username: reviewer71456@ebi.ac.uk, password: B5YuYNx8) and PXD017832 [analysis of C. fetus fetus pgl enzymes in the heterologous E. coli glycosylation system (Supplementary MS Data 2) accessible using the username: reviewer23740@ebi.ac.uk password: PHKlhnSp].
Glycopeptide Data Analysis
Glycopeptides were identified by manually interrogating possible glycopeptide scans based on the presence of the diagnostic oxonium ion (204.09 m/z) of HexNAc. To facilitate glycopeptide assignments from HCD scans, the ions below the mass of the predicted deglycosylated peptides were extracted with Xcalibur v2.2 using the Spectrum list function. Ions with a deconvoluted mass above that of the deglycosylated peptide and ions corresponding to known carbohydrate oxoniums were removed in a similar approach to post-spectral processing of ETD data and then searched with Mascot1. Searches were carried out using semi-trypsin specificity, carbamidomethylation of cysteine as a fixed modification, and oxidation (M) as a variable modification. A precursor and product tolerance of 20 ppm was used, and the taxonomy restricted to “Other Proteobacteria.” All spectra were searched with the decoy option enabled with all peptides passing a 1% FDR. Identified glycopeptide spectra were manually inspected and spectra annotated according to the nomenclature of Roepstorff and Fohlman (1984) for peptides as well as Domon and Costello (1988) for glycans.
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (1) partner repository with the dataset identifiers PXD014538 and PXD017832.
Author Contributions
JD, HN, and CS designed the experiments, interpreted the results, and wrote the manuscript. BB and CF constructed expression plasmids and Cf-pgl mutants. SB and RM performed the hydrogenase activity and assisted with analysis and interpretation of hydrogenase and AAS data. NS performed all mass spectrometry and LFQ analysis. DW and DL made pgl expression constructs for E. coli and assisted in data analysis and interpretation. All authors read and approved of the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the Melbourne Mass Spectrometry and Proteomics Facility of the Bio21 Molecular Science and Biotechnology Institute at The University of Melbourne for mass spectrometry analysis. We also thank Susan Sanchez and the Athens Veterinary Diagnostic Laboratory for help with the antibiotic sensitivity assay.
Nomenclature
- Cf
Campylobacter fetus
- Cff
Campylobacter fetus subsp. fetus
- Cft
Campylobacter fetus subsp. testudinum
- Cfv
Campylobacter fetus subsp. venerealis
- Cj
Campylobacter jejuni
- diNAcBac
2,4-diacetamido-2,4,6-trideoxyglucopyranose
- fOS
free oligosaccharides
- GalNAc
N-acetyl-galactosamine
- Glc
glucose
- GlcNAc
N-acetyl-glucosamine
- GTase
Glycosyltransferase
- Hex
hexose
- HexNAc
N-acetyl-hexosamine
- LLO
lipid-linked oligosaccharide
- MS
mass spectrometry.
Funding. JD was previously supported by the Alberta Glycomics Centre and subsequently the National Institute of General Medical Sciences Training Grant Award Number T32GM107004. NS was supported by the National Health and Medical Research Council of Australia (NHMRC) project grant (APP1100164) and by an Overseas (Biomedical) Fellowship (APP1037373). SB and RM received support from the University of Georgia Foundation. CS was an Alberta Innovates Strategic Chair in Bacterial Glycomics. DW was supported by a Society for Applied Microbiology Studentship.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01191/full#supplementary-material
References
- Abouelhadid S., North S. J., Hitchen P., Vohra P., Chintoan-Uta C., Stevens M., et al. (2019). Quantitative analyses reveal novel roles for N-glycosylation in a major enteric bacterial pathogen. mBio 10:e00297-19. 10.1128/mBio.00297-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiba M., Lin J., Barton Y. W., Zhang Q. (2006). Interaction of CmeABC and CmeDEF in conferring antimicrobial resistance and maintaining cell viability in Campylobacter jejuni. J. Antimicrob. Chemother. 57 52–60. 10.1093/jac/dki419 [DOI] [PubMed] [Google Scholar]
- Alagesan K., Hinneburg H., Seeberger P. H., Silva D. V., Kolarich D. (2019). Glycan size and attachment site location affect electron transfer dissociation (ETD) fragmentation and automated glycopeptide identification. Glycoconj J. 36 487–493. 10.1007/s10719-019-09888-w [DOI] [PubMed] [Google Scholar]
- Alaimo C., Catrein I., Morf L., Marolda C. L., Callewaert N., Valvano M. A., et al. (2006). Two distinct but interchangeable mechanisms for flipping of lipid-linked oligosaccharides. EMBO J. 25 967–976. 10.1038/sj.emboj.7601024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baillon M. L., Van Vliet A. H., Ketley J. M., Constantinidou C., Penn C. W. (1999). An iron-regulated alkyl hydroperoxide reductase (AhpC) confers aerotolerance and oxidative stress resistance to the microaerophilic pathogen Campylobacter jejuni. J. Bacteriol. 181 4798–4804. 10.1128/jb.181.16.4798-4804.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit S. L., Maier R. J. (2018). Site-directed mutagenesis of Campylobacter concisus respiratory genes provides insight into the pathogen’s growth requirements. Sci. Rep. 8:14203. 10.1038/s41598-018-32509-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit S. L., Maier R. J., Sawers R. G., Greening C. (2020). Molecular hydrogen metabolism: a widespread trait of pathogenic bacteria and protists. Microbiol. Mol. Biol. Rev. 84:e00092-19. 10.1128/MMBR.00092-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cain J. A., Dale A. L., Niewold P., Klare W. P., Man L., White M. Y., et al. (2019). Proteomics reveals multiple phenotypes associated with N-linked glycosylation in Campylobacter jejuni. Mol. Cell. Proteomics 18 715–734. 10.1074/mcp.RA118.001199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M. M., Glover K. J., Imperiali B. (2007). From peptide to protein: comparative analysis of the substrate specificity of N-linked glycosylation in C. jejuni. Biochemistry 46 5579–5585. 10.1021/bi602633n [DOI] [PubMed] [Google Scholar]
- Cid E., Gomis R. R., Geremia R. A., Guinovart J. J., Ferrer J. C. (2000). Identification of two essential glutamic acid residues in glycogen synthase. J. Biol. Chem. 275 33614–33621. 10.1074/jbc.M005358200 [DOI] [PubMed] [Google Scholar]
- Coutinho P. M., Deleury E., Davies G. J., Henrissat B. (2003). An evolving hierarchical family classification for glycosyltransferases. J. Mol. Biol. 328 307–317. 10.1016/s0022-2836(03)00307-3 [DOI] [PubMed] [Google Scholar]
- Cox J., Hein M. Y., Luber C. A., Paron I., Nagaraj N., Mann M. (2014). Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics 13 2513–2526. 10.1074/mcp.M113.031591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J., Mann M. (2008). MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26 1367–1372. 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- Domon B., Costello C. E. (1988). A systematic nomenclature for carbohydrate fragmentations in FAB-MS/MS spectra of glycoconjugates. Glycoconjugate J. 5 397–409. 10.1007/BF01049915 [DOI] [Google Scholar]
- Dosanjh N. S., West A. L., Michel S. L. (2009). Helicobacter pylori NikR’s interaction with DNA: a two-tiered mode of recognition. Biochemistry 48 527–536. 10.1021/bi801481j [DOI] [PubMed] [Google Scholar]
- Dubb R. K., Nothaft H., Beadle B., Richards M. R., Szymanski C. M. (2020). N-glycosylation of the CmeABC multidrug efflux pump is needed for optimal function in Campylobacter jejuni. Glycobiology 30 105–119. 10.1093/glycob/cwz082 [DOI] [PubMed] [Google Scholar]
- Duncan J. S., Leatherbarrow A. J., French N. P., Grove-White D. H. (2014). Temporal and farm-management-associated variation in faecal-pat prevalence of Campylobacter fetus in sheep and cattle. Epidemiol. Infect. 142 1196–1204. 10.1017/S0950268813002379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi R., Nothaft H., Reiz B., Whittal R. M., Szymanski C. M. (2013). Generation of free oligosaccharides from bacterial protein N-linked glycosylation systems. Biopolymers 99 772–783. 10.1002/bip.22296 [DOI] [PubMed] [Google Scholar]
- Ernst F. D., Kuipers E. J., Heijens A., Sarwari R., Stoof J., Penn C. W., et al. (2005). The nickel-responsive regulator NikR controls activation and repression of gene transcription in Helicobacter pylori. Infect. Immun. 73 7252–7258. 10.1128/IAI.73.11.7252-7258.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman M. F., Wacker M., Hernandez M., Hitchen P. G., Marolda C. L., Kowarik M., et al. (2005). Engineering N-linked protein glycosylation with diverse O antigen lipopolysaccharide structures in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 102 3016–3021. 10.1073/pnas.0500044102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald C., Tu Z. C., Patrick M., Stiles T., Lawson A. J., Santovenia M., et al. (2014). Campylobacter fetus subsp. testudinum subsp. nov., isolated from humans and reptiles. Int. J. Syst. Evol. Microbiol. 64 2944–2948. 10.1099/ijs.0.057778-0 [DOI] [PubMed] [Google Scholar]
- Glover K. J., Weerapana E., Chen M. M., Imperiali B. (2006). Direct biochemical evidence for the utilization of UDP-bacillosamine by PglC, an essential glycosyl-1-phosphate transferase in the Campylobacter jejuni N-linked glycosylation pathway. Biochemistry 45 5343–5350. 10.1021/bi0602056 [DOI] [PubMed] [Google Scholar]
- Glover K. J., Weerapana E., Imperiali B. (2005). In vitro assembly of the undecaprenylpyrophosphate-linked heptasaccharide for prokaryotic N-linked glycosylation. Proc. Natl. Acad. Sci. U.S.A. 102 14255–14259. 10.1073/pnas.0507311102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden N. J., Acheson D. W. (2002). Identification of motility and autoagglutination Campylobacter jejuni mutants by random transposon mutagenesis. Infect. Immun. 70 1761–1771. 10.1128/iai.70.4.1761-1771.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B., Lin J., Reynolds D. L., Zhang Q. (2010). Contribution of the multidrug efflux transporter CmeABC to antibiotic resistance in different Campylobacter species. Foodborne Pathog. Dis. 7 77–83. 10.1089/fpd.2009.0354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrixson D. R., DiRita V. J. (2004). Identification of Campylobacter jejuni genes involved in commensal colonization of the chick gastrointestinal tract. Mol. Microbiol. 52 471–484. 10.1111/j.1365-2958.2004.03988.x [DOI] [PubMed] [Google Scholar]
- Holst E., Wathne B., Hovelius B., Mardh P. A. (1987). Bacterial vaginosis: microbiological and clinical findings. Eur. J. Clin. Microbiol. 6 536–541. 10.1007/bf02014242 [DOI] [PubMed] [Google Scholar]
- Ielmini M. V., Feldman M. F. (2011). Desulfovibrio desulfuricans PglB homolog possesses oligosaccharyltransferase activity with relaxed glycan specificity and distinct protein acceptor sequence requirements. Glycobiology 21 734–742. 10.1093/glycob/cwq192 [DOI] [PubMed] [Google Scholar]
- Iraola G., Forster S. C., Kumar N., Lehours P., Bekal S., Garcia-Pena F. J., et al. (2017). Distinct Campylobacter fetus lineages adapted as livestock pathogens and human pathobionts in the intestinal microbiota. Nat. Commun. 8:1367. 10.1038/s41467-017-01449-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihama Y., Rappsilber J., Mann M. (2006). Modular stop and go extraction tips with stacked disks for parallel and multidimensional Peptide fractionation in proteomics. J. Proteome Res. 5 988–994. 10.1021/pr050385q [DOI] [PubMed] [Google Scholar]
- Jervis A. J., Butler J. A., Lawson A. J., Langdon R., Wren B. W., Linton D. (2012). Characterization of the structurally diverse N-linked glycans of Campylobacter species. J. Bacteriol. 194 2355–2362. 10.1128/JB.00042-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jervis A. J., Langdon R., Hitchen P., Lawson A. J., Wood A., Fothergill J. L., et al. (2010). Characterization of N-linked protein glycosylation in Helicobacter pullorum. J. Bacteriol. 192 5228–5236. 10.1128/JB.00211-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly J., Jarrell H., Millar L., Tessier L., Fiori L. M., Lau P. C., et al. (2006). Biosynthesis of the N-linked glycan in Campylobacter jejuni and addition onto protein through block transfer. J. Bacteriol. 188 2427–2434. 10.1128/JB.188.7.2427-2434.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein B. S., Vergeront J. M., Blaser M. J., Edmonds P., Brenner D. J., Janssen D., et al. (1986). Campylobacter infection associated with raw milk. An outbreak of gastroenteritis due to Campylobacter jejuni and thermotolerant Campylobacter fetus subsp fetus. JAMA 255 361–364. 10.1001/jama.255.3.361 [DOI] [PubMed] [Google Scholar]
- Kowarik M., Young N. M., Numao S., Schulz B. L., Hug I., Callewaert N., et al. (2006). Definition of the bacterial N-glycosylation site consensus sequence. EMBO J. 25 1957–1966. 10.1038/sj.emboj.7601087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhns L. G., Benoit S. L., Bayyareddy K., Johnson D., Orlando R., Evans A. L., et al. (2016). Carbon fixation driven by molecular hydrogen results in chemolithoautotrophically enhanced growth of Helicobacter pylori. J. Bacteriol. 198 1423–1428. 10.1128/JB.00041-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J., Michel L. O., Zhang Q. (2002). CmeABC functions as a multidrug efflux system in Campylobacter jejuni. Antimicrob. Agents Chemother. 46 2124–2131. 10.1128/aac.46.7.2124-2131.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linton D., Dorrell N., Hitchen P. G., Amber S., Karlyshev A. V., Morris H. R., et al. (2005). Functional analysis of the Campylobacter jejuni N-linked protein glycosylation pathway. Mol. Microbiol. 55 1695–1703. 10.1111/j.1365-2958.2005.04519.x [DOI] [PubMed] [Google Scholar]
- Liu X., Mcnally D. J., Nothaft H., Szymanski C. M., Brisson J. R., Li J. (2006). Mass spectrometry-based glycomics strategy for exploring N-linked glycosylation in eukaryotes and bacteria. Anal. Chem. 78 6081–6087. 10.1021/ac060516m [DOI] [PubMed] [Google Scholar]
- Ma Z., Jacobsen F. E., Giedroc D. P. (2009). Coordination chemistry of bacterial metal transport and sensing. Chem. Rev. 109 4644–4681. 10.1021/cr900077w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macomber L., Hausinger R. P. (2011). Mechanisms of nickel toxicity in microorganisms. Metallomics 3 1153–1162. 10.1039/c1mt00063b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier R. J., Benoit S. L. (2019). Role of nickel in microbial pathogenesis. Inorganics 7:80 10.3390/inorganics7070080 [DOI] [Google Scholar]
- Maier R. J., Fu C., Gilbert J., Moshiri F., Olson J., Plaut A. G. (1996). Hydrogen uptake hydrogenase in Helicobacter pylori. FEMS Microbiol. Lett. 141 71–76. 10.1111/j.1574-6968.1996.tb08365.x [DOI] [PubMed] [Google Scholar]
- Marchand-Senecal X., Bekal S., Pilon P. A., Sylvestre J. L., Gaudreau C. (2017). Campylobacter fetus cluster among men who have sex with men, montreal, quebec, Canada, 2014-2016. Clin. Infect. Dis. 65 1751–1753. 10.1093/cid/cix610 [DOI] [PubMed] [Google Scholar]
- Miller C. E., Williams P. H., Ketley J. M. (2009). Pumping iron: mechanisms for iron uptake by Campylobacter. Microbiology 155 3157–3165. 10.1099/mic.0.032425-0 [DOI] [PubMed] [Google Scholar]
- Mills D. C., Jervis A. J., Abouelhadid S., Yates L. E., Cuccui J., Linton D., et al. (2016). Functional analysis of N-linking oligosaccharyl transferase enzymes encoded by deep-sea vent Proteobacteria. Glycobiology 26 398–409. 10.1093/glycob/cwv111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintmier B., Mcgarry J. M., Sparacino-Watkins C. E., Sallmen J., Fischer-Schrader K., Magalon A., et al. (2018). Molecular cloning, expression and biochemical characterization of periplasmic nitrate reductase from Campylobacter jejuni. FEMS Microbiol. Lett. 365:fny151. 10.1093/femsle/fny151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraoka W. T., Zhang Q. (2011). Phenotypic and genotypic evidence for L-fucose utilization by Campylobacter jejuni. J. Bacteriol. 193 1065–1075. 10.1128/JB.01252-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S., Takaki Y., Shimamura S., Reysenbach A. L., Takai K., Horikoshi K. (2007). Deep-sea vent epsilon-Proteobacterial genomes provide insights into emergence of pathogens. Proc. Natl. Acad. Sci. U.S.A. 104 12146–12150. 10.1073/pnas.0700687104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothaft H., Liu X., Mcnally D. J., Li J., Szymanski C. M. (2009). Study of free oligosaccharides derived from the bacterial N-glycosylation pathway. Proc. Natl. Acad. Sci. U.S.A. 106 15019–15024. 10.1073/pnas.0903078106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothaft H., Liu X., Mcnally D. J., Szymanski C. M. (2010). N-linked protein glycosylation in a bacterial system. Methods Mol. Biol. 600 227–243. 10.1007/978-1-60761-454-8_16 [DOI] [PubMed] [Google Scholar]
- Nothaft H., Scott N. E., Vinogradov E., Liu X., Hu R., Beadle B., et al. (2012). Diversity in the protein N-glycosylation pathways among Campylobacter species. Mol. Cell Proteomics 11 1203–1219. 10.1074/mcp.M112.021519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothaft H., Szymanski C. M. (2010). Protein glycosylation in bacteria: sweeter than ever. Nat. Rev. Microbiol. 8 765–778. 10.1038/nrmicro2383 [DOI] [PubMed] [Google Scholar]
- Nothaft H., Szymanski C. M. (2013). Bacterial protein N-glycosylation: new perspectives and applications. J. Biol. Chem. 288 6912–6920. 10.1074/jbc.R112.417857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson J. W., Maier R. J. (2002). Molecular hydrogen as an energy source for Helicobacter pylori. Science 298 1788–1790. 10.1126/science.1077123 [DOI] [PubMed] [Google Scholar]
- Palyada K., Threadgill D., Stintzi A. (2004). Iron acquisition and regulation in Campylobacter jejuni. J. Bacteriol. 186 4714–4729. 10.1128/JB.186.14.4714-4729.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick M. E., Gilbert M. J., Blaser M. J., Tauxe R. V., Wagenaar J. A., Fitzgerald C. (2013). Human infections with new subspecies of Campylobacter fetus. Emerg. Infect. Dis. 19 1678–1680. 10.3201/eid1910.130883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez A. S., Boilevin J., Mehdipour A. R., Hummer G., Darbre T., Reymond J. L., et al. (2018). Structural basis of the molecular ruler mechanism of a bacterial glycosyltransferase. Nat. Commun. 9:445. 10.1038/s41467-018-02880-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J., Mann M., Ishihama Y. (2007). Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2 1896–1906. 10.1038/nprot.2007.261 [DOI] [PubMed] [Google Scholar]
- Roepstorff P., Fohlman J. (1984). Proposal for a common nomenclature for sequence ions in mass spectra of peptides. Biomed. Mass Spectrom. 11:601. 10.1002/bms.1200111109 [DOI] [PubMed] [Google Scholar]
- Scott N. E., Nothaft H., Edwards A. V., Labbate M., Djordjevic S. P., Larsen M. R., et al. (2012). Modification of the Campylobacter jejuni N-linked glycan by EptC protein-mediated addition of phosphoethanolamine. J. Biol. Chem. 287 29384–29396. 10.1074/jbc.m112.380212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott N. E., Parker B. L., Connolly A. M., Paulech J., Edwards A. V., Crossett B., et al. (2011). Simultaneous glycan-peptide characterization using hydrophilic interaction chromatography and parallel fragmentation by CID, higher energy collisional dissociation, and electron transfer dissociation MS applied to the N-linked glycoproteome of Campylobacter jejuni. Mol. Cell. Proteomics 10:M000031-MCP201. 10.1074/mcp.M000031-MCP201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski C. M., Yao R., Ewing C. P., Trust T. J., Guerry P. (1999). Evidence for a system of general protein glycosylation in Campylobacter jejuni. Mol. Microbiol. 32 1022–1030. 10.1046/j.1365-2958.1999.01415.x [DOI] [PubMed] [Google Scholar]
- Troutman J. M., Imperiali B. (2009). Campylobacter jejuni PglH is a single active site processive polymerase that utilizes product inhibition to limit sequential glycosyl transfer reactions. Biochemistry 48 2807–2816. 10.1021/bi802284d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Z. C., Zeitlin G., Gagner J. P., Keo T., Hanna B. A., Blaser M. J. (2004). Campylobacter fetus of reptile origin as a human pathogen. J. Clin. Microbiol. 42 4405–4407. 10.1128/JCM.42.9.4405-4407.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M. Y., Geiger T., et al. (2016). The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13 731–740. 10.1038/nmeth.3901 [DOI] [PubMed] [Google Scholar]
- Vizcaino J. A., Csordas A., Del-Toro N., Dianes J. A., Griss J., Lavidas I., et al. (2016). 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44:11033. 10.1093/nar/gkw880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacker M., Linton D., Hitchen P. G., Nita-Lazar M., Haslam S. M., North S. J., et al. (2002). N-linked glycosylation in Campylobacter jejuni and its functional transfer into E. coli. Science 298 1790–1793. 10.1126/science.298.5599.1790 [DOI] [PubMed] [Google Scholar]
- Wagenaar J. A., Van Bergen M. A., Blaser M. J., Tauxe R. V., Newell D. G., Van Putten J. P. (2014). Campylobacter fetus infections in humans: exposure and disease. Clin. Infect. Dis. 58 1579–1586. 10.1093/cid/ciu085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Taylor D. E. (1990). Natural transformation in Campylobacter species. J. Bacteriol. 172 949–955. 10.1128/jb.172.2.949-955.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A. M., Procter J. B., Martin D. M., Clamp M., Barton G. J. (2009). Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics 25 1189–1191. 10.1093/bioinformatics/btp033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weerakoon D. R., Borden N. J., Goodson C. M., Grimes J., Olson J. W. (2009). The role of respiratory donor enzymes in Campylobacter jejuni host colonization and physiology. Microb. Pathog. 47 8–15. 10.1016/j.micpath.2009.04.009 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (1) partner repository with the dataset identifiers PXD014538 and PXD017832.