Key Points
MHC class II specificity was assessed by high-density peptide arrays.
A novel method is proposed to expand and update MHC class II prediction algorithms.
Abstract
The ability to predict and/or identify MHC binding peptides is an essential component of T cell epitope discovery, something that ultimately should benefit the development of vaccines and immunotherapies. In particular, MHC class I prediction tools have matured to a point where accurate selection of optimal peptide epitopes is possible for virtually all MHC class I allotypes; in comparison, current MHC class II (MHC-II) predictors are less mature. Because MHC-II restricted CD4+ T cells control and orchestrated most immune responses, this shortcoming severely hampers the development of effective immunotherapies. The ability to generate large panels of peptides and subsequently large bodies of peptide–MHC-II interaction data are key to the solution of this problem, a solution that also will support the improvement of bioinformatics predictors, which critically relies on the availability of large amounts of accurate, diverse, and representative data. In this study, we have used rHLA-DRB1*01:01 and HLA-DRB1*03:01 molecules to interrogate high-density peptide arrays, in casu containing 70,000 random peptides in triplicates. We demonstrate that the binding data acquired contains systematic and interpretable information reflecting the specificity of the HLA-DR molecules investigated, suitable of training predictors able to predict T cell epitopes and peptides eluted from human EBV-transformed B cells. Collectively, with a cost per peptide reduced to a few cents, combined with the flexibility of rHLA technology, this poses an attractive strategy to generate vast bodies of MHC-II binding data at an unprecedented speed and for the benefit of generating peptide–MHC-II binding data as well as improving MHC-II prediction tools.
Introduction
The binding of peptides to MHC class II (MHC-II) molecules is one of the most selective events in Ag presentation. Expressed by professional APC and presenting exogenous peptides to CD4+ T cells, MHC-II is central to adaptive immunity. Because CD4+ T cells, restricted to MHC-II, are recognized for their orchestrating role in shaping both humoral and cellular immune responses, much effort has been put into understanding the specificity of MHC-II. The human MHC HLA loci is one of the most polymorphic gene families known (1), and as of September 2019, the number of HLA class II (HLA-II) alleles amounts to 7065 according to hla.alleles.org, not considering the additional diversity created by the combinatorial HLA-DPA/DPB and HLA-DQA/DQB pairing. Furthermore, considering the number of peptides that can be presented and the huge diversity of TCRs that can recognize peptide–HLA-II (pHLAII) complexes, the combinations of peptides, HLA-I,I and TCRs that should be evaluated to characterize this interaction system are staggering. This calls for high-throughput and bioinformatics-based approaches. Indeed, bioinformatics has been a driving force in describing MHC-II specificity. Based on binding and affinity data, several different algorithms have been developed capable of predicting peptide–MHC-II interactions (e.g., the NetMHC suite) as an efficient approach to address the vast number of peptides and MHC allotypes. Prediction algorithms powered by artificial neural networks (ANNs) are dependent on large bodies of data which are costly to generate because each peptide conventionally needs to be individually synthesized and handled before conducting binding experiments. More recently, proteomics-driven, high-throughput identification of peptides eluted off MHC-II molecules has become possible and used to improve the performance of MHC-II prediction algorithms (2).
Although the prediction tools available today have improved significantly over the years, they are still impaired by a high false positive rate (3), making it difficult to select true T cell epitopes. This is recognized as a major current problem in the development of personalized medicine and immunotherapies, in particular those modes that are based on real-time discovery of tumor neoantigens (4, 5). The recent advances within the field of cancer immunotherapy have suggested that immunization with tumor-derived neoantigens is a very promising strategy, and growing evidence suggests that MHC-I–-restricted neoantigens should be included (4, 6, 7). Hence, there is an increasing demand for more accurate and efficient predictors of peptide–MHC-II interactions, underscored by the pharmaceutical industry now focusing on developing MHC-II prediction tools (8).
We have for years chosen a biochemical approach to study peptide-MHC interactions (9). This has resulted in the generation of a large collection of rHLA molecules, which have been used to generate tens of thousands of data points subsequently used to develop the NetMHC suite of prediction algorithms (10–12), which continues to be updated. Whereas high-throughput assays have been developed to study peptide-MHC interactions, classical orthogonal peptide synthesis remains a very costly and time-consuming bottleneck. However, within the last decade, high-density peptide microarray platforms, potentially containing up to millions of individual peptides/array, have become available at a cost as low as a few cents per peptide. We have recently developed and used high-density peptide microarrays to characterize linear sequence motifs such as enzyme cleavage sites and B cell epitopes in great detail (13–18). Such peptide microarrays containing C-terminally anchored 15-mer peptides should, in theory, be able to interact with MHC-II molecules because of its open-ended peptide-binding cleft allowing peptides to protrude through the ends of the cleft; something that should allow C-terminal tethering.
The ability to acquire millions of peptide–MHC-II binding data in a single experiment could potentially transform CD4+ T cell epitope discovery. In particular, the combination of such data with machine-learning methods has the potential to transform the development of peptide–MHC-II predictors. We have previously produced rHLA-DRA1*01:01/HLA-DRB1*01:01 and HLA-DRA1*01:01/HLA-DRB1*03:01 and successfully used these molecules to demonstrate specific staining of CD4+ T cells with peptide–HLA-II tetramers (19). In this study, we have investigated the binding of HLA-DRA1*01:01/HLA-DRB1*01:01 and HLA-DRA1*01:01/HLA-DRB1*03:01 to high-density peptide microarrays containing ∼70,000 random peptides in triplicate, used the binding data to train ANNs, validated these against peptides eluted off HLA-DRA1*01:01/HLA-DRB1*01:01 and HLA-DRA1*01:01/HLA-DRB1*03:01, and compared the prediction power to ANNs trained on publicly available binding affinity data from the Immune Epitope Database (IEDB) (20).
Materials and Methods
Production of rHLA-DR molecules HLA-DRA1*01:01, -DRB1*01:01, and -DRB1*03:01
HLA-DR molecules were produced and purified according to (21). Briefly, this entailed separate Escherichia coli expressions of each rHLA-DR chain followed by multiple liquid chromatography purification steps. The purified HLA-DR molecules were stored in 8 M urea, 25 mM Tris, and 15 mM NaCl at −80°C.
High-density peptide arrays
The conformational propensity of amino acids extending out of the peptide-binding groove of MHC-II is known to affect their peptide binding drastically (22). To ensure a hydrophilic peptide-binding environment. Nexterion-E microscope slides (Schott, Jena, Germany) were amino-functionalized with amino-dextran 250 (Fina Biosolutions, Rockville, MD) and used as substrate for solid-phase peptide synthesis. The ε-amino-caproic acid was inserted between the dextran-coat and the synthetic peptide as a spacer of a nonamino acid nature. High-density peptide arrays were then synthesized on the surface of the amino-functionalized microscope glass slides using a principle of maskless photolithographic synthesis (17, 23) combined with standard Fmoc peptide synthesis modified with the photolabile NPPOC (2-(2-nitrophenyl)propyl oxycarbonyl). A detailed description of the peptide synthesis has been described elsewhere (13).
High-density peptide array design
The array designs were all made with a proprietary software (PepArray, Schafer-N, Denmark) by importing peptide sequences and randomly distributing the sequences across 12 virtual sectors. A design to analyze the effect of peptide length on MHC-II binding ∼2300 15-mer peptides with random sequences were generated in silico and subsequently chopped into overlapping 8–14 mers and randomly distributed across the peptide array.
For the generation of peptide–MHC-II binding data, 500,000 13-mer peptides were generated from a list of pathogen proteins from which 72,000 peptide sequences were randomly sampled and distributed across the peptide array in triplicates.
Binding of HLA-II molecules to peptide microarray
The peptide arrays synthesized on microscope glass slides were hydrated in Lockmailer microscope jars containing 15 ml of PBS prior to incubation with HLA-DR molecules.
HLA-DR molecules (DRA1*01:01, DRB1*01:01, DRB1*03:01; immunAware, Copenhagen, Denmark) dissolved in storage buffer (8 M urea, 25 mM Tris [pH 8], 25 mM NaCl) were diluted in 12 ml of refolding buffer (PBS [pH 7.4] supplemented with 0.01% Lutrol F68, glycerol 20% [v/v], TPCK, TLCK, PMSF, TCEP) to a final concentration of 500 nM HLA-DRA and HLA-DRB and transferred to the incubation jar containing the hydrated peptide array and allowed to (de novo) refold at 18°C for 48 h. Following incubation, the incubation jars were drained, and the peptide arrays washed five times in PBS-T (PBS [pH 7.4], 1% Tween-20) followed by incubation with 1 μg/ml of monoclonal mouse anti–HLA-DR Ab (clone #L243) in PBS-T for 2 h. The peptide arrays were washed five times in 15 ml of PBS-T before a final incubation with 1 μg/ml Cy3-conjugated goat anti-mouse IgG (Bethyl Laboratory, Montgomery, Texas) in PBS-T. A final wash (five times) with PBS-T was conducted before transferring the peptide array to PBS for 20–30 min before drying the microscope slide in a spin centrifuge dryer.
Data acquisition
The peptide arrays were scanned in a microscope slides laser scanner (INNOSCAN 900; Innopsys, Carbonne, France) at 534 nm, resolution 1 μm and stored as TIFF-format images. Prior to qualification of the image scans, each image was cropped to include only the peptide array area surrounded by a thin border; subsequently, the cropped images were rescaled to (10,000 × 6,000 pixels) and saved in PNG format.
Quantification of signals (proprietary software, Schafer-N)
The processed images were quantified with a proprietary software (PepArray) by positioning a virtual grid corresponding to the peptide fields on top of the scanned images and quantifying each peptide field. The quantified signals (eight-bit) were stored in a text-file format containing corresponding peptide sequence information, peptide field identification, and quantified signal, row, column, and sector information [deposited at https://doi.org/10.5061/dryad.tqjq2bvvv (24)]. The PepArray software can be obtained from Schafer-N upon reasonable request. The quantified data were prepared for ANN (NNAlign) training by calculating the mean value SD and coefficient of variance (cv) (based on triplicate measurements). Inclusion criteria for ANN training were set at a threshold of cv < 0.5. The NNAlign server (2.1) https://services.healthtech.dtu.dk/service.php?NNAlign-2.1 accepts columns of a text strings and corresponding numerical values [0–1] with a limit of 50,000 inputs, hence the datasets were reduced to 50,000 by selecting, based on signal strength, the top 2000 peptides and random sampling of the remaining peptides.
Data transformation
The data were transformed from the eight-bit format [0–255] to [0–1] using a log transformation optimized by a BoxCox algorithm written in R using the EnvStats package (25).
ANN training
Log-transformed data with corresponding peptide sequences (one letter code) were uploaded to the NNAling 2.1 server and trained with the parameters indicated in Table I.
Table I. ANN summary.
| Neural Network Architecture | |
|---|---|
| Motif length | 9 |
| Peptide flanking region size | 3 |
| Peptide flanking region length encoding type | 0 |
| Peptide flanking region encoding | SPARSE OR BLOSUM |
| Number of hidden neurons | 2, 10, 20 |
| Burn-in period without insertions or deletions | 10 |
| Impose amino acid preference during burn-in | ILVMFYW |
| Amino acid numerical encoding | SPARSE OR BLOSUM |
| Number of training cycles | 500 |
| Number of random seeds | 10 |
| Number of networks in final ensemble | 150 |
| Cross-Validation | |
|---|---|
| Folds for cross-validation | 5 |
| Method to create subsets | Common motif with maximum overlap of 9 aa |
| Cross-validation setup | Simple |
| Stop training on best test-set performance | No |
Summary of NNAlign architecture and encoding applied to all four models. Datasets were reduced to 50,000 peptide sequences according to the maximum accepted number of inputs by the server.
ANN model evaluation
Peptide sequences used to evaluate the ANN models were obtained by selecting IEDB epitopes restricted to HLA-DRB1*01:01 or HLA-DRB1*03:01 and selecting only peptide sequences with a positive multimer/tetramer (MMR/TMR) assay (see list of filters applied in Table III Evaluation data). The remaining epitopes were further filtered for inconsistencies between the peptides sequence and reported source Ag and subsequent in silico digested into 13-mer peptide sequences. The source Ag for each epitope were also retrieved based on the Genbank identification and in silico chopped into peptides with the length of the relevant epitope in question; each of these peptides were further digested into 13-mer peptides where the highest predicted score of each derived overlapping 13-mer were assigned to the epitope or source Ag peptide. The applied filters and number of epitopes and Ags are summarized in Table III.
Table III. Evaluation data.
| Source | HLA-DRB1 | Applied Filters | No. of Epitopes | No. of Epitopes Curated | No. of Source Ags |
|---|---|---|---|---|---|
| IEDB | 01:01 | Positive assays only | 266 | 235 | 80 |
| Qualitative binding (MMR/TMR) | |||||
| No B cell assays | |||||
| No MHC ligand assays | |||||
| MHC restriction-MRO_0001279 HLA-DRB1*01:01 | |||||
| IEDB | 03:01 | Positive assays only | 114 | 105 | 49 |
| Qualitative binding (MMR/TMR) | |||||
| No B cell assays | |||||
| No MHC ligand assays | |||||
| MHC restriction-MRO_0001284 HLA-DRB1*03:01 |
Evaluation datasets (epitopes) extracted from IEDB after selecting positive assays (tetramer/multimer staining).
Isolation of HLA-DR–bound peptides
Cell pellets from International Histocompatibility Workshop B-LCLs 9022 (COX: DRA*01:02-DRB1*03:01:01) and 9087 (STEINLEN: HLA-DRA*01:02-DRB1*03:01:01) (26) were lysed in a mild detergent. Following lysis, peptide-HLA complexes were affinity purified by anti–HLA-DR Ab (LB3.1). Affinity-purified HLA molecules and their peptide cargo were separated using reversed-phase chromatography, and peptides were subsequently analyzed using mass spectrometry as described (27). In brief, cells were expanded in RPMI 1640–10% FCS, and pellets of 109 cells were snap frozen in liquid nitrogen. Cells were ground under cryogenic conditions and resuspended in lysis buffer (0.5% IGEPAL, 50 mM Tris [pH 8], 150 mM NaCl, and protease inhibitors) and cleared lysates passed over a protein A precolumn followed by an affinity column cross-linked with a mAb specific for HLA-DR (LB3.1). Peptide-MHC complexes were eluted from the column by acidification with 10% acetic acid. Peptides were isolated using reversed-phase HPLC (Chromolith C18 Speed Rod; Merck) on an Akta Ettan HPLC system (GE Healthcare). Fractions were concentrated and run on an AB SCIEX 5600+ TripleTOF High-Resolution Mass Spectrometer. Acquired data were searched against the human proteome (Uniprot/Swissprot v2012_7) using ProteinPilot (v5; SCIEX) using the following parameters: database, human proteins from UniProt/SwissProt v2016_12; no cysteine alkylation; no enzyme digestion (considers all peptide bond cleavages); instrument-specific settings for TripleTOF 5600+ (mass spectrometry tolerance, 0.05 Da; tandem mass spectrometry tolerance, 0.1 Da; charge state, +2 to +5); biological modification probabilistic features on; thorough ID algorithm; detected protein threshold, 0.05. The resulting peptide identities were subject to strict bioinformatic criteria, including the use of a decoy database to calculate the false discovery rate. A 5% false discovery rate cut-off was applied, and the filtered dataset was further analyzed manually to exclude redundant peptides and known contaminants.
Results
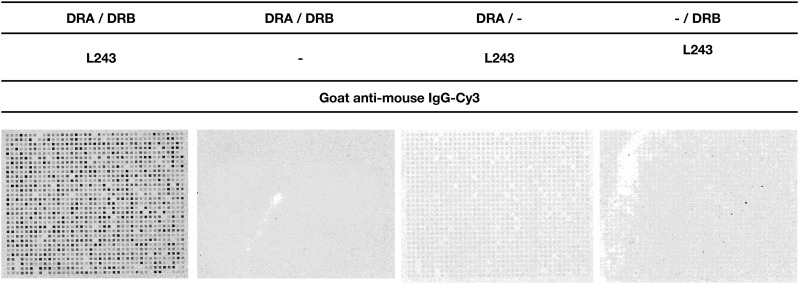
We investigated the binding of HLA-DR molecules to peptides, synthesized in situ on microscope glass slides (17) by de novo folding of rHLA-DR molecules (21) followed by staining with a monoclonal mouse anti–HLA-DR Ab known to react with a conformational HLA-DR α epitope, exclusively expressed by correctly folded HLA-DR α-β heterodimers (28), and finally stained with Cy3-labeled polyclonal goat anti-mouse IgG. Only in the presence of both HLA-DRA and HLA-DRB did we obtain a staining pattern corresponding to the location, geometry, and size of relevant peptides synthesized in situ (Fig. 1), confirming that conformationally intact peptide–HLA-DR complexes were obtained. We found no binding of the Cy3-goat anti-mouse IgG to the peptide arrays in absence of HLA-DRA, HLA-DRB, or L243 (Fig. 1), showing that the signals obtained was dependent on the presence of on chip–generated peptide–HLA-DR complexes.
FIGURE 1.
Peptide microarray incubated with HLA-DRA1*01:01/HLA-DRB1*03:01 stained with monoclonal anti–HLA-DR (L243) and Cy3-conjugated goat anti-mouse in combinations indicated in the figure heading. A fluorescent signal was only detectable in presence of HLA-DRA, HLA-DRB, mouse anti HLA-DR, and Cy3-goat anti-mouse, suggesting that the binding assay reveals HLA-DR molecules bound to specific peptides.
We determined the optimal peptide length to be synthesized on these peptide microarrays by synthesizing peptides with lengths from 9–15 aa residues and using the binding data to train ANN. Initially, NNAlign-1.4, was used to evaluate the effect of peptide length on network performance expressed as Pearson correlation coefficient (PCC) and root mean square error (RMSE) with an optimal performance identified as maximizing the PCC in combination with the lowest RMSE. This identified a peptide length of 13 aa being optimal for developing efficient predictors with a combination of high PPC and low RMSE. It should be noted that later versions of the algorithm such as NNAlign-2.1, which allows the incorporation of insertions and deletions in the analysis, can handle longer length (at least up to 15 aa) without loss of performance (Supplemental Fig. 1).
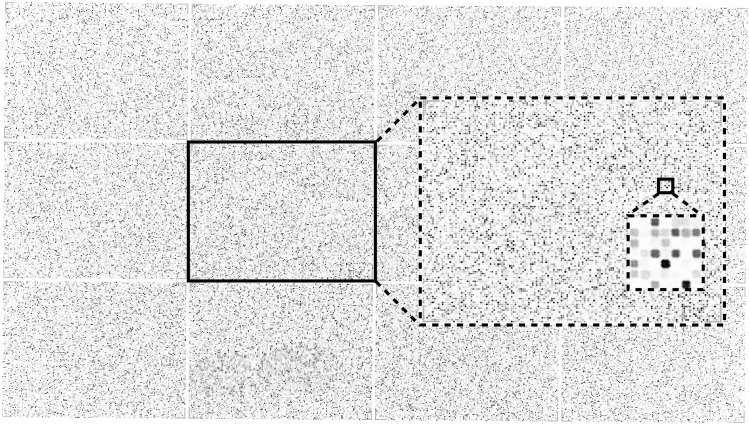
A list containing 63,802 peptides was generated from randomly sampling 13-mer peptides originating from an in-house database of human pathogens and synthesized in triplicate on a microscope slide with a randomly distributed localization. The microscope slide was incubated with HLA-DRA1*01:01/HLA-DRB1*03:01 and stained with L243/Cy3-goat anti-mouse IgG before obtaining a laser-scanning image at 1 μm resolution (Fig. 2). The individual peptide fields (20 × 20 μm) are clearly distinct in the zoomed section (Fig. 2, insert) and present themselves with varying intensities reflecting the amounts of peptide–HLA-DR complexes present on each field.
FIGURE 2.
A full view of a high-density peptide array with 217,000 individual peptides (area 20 × 11 mm) divided in 12 virtual sectors. Zoomed inserts of sector 6 and a further zoom of a subsection of sector 6 with individual peptide fields visible. The size of each peptide field is 20 × 20 μm with a 10-μm pitch scanned with a resolution of 1 μm per pixel.
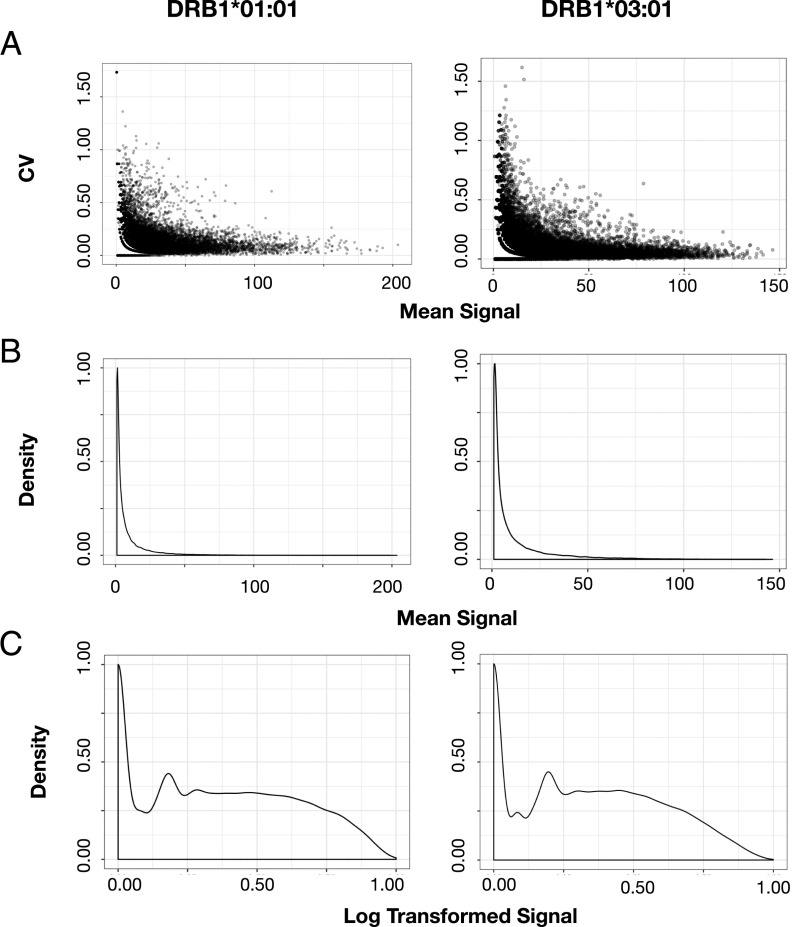
Each individual peptide field was quantified with a proprietary software (PepArray), and the mean signal and the cv based on triplicate values was calculated for each peptide. For both HLA-DRB1*01:01 and HLA-DRB1*03:01, we found a high concordance between triplicate signal values generally showing cv < 0.25 for peptides with mean signal values >50% of the maximal signal. Peptides with mean signal in the lower 10% of maximal value showed a marked increase in cv, suggesting that these lower signals approached the noise level of detection (Fig. 3A). Inclusion criteria for downstream ANN training was chosen at cv < 0.5, which almost exclusively filtered away weak and nonbinding signals. The mean signal of the remaining (cv < 0.5) peptides were, for both HLA-DR allotypes, displaying a log-normal distribution (Fig. 3B); for ANN training, signal values were log-transformed to bring the signal in the range [0–1] and reduce the distribution skewness Fig. 3C.
FIGURE 3.
(A) Raw data, mean signal versus cv calculated from triplicate signal values. (B) Signal distribution of the calculated mean signal. (C) Signal distribution after log transformation of the mean signal calculated after optimization with a BoxCox transformation (R package EnvStats Version 2.3.1).
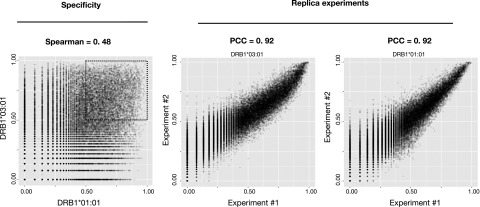
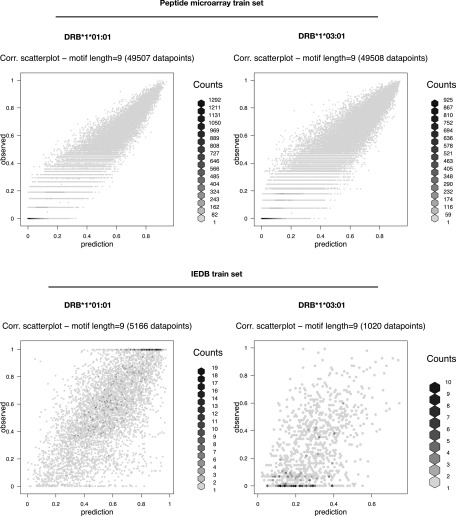
By comparing the log-transformed signals of HLA-DRB1*01:01 against HLA-DRB1*03:01 peptide by peptide, we found, as expected, the two molecules to bind to very different peptides (Fig. 4, left), confirming that a particular HLA-DR molecule has preference toward a different subset of peptides. This is further confirmed when considering only subsets of data with log-transformed signal >0.5 (Fig. 4, left black dashed square). In this study, the Spearman rank correlation (SRC) = 0.11, suggesting a very limited specificity overlap between the two molecules for strong binding peptides. The reproducibility of the HLA-II/peptide microarray binding assay was examined by repeating the peptide array synthesis (i.e., the peptide design and layout was identical to the first peptide microarray) and repeating the binding of DRA/HLA-DRB1*01:01 and DRA/HLA-DRB1*03:01. Plotting the log-transformed signals from duplicate binding experiments against each other by HLA, we found a PCC (PCC = 0.92) for both HLA-DRB1*01:01 and HLA-DRB1*03:01(Fig. 4, middle and right), suggesting that the binding of each HLA-DR molecule was highly reproducible. To assess whether the binding data possessed peptide sequence-based information of a quality sufficient to support the development of ANN predictors, we submitted the datasets from HLA-DRB1*01:01 and HLA-DRB1*03:01 to the publicly available NNAlign server with the network architecture parameters specified in Table I. As a reference and comparison with the peptide microarray data, we used the original datasets (originating from IEDB binding data) used to train the NetMHCII prediction algorithms (available at https://services.healthtech.dtu.dk/service.php?NetMHCII-2.3) and subjected them to NNAlign with the exact same network architecture as the peptide array datasets (Table I). For both HLA-DRB1*01:01 and HLA-DRB1*03:01, ANN training with the relevant peptide microarray datasets returned prediction models with a very high internal correlation (PCC > 0.95) between predicted and observed data (Fig. 5). The results summarized in Table II suggests that the peptide microarray dataset of the respective HLA-DR molecules contains a distinct and recognizable pattern that can be extracted by the NNAlign algorithm.
FIGURE 4.
Left, Log-transformed signals of DRB1*01:01 plotted against DRB1*03:01 by peptide sequence show a weak correlation (PCC = 0.48). Considering only peptides with a predicted score >0.5 returns a Spearman rank correlation (SRC) = 0.11, suggesting HLA-DR specificity. Middle and right, Log-transformed signals obtained by replica experiments of DRB1*03:01 and DRB1*01:01, respectively, correlate well (PCC = 0.92), conferring an acceptable reproducibility.
FIGURE 5.
Scatter plots of predicted versus observed values (log-transformed signals) of NNAlign-2.1 models trained on ∼50,000 peptide sequences obtained from peptide microarrays with SPARSE encoding. IEDB train sets (downloaded as pairs of peptide sequences and log-transformed binding data from https://services.healthtech.dtu.dk/service.php?NetMHCII-2.3) encoded with SPARSE. The internal model performances are listed in Table II. BLOSUM encoding rendered network ensembles with comparable performances (data not shown). The neural network architecture is disseminated in Table I.
Table II. Prediction performances.
| Peptide Microarray Data | IEDB Data | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Allotype | ANN Performance | TMR+ Evaluation | Eluted Peptides Evaluation | ANN Performance | TMR+ Evaluation | Eluted Peptides Evaluation | ||||
| No. of Data Points | PCC | RMSE | AUC | AUC | No. of Data Points | PCC | RMSE | AUC | AUC | |
| DRB1*01:01 | 49,507 | 0.954 | 0.065 | 0.722 | 0.832 | 5166 | 0.671 | 0.199 | 0.821 | 0.923 |
| DRB1*03:01 | 49,508 | 0.952 | 0.086 | 0.817 | 0.835 | 1020 | 0.511 | 0.204 | 0.753 | 0.845 |
Summary of the final network ensemble for all models listing the number of data points and prediction performance reported as PCC and RMSE. The mean area under the curve (AUC) values are reported by a receiver-operating characteristic analysis of predicted scores of IEDB epitopes versus epitope source Ags.
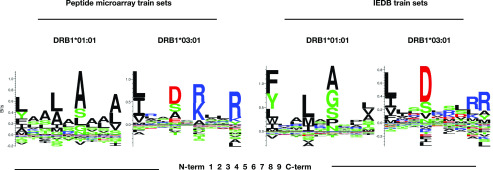
The sequence logo representations of the final NNAlign network ensembles trained on peptide microarray datasets revealed motifs with distinct anchor residues in positions P1, P4, P6, and P9 for both HLA-DRB1*01:01 and HLA-DRB1*03:01 (Fig. 6). Although P1 in HLA-DRB1*01:01 did not appear to be as prominent an anchor position (reflected by a relatively low bit score) as normally observed for P1 in many HLA-II motifs, we found the motifs and anchor positions to be comparable to the commonly accepted motifs of the respective molecules. A head to head comparison of the final DRB1*01:01 ANN network ensembles trained on peptide microarray versus network ensembles trained on IEDB data, showed overall that the peptide microarray generated motif appeared more blurred with less distinct anchor positions (P1 in particular). In contrast, the motif for HLA-DRB1*03:01 as derived from the microarray data appeared sharper and with higher information context compared with the motif derived from the IEDB data. It should be noted that the P1 anchor is the one most sensitive to the peptide quality, which affects in particular the N terminus because of the accumulation of errors in the peptide elongation process as synthesis proceeds toward the N terminus.
FIGURE 6.
LOGO representation of the final NNAlign network ensemble based on the peptide microarray dataset and IEDB datasets trained with the neural network architecture listed in Table I.
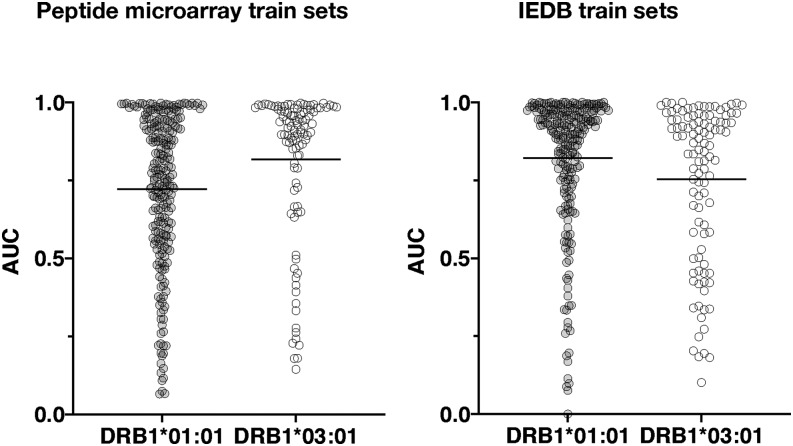
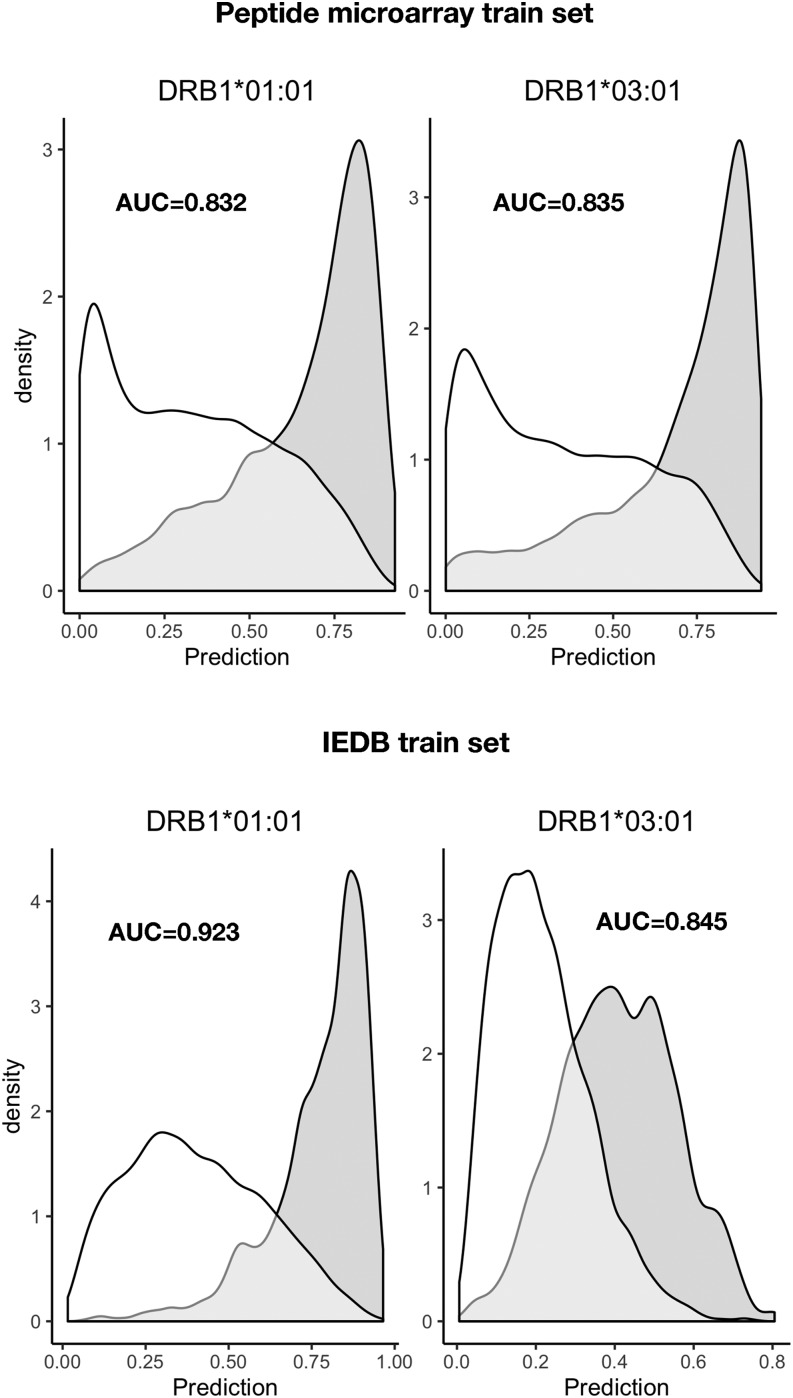
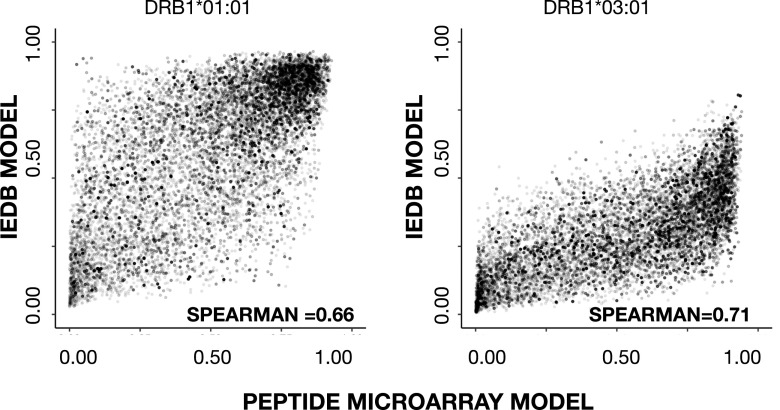
To benchmark the prediction power of the final network ensembles, we evaluated the prediction of HLA-DRB1*01:01 and HLA-DRB1*03:01 epitopes extracted from IEDB with selection criteria outlined in Table III (note that epitopes were curated for cases in which the epitope sequence not being present in the corresponding source Ag). To benchmark the peptide microarray–driven models, which operate in a 13-mer peptide space, each source Ag was in silico digested into overlapping peptides with the length of the epitope; within each of these peptides, the highest predicted score of the underlying overlapping 13-mers was assigned to the peptide. Similarly, for each epitope, the highest predicted 13-mer within each epitope was assigned to the epitope. The IEDB dataset–driven models were trained on peptides with varying lengths, hence predictions were made on the epitope sequence, and overlapping peptides from the source Ag with same lengths of the epitope. By comparing the prediction of each epitope against the predictions of the overlapping source Ag peptides, an area under the curve (AUC) was calculated for each epitope; AUC values of each epitope and models are shown in Fig. 7. The overall performance (mean AUC values) of each model, summarized in Table II, shows that the prediction power of IEDB data–driven networks is higher (p < 0.05), AUC = 0.821, as compared with AUC = 0.722 of peptide microarray–driven networks, with respect to predicting HLA-DRB1*01:01 epitopes. In contrast was the power to predict HLA-DRB1*03:01 higher epitopes (p < 0.05) by the peptide microarray–driven networks AUC = 0.817 compared with IEDB-driven networks predicting with an AUC = 0.753. As a further validation, we evaluated the prediction of peptides eluted off HLA-DRB1*01:01 or HLA-DRB1*03:01 and sequenced by mass spectrometry (29), as described in this study (eluted peptide sequences provided in Supplemental Table I). Peptides (5515) of different lengths eluted off HLA-DRB1*01:01 were in silico digested to a length of 13 aa and subjected to prediction, the highest predicted 13-mer within each eluted peptide were used in the evaluation against 5000 randomly generated 13-mer peptides. An identical strategy was applied for the (5713) eluted HLA-DRB1*03:01 peptides evaluated against the same 5000 randomly generated peptides. Density plots (Fig. 8) of the predicted scores confirmed that all four network ensembles are assigning higher scores to peptides eluted off relevant MHC-II allotypes than to random peptides, resulting in a clear skewness of the score distribution. The AUC values, summarized in Table II, of the evaluation of eluted peptides against random peptides were comparable to AUC values obtained for the evaluation of epitopes. Using a Student t test to compare the performances of the peptide microarray–driven models against the IEDB-driven models, we found no statistical difference (p = 0.17, p = 0.31, HLA-DRB1*01:01 and HLA-B1*03:01, respectively) of the combined mean AUC of epitopes and eluted peptides. A direct comparison of the prediction scores of eluted peptides between the peptide microarray–driven models and IEDB models (Fig. 9) reveals that the peptide microarray and IEDB model performances are comparable, with SRC = 0.66 (HLA-DRB1*01:01) and SRC = 0.71 (HLA-DRB1*03:01). There is a tendency toward agreement between the models in particular for peptides with high prediction scores. It is possible that the nature of the binding data, which is obtained from two different assays, chip binding and traditional affinity data (IEDB), contributes to the differences. For all datasets, the peptide with the strongest measured binding is transformed to a value of 1 in the training sets. Hence, the differences in distribution of the transformed data originating from the different datasets may, in part, explain some of the differences observed for prediction scores <0.8, which, in any case, translates into weak to nonbinders, and thus less relevant.
FIGURE 7.
AUC values of HLA-DRB1*01:01 (light gray) and HLA-DRB1*03:01 (white) epitopes based on predictions by network ensembles trained on peptide microarray data (left) and IEDB binding data (right). Neural network architecture is listed in Table I and the mean AUC values are listed in Table II. In general, all four models assign high prediction scores to peptide sequences that can be identified as HLA-II tetramer–validated epitopes as expected if the input data reflects the peptide–HLA-DR interaction. The prediction performances reported as AUC values are comparable between the models with respect to allotype. This indicates that the data obtained by peptide microarray binding studies contain specific peptide–HLA-DR information comparable to traditional binding data and suitable to ANN training.
FIGURE 8.
Predicted scores of peptides eluted off HLA-DRB1*01:01 and HLA-DRB1*03:01 and compared with random peptides. Top, the peptide microarray model prediction of eluted peptides (gray density) are clearly skewed toward high scores whereas random peptides (white density) are primarily skewed toward low scores for both HLA-DR allotypes. Bottom, predicted scores by the models trained in IEDB data presents a similar pattern of eluted peptides (gray density) and random peptides (white density). The model performances are summarized in Table II, showing that HLA-DRB1*03:01 models performed at par, whereas the HLA-DRB1*01:01 model, based on IEDB datasets, had a better prediction power compared with the model trained on peptide microarray data.
FIGURE 9.
Scatter plots of eluted peptide scores (left, HLA-DRB1*01:01; right, HLA-DRB1*03:01) predicted by peptide microarray models versus IEDB models. The SRC of the respective dataset are inserted.
Collectively, the high internal performance of the peptide microarray–driven models and a prediction power comparable to prediction models based on traditional binding data (IEDB) suggests that high-density peptide microarrays can be used to generate relevant pHLAII binding data. As a final examination of the prediction models, we calculated the distance between the models one by one and their individual distances to NetMHC-2.3, summarized in Table IV. Briefly, the distance is calculated as 1-SRC2, where SRC is found by comparing prediction scores of 100,000 random peptides from two prediction models. We found the distances of network ensembles to be closer within allotypes (HLA-DRB1*01:01 distance = 0.31 and HLA-DRB1*03:01 distance = 0.43) than between allotypes (0.49 < distance < 0.85), underscoring that the training data contains allotype-specific binding data.
Table IV. Distances between prediction models.
| DRB1*01:01 |
DRB1*03:01 |
|||||
|---|---|---|---|---|---|---|
| Model |
Microarray | IEDB | Microarray | IEDB | ||
| DRB1*01:01 | Microarray | 0.00 | 0.31 | 0.49 | 0.85 | |
| IEDB | — | 0.00 | 0.68 | 0.68 | ||
| DRB1*03:01 | Microarray | — | — | 0.00 | 0.43 | |
| IEDB | — | — | — | 0.00 | ||
Table of calculated distances between the models trained on peptide microarray data, IEDB binding data, and NetMHC-2.3. The distance is calculated as 1-R2, with SRC calculated by the prediction scores of 100,000 random peptides.
In conclusion, the binding data obtained from HLA-II–peptide microarray assays produced comparable HLA-II–peptide prediction models compared with models trained on pHLAII affinity data available at IEDB, and as such, represents an attractive way forward to generate large amounts of peptide–MHC-II binding data and to improve existing prediction models.
Discussion
CD4+ T cells arguably perform the most important function of the immune system; by controlling and coordinating the responses of most immune cells, they essentially orchestrate the overall specificity and reactivity of the immune system. To this end, they survey peptide–MHC-II complexes and determine whether these peptides are of self or nonself origin. This essential process of peptide sampling and display is diversified immensely by the polymorphism of the MHC system, which assures that no two individuals will perform self-nonself–discrimination in an identical manner, thereby avoiding the evolution of pathogen variants that can escape the immune systems of the entire species. An in-depth understanding of which peptides bind to MHC-II and how this interaction is affected by MHC polymorphism is of paramount importance for our ability to understand and manipulate the immune system. Many autoimmune diseases (e.g., type I diabetes, multiple sclerosis, rheumatoid arthritis, narcolepsy, and many more) are tightly associated with specific MHC-II allotypes, which obviously make these allotypes interesting in their own right. More recently, the field of personalized immunotherapy (emerging as promising cancer treatments) increasingly rely on a better understanding and prediction of immune specificities in a much broader representative coverage of the “MHC space” underscoring the relevance of panspecific predictors.
The need for experimental coverage became apparent when panspecific predictors were applied to MHC allotypes lacking experimental data, and it could be shown that these pan-predictors are more accurate when they have been trained on experimental data from a closely related allotype rather than a more distant allotype (30). The development of these bioinformatics methods requires large bodies of data that are representative of peptide–MHC-II interactions. In other words, it requires access to large panels of natural and/or synthetic peptides representing peptide–MHC-II binding events. By combining recombinant MHC-II molecules with high-density peptide array technology, it is be possible to generate the large bodies of binding data representing many different allotypes, and do so at significant cost reductions compared with generating the same amount of data using more traditional binding experiments.
In this study, we have demonstrated that MHC-II can interact with in situ–synthesized peptides in a high-density array format; that the peptide–MHC-II interaction is specific; that there is a systematic binding pattern that is interpretable by ANNs, in this study exemplified by NNAlign training (31, 32); furthermore, the resulting prediction models are able to predict peptide–MHC-II binding at a level comparable to that of models trained on traditional binding data deposited at the IEDB, which have been used to develop NetMHCII models (32). We also found the binding motifs revealed by the models trained on peptide microarray data comparable with the motifs obtained by training on IEDB binding data.
The capacity of this high-density peptide microarray technology should enable large-scale analyses of any MHC-II allotype of interest, even including all posttranslational modifications that are available for solid-phase peptide synthesis. Without the limitations inherent to conventional solid-phase peptide synthesis, peptides could be selected entirely based on the scientific and/or practical merits of the question at hand. A completely random selection of peptides, as applied in this study, would ensure a truly unbiased approach to define MHC-II specificity. Alternatively, one could randomly extract peptides from the proteomes of organism(s) of interest (microbiomes for infectious diseases, the human proteome for autoimmune disease, oncoproteomes for cancer, etc.). Other “peptide-intensive” strategies could also be pursued (e.g., using an initial random screen to generate a primary predictor and use that to enrich for binders in a secondary experiment). In contrast, the current cost of obtaining peptides for such mapping projects tend to be prohibitive and may lead to the use of existing prediction tools to downsize sampling with the inherent risk of introducing bias leading to an incomplete representation of the relevant sequence space.
The prediction models developed using peptide microarray binding data are at performance levels comparable to prediction models trained on traditional binding (affinity) data. Thus, peptide microarrays could be used to develop large bodies of peptide–MHC-II binding data and/or to complement other methods capable of generating large amounts of binding data (e.g., peptide elution and mass spectrometry) (2, 30). This could even include nonstandard amino acids representing posttranslational modifications that otherwise are scarcer and/or more difficult to obtain. The massive amounts of representative peptide–MHC-II data that can be generated should allow an exhaustive scanning of both the peptide and MHC spaces. This should support the generation of improved pan–MHC-II predictors, which should allow peptide–MHC-II interactions to be evaluated in silico cheaper, faster, and at an even higher capacity than any experimental approach, even one enabled by the high-density peptide microarray technology could do. One area may always benefit from an experimental approach: the binding of epitopes derived from cancer neoepitopes identified by genomics or proteomics sequencing of tumor cells versus normal cells. In this study, all mutations and corresponding wildtype sequences could potentially be synthesized in multiple length variants and tested experimentally against all of the MHC-II allotypes of a given patient.
One current disadvantage of the present peptide microarray technology is the inability (at least with current technologies) to validate the identity and quality of the peptides. Thus, some of the experimental peptide–MHC-II data points might be erroneous. We surmise that as long as such errors are the exception rather than the rule, then the predictors should ignore these experimental errors. Notwithstanding, it would be a major advance if the quality of the individual peptides on a microarray could be validated through independent physical means (e.g., by mass spectrometry).
Other recent technologies have emerged as possible ways of generating large-scale peptide–MHC-II data. One approach is phage expression of peptide libraries followed by MHC-II selection and sequencing (33). This technology does not offer the same degree of control of the peptides expressed; it requires DNA sequencing to identify the peptides involved rather than the simple “identification by look-up” used by the peptide microarray technology; it does not readily allow the incorporation of posttranslational modifications; nor does it allow the identification of nonbinders, which are important for ANN training. Another recent approach, “immunopeptidomics,” involves sequencing of natural peptides eluted of MHC-II by liquid chromatography followed by mass spectrometry (34). Sequencing natural peptides has the huge qualitative advantage that it includes events of Ag processing and includes posttranslational modification. Otherwise, it shares some of the same disadvantages of the phage display approach: lack of control of which peptides and posttranslational modification are interrogated and lack of nonbinders. Ideally, one should include data obtained from both synthetic and natural peptide and generate immune-bioinformatics predictors incorporating both kinds of input.
In conclusion, we have demonstrated that high-density peptide microarrays can be used to generate very large numbers of discrete peptide–MHC-II interaction data, something that represents a major advance in the analysis of peptide–MHC-II interactions and should become instrumental in developing improved bioinformatics methods representing these important interactions.
Supplementary Material
This work was supported by The Danish Council for Independent Research (DFF – 6110-00644), Scleroseforeningen (A31444), the European Commission (278832), and the Department of Health, National Health and Medical Research Council (1165490).
The online version of this article contains supplemental material.
- ANN
- artificial neural network
- AUC
- area under the curve
- cv
- coefficient of variance
- HLA-II
- HLA class II
- IEDB
- Immune Epitope Database
- MHC-II
- MHC class II
- MMR/TMR
- multimer/tetramer
- PBS-T
- PBS (pH 7.4), 1% Tween-20
- PCC
- Pearson correlation coefficient
- pHLAII
- peptide–HLA-II
- RMSE
- root mean square error
- SRC
- Spearman rank correlation.
Disclosures
T.O. and S.B. are cofounders of immunAware Aps, a provider of MHC class I and II reagents. C.S.-N. is the owner of Schafer-N, a provider of high density peptide arrays. The other authors have no financial conflicts of interest.
References
- 1.Reche P. A., Reinherz E. L. 2003. Sequence variability analysis of human class I and class II MHC molecules: functional and structural correlates of amino acid polymorphisms. J. Mol. Biol. 331: 623–641. [DOI] [PubMed] [Google Scholar]
- 2.Garde C., Ramarathinam S. H., Jappe E. C., Nielsen M., Kringelum J. V., Trolle T., Purcell A. W. 2019. Improved peptide-MHC class II interaction prediction through integration of eluted ligand and peptide affinity data. Immunogenetics 71: 445–454. [DOI] [PubMed] [Google Scholar]
- 3.Lin H. H., Zhang G. L., Tongchusak S., Reinherz E. L., Brusic V. 2008. Evaluation of MHC-II peptide binding prediction servers: applications for vaccine research. BMC Bioinformatics 9(Suppl. 12): S22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sahin U., Türeci Ö. 2018. Personalized vaccines for cancer immunotherapy. Science 359: 1355–1360. [DOI] [PubMed] [Google Scholar]
- 5.2017. The problem with neoantigen prediction. Nat. Biotechnol. 35: 97. [DOI] [PubMed] [Google Scholar]
- 6.Kreiter S., Vormehr M., van de Roemer N., Diken M., Löwer M., Diekmann J., Boegel S., Schrörs B., Vascotto F., Castle J. C., et al. 2015. Mutant MHC class II epitopes drive therapeutic immune responses to cancer. [Published erratum appears in 2015 Nature 523: 370.] Nature 520: 692–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tran E., Turcotte S., Gros A., Robbins P. F., Lu Y. C., Dudley M. E., Wunderlich J. R., Somerville R. P., Hogan K., Hinrichs C. S., et al. 2014. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science 344: 641–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abelin J. G., Harjanto D., Malloy M., Suri P., Colson T., Goulding S. P., Creech A. L., Serrano L. R., Nasir G., Nasrullah Y., et al. 2019. Defining HLA-II ligand processing and binding rules with mass spectrometry enhances cancer epitope prediction. Immunity 51: 766–779.e17. [DOI] [PubMed] [Google Scholar]
- 9.Buus S. 1999. Description and prediction of peptide-MHC binding: the ‘human MHC project’. Curr. Opin. Immunol. 11: 209–213. [DOI] [PubMed] [Google Scholar]
- 10.Buus S., Lauemøller S. L., Worning P., Kesmir C., Frimurer T., Corbet S., Fomsgaard A., Hilden J., Holm A., Brunak S. 2003. Sensitive quantitative predictions of peptide-MHC binding by a ‘query by committee’ artificial neural network approach. Tissue Antigens 62: 378–384. [DOI] [PubMed] [Google Scholar]
- 11.Nielsen M., Lundegaard C., Blicher T., Lamberth K., Harndahl M., Justesen S., Røder G., Peters B., Sette A., Lund O., Buus S. 2007. NetMHCpan, a method for quantitative predictions of peptide binding to any HLA-A and -B locus protein of known sequence. PLoS One 2: e796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nielsen M., Lundegaard C., Lund O. 2007. Prediction of MHC class II binding affinity using SMM-align, a novel stabilization matrix alignment method. BMC Bioinformatics 8: 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hansen L. B., Buus S., Schafer-Nielsen C. 2013. Identification and mapping of linear antibody epitopes in human serum albumin using high-density peptide arrays. PLoS One 8: e68902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zandian A., Forsström B., Häggmark-Månberg A., Schwenk J. M., Uhlén M., Nilsson P., Ayoglu B. 2017. Whole-proteome peptide microarrays for profiling autoantibody repertoires within multiple sclerosis and narcolepsy. J. Proteome Res. 16: 1300–1314. [DOI] [PubMed] [Google Scholar]
- 15.Hecker M., Fitzner B., Wendt M., Lorenz P., Flechtner K., Steinbeck F., Schröder I., Thiesen H. J., Zettl U. K. 2016. High-density peptide microarray analysis of IgG autoantibody reactivities in serum and cerebrospinal fluid of multiple sclerosis patients. Mol. Cell. Proteomics 15: 1360–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hecker M., Lorenz P., Steinbeck F., Hong L., Riemekasten G., Li Y., Zettl U. K., Thiesen H. J. 2012. Computational analysis of high-density peptide microarray data with application from systemic sclerosis to multiple sclerosis. Autoimmun. Rev. 11: 180–190. [DOI] [PubMed] [Google Scholar]
- 17.Buus S., Rockberg J., Forsström B., Nilsson P., Uhlen M., Schafer-Nielsen C. 2012. High-resolution mapping of linear antibody epitopes using ultrahigh-density peptide microarrays. Mol. Cell. Proteomics 11: 1790–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andreatta M., Schafer-Nielsen C., Lund O., Buus S., Nielsen M. 2011. NNAlign: a web-based prediction method allowing non-expert end-user discovery of sequence motifs in quantitative peptide data. PLoS One 6: e26781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Braendstrup P., Mortensen B. K., Justesen S., Osterby T., Rasmussen M., Hansen A. M., Christiansen C. B., Hansen M. B., Nielsen M., Vindeløv L., et al. 2014. Identification and HLA-tetramer-validation of human CD4+ and CD8+ T cell responses against HCMV proteins IE1 and IE2. PLoS One 9: e94892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vita R., Mahajan S., Overton J. A., Dhanda S. K., Martini S., Cantrell J. R., Wheeler D. K., Sette A., Peters B. 2019. The immune epitope database (IEDB): 2018 update. Nucleic Acids Res. 47: D339–D343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Justesen S., Harndahl M., Lamberth K., Nielsen L. L., Buus S. 2009. Functional recombinant MHC class II molecules and high-throughput peptide-binding assays. Immunome Res. 5: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sette A., Lamont A., Buus S., Colon S. M., Miles C., Grey H. M. 1989. Effect of conformational propensity of peptide antigens in their interaction with MHC class II molecules. Failure to document the importance of regular secondary structures. J. Immunol. 143: 1268–1273. [PubMed] [Google Scholar]
- 23.Singh-Gasson S., Green R. D., Yue Y., Nelson C., Blattner F., Sussman M. R., Cerrina F. 1999. Maskless fabrication of light-directed oligonucleotide microarrays using a digital micromirror array. Nat. Biotechnol. 17: 974–978. [DOI] [PubMed] [Google Scholar]
- 24.Osterbye T. 2020. Data from: HLA-class II specificity assessed by high-density peptide microarray interactions, v3, Dryad, Dataset, 10.5061/dryad.tqjq2bvvv [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Millard S. P. 2013. EnvStats: An R Package for Environmental Statistics. Springer, Berlin, Germany. [Google Scholar]
- 26.Robinson J., Barker D. J., Georgiou X., Cooper M. A., Flicek P., Marsh S. G. E. 2020. IPD-IMGT/HLA database. Nucleic Acids Res. 48: D948–D955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Purcell A. W., Ramarathinam S. H., Ternette N. 2019. Mass spectrometry-based identification of MHC-bound peptides for immunopeptidomics. Nat. Protoc. 14: 1687–1707. [DOI] [PubMed] [Google Scholar]
- 28.Moro M., Cecconi V., Martinoli C., Dallegno E., Giabbai B., Degano M., Glaichenhaus N., Protti M. P., Dellabona P., Casorati G. 2005. Generation of functional HLA-DR*1101 tetramers receptive for loading with pathogen- or tumour-derived synthetic peptides. BMC Immunol. 6: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ooi J. D., Petersen J., Tan Y. H., Huynh M., Willett Z. J., Ramarathinam S. H., Eggenhuizen P. J., Loh K. L., Watson K. A., Gan P. Y., et al. 2017. Dominant protection from HLA-linked autoimmunity by antigen-specific regulatory T cells. Nature 545: 243–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Racle J., Michaux J., Rockinger G. A., Arnaud M., Bobisse S., Chong C., Guillaume P., Coukos G., Harari A., Jandus C., et al. 2019. Robust prediction of HLA class II epitopes by deep motif deconvolution of immunopeptidomes. Nat. Biotechnol. 37: 1283–1286. [DOI] [PubMed] [Google Scholar]
- 31.Nielsen M., Andreatta M. 2017. NNAlign: a platform to construct and evaluate artificial neural network models of receptor-ligand interactions. Nucleic Acids Res. 45: W344–W349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nielsen M., Lund O. 2009. NN-align. An artificial neural network-based alignment algorithm for MHC class II peptide binding prediction. BMC Bioinformatics 10: 296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Somasundaram R., Satyamoorthy K., Caputo L., Yssel H., Herlyn D. 2004. Detection of HLA class II-dependent T helper antigen using antigen phage display. Clin. Exp. Immunol. 135: 247–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dudek N. L., Croft N. P., Schittenhelm R. B., Ramarathinam S. H., Purcell A. W. 2016. A systems approach to understand antigen presentation and the immune response. Methods Mol. Biol. 1394: 189–209. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.