Abstract
Calypsoinae is a small subtribe in Orchidaceae (Epidendroideae) characterized by diverse trophic strategies and morphological characters. Calypsoinae includes 13 genera, four of which are leafless and mycoheterotrophic. Mycoheterotrophic species in the leafless genus Corallorhiza are well suited to studies of plastome evolution. However, the lack of plastome sequences for other genera in Calypsoinae limits the scope of comparative and phylogenetic analyses, in particular our understanding of plastome evolution. To understand plastid genome evolution in Calypsoinae, we newly sequenced the plastomes of 12 species in the subtribe, including representatives of three mycoheterotrophic genera as well as five autotrophic genera. We detected two parallel photosynthetic losses in Corallorhiza. Evolutionary analyses indicated that the transition to obligate mycoheterotrophy leads to the relaxation of selection in a highly gene-specific pattern.
Keywords: Corallorhiza, Calypsoinae, Danxiaorchis, plastome, Risleya, mycoheterotrophy
Introduction
Mycoheterotrophic species have received substantial attention owing to their unique lifestyle (Delannoy et al. 2011; Schelkunov et al. 2015; Barrett et al. 2018; Tsukaya 2018; Kim et al. 2019), in which they obtain carbon from fungi instead of photosynthesis (Merckx, Freudenstein, et al. 2013). At least, 47 independent origins of full mycoheterotrophy in land plants have been reported (Merckx, Mennes, et al. 2013).
There were about 25 independent origins of mycoheterotrophy in the family Orchidaceae (Merckx, Mennes, et al. 2013). The subtribe Calypsoinae (Epidendreae, Epidendroideae) includes 13 genera, four of which are mycoheterotrophic (Chase et al. 2015; Freudenstein et al. 2017). In Calypsoinae, the leafless genus Corallorhiza has been established as an ideal system for studies of plastome evolution in heterotrophic lineages (Barrett and Davis 2012; Barrett et al. 2014, 2018). However, the plastomes of Calypsoinae species published to date are limited to the genus Corallorhiza, emphasizing the need for sequence data for other genera, especially closely related autotrophic clades.
To characterize plastome evolution across Calypsoinae, we newly sequenced 12 Calypsoinae plastomes, including representative species of three mycoheterotrophic genera (Corallorhiza, Danxiaorchis, and Risleya) and five autotrophic genera.
Materials and Methods
DNA Extraction and Sequencing
Twelve species from eight Calypsoinae genera were sampled, including Calypso bulbosa, Changnienia amoena, Corallorhiza trifida, Cremastra appendiculata, Danxiaorchis singchiana, and Risleya atropurpurea, four Oreorchis species, and two Tipularia species (supplementary table S1, Supplementary Material online). Agrostophyllum callosum (Agrostophyllinae) was sampled as an autotrophic outgroup for comparative analyses. Fourteen previously published plastomes obtained from the NCBI database were included in the analyses (supplementary table S1, Supplementary Material online).
Total genomic DNA was extracted from silica-dried materials using a modified cetyltrimethylammonium bromide (CTAB) method (Li et al. 2013). DNA with a concentration of >100 ng/ml was sheared to fragments of ∼400–600 bp using Covaris M220. The NEBNext Ultra DNA Library Prep Kit (New England Biolabs, USA) was used to prepare DNA libraries for sequencing according to the manufacturer’s protocol. Paired-end sequencing (100- or 150-bp read lengths) was performed on the Illumina HiSeq 2500 platform at the Institute of Botany, Chinese Academy of Sciences, which generated ∼10 GB of raw data for heterotrophic species and at least 2 GB for autotrophic species.
Plastome Assembly and Annotation
Plastome assembly was performed following previously described methods (Feng et al. 2016). In brief, raw reads were trimmed and filtered using NGSQCTOOLKIT v. 2.3.3 (Patel and Jain 2012), and bases with PHRED quality scores of lower than 20 were trimmed. The filtered paired reads were mapped to the plastome of Calanthe triplicata (NC_024544.1) using Geneious v. 10.1.2 to filter reads matching the reference genome. De novo assemblies were constructed using VELVET (Zerbino and Birney 2008) with K-mer values from 37 to 45. Contigs from consensus sequences were merged and combined into scaffolds using Geneious with default parameters. Scaffolds were extended by mapping reads using Geneious. The assembly steps were repeated to obtain the draft plastome. Assembly errors were corrected by mapped all sequencing reads to draft plastome. IR boundaries for each plastome were confirmed by BLAST and the reverse IR was added to complete plastome by hand.
Completed plastomes were annotated using PGA (Qu et al. 2019) with the annotated plastome of Calanthe triplicata as a reference, followed by manual checking and adjustment of gene or exon boundaries using Geneious v. 10.1.2. The initiation codon, termination codon, and other annotation errors for each gene were revised using Sequin, and results were exported as GenBank files.
Phylogenetic Analysis
To reconstruct phylogenetic relationships, all protein-coding sequences (supplementary table S4, Supplementary Material online) were exported from plastomes using Geneious v. 10.1.2. Each single gene matrix was aligned using MAFFT under the automatic model selection option (Katoh and Standley 2013) with manual adjustments in BioEdit (Hall 1999). These were subsequently combined into a single plastome supermatrix using PhyloSuite (Zhang et al. 2020). The concatenated sequences were used for a phylogenetic analysis by the maximum likelihood (ML) method using IQ-TREE (Nguyen et al. 2015) with the best-fit model TVM + F + R3 was automatically selected by ModelFinder (Kalyaanamoorthy et al. 2017). Branch support was evaluated by 1,000 bootstrap replicates (-bb 1000).
Molecular Evolutionary Analyses
Plastomes of mycoheterotrophic Calypsoinae are highly reduced (see Results and Discussion), there are 25 common protein-coding genes (CDS) in all sampled plastomes (see table 1 and supplementary table S4, Supplementary Material online). These CDS were aligned at the codon level with the option “-codon” using MUSCLE in MEGA v. 7.0.2 (Kumar et al. 2016). Stop codons were removed from the sequences prior to alignment. The output topology from the ML phylogenetic analysis based on all of the protein-coding sequences was used for the evolutionary analysis. CODEML in the PAML software package (Yang 2007) was used to calculate branch lengths of dS and dN of the 25 concatenated protein-coding genes.
Table 1.
The Number of Genes, Length, and GC Content of the Newly Sequenced Plastid Genomes in This Study
| Species | Number of Genes |
Length (bp) |
GC Content (%) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Protein Coding | tRNA | rRNA | Total | LSC | IR | SSC | Total | LSC | IR | SSC | |
| Agrostophyllum callosum | 79 | 30 | 4 | 158,365 | 86,349 | 26,743 | 18,530 | 37.0 | 34.8 | 43.2 | 29.9 |
| Calypso bulbosa | 69 | 30 | 4 | 149,451 | 83,800 | 25,654 | 14,343 | 37.1 | 34.5 | 43.5 | 29.1 |
| Changnienia amoena | 79 | 30 | 4 | 156,828 | 84,856 | 26,915 | 18,142 | 37.1 | 34.8 | 43.2 | 29.8 |
| Corallorhiza trifida_14268 | 68 | 30 | 4 | 149,408 | 83,171 | 25,920 | 14,397 | 37.2 | 34.5 | 43.7 | 28.8 |
| Cremastra appendiculata | 79 | 30 | 4 | 159,493 | 86,948 | 27,089 | 18,367 | 37.0 | 34.6 | 43.3 | 29.9 |
| Danxiaorchis singchiana | 29 | 22 | 4 | 87,910 | 42,494 | 13,763 | 17,890 | 34.6 | 31.1 | 37.0 | 39.0 |
| Oreorchis angusta | 79 | 30 | 4 | 158,654 | 86,294 | 27,024 | 18,312 | 37.0 | 34.6 | 43.2 | 29.8 |
| Oreorchis foliosa | 75 | 30 | 4 | 158,496 | 86,061 | 27,037 | 18,361 | 36.9 | 34.6 | 43.2 | 29.6 |
| Oreorchis indica | 75 | 30 | 4 | 158,599 | 86,109 | 27,437 | 17,616 | 36.9 | 34.6 | 43 | 29.6 |
| Oreorchis patens | 79 | 30 | 4 | 158,256 | 86,253 | 26,831 | 18,341 | 37.0 | 34.6 | 43.3 | 29.6 |
| Risleya atropurpurea | 25 | 15 | 4 | 77,821 | 20,656 | 26,260 | 4,645 | 33.3 | 25.7 | 37.5 | 19.2 |
| Tipularia josephii | 68 | 30 | 4 | 146,816 | 82,881 | 25,977 | 11,981 | 37.4 | 34.9 | 43.5 | 28.2 |
| Tipularia szechuanica | 68 | 30 | 4 | 142,799 | 82,378 | 24,444 | 11,533 | 37.4 | 34.9 | 43.6 | 28.7 |
The selection intensity parameter (k) was calculated using RELAX (Wertheim et al. 2015) within the Datamonkey server (Delport et al. 2010). In particular, test branches (mycoheterotrophic species) under relaxed (k < 1) or intensified (k > 1) selection relative to the reference branches (autotrophic species) were examined. The significance of the k parameter was determined by likelihood ratio tests.
Results and Discussion
Phylogenetic Relationships and Photosynthetic Losses of Calypsoinae
Plastome data provided better understanding of phylogenetic relationships and losses of photosynthesis within Calypsoinae (supplementary fig. S1, Supplementary Material online). Our phylogenetic analysis of interrelationships of Calypsoinae is generally consistent with previous studies (Chase et al. 2015; Freudenstein et al. 2017; Li et al. 2019) but provides new insight into relationships among taxa. Corallorhiza was not monophyletic, including two groups nested within Oreorchis lineages. Danxiaorchis and Cremastra were sister groups. We detected four independent losses of photosynthesis in Calypsoinae, two of which were parallel losses in Corallorhiza. Parallel photosynthetic losses have been observed in Hexalectris (Barrett et al. 2019), and our results indicated that parallel photosynthetic losses may be more common in Orchidaceae than expected.
Genome Size, Gene Content, and Structure of Calypsoinae Plastids
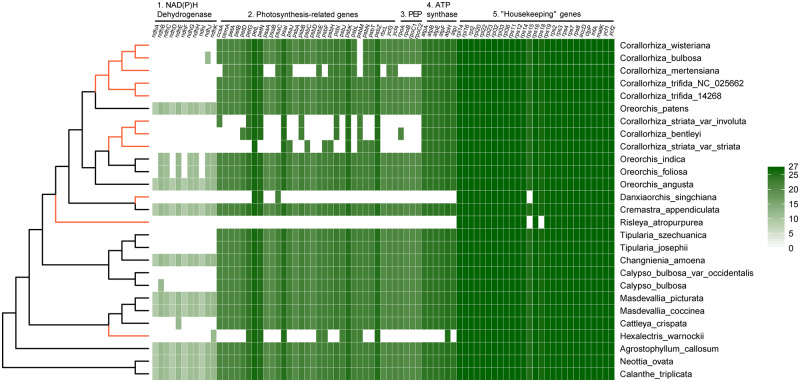
Plastid genomes of sampled Calypsoinae species ranged in size from 77,821 bp in R. atropurpurea to 159,493 bp in Cremastra appendiculata (table 1). In mycoheterotrophic species, we observed high variation in genome size, from 77,821 to 149,384 bp. The fully mycoheterotrophic R. atropurpurea plastome (77,821 bp) was the most highly reduced among the plastomes of Calypsoinae species sequenced to date (table 1). The plastid genome of the mycoheterotrophic species D. singchiana also showed a dramatic reduction to 87,910 bp (table 1). The loss or pseudogenization of rps genes, such as rps15 in D. singchiana and rps15 and rps18 in R. atropurpurea (fig. 1), is consistent with the final stage of the plastome degradation model proposed by Barrett and Davis (2012).
Fig. 1.
—Summary of retained genes in the plastomes of sampled species. Intact genes per species are indicated by green boxes, whereas the white boxes mark functional and or physical losses. The shading boxes (0–27) indicate the number of species in this study retaining the genes. Orange branch indicates leafless and mycoheterotrophic species.
The highly reduced plastomes of R. atropurpurea and D. singchiana still had a quadripartite structure but exhibited a shift in the IR/SC region boundary. The plastome of R. atropurpurea was basically collinear with those of autotrophic relatives (supplementary fig. S2, Supplementary Material online), but rpl36, infA, rps8, rpl14, rpl16, and rps3, which are usually present in the large single copy region, were translocated to the IR region. In plastome of D. singchiana, all rrn genes were translocated from the IR region to the SC region. In addition, we detected two inversions in the plastome of D. singchiana (supplementary fig. S2, Supplementary Material online).
Rate of Evolution and Selection
Mycoheterotrophic species exhibited a higher substitution rate and thereby longer branches relative to those of autotrophic lineages (supplementary fig. S3, Supplementary Material online). Both synonymous and nonsynonymous substitution rates of R. atropurpurea were higher than those of other Calypsoinae species (supplementary fig. S3, Supplementary Material online). The RELAX model predicted that the relative strength of selection (relaxation or intensification) varied among plastid protein-coding genes in mycoheterotrophic species. In particular, 15 of 25 common genes displayed evidence of relaxed selective constraint associated with the loss of photosynthesis, including a significant relaxation of selective constraint on five genes, accD, rpl23, rps11, rps16, and ycf1 (P < 0.05; supplementary table S2, Supplementary Material online). Analyses of concatenated gene sets for each functional class revealed evidence for significantly relaxed selection in ACIM (accD, clpP, infA, and matK) and the rps complex (P < 0.05; supplementary table S3, Supplementary Material online).
Based on previous molecular dating, the estimated crown age of Corallorhiza is ∼9 Myr (Barrett et al. 2018), whereas Danxiaorchis split from its autotrophic sister group 11–12 Ma (Barrett et al. 2018; Li et al. 2019) and Risleya diverged about 24 Ma (Li et al. 2019). Based on these estimates and the degree of plastome reduction in the three lineages (fig. 1), the time since the shift to mycoheterotrophy might be the main determinant of the degree of plastome reduction and elevated evolutionary rate in heterotrophic lineages (Barrett et al. 2019).
Supplementary Material
Data deposition: This project has been deposited at NCBI under the accessions MN990431–MN990443.
Literature Cited
- Barrett CF, Davis JI.. 2012. The plastid genome of the mycoheterotrophic Corallorhiza striata (Orchidaceae) is in the relatively early stages of degradation. Am J Bot. 99(9):1513–1523. [DOI] [PubMed] [Google Scholar]
- Barrett CF, Sinn BT, Kennedy AH.. 2019. Unprecedented parallel photosynthetic losses in a heterotrophic orchid genus. Mol Biol Evol. 36(9):1884–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett CF, Wicke S, Sass C.. 2018. Dense infraspecific sampling reveals rapid and independent trajectories of plastome degradation in a heterotrophic orchid complex. New Phytol. 218(3):1192–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett CF, et al. 2014. Investigating the path of plastid genome degradation in an early-transitional clade of heterotrophic orchids, and implications for heterotrophic angiosperms. Mol Biol Evol. 31(12):3095–3112. [DOI] [PubMed] [Google Scholar]
- Chase MW, et al. 2015. An updated classification of Orchidaceae. Bot J Linn Soc. 177(2):151–174. [Google Scholar]
- Delannoy E, Fujii S, Colas des Francs-Small C, Brundrett M, Small I.. 2011. Rampant gene loss in the underground orchid Rhizanthella gardneri highlights evolutionary constraints on plastid genomes. Mol Biol Evol. 28(7):2077–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delport W, Poon AF, Frost SD, Kosakovsky Pond SL.. 2010. Datamonkey 2010: a suite of phylogenetic analysis tools for evolutionary biology. Bioinformatics 26(19):2455–2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y-L, et al. 2016. Lineage-specific reductions of plastid genomes in an orchid tribe with partially and fully mycoheterotrophic species. Genome Biol Evol. 8(7):2164–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freudenstein JV, Yukawa T, Luo Y-B.. 2017. A reanalysis of relationships among Calypsoinae (Orchidaceae: Epidendroideae): floral and vegetative evolution and the placement of Yoania. Syst Bot. 42(1):17–25. [Google Scholar]
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 41(41):95–98. [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler AV, Jermiin LS.. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YK, et al. 2019. Extensive losses of photosynthesis genes in the plastome of a mycoheterotrophic orchid, Cyrtosia septentrionalis (Vanilloideae: Orchidaceae). Genome Biol Evol. 11(2):565–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K.. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J-L, Wang S, Jing Y, Wang L, Zhou S-L.. 2013. A modified CTAB protocol for plant DNA extraction. Chin Bull Bot. 48(1):72–78. [Google Scholar]
- Li Y-X, et al. 2019. Phylogenomics of Orchidaceae based on plastid and mitochondrial genomes. Mol Phylogenet Evol. 139:106540. [DOI] [PubMed] [Google Scholar]
- Merckx VSFT, Freudenstein JV, et al. 2013. Taxonomy and classification In: Merckx VSFT, editor. Mycoheterotrophy: the biology of plants living on fungi. New York: Springer; p. 19–101. [Google Scholar]
- Merckx VSFT, Mennes CB, Peay KG, Geml J.. 2013. Evolution and diversification In: Merckx VSFT, editor. Mycoheterotrophy: the biology of plants living on fungi. New York: Springer; p. 215–244. [Google Scholar]
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel RK, Jain M.. 2012. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One 7(2):e30619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X-J, Moore MJ, Li D-Z, Yi T-S.. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schelkunov MI, et al. 2015. Exploring the limits for reduction of plastid genomes: a case study of the mycoheterotrophic orchids Epipogium aphyllum and Epipogium roseum. Genome Biol Evol. 7(4):1179–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukaya H. 2018. How have leaves of mycoheterotrophic plants evolved—from the view point of a developmental biologist. New Phytol. 217(4):1401–1406. [DOI] [PubMed] [Google Scholar]
- Wertheim JO, Murrell B, Smith MD, Kosakovsky Pond SL, Scheffler K.. 2015. RELAX: detecting relaxed selection in a phylogenetic framework. Mol Biol Evol. 32(3):820–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z-H. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
- Zerbino DR, Birney E.. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18(5):821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, et al. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.