Abstract
Cold seeps, characterized by the methane, hydrogen sulfide, and other hydrocarbon chemicals, foster one of the most widespread chemosynthetic ecosystems in deep sea that are densely populated by specialized benthos. However, scarce genomic resources severely limit our knowledge about the origin and adaptation of life in this unique ecosystem. Here, we present a genome of a deep-sea limpet Bathyacmaea lactea, a common species associated with the dominant mussel beds in cold seeps. We yielded 54.6 gigabases (Gb) of Nanopore reads and 77.9-Gb BGI-seq raw reads, respectively. Assembly harvested a 754.3-Mb genome for B. lactea, with 3,720 contigs and a contig N50 of 1.57 Mb, covering 94.3% of metazoan Benchmarking Universal Single-Copy Orthologs. In total, 23,574 protein-coding genes and 463.4 Mb of repetitive elements were identified. We analyzed the phylogenetic position, substitution rate, demographic history, and TE activity of B. lactea. We also identified 80 expanded gene families and 87 rapidly evolving Gene Ontology categories in the B. lactea genome. Many of these genes were associated with heterocyclic compound metabolism, membrane-bounded organelle, metal ion binding, and nitrogen and phosphorus metabolism. The high-quality assembly and in-depth characterization suggest the B. lactea genome will serve as an essential resource for understanding the origin and adaptation of life in the cold seeps.
Keywords: deep-sea adaptation, cold seeps, limpet, Bathyacmaea, mollusk genome, Nanopore
Introduction
Cold seeps are the deep-sea areas where methane, hydrogen sulfide, and other hydrocarbon-rich fluid seepages occur (Joseph 2017). Cold-seep animals not only have to adapt the harsh deep-sea environments including high hydrostatic pressure, darkness, and low temperatures, but also should cope with hypoxia and the rich reducing chemicals that are toxic to most animals (Levin 2005; Hourdez and Lallier 2006). Despite inhospitable, cold seeps support huge chemosynthesis-based ecosystems characterized by abundant specialized benthos (Van Dover et al. 2002; Niu et al. 2017; Åström et al. 2019). In a typical cold-seep ecosystem, certain invertebrates of mussels, clams, tubeworms, shrimps, and gastropods are the common species (Levin 2005; Mazumdar et al. 2019), most of them perform symbioses with chemosynthetic bacteria (Petersen and Dubilier 2009; Kiel 2010), whereas some gastropods feed on bacteria and other animals (Zande and Carney 2001; Amano 2003). Biologists have long wondered the origin and adaptations of these specialized organisms in such unique environments. To date, genomes of symbiotic Bathymodiolus platifrons (Sun et al. 2017) and Lamellibrachia luymesi (Li et al. 2019) in cold seeps have been published, which have well revealed the mechanisms of symbioses. However, our knowledge about the underlying adaptations of them to the cold-seep environment is still lagging. This is particularly true for nonsymbiotic animals. Thus, the genomes of cold-seep animals with different nutrition modes are vital for a comprehensive understanding to the origin and adaptation of cold-seep life.
As important members of deep-sea gastropods, Pectinodontid limpets are often associated with the dominant bathymodioline mussel beds (Feng et al. 2018; Mazumdar et al. 2019). Previous investigations reported that some limpets (e.g., Bathyacmaea lactea) feed on bacteria and decomposing the periostracum of Bathymodiolus shells (Zande and Carney 2001; Zhang et al. 2016; Feng et al. 2018; Linse et al. 2019; Mazumdar et al. 2019), which makes deep-sea limpets different from the well-studied symbionts. Meanwhile, a large number of reports (Nakano and Ozawa 2007) accompanied by complete fossil records (Jenkins et al. 2017) make the Pectinodontid limpets ideal for studying phylogeography and paleoecology.
Here, we present a genome assembly of cold-seep limpet B. lactea (NCBI: txid2049241; Gastropoda, Pectinodontidae) by Nanopore and BGI-seq sequencing. To the best of our knowledge, this is the first genome of gastropods in the deep-sea cold seeps. We believe that high-quality assembly and annotation will provide a solid foundation in answering the fundamental issues about the origin and adaptation of cold-seep life.
Materials and Methods
Sampling and sequencing
A total of eight specimens of deep-sea limpet B. lactea were obtained by the HOV Shenhai Yongshi at a depth of ∼1,388 m in the Haima cold seeps in the South China Sea (supplementary fig. S1, Supplementary Material online). These samples were clean and immediately refrigerated by liquid nitrogen. The whole-genome DNA of a specimen (sample No. MBSQW58) was extracted by Qiagen Genomic DNA extraction kit (Cat#13323, Qiagen, Hilden, Germany) and then subjected to construct genome BGI-seq library using an MGIEasy Library Prep Kit (v1.1, MGI Tech). We conducted 100-bp paired-end single-indexed sequencing on the MGISEQ-2000 platform (BGI, Shenzhen, China). Nanopore libraries with insertions >20 kb were prepared and sequenced on one flow cell using a PromethION DNA sequencer (v24, Oxford Nanopore, Oxford, UK).
Genome Assembly and Assessment
For the BGI-seq data, both low-quality reads and adaptor sequences were removed by fastp (v0.20, Chen et al. 2018) with default parameters. We estimated the genome size via a 23-mer frequency distribution analysis using the formula: G = K_num/K_depth. The K_num and K_depth represented the total number and the expected depth of 23-mer analysis, respectively.
For the Nanopore data, raw reads were filtered by program ontbc (https://github.com/FlyPythons/ontbc) with parameters “–min_score 7 –min_length 1000.” The filtered Nanopore reads were first corrected the base errors by NextDenovo (https://github.com/Nextomics/NextDenovo) with default settings, and then assembled into contigs by program wtdbg (v2.4, Ruan and Li 2019) with parameters “-L 5000 -k 0 -p 21 -S 1.” The filtered BGI-seq reads were mapped to assembled contigs using Burrows–Wheeler Aligner (BWA, Li and Durbin 2010) and polished twice with Pilon (v1.21, Walker et al. 2014) under default settings.
We employed Benchmarking Universal Single-Copy Orthologs (BUSCO v3.0, Simão et al. 2015) to evaluate the completeness of the genome assembly and annotation using the “metazoa_odb9” database. To further evaluate the accuracy, we aligned the clean BGI-seq reads against the genome assembly using BWA (Li and Durbin 2010). Then, SAMtools/BCFtools package (v1.3.1, Li et al. 2009) was employed to investigate the reads coverage of the final assembly and calculate the base error rate of it.
Genome Annotation
We carried out annotations for repeats before predicting the structure of genes. Firstly, we build a de novo repeat library by RepeatModeler (v1.0.11, Tarailo-Graovac and Chen 2009), then we employed RepeatMasker (v3.3.0, Tarailo-Graovac and Chen 2009) to identify homolog repeats in B. lactea. Moreover, RepeatMasker (v3.3.0, Tarailo-Graovac and Chen 2009) and RepeatProteinMask (v3.3.0, a package in RepeatMasker) were employed to search previously reported repeats in the Repbase. Further, we used Tandem Repeat Finder (v4.04, Benson 1999) to identify tandem repeats with following settings: Match = 2, Mismatch = 7, Delta = 7, PM = 80, PI = 10, Minscore = 50.
The location and structure of genes were predicted by a combination of ab initio and homology-based approaches. Before homology-based annotation, the genomes of six mollusk species were retrieved from NCBI (supplementary table S1, Supplementary Material online). Transcript of each gene with the longest length and no premature termination site was picked out and translated into amino acids (Wang, Xu, et al. 2019). These protein sequences were aligned to the repeats soft-masked genome by TBlastN (v2.7.1, Altschul et al. 1990) with a cut-off value 1e-5. Then, identified homologous sequences were subjected to GeneWise (Birney et al. 2004) to define gene models. For the ab initio annotation, we used AUGUSTUS (v3.2.1, Stanke et al. 2008) to predict gene models based on the “seahare” training set. All gene models were integrated into a nonredundant gene set by EvidenceModeler (EVM, v1.1.1, Haas et al. 2008).
To determine the function of those predicted protein-coding genes, we employed InterProScan (v5.30-69.0, Jones et al. 2014) to searched for the best hits of these proteins against all incorporated databases.
Phylogenetic Analysis and Divergence Time Analysis
To assess the phylogenetic position of B. lactea, genomic data of another seven lophotrochozoa species (see supplementary table S1, Supplementary Material online) were retrieved from NCBI and preprocessed in the same way as those six mollusk species described above. Protein set of B. lactea and these thirteen species were clustered into families by running reciprocal BLAST analysis in OrthoFinder (v2.3.1, Emms and Kelly 2015). The 580 produced single-copy orthologues were aligned using MUSCLE (v3.8.1551, Edgar 2004) with default parameters. Low-quality alignments were trimmed by TrimAl (Capella-Gutiérrez et al. 2009) using an automated1 algorithm. Subsequently, the remaining alignments were concatenated into a supergene and used for phylogeny reconstruction. Finally, a maximum likelihood (ML) tree was built by RAxML (v8.2.12, Stamatakis 2014) with the GTRGAMMA model and 1,000 bootstraps.
Divergence time among species was estimated via the MCMCtree program in PAML (v4.9, Yang 2007). Five fossil-based calibration points were used as time priors (Benton et al. 2009; Plazzi and Passamonti 2010): The most recent common ancestor of lophotrochozoa (550.3–636.1 Ma); the most recent common ancestor of molluscs (532–549 Ma); Aplysia californica (or Biomphalaria glabrata)–Lottia gigantea (470.2–531.5 Ma); A. californica–B. glabrata (168.6–473.4 Ma); Crassostrea gigas–Crassostrea virginica (63–83 Ma).
Substitution Rate, Demographic History, and TE Activity
The branch-length calibrated ML tree was subjected to r8s (v1.81, Sanderson 2003) to investigate the substitution rate of each species under the penalized likelihood method. We explored demographic histories using the Pairwise Sequentially Markovian Coalescence model (PSMC, v0.6.5-r67, Li and Durbin 2011) based on heterozygous sites. The filtered BGI-seq reads were mapped to the final genome assembly using BWA-mem (Li and Durbin 2010). Subsequently, SAMtools (v1.3.1, Li et al. 2009) with the parameters “mpileup -q 20 -Q 20” were employed to extract heterozygous sites. The substitution rate was set according to the r8s (Sanderson 2003) results. Finally, the PSMC model (Li and Durbin 2011) was analyzed using parameters: -N25 -t15 -r5 -b -p “4 + 25*2 + 4 + 6.”
We further used a custom Perl script parseRM.pl (available at https://github.com/4ureliek/Parsing-RepeatMasker-Outputs; Kapusta et al. 2017) to parse the TE activities in B. lactea based on alignment outputs from RepeatMasker (v3.3.0, Tarailo-Graovac and Chen 2009). The substitution rate was set as 0.00121 per site per million years, according to the results of r8s (v1.81, Sanderson 2003). The analysis result was packed into bin per 3 Ma.
Gene Family Analysis and Rapidly Evolving Gene Ontology Categories
Based on the OrthoFinder analysis, we investigated significant expansion and contraction of gene families with CAFÉ (v4.0, Berger and Young 2006) along the time-calibrated tree (Viterbi P-values < 0.01).
Single-copy orthologues from the OrthoFinder analysis were realigned with PRANK (v170427, Löytynoja 2014) and filtered by Gblocks (v0.91b, Castresana 2000) using default settings to remove the ambiguously aligned sequences. We used the free-ratio model in the “Codeml” module of the PAML (v4.9, Yang 2007) to estimate the Ka/Ks values of species for each gene. According to Gene Ontology (GO) annotations, these genes were assigned to the corresponding GO categories. Any GO comprised <10 orthologues was excluded from subsequent analysis. Following the method of Wang, Shen, et al. (2019), we counted the Ka and Ks values of B. lactea and it is shallow-water relative L. gigantea in each GO and set the nonsynonymous substitution probability of L. gigantea corrected by substitutions of these two genomes as the background expectation. By a one-sided binomial test, those with a significantly higher Ka/Ks level in B. lactea than expected were considered as rapidly evolving GO categories (FDR < 0.05).
Results and Discussion
Genome Assembly and Assessment
In total, we generated a 77.9 Gb (98.3×) raw data set of BGI-seq and a 54.6 Gb (68.9×, reads N50 of 18.3 kb) raw data set of Nanopore, respectively (supplementary table S2, Supplementary Material online). After filtering, 77.0 Gb (97.1×) BGI reads and 46.2 Gb (58.3×, N50 of 18.7 kb) passed Nanopore reads were harvested (supplementary table S2, Supplementary Material online). The 23-mer survey suggested an estimated genome size of ∼792.8 Mb for B. lactea (supplementary fig. S2 and table S3, Supplementary Material online). A slight subpeak was observed at half the value of the expected depth (supplementary fig. S2, Supplementary Material online), indicating a moderately heterozygous genome.
After assembly and polishing, we obtained a final genome of 754.3 Mb, with 3,720 contigs and a contig N50 of 1.57 Mb (table 1). The completeness assessment showed that the assembly included complete matches for 922 of 978 metazoan BUSCOs (94.3%), which represented a high-quality assembly compared with other gastropod genomes (supplementary table S4, Supplementary Material online). Furthermore, we found that 98.1% of the BGI-seq reads were reliably aligned to the genome assembly, and 93.9% of the reads were properly aligned to the genome with their mates. The genome-level base error rate was as low as 2.3 bp per 100,000 bases. These results suggested a high-quality assembly of the B. lactea genome.
Table 1.
Assembly Statistics of the Bathyacmaea lactea Genome
| Contig |
||
|---|---|---|
| Size (bp) | Number | |
| N90 | 111,041 | 755 |
| N50 | 1,568,604 | 125 |
| Longest | 8,382,764 | — |
| Shortest | 1,973 | — |
| Total size | 754,256,772 | 3,720 |
Genome Annotations
Totally, ∼463.4 Mb of repeat sequences that accounted for 61.4% of the genome were identified (supplementary table S5, Supplementary Material online). This proportion was quite high compared with that of other mollusk genomes (20.5–61.1%, Sun et al. 2017; Cai et al. 2019). DNA transposons (10.4%) and long interspersed nuclear elements (LINEs; 6.5%) were the most abundant TEs in the B. lactea genome (supplementary table S6, Supplementary Material online).
We finally generated a gene set of 23,574 protein-coding genes with an average of 6.8 exons and 1,417-bp coding region per gene, which was comparable to that of other gastropod genomes (supplementary fig. S3 and table S7, Supplementary Material online). BUSCO assessment indicated that 95.7% of the metazoa conserved genes were completely detected in the B. lactea gene set. This result was better than most of the published gastropod genomes (supplementary table S4, Supplementary Material online). As for function annotation, 21,136 of the 23,574 genes were annotated by at least one database, representing 89.7% of the total genes (supplementary table S8, Supplementary Material online).
Phylogenetics and Divergence Time Analysis
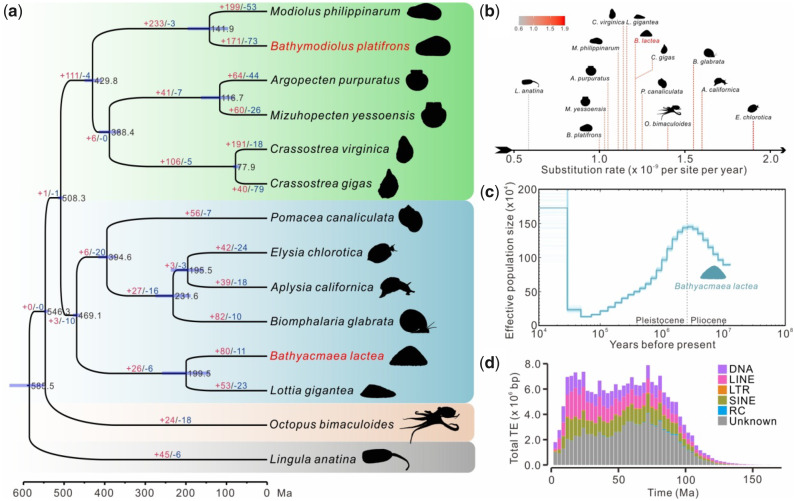
Totally, 580 single-copy orthologues were identified among fourteen lophotrochozoa species. Based on the single-copy orthologues, ML analysis recovered a well-resolved phylogeny of lophotrochozoan and supported B. lactea was most closely related to limpet L. gigantea. They diverged from a common ancestor at ∼199.5 Ma (fig. 1a), with a 95% confidence interval of 141.4–258.7 Ma.
Fig. 1.
Evolutionary analysis of Bathyacmaea lactea. (a) Phylogeny and divergence time (Ma) of fourteen Lophotrochozoa species, labeled with the 95% confidence interval (purple bars). Numbers on branches indicate the event of gene family expansions (red) and contractions (blue). (b) Comparison of substitution rates among fourteen Lophotrochozoa species. (c) Demographic history of B. lactea estimated by PSMC. (d) The landscape of TE accumulation along the timeline. The result was exhibited by bins of 3 Ma.
Substitution Rate, Demographic History, and TE Activity
The r8s analysis showed that gastropods tended to have a higher substitution rate than bivalves (fig. 1b). The substitution rate of B. lactea was ∼1.21 × 10−9 per site per year, which was slightly higher than that of its shallow-sea relative L. gigantea (fig. 1b).
Demographic history analysis showed that the effective population size of B. lactea reached a peak of ∼2.6 Ma and then experienced a long-term recession until it recovered again at ∼21,000 years ago (21.0 ka, fig. 1c). The rise and fall of population of B. lactea coincided with the great events in ancient times, such as the marine life extinction around the Pliocene-Pleistocene boundary (∼2.58 Ma, Pimiento et al. 2017) and the Last Glacial Maximum (26.5–19.0 ka, Clark et al. 2009). These results indicate the deep-sea ecosystems may also be influenced by paleoclimate. Furthermore, we found that B. lactea had undergone a long-lasting TE activity until the last 10 Ma, including two concentrated TE expansions (fig. 1d and supplementary fig. S4, Supplementary Material online). It has been reported that TEs could play important roles in response to environmental challenges (Casacuberta and González 2013). Hence, whether and how TEs facilitate the adaptability of deep-sea limpet to cold seeps also deserve the attention of further studies.
Gene Family Analysis and Rapidly Evolving GO Categories
We detected 80 expanded gene families and 11 contracted gene families in the B. lactea lineage (fig. 1a). The expanded gene families were enriched in 97 GO categories, and many of them were associated with metal ion binding, heterocyclic compound metabolism, apoptotic process, nitrogen compound metabolism, and membranes (supplementary table S9, Supplementary Material online). Meanwhile, gene families related to peptidase inhibitor activity were contracted (supplementary table S10, Supplementary Material online).
Further analysis harvested 87 rapidly evolving GOs (supplementary table S11, Supplementary Material online), most of them were involved in heterocyclic compound metabolism, membrane-bounded organelle, metal ion binding, and nitrogen and phosphorus metabolism (supplementary fig. S5, Supplementary Material online). These expanded and rapidly evolving genes depict the holistic adaptive changes within the genome. Positive changes of genes associated with heterocyclic and nitrogen compound metabolism hinted the genetic basis of B. lactea adapting to the chemosynthesis-based and highly toxic reducing environments in cold seeps (Berry, et al. 1987; Xue and Warshawsky 2005). Moreover, changes in membranes and metal ion binding may facilitate lives survive and thrive in the inhospitable conditions of cold seeps including high pressures, coldness, and hypoxia (Hourdez and Lallier 2006; Wang, Xu, et al. 2019).
Although many works remain to be done to disentangle the origin and adaptation of life in cold seeps, we believe that the high-quality genome and numerous data created in this study will serve as important resources in answering these issues.
Supplementary Material
Acknowledgments
The authors want to thank the crews of the vessel Tansuo 1 and the pilots of the HOV Shenhai Yongshi. We appreciate Prof. Qiang Qiu (Northwestern Polytechnical University, China), Dr Chong Chen (Japan Agency for Marine-Earth Science and Technology, Japan), and Dr Ping Zheng (Fujian Agriculture and Forestry University, China) for their helpful advice. This study was supported by the National Key Research and Development Program of China (2016YFC0304905 and 2017YFC0306604 to H.Z.), and the Major scientific and technological projects of Hainan Province (ZDKJ2019011 to H.Z.), the 1000 Talent Project of Shaanxi Province to K.W., and Research Funds for Interdisciplinary subject, NWPU (19SH030408 to K.W.).
Data deposition: This project has been deposited at GenBank under the BioProject PRJNA588374 (BioSamples SAMN13255183, SRA of Nanopore reads: SRR10425370, SRA of Illumina data: SRR10425354–SRR10425369).
Literature Cited
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ.. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410. [DOI] [PubMed] [Google Scholar]
- Amano K. 2003. Predatory gastropod drill holes in Upper Miocene cold seep bivalves, Hokkaido, Japan . Veiliger 46:90–96. [Google Scholar]
- Åström EK, et al. 2019. Chemosynthesis influences food web and community structure in high-Arctic benthos. Mar Ecol Prog Ser. 629:19–42. [Google Scholar]
- Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27(2):573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton MJ, Donoghue PC, Asher RJ.. 2009. Calibrating and constraining molecular clocks. In: Hedges SB Kumar S, editors. The timetree of life. Vol. 35. Oxford: Oxford University Press. p. 86. [Google Scholar]
- Berger MS, Young CM.. 2006. Physiological response of the cold-seep mussel Bathymodiolus childressi to acutely elevated temperature. Mar Biol. 149(6):1397–1402. [Google Scholar]
- Berry DF, Francis AJ, Bollag J-M.. 1987. Microbial metabolism of homocyclic and heterocyclic aromatic compounds under anaerobic conditions. Microbiol Rev. 51(1):43–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Clamp M, Durbin R.. 2004. GeneWise and genomewise. Genome Res. 14(5):988–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H, et al. 2019. A draft genome assembly of the solar-powered sea slug Elysia chlorotica. Sci Data. 6(1):190022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T.. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25(15):1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casacuberta E, González J.. 2013. The impact of transposable elements in environmental adaptation. Mol Ecol. 22(6):1503–1517. [DOI] [PubMed] [Google Scholar]
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17(4):540–552. [DOI] [PubMed] [Google Scholar]
- Chen S, Zhou Y, Chen Y, Gu J.. 2018. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34(17):i884–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark PU, et al. 2009. The last glacial maximum. Science 325(5941):710–714. [DOI] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms DM, Kelly S.. 2015. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16(1):157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng D, et al. 2018. Cold seep systems in the South China Sea: an overview. J Asian Earth Sci. 168:3–16. [Google Scholar]
- Haas BJ, et al. 2008. Automated eukaryotic gene structure annotation using EVidenceModeler and the program to assemble spliced alignments. Genome Biol. 9(1):R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hourdez S, Lallier FH.. 2006. Adaptations to hypoxia in hydrothermal-vent and cold-seep invertebrates. In: Life in extreme environments. Dordrecht: Springer. p. 297–313.
- Jenkins R, et al. 2017. Antiquity of the substrate choice among acmaeid limpets from Late Cretaceous chemosynthesis-based communities. Acta Palaeontol Polon. 62:369–373. [Google Scholar]
- Jones P, et al. 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 30(9):1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph A. 2017. Investigating seafloors and oceans: from mud volcanoes to giant squid, foreword. Amsterdam: Elsevier. p. ix–xxvi. doi: 10.1016/B978-0-12-809357-3.09995-1. [DOI]
- Kapusta A, Suh A, Feschotte C.. 2017. Dynamics of genome size evolution in birds and mammals. Proc Natl Acad Sci USA. 114(8):E1460–E1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiel S. 2010. The vent and seep biota: aspects from microbes to ecosystems. Berlin: Springer Science & Business Media.
- Levin LA. 2005. Ecology of cold seep sediments: interactions of fauna with flow, chemistry and microbes In: Oceanography and marine biology. Boca Raton: CRC Press. p. 11–56. [Google Scholar]
- Li H, Durbin R.. 2010. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26(5):589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2011. Inference of human population history from individual whole-genome sequences. Nature 475(7357):493–U484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, et al. 2019. Genomic adaptations to chemosymbiosis in the deep-sea seep-dwelling tubeworm Lamellibrachia luymesi. BMC Biol. 17(1):91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linse K, Roterman C, Chen C.. 2019. A new vent limpet in the genus Lepetodrilus (Gastropoda: Lepetodrilidae) from Southern Ocean hydrothermal vent fields showing high phenotypic plasticity. Front Mar Sci. 6: 381. [Google Scholar]
- Löytynoja A. 2014. Phylogeny-aware alignment with PRANK In: Multiple sequence alignment methods. Dordrecht: Springer. p. 155–170. [DOI] [PubMed] [Google Scholar]
- Mazumdar A, et al. 2019. The first record of active methane (cold) seep ecosystem associated with shallow methane hydrate from the Indian EEZ. J Earth Syst Sci. 128(1):18. [Google Scholar]
- Nakano T, Ozawa T.. 2007. Worldwide phylogeography of limpets of the order Patellogastropoda: molecular, morphological and palaeontological evidence. J Molluscan Stud. 73(1):79–99. [Google Scholar]
- Niu M, Fan X, Zhuang G, Liang Q, Wang F.. 2017. Methane-metabolizing microbial communities in sediments of the Haima cold seep area, northwest slope of the South China Sea. FEMS Microbiol Ecol. 93:fix101. [DOI] [PubMed] [Google Scholar]
- Petersen JM, Dubilier N.. 2009. Methanotrophic symbioses in marine invertebrates. Environ Microbiol Rep. 1(5):319–335. [DOI] [PubMed] [Google Scholar]
- Pimiento C, et al. 2017. The Pliocene marine megafauna extinction and its impact on functional diversity. Nat Ecol Evol. 1(8):1100–1106. [DOI] [PubMed] [Google Scholar]
- Plazzi F, Passamonti M.. 2010. Towards a molecular phylogeny of Mollusks: bivalves’ early evolution as revealed by mitochondrial genes. Mol Phylogenet Evol. 57(2):641–657. [DOI] [PubMed] [Google Scholar]
- Ruan J, Li H.. 2019. Fast and accurate long-read assembly with wtdbg2. Nat Methods 17:155–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson MJ. 2003. r8s: inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics 19(2):301–302. [DOI] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM.. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31(19):3210–3212. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, Diekhans M, Baertsch R, Haussler D.. 2008. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 24(5):637–644. [DOI] [PubMed] [Google Scholar]
- Sun J, et al. 2017. Adaptation to deep-sea chemosynthetic environments as revealed by mussel genomes. Nat Ecol Evol. 1(5):121. [DOI] [PubMed] [Google Scholar]
- Tarailo-Graovac M, Chen N.. 2009. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr Protoc Bioinform. 25(1):4.10.11–14.10.14. [DOI] [PubMed] [Google Scholar]
- Van Dover CL, German C, Speer KG, Parson L, Vrijenhoek R.. 2002. Evolution and biogeography of deep-sea vent and seep invertebrates. Science 295(5558):1253–1257. [DOI] [PubMed] [Google Scholar]
- Walker BJ, et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9(11):e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Shen Y, et al. 2019. Morphology and genome of a snailfish from the Mariana Trench provide insights into deep-sea adaptation. Nat Ecol Evol. 3(5):823–833. [DOI] [PubMed] [Google Scholar]
- Wang X, Xu W, et al. 2019. Nanopore sequencing and de novo assembly of a black-shelled Pacific oyster (Crassostrea gigas) genome. Front Genet. 10:1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W, Warshawsky D.. 2005. Metabolic activation of polycyclic and heterocyclic aromatic hydrocarbons and DNA damage: a review. Toxicol Appl Pharmacol. 206(1):73–93. [DOI] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
- Zande JM, Carney RS.. 2001. Population size structure and feeding biology of Bathynerita naticoidea Clarke 1989 (Gastropoda: Neritacea) from Gulf of Mexico hydrocarbon seeps. Gulf Mexico Sci. 19(2):4. [Google Scholar]
- Zhang S, Zhang J, Zhang S.. 2016. A new species of Bathyacmaea (Gastropoda: Pectinodontidae) from a methane seep area in the South China Sea. Nautilus 130:1–4. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.