Abstract
Odorant receptors represent the largest family of mammalian G protein-coupled receptors. Phylogenetically, they are split into two classes (I and II). By analyzing the entire subclass I odorant receptors sequences, we identified two class I-specific and highly conserved motifs. These are predicted to face each other at the extra-cellular portion of the transmembrane domain, forming a vestibular site at the entrance to the orthosteric-binding cavity. Molecular dynamics simulation combined with site-directed mutagenesis and in vitro functional assays confirm the functional role of this vestibular site in ligand-driven activation. Mutations at this part of the receptor differentially affect the receptor response to four agonists. Since this vestibular site is involved in ligand recognition, it could serve ligand design that targets specifically this sub-genome of mammalian odorant receptors.
Electronic supplementary material
The online version of this article (10.1007/s00018-018-2996-4) contains supplementary material, which is available to authorized users.
Keywords: Olfactory, GPCR, Molecular modeling, Mechanism, Binding, Mutant
Introduction
Mammals sense their chemical environment when volatile molecules activate the odorant receptors (ORs) embedded in the membrane of their olfactory sensory neurons [1]. ORs belong to the class A G protein-coupled receptors (GPCRs) and represent more than 3% of all protein-coding genes [2]. Structurally, class A GPCRs are made up of seven transmembrane helices (TM1–TM7) forming a bundle within the cell membrane, connected by intra- and extra-cellular loops. Class A GPCRs show a remarkably conserved fold and, from a sequence point of view, they also share typical hallmark motifs that are distributed at various locations within their sequences [3].
In the absence of any OR experimental structure, molecular modeling has been the tool of choice for studying structure–function relationships in ORs. Comparative modeling benefits from the high level of conservation in the structural fold of class A GPCRs. Combined with in vitro site-directed mutagenesis and functional assays, it has proven extremely powerful for predicting various OR features, such as ligand binding and selectivity, and selective activation by agonist bound within their orthosteric-binding site [4–9].
In ORs, the orthosteric-binding cavity exhibits the greatest variability [10], endowing mammals with an extraordinarily broad chemical discriminatory power. Phylogenetic classification of the mammalian olfactory receptor genome splits this family of receptors into two distinct classes [11–13]. In humans, the 58 class I ORs represent ~ 15% of the 397 receptors that are considered functional. Class I ORs are also referred to as “fish-like”, since they resemble the family that was first identified in aquatic animals such as fish and frog. They were considered to be ancestral ORs that were maintained in mammals [12]. The second class of ORs (class II or “tetrapod”) is thought to have appeared during tetrapod terrestrial evolution. However, the diversity of mammalian ORs remains much smaller than that found in fishes, where many more classes have been identified [14]. Interestingly, during evolution, class I ORs were maintained in a single cluster as opposed to class II OR genes which spread over most chromosomes [15]. Class I odorant receptor genes also share a specific and conserved genetic mechanism regulating the allelic exclusion during expression [16].
In mammals, class I and class II, despite sharing all the typical OR sequence-specific hallmarks, can be clearly distinguished by some characteristic features that are highly conserved within their sequences [13].The extra-cellular part of TM6 has been suggested to play an role in the differential ligand recognition between class I and class II ORs [17], although the associated structural features are yet to be uncovered. Between the two classes, no difference could be identified concerning the breadth of tuning, the number of agonists, or the sensitivity [18].
Extra-cellular allosteric sites have been identified in numerous class A GPCRs [19], such as the purinoceptor 1 (P2Y1 [20]), sphingosine-1-phosphate receptor 1 (S1P1 [21]), β2-adrenergic receptor [22], and the muscarinic acetylcholine receptors [23]. More specifically, a so-called “vestibular-binding site” has been identified at the top of the transmembrane domain in the muscarinic acetylcholine receptors [23], in the β2 adrenergic receptor [22] experimental structures, as well as in molecular models of the δ-opioid receptor [24]. Focusing on chemosensory receptors, such a site was reported in the bitter taste receptor TAS2R46 [25] and in the trace amine associated receptor TAAR13c [26]. These discoveries laid the groundwork for the design of highly selective allosteric modulators. This type of vestibular-binding site has never been identified in any OR.
In this article, we report the identification of two class I OR-specific motifs that face each other at the extra-cellular extremities of TM5 and TM6. These motifs are conserved in class I ORs but not in the class II. Molecular dynamics simulations showed that these motifs form a vestibular site at the entrance to the orthosteric-binding cavity. Site-directed mutations at the vestibular-binding site affected the receptors’ response to odorant stimulation in vitro, confirming the functional role of this vestibule.
Results
ORs show distinct specific sequence motifs
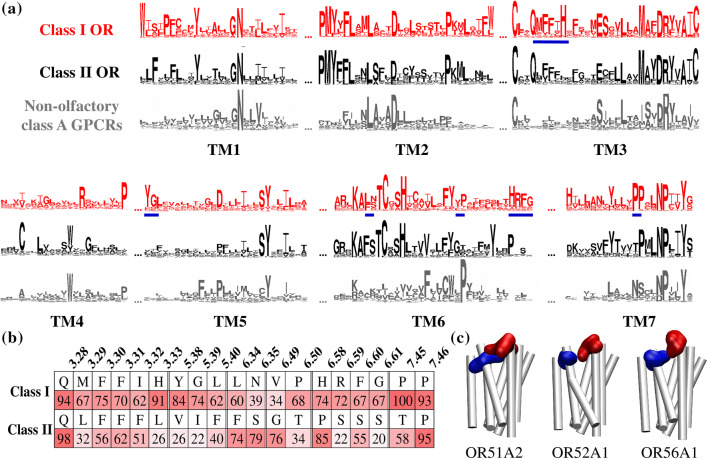
Figure 1 reports the conservation analysis of human GPCR sequences. Here, 397 human ORs were compared with 204 non-olfactory class A GPCRs. As observed previously in mice [13], both OR classes show identical conserved regions such as the PMYxFL motif in TM2, MAYDRYVAIC in TM3, SY residues in TM5, RxKAxxTCxSH, and FY in TM6. Other motifs considered to be the hallmarks of the GPCR class A family (GN in TM1, DRY in TM3, KA in TM6, and NPxxY in TM7) can also be identified (Fig. 1a).
Fig. 1.
Conserved motifs in class I and class II human ORs and comparison with non-olfactory class A GPCRs. a Logos summarize the conservation amongst 397 human OR sequences (58 are class I ORs and 339 are class II) and 204 non-olfactory class A GPCRs [10, 27]. Major differences are underlined in blue. b Highest conservation of class-specific motifs in TM3, TM5, TM6, and TM7. c Three class I human OR structures (representative of each class I subgroup) show the position of conserved motifs in YGLTM5 (blue) and HRFGTM6 (in red). Both motifs are predicted to face each other; in these representations, intra- and extra-cellular loops are omitted for image clarity
Once the typical class A GPCRs motifs are excluded, the hallmarks of class I and class II ORs become quite distinct (Files S3 and S4). Specific conserved motifs can be distinguished in both classes (Fig. 1b, c). As a general rule, TM3 of class A GPCRs is a structural and functional hub [3]. Class I ORs show a highly conserved QM3.29FFxH3.33 motif (superscripts refer to the Ballesteros–Weinstein notation) [28] where the polar methionine and histidine residues are predicted to point towards the inside of the binding cavity of class I ORs [10, 29]. These two conserved and hydrophilic residues (M3.29 and H3.33, respectively, conserved at 67% and 91%, Fig. 1b) are not conserved in class II ORs (where only a conserved QxFFxx signature is found).
In TM5, a YGL5.40 motif is specific to the extra-cellular side of class I ORs and is conserved at more than 60% for each residue, whereas, in class II ORs, the conservation rate at each position drops to less than 40% (Fig. 1b). TM6 also presents notable differences in conservation in the extra-cellular side of the receptors between both classes. A HRFG6.61 motif, where each residue is conserved at about 70%, is specific to class I, but this motif is replaced by a conserved PxSx motif in class II.
Focusing on the middle of TM6, the highly conserved FY6.48XX motif was shown to act as a toggle switch in ORs [6] and is conserved as an FYxP motif in class I ORs. At the N-terminal part of TM6 (RxKAFSTCxSH motif), a highly conserved FS motif in class II ORs is replaced by an LN motif in class I. Note that this motif was shown to be crucial in receptor activation and dynamics [4, 30]. Finally, as previously reported in mice [13], three conserved proline residues in TM7 are present in class I ORs, while only two can be found in class II. TM4 and TM5 are connected by the extra-cellular loop 2 (ECL2). Although this structure surely plays an active role in ligand recognition [31], it may not be specific to class I or class II ORs given its poor conservation amongst mammalian ORs.
The conserved motifs YGLTM5 and HRFGTM6 form a vestibular site
In human, class I ORs are phylogenetically split into three sub-groups, OR51, OR52, and OR56 [12]. YGLTM5 and HRFGTM6 are conserved for the two ORs that are representative of the two first groups (OR51A2 and OR52A1). In the third group (OR56 sub-family comprises 6 ORs), the typical OR56A1 receptor shows slightly different motifs in TM5 and TM6 (YQFTM5 and NLARTM6, see File S1), which, nonetheless, face each other, as well. Above all, this highlights that the structural feature made up of these facing motifs is observed amongst the whole class I OR sub-genome (Fig. 1c). We focused specifically on understanding the functional role of such conserved and class-specific motifs in TM5 and TM6 extra-cellular regions. OR51E1 was considered prototypical, because its sequence shows all the signatures of class I ORs and it has already been thoroughly studied, both in vitro and in silico [18, 29, 32]. The model was built using a previous protocol, where homology modeling was confirmed to fulfill constraints obtained by functional in vitro assays [29]. In total, 141 mutated positions on mammalian ORs were gathered [4], covering 35% of the whole sequence of the receptor. As expected, YGLTM5 and HRFGTM6 form the last extra-cellular helical turn of TM5 and TM6, respectively, and face each other.
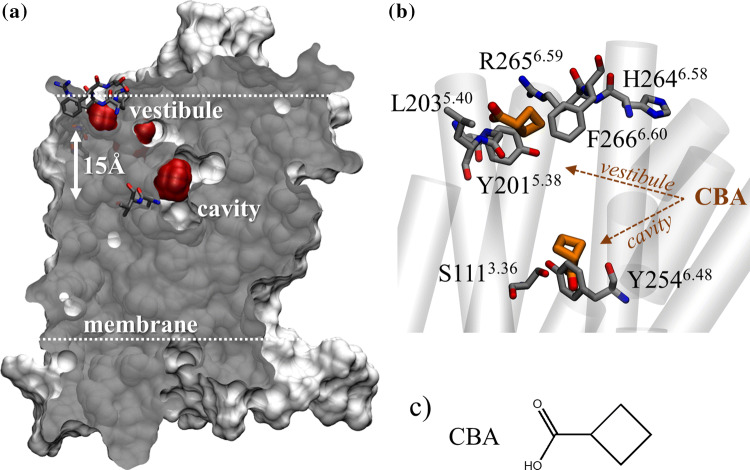
The two motifs are located at the solvent/membrane interface. The vestibule is predicted to be 2.5 helical turns (approximately 15 Å) above the orthosteric-binding pocket (Fig. 2a). Four known agonists of OR51E1, cyclobutanoic acid (CBA), butyric acid (BA), isovaleric acid (IVA), and nonanoic acid (NA) were docked into the orthosteric-binding site and the vestibule. The four acids with varied chemical propertied showed comparable affinities for both sites (Table S1 and Fig. 2b), suggesting that the vestibule is likely to interact with agonists.
Fig. 2.
OR51E1 conserved motifs form a vestibular-binding site visited by ligands. a Cross section of OR51E1 van der Waals volume (gray). Extra-cellular loops were modeled but are omitted for image clarity. A vestibular-binding site and the orthosteric-binding cavity (red) are detected by a cavity detector. Y2546.48 and V2556.49 at the orthosteric cavity and HRFGTM5 and YGLTM6 forming the vestibule are shown in licorice. b Superpositions of typical positions of CBA (shown in brown) at the cradle of the orthosteric-binding cavity (S1113.36 and Y2546.48) and at the vestibular site. c Chemical structure of CBA
At the orthosteric cavity, the agonists established contacts with S1113.36, I2065.44, and H1083.33, as we have previously shown [29]. The ligands locations are consistent with the binding poses predicted by several ligand–OR interaction studies [4, 7–10, 29, 33].
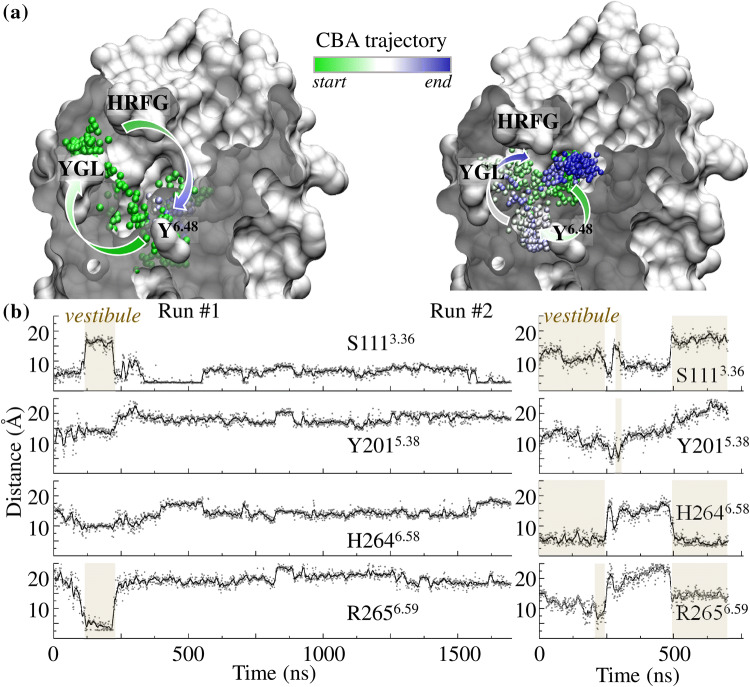
The dynamics of the four systems was investigated through molecular dynamics (MD) simulations. In three independent MD simulations, CBA visited both sites during the same run (Figs. 2, 3, and S1). The same behavior is observed for the three other agonists (Fig. S1), confirming the connection between the vestibular site and the orthosteric cavity.
Fig. 3.
Vestibule is connected to the orthosteric-binding cavity. a Locations of CBA (as shown by a sphere corresponding to its carboxylic atom) during two independent MD trajectories initiated either at the orthosteric cavity (Run #1, left) or at the vestibule (Run #2, right). Green-to-blue colors indicate simulation time evolution. b Distance analysis between a CBA oxygen atom and donor/acceptor groups of S111 (bottom of the cavity), and Y201, H264, and R265 (forming the vestibule). In both simulations, CBA engages opportunistic hydrogen bonds with residues forming the vestibule. Periods during which CBA visits the vestibule are highlighted
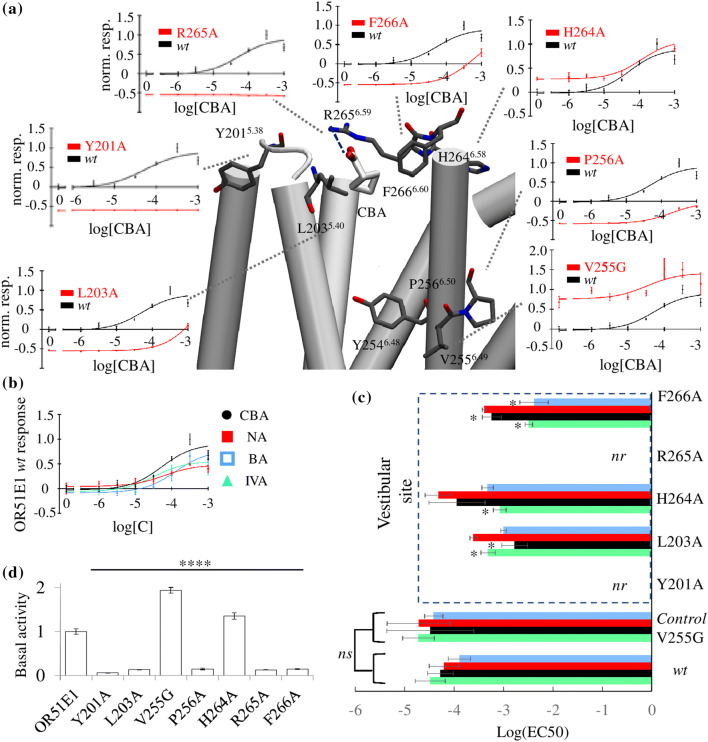
We have previously found that S1113.36 was involved in agonist binding to OR51E1 [29]. When the agonist are hydrogen-bonded with S1113.36, they are close to the conserved motif (FY6.48VP in class I or FY6.48GT in class II ORs) acting as the activation toggle switch of the receptor [6]. Although residues 2556.49 and 2566.50 do not strongly interact with the ligands (Fig. S1), structural perturbations at these positions affect the general ability of the receptor to sense agonists and to activate [5, 6]. The V2556.49G and P2566.50A mutations consistently modified the receptor’s basal activity and decreased its efficacy (Fig. 4a, d).
Fig. 4.
In vitro assessment of the functional role of the vestibule and the binding cavity. a Snapshot where CBA bound at the vestibular site can form a hydrogen bond with R2656.59 (blue dashed line) and hydrophobic contacts with L2035.40 and F2666.60. In this representation, extra-cellular loops are omitted for image clarity. For each residue near the agonist, normalized dose–response curves of the response of OR51E1 mutants (red) in comparison with the wt (black) to CBA stimulation are shown. The y-axis represents the firefly luminescence normalized to Renilla firefly and normalized to the wt response. b Normalized dose–response curves (error bars represent SD; n = 3) for OR51E1 wt to nonanoic acid (NA), isovaleric acid (IVA), and butyric acid (BA). c Log(EC50) values for the four agonists. Comparison between the wt and the mutant ORs at the position V2556.49 and at the vestibular site. nr non-responsive. Mutation V2556.49G, which does not interact with agonists, has no statistically significant (ns) effect on the EC50 for any agonist. Mutations at the vestibular site differentially affect potency. A ‘asterisk’ indicates that the 95% EC50 confidence intervals do not intersect (Table S3). The H2646.58A mutation only affects IVA binding, while the F2666.60A mutation affects all ligands except NA. The L203A5.40 mutation mostly decreases both CBA and IVA potencies. d Normalized basal activity for all mutant ORs normalized to wt. All mutants’ basal activity is significantly different than the wt (****p < 0.001; ANOVA Dunnett’s multiple comparisons test)
In general, mutating the residues identified either at the orthosteric cavity or at the vestibule affected the basal activity (Fig. 4) without apparent impact on the surface expression (Fig. S2). This highlights the role of the vestibular site in the activation mechanism.
During the course of the MD simulations, the agonists formed transitory hydrophilic or hydrophobic contacts with residues belonging to the orthosteric cavity and the vestibule, whatever the starting point of the ligand (Figs. 3, S1). This suggests an active role of these residues in ligand recognition. In the vestibule, transitory hydrogen bonds could be observed between the agonists and R2656.59, H2646.58, or Y2015.38, as illustrated for CBA in Fig. 3.
Mutations at the vestibular site differentially affect agonists’ recognition in vitro
The opportunistic and transitory agonist–vestibule interactions observed during the MD simulations underline the role of the vestibule in agonist recognition. R265A6.59 as well as Y2015.38A mutant ORs were no longer responsive to the agonists in vitro (Fig. 4a), confirming the crucial roles of these residues in ligand-driven receptor activation.
The EC50 confidence intervals for each ligand and each mutant (Tables S2 and S3) are compared in Fig. 4c to examine the changes in potency upon mutation. The V2556.49G mutation has a negligible effect on the potency of all the agonists, consistent with the orientation of V2556.49 towards the membrane. However, different modulations are observed when mutating residues located at the vestibule, namely L2035.40, H2646.58, and F2666.60. All mutant ORs are less responsive to agonists, but each mutation affects each ligand in a different manner. This suggests that the vestibular residues do interact with the ligands. Each residue of the vestibules, thus, plays a role in the ligand recognition process, which naturally depends on the chemical property of the stimulating odorant (Fig. S3 and Table S2).
Discussion: class I odorant receptor sub-genome exhibits a vestibular cavity
Sequence analysis of human class I and class II ORs showed that both classes have distinct signatures in their sequences, as previously observed in mice [13]. Motifs within the ECL3 were considered to play an active role in ligand recognition by class I ORs [17]. Interestingly, we identified two motifs in the extra-cellular domain of class I ORs that face towards each other and are not conserved amongst class II ORs. Site-directed mutagenesis and functional assays determined that these motifs form a functional allosteric extra-cellular binding cavity, 15 Å removed from the canonical ligand cavity of ORs. This structural feature is likely to be observed within the whole class I sub-genome.
Allosteric-binding sites have been identified in many GPCRs from distinct classes [19]. In ORs, notably, some residues located in helix eight were reported to play a functional role in response to ligand binding [34]. In the case of extra-cellular allosteric sites and more specifically in muscarinic acetylcholine receptors, the vestibular site has been extensively studied, since these receptors have strictly conserved binding sites, while the vestibule is an allosteric pocket that acts as a potential subtype selectivity filter [35]. Class I ORs show the opposite mechanism, where the vestibule appears to be highly conserved, while the binding pocket (either in class I or class II) is strongly divergent. Coming back to the vestibular-binding site, class II ORs do not show any conservation at these positions. This could be connected with observed differences in ligand properties within the two classes.
From a sequence point of view, the most comparable vestibular motifs to those reported herein are found in the β2-adrenergic receptor (β2AR) vestibular entry site [22]. In the β2AR, the OR-equivalent motif of YGLTM5-OR is YAITM5-β2AR, while the HRFGTM6-OR equivalent is HVIQTM6-β2AR. These residues, as well as others located in the extra-cellular domain, interact with the agonists in the very first steps of the binding of alprenolol to the receptor [22]. Namely, A2005.39, H2966.58, and V2676.59 interact with the ligand through hydrophobic interaction. An analysis of the structures of class II ORs studied by us previously did not reveal the presence of any vestibular cavity [4, 33, 36]. Hence, the presence of a vestibule in the class I olfactory sub-genome suggests that the ligand-binding mechanism of class I ORs lies between non-olfactory class A GPCRs (where vestibules can be identified) and class II ORs.
Molecular dynamics simulations of OR51E1, a prototypical class I OR, showed that the motifs facing each other in the three-dimensional structure constitute a similar vestibular-binding site as those found in the β2AR. In the simulation, the ligands transiently visited the vestibule highlighting the connection that exists between the vestibule and the orthosteric-binding site. Docking experiments suggested that the vestibular-binding site had a comparable binding affinity for the agonist with respect to the orthosteric-binding cavity. The functionality of the vestibule was assessed through in vitro experiments. Here, we show that mutation of the residues identified as being in contact with the ligand during molecular dynamics simulations strongly affected the response of the receptor. The residues constituting this vestibular pocket play an important role in agonists’ recognition.
Class I receptors have already been shown to bind more hydrophilic compounds than class II ORs [18]. We have revisited and updated all ligands considered to be agonists for any human OR (File S3). A statistical analysis of the agonist chemical properties shows that class I and class II ORs agonists have noticeably different properties (Figs. S4 and S5; Tables S4 and S5) [17, 18, 37]. Typically, class I OR agonists are more hydrophilic than the class II. Counterintuitively, an analysis of the residues lining the orthosteric-binding cavities of class I ORs shows that they are less hydrophilic than those of class II ORs (Fig. S6). The vestibular site could act as a molecular sieve in class I ORs to favor hydrophilic ligands.
Beyond providing insight into the binding process of class I OR ligands, the identification of these TM5 and TM6 conserved motifs lays the groundwork for the rational design of allosteric modulators for the entire olfactory class I OR sub-genome. Such modulators will be of broad interest beyond olfaction, since this class of receptors is expressed not only in the nose. They are found in numerous non-olfactory tissues, although their functional roles and expression levels remain to be established in several cases. More specifically, six class I ORs were shown to be the amongst the 40 most highly expressed ORs outside of the nose. These include OR51E1, OR51E2, OR52N4, OR52B6, OR52D1, and OR51B5 [38]. In addition, 53 class I human ORs transcripts were found to be expressed in polymorphonuclear leukocytes, B and T cells, whereas class II ORs were not detected in these cell types [39]. This wide-spread expression and evidence for important regulatory roles in various diseases makes class I ORs interesting and novel pharmacological targets. The design of ligands with specific interactions with this allosteric cavity would thus be extremely useful.
Materials and methods
Chemical space analysis
To examine the characteristics of the molecules that class I and class II ORs respond to, all the data concerning deorphanization of human ORs were gathered. All references regarding OR deorphanization data and used in this study can be found in the Supplementary Material (File S3). Only molecules eliciting agonist activity on ORs were considered for further analysis: 92 were identified as agonists for class I ORs and 189 as agonists for class II ORs. We calculated 4884 chemical and topological descriptors using the Dragon Software [40] on this set of molecule. We extracted eight descriptors that are of interest due to their pharmacological importance (they relate to the well-known Lipinski’s rule of five) (File S4). The descriptors were normalized and averaged to create the spider plot. An average was identified as significantly different with a one-way ANOVA test, using p < 0.05.
Sequence logos
Multiple sequence alignment was performed using Jalview [41]. 397 sequences of human ORs were gathered and aligned as well as 204 non-olfactory class A GPCR sequences. The alignment was then split into three: 58 sequences were used for generating the class I sequence logo, 339 sequences were used to generate class II sequence logo, and 204 sequences for other class A GPCRs. Phylogenetic trees and secondary structure prediction were performed using the Jalview built-in Web Service application. Logos were generated using WebLogo. Sequence alignments for class I and class II ORs are provided in separate fasta files (Files S1 and S2).
Conservation analysis
Information about the conservation rate of each amino acid making up underlined motifs is given in Fig. 1. These conservation rates were obtained using Jalview [41], and the most conserved residue and its associated value are reported.
Human OR51E1 3D model
The protocol follows a previously published method [10]. Briefly, all 397 human OR sequences were aligned to the sequence of GPCRs for which the experimental structure is known. Manual adjustments were performed to be consistent with the data from the 141 mutants previously described in the literature. A homology model was obtained using the crystal structures of bovine rhodopsin receptor (PDB id: 1U19), CXCR4 chemokine receptor (3ODU), human adenosine A2A receptor (2YDV), and human chemokine CXCR1 receptor (2LNL) as structural templates using Modeler [42]. The N-terminal structure was omitted to avoid perturbing the modeling protocol. Five models were obtained, and the one that was consistent with the in vitro data and several structural constraints [no large folded structure in extra-cellular loops, all trans-membranes helices (TMs) folded as α-helices, and a tiny α-helix 8 at the C-terminal extremity] was kept.
Cyclobutanoic acid (CBA), nonanoic acid (NA), isovaleric acid (IVA), and butyric acid (BA) structures parameters were prepared with the antechamber module of AMBER with AM1-BCC charges. They were docked into the receptor cavity, using flexible docking parameters on residues H1083.33, Y2546.48, and F2576.51 with Autodock Vina for the docking in the orthosteric-binding cavity and Y2015.38, H2646.58, and R2656.59 for docking in the vestibular-binding site [43]. In each site, all docking poses were similar, and we considered the one with the lowest binding free energy for simulations.
Ligand-binding poses considered for the MD simulations are given in Table S6. The cavity volumes were analyzed with MDPocket [44].
Molecular dynamics
The OR51E1 model was embedded into a model membrane. Its orientation in the membrane was determined using the OPM server [45]. The simulation box is made of POPC lipids solvated using TIP3P water molecules in Maestro [46]. The total system is made up of ~ 36,000 atoms in a periodic bow of 74*59*89 Å3. Molecular dynamics simulations were performed with sander and pmemd.cuda modules of AMBER16 with the ff14SB force field for the protein, and the lipid14 for the membrane, and the gaff2 force field for the ligands. Bonds involving hydrogen atoms are constrained using the SHAKE algorithm and long-range electrostatic interactions are handled with Particle Mesh Ewald (PME). The cut-off for non-bonded interactions is set to 8 Å. With CBA, an alternative run (Run #3) with a cut-off set to 10 Å was also run for comparison purposes. Similar findings were obtained, i.e., a visit of both the vestibule and the orthosteric cavity (CBA run #3 behavior is shown in Fig. S1). MD simulations were stopped once the ligand sampled both the orthosteric cavity and the vestibule.
Temperature is kept constant in the system using a Langevin thermostat with a collision frequency of 2 ps−1. In addition, a weak anisotropic algorithm with a relaxation time of 1 ps−1 is applied to keep a constant pressure. Snapshots are saved every 20 ps. The workflow used for energy minimization, thermalization, equilibration, and production of molecular dynamics simulations is detailed in Fig. S7. The total simulation time for this study is 5.6 µs. The RMS deviations of OR51E1 during all molecular dynamics simulations are shown in Fig. S8. The distance analysis was performed considering heteroatoms of each agonist and the closest H-bond donor/acceptor heteroatom of residue S111, Y201, H264, and R265.
Site-directed mutagenesis
The coding sequence of OR51E1 was cloned into a pCI vector (Promega) and tagged at the N-terminal with the 20 first amino acids of rhodopsin. Site-directed mutagenesis was performed by Phusion DNA polymerase (NEB) [29]. The sequence of all plasmids was validated using the BigDye Terminator Sequencing Kit (Applied Biosystem).
Dual-luciferase reporter gene assay
The dual-luciferase reported gene assay was used to evaluate the functionality of wild-type and mutant clones of OR51E1 in an in vitro system [29, 47]. Hana3A cells have been cultured and plated the day before transfection at 1/10 of a 100% confluence 100 mm plate into 96-well plates coated with poly D lysine. After overnight incubation, the required genes were transfected in triplicate using, for each plate, 5 ng SV40-RL, 10 ng CRE-Luc, 5 ng human RTP1S [48], 2.5 ng M3 receptor [49], and 5 ng of receptor (OR51E1 wt or mutant) plasmid. After around 18 h of transfection, cells were stimulated during 3.5 h by 25 µL of odorant diluted in CD293 + glutamine + CuCl2. The luminescence of Firefly (Luc) and Renilla (Rluc) luciferase was then sequentially monitored by injecting the corresponding substrate. The activity in each well was normalized as (Luc-400)/(Rluc-400), and mean and standard deviation were determined for each triplicate. The response of a receptor was also normalized to its basal activity as (NLX/NL0)-1, where NL0 is the normalized luminescence value at 0 µM of odorant and NLX is the value at X µM.
Cell surface expression
Fluorescent-Activated Cell Sorting (FACS) was conducted to evaluate cell surface expression of OR51E1 and mutants. Hana3A were seeded in a 35 mm dish (Corning) in Minimum Essential Medium containing 10% FBS (M10). Lipofectamine2000 (Invitrogen) was used for transfection of plasmid OR and RTP1s DNA. At the time of transfection, green fluorescent protein (GFP) expression vector and RTP1s were co-transfected to monitor and improve the transfection efficiency. About 24 h post-transfection, cells were incubated 30 min with PBS containing anti Rho-tag antibody 4D2 (gift from R. Molday), 15 mM NaN3, and 2% FBS, and then washed and incubated 30 min with phycoerythrin (PE)-conjugated donkey anti-mouse IgG (Jackson Immunologicals). 7-amino-actinomycin D (7-AAD; Calbiochem), a fluorescent, cell-impermeant DNA-binding agent, was added before flow cytometry to eliminate dead cells from analysis as 7-AAD selectively stains dead cells. The intensity of PE signal among the GFP-positive population was measured and plotted to evaluate the OR expression to the plasma membrane.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work is supported by Grants from the National Institute on Deafness and Other Communication Disorders, National Institute of Health Grants DC014423 and DC016224, National Science Foundation (NSF) Grants 1515801 and 1556207 (to H.M.) and from Agence Nationale de la Recherche (Neurolf project to J.G.) as part of NSF/NIH/ANR Collaborative Research in Computational Neuroscience and from the Fondation Roudnitska under the aegis of Fondation de France to C.B. This work is also funded by Université Côte d’Azur IDEX JEDI to J.G. C.B. thanks GIRACT and the GEN foundation for a Ph.D. bursary.
Author contributions
CB: performed numerical modeling, analyzed the data, and wrote the paper. CADM: performed in vitro experiments and analyzed the data. JT: performed numerical modeling and analyzed the data. MD: performed in vitro experiments. HM: conceived the study and analyzed the data. JG: conceived the study, analyzed the data, and wrote the paper.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Caroline Bushdid and Claire A. de March are co-first authors.
Contributor Information
Hiroaki Matsunami, Email: hiroaki.matsunami@duke.edu.
Jérôme Golebiowski, Email: jerome.golebiowski@unice.fr.
References
- 1.Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65(1):175–187. doi: 10.1016/0092-8674(91)90418-X. [DOI] [PubMed] [Google Scholar]
- 2.Niimura Y. Olfactory receptor multigene family in vertebrates: from the viewpoint of evolutionary genomics. Curr Genom. 2012;13(2):103–114. doi: 10.2174/138920212799860706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Venkatakrishnan AJ, Deupi X, Lebon G, Tate CG, Schertler GF, Babu MM. Molecular signatures of G-protein-coupled receptors. Nature. 2013;494(7436):185–194. doi: 10.1038/nature11896. [DOI] [PubMed] [Google Scholar]
- 4.de March CA, Topin J, Bruguera E, Novikov G, Ikegami K, Matsunami H, Golebiowski J. Odorant receptor 7D4 activation dynamics. Angew Chem Int Ed Engl. 2018;57(17):4554–4558. doi: 10.1002/anie.201713065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu Y, de March CA, Ni MJ, Adipietro KA, Golebiowski J, Matsunami H, Ma M. Responsiveness of G protein-coupled odorant receptors is partially attributed to the activation mechanism. Proc Natl Acad Sci USA. 2015;112(48):14966–14971. doi: 10.1073/pnas.1517510112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de March CA, Yu Y, Ni MJ, Adipietro KA, Matsunami H, Ma M, Golebiowski J. Conserved residues control activation of mammalian G protein-coupled odorant receptors. J Am Chem Soc. 2015;137(26):8611–8616. doi: 10.1021/jacs.5b04659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geithe C, Protze J, Kreuchwig F, Krause G, Krautwurst D. Structural determinants of a conserved enantiomer-selective carvone binding pocket in the human odorant receptor OR1A1. Cell Mol Life Sci CMLS. 2017;74(22):4209–4229. doi: 10.1007/s00018-017-2576-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahmed L, Zhang Y, Block E, Buehl M, Corr MJ, Cormanich RA, Gundala S, Matsunami H, O’Hagan D, Ozbil M, Pan Y, Sekharan S, Ten N, Wang M, Yang M, Zhang Q, Zhang R, Batista VS, Zhuang H. Molecular mechanism of activation of human musk receptors OR5AN1 and OR1A1 by (R)-muscone and diverse other musk-smelling compounds. Proc Natl Acad Sci USA. 2018;115(17):E3950–E3958. doi: 10.1073/pnas.1713026115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gelis L, Wolf S, Hatt H, Neuhaus EM, Gerwert K. Prediction of a ligand-binding niche within a human olfactory receptor by combining site-directed mutagenesis with dynamic homology modeling. Angew Chem Int Ed Engl. 2012;51(5):1274–1278. doi: 10.1002/anie.201103980. [DOI] [PubMed] [Google Scholar]
- 10.de March CA, Kim SK, Antonczak S, Goddard WA, 3rd, Golebiowski J. G protein-coupled odorant receptors: from sequence to structure. Protein Sci Publ Protein Soc. 2015;24(9):1543–1548. doi: 10.1002/pro.2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freitag J, Krieger J, Strotmann J, Breer H. Two classes of olfactory receptors in Xenopus laevis. Neuron. 1995;15(6):1383–1392. doi: 10.1016/0896-6273(95)90016-0. [DOI] [PubMed] [Google Scholar]
- 12.Glusman G, Bahar A, Sharon D, Pilpel Y, White J, Lancet D. The olfactory receptor gene superfamily: data mining, classification, and nomenclature. Mamm Genome Off J Int Mamm Genome Soc. 2000;11(11):1016–1023. doi: 10.1007/s003350010196. [DOI] [PubMed] [Google Scholar]
- 13.Zhang X, Firestein S. The olfactory receptor gene superfamily of the mouse. Nat Neurosci. 2002;5(2):124–133. doi: 10.1038/nn800. [DOI] [PubMed] [Google Scholar]
- 14.Niimura Y, Nei M. Evolutionary dynamics of olfactory receptor genes in fishes and tetrapods. Proc Natl Acad Sci USA. 2005;102(17):6039–6044. doi: 10.1073/pnas.0501922102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Niimura Y, Nei M. Evolution of olfactory receptor genes in the human genome. Proc Natl Acad Sci USA. 2003;100(21):12235–12240. doi: 10.1073/pnas.1635157100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iwata T, Niimura Y, Kobayashi C, Shirakawa D, Suzuki H, Enomoto T, Touhara K, Yoshihara Y, Hirota J. A long-range cis-regulatory element for class I odorant receptor genes. Nat Commun. 2017;8(1):885. doi: 10.1038/s41467-017-00870-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Freitag J, Ludwig G, Andreini I, Rossler P, Breer H. Olfactory receptors in aquatic and terrestrial vertebrates. J Comp Physiol A. 1998;183(5):635–650. doi: 10.1007/s003590050287. [DOI] [PubMed] [Google Scholar]
- 18.Saito H, Chi Q, Zhuang H, Matsunami H, Mainland JD. Odor coding by a mammalian receptor repertoire. Sci Signal. 2009;2(60):ra9. doi: 10.1126/scisignal.2000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thal DM, Glukhova A, Sexton PM, Christopoulos A. Structural insights into G-protein-coupled receptor allostery. Nature. 2018;559(7712):45–53. doi: 10.1038/s41586-018-0259-z. [DOI] [PubMed] [Google Scholar]
- 20.Yuan X, Raniolo S, Limongelli V, Xu Y. The molecular mechanism underlying ligand binding to the membrane-embedded site of a G-protein-coupled receptor. J Chem Theory Comput. 2018;14(5):2761–2770. doi: 10.1021/acs.jctc.8b00046. [DOI] [PubMed] [Google Scholar]
- 21.Stanley N, Pardo L, Fabritiis GD. The pathway of ligand entry from the membrane bilayer to a lipid G protein-coupled receptor. Sci Rep. 2016;6:22639. doi: 10.1038/srep22639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dror RO, Pan AC, Arlow DH, Borhani DW, Maragakis P, Shan Y, Xu H, Shaw DE. Pathway and mechanism of drug binding to G-protein-coupled receptors. Proc Natl Acad Sci USA. 2011;108(32):13118–13123. doi: 10.1073/pnas.1104614108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kruse AC, Hu J, Pan AC, Arlow DH, Rosenbaum DM, Rosemond E, Green HF, Liu T, Chae PS, Dror RO, Shaw DE, Weis WI, Wess J, Kobilka BK. Structure and dynamics of the M3 muscarinic acetylcholine receptor. Nature. 2012;482(7386):552–556. doi: 10.1038/nature10867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Provasi D, Bortolato A, Filizola M. Exploring molecular mechanisms of ligand recognition by opioid receptors with metadynamics. Biochemistry. 2009;48(42):10020–10029. doi: 10.1021/bi901494n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sandal M, Behrens M, Brockhoff A, Musiani F, Giorgetti A, Carloni P, Meyerhof W. Evidence for a transient additional ligand binding site in the TAS2R46 bitter taste receptor. J Chem Theory Comput. 2015;11(9):4439–4449. doi: 10.1021/acs.jctc.5b00472. [DOI] [PubMed] [Google Scholar]
- 26.Sharma K, Balfanz S, Baumann A, Korsching S. Full rescue of an inactive olfactory receptor mutant by elimination of an allosteric ligand-gating site. Sci Rep. 2018;8(1):9631. doi: 10.1038/s41598-018-27790-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cvicek V, Goddard WA, 3rd, Abrol R. Structure-based sequence alignment of the transmembrane domains of all human GPCRs: phylogenetic, structural and functional implications. PLoS Comput Biol. 2016;12(3):e1004805. doi: 10.1371/journal.pcbi.1004805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ballesteros JA, Weinstein H. Integrated methods for the construction of three-dimensional models and computational probing of structure–function relations in G protein-coupled receptors. Methods Neurosci. 1995;25:366–428. doi: 10.1016/s1043-9471(05)80049-7. [DOI] [Google Scholar]
- 29.Bushdid C, de March CA, Fiorucci S, Matsunami H, Golebiowski J. Agonists of G-protein-coupled odorant receptors are predicted from chemical features. J Phys Chem Lett. 2018;9(9):2235–2240. doi: 10.1021/acs.jpclett.8b00633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Katada S, Hirokawa T, Oka Y, Suwa M, Touhara K. Structural basis for a broad but selective ligand spectrum of a mouse olfactory receptor: mapping the odorant-binding site. J Neurosci Off J Soc Neurosci. 2005;25(7):1806–1815. doi: 10.1523/JNEUROSCI.4723-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wheatley M, Wootten D, Conner MT, Simms J, Kendrick R, Logan RT, Poyner DR, Barwell J. Lifting the lid on GPCRs: the role of extracellular loops. Br J Pharmacol. 2012;165(6):1688–1703. doi: 10.1111/j.1476-5381.2011.01629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Audouze K, Tromelin A, Le Bon AM, Belloir C, Petersen RK, Kristiansen K, Brunak S, Taboureau O. Identification of odorant-receptor interactions by global mapping of the human odorome. PLoS One. 2014;9(4):e93037. doi: 10.1371/journal.pone.0093037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Charlier L, Topin J, Ronin C, Kim SK, Goddard WA, 3rd, Efremov R, Golebiowski J. How broadly tuned olfactory receptors equally recognize their agonists. Human OR1G1 as a test case. Cell Mol Life Sci CMLS. 2012;69(24):4205–4213. doi: 10.1007/s00018-012-1116-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sato T, Kawasaki T, Mine S, Matsumura H. Functional role of the C-terminal amphipathic helix 8 of olfactory receptors and other G protein-coupled receptors. Int J Mol Sci. 2016 doi: 10.3390/ijms17111930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Granier S, Kobilka B. A new era of GPCR structural and chemical biology. Nat Chem Biol. 2012;8(8):670–673. doi: 10.1038/nchembio.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Topin J, de March CA, Charlier L, Ronin C, Antonczak S, Golebiowski J. Discrimination between olfactory receptor agonists and non-agonists. Chemistry. 2014;20(33):10227–10230. doi: 10.1002/chem.201402486. [DOI] [PubMed] [Google Scholar]
- 37.Dunkel A, Steinhaus M, Kotthoff M, Nowak B, Krautwurst D, Schieberle P, Hofmann T. Nature’s chemical signatures in human olfaction: a foodborne perspective for future biotechnology. Angew Chem Int Ed Engl. 2014;53(28):7124–7143. doi: 10.1002/anie.201309508. [DOI] [PubMed] [Google Scholar]
- 38.Massberg D, Hatt H. Human olfactory receptors: novel cellular functions outside of the nose. Physiol Rev. 2018;98(3):1739–1763. doi: 10.1152/physrev.00013.2017. [DOI] [PubMed] [Google Scholar]
- 39.Malki A, Fiedler J, Fricke K, Ballweg I, Pfaffl MW, Krautwurst D. Class I odorant receptors, TAS1R and TAS2R taste receptors, are markers for subpopulations of circulating leukocytes. J Leukoc Biol. 2015;97(3):533–545. doi: 10.1189/jlb.2A0714-331RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dragon TS (2014) Software for Molecular descriptor calculation. 6.0, Talete Srl editor. http://www.talete.mi.it
- 41.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25(9):1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, Pieper U, Sali A. Comparative protein structure modeling using modeller. Curr Protoc Bioinform. 2006 doi: 10.1002/0471250953.bi0506s15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmidtke P, Bidon-Chanal A, Luque FJ, Barril X. MDpocket: open-source cavity detection and characterization on molecular dynamics trajectories. Bioinformatics. 2011;27(23):3276–3285. doi: 10.1093/bioinformatics/btr550. [DOI] [PubMed] [Google Scholar]
- 45.Lomize MA, Pogozheva ID, Joo H, Mosberg HI, Lomize AL. OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res. 2012;40(Database issue):D370–D376. doi: 10.1093/nar/gkr703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maestro (2018) Schrödinger Release 2018-1: Maestro S, LLC, New York. https://www.schrodinger.com
- 47.Zhuang H, Matsunami H. Evaluating cell-surface expression and measuring activation of mammalian odorant receptors in heterologous cells. Nat Protoc. 2008;3(9):1402–1413. doi: 10.1038/nprot.2008.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhuang H, Matsunami H. Synergism of accessory factors in functional expression of mammalian odorant receptors. J Biol Chem. 2007;282(20):15284–15293. doi: 10.1074/jbc.M700386200. [DOI] [PubMed] [Google Scholar]
- 49.Li YR, Matsunami H. Activation state of the M3 muscarinic acetylcholine receptor modulates mammalian odorant receptor signaling. Sci Signal. 2011;4(155):ra1. doi: 10.1126/scisignal.2001230. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.