Abstract
Resolvins D1 and D2 (RvDs) are structural isomers and metabolites of docosahexaenoic acid, an omega-3 fatty acid, enzymatically produced in our body in response to acute inflammation or microbial invasion. Resolvins have been shown to play an essential role in the resolution of inflammation, tissue repair, and return to homeostasis, and thus are actively pursued as potential therapeutics in treating inflammatory disorders and infectious diseases. However, effective in vivo delivery of RvDs continues to be a challenging task. Recent studies demonstrated that RvD1 or RvD2 loaded in cell membrane-derived nanovesicles significantly increased therapeutic efficacy in treating murine peritonitis and ischemic stroke, respectively. The mechanistic details of how the subtle structural difference between RvD1 and RvD2 alters their molecular interactions with the membrane lipids of the nanovesicles, and thus affects the loading efficiency, remain unknown. Here we report the encapsulation profiles of the neutral and ionized species of both RvD1 and RvD2 determined with the cell membrane-derived nanovesicles at pH values 5.4 and 7.4, respectively. Also, we performed microsecond time-scale all-atom molecular dynamics (MD) simulations in explicit water to elucidate the molecular interactions of both neutral and ionized species of RvD1 and RvD2 with the lipid bilayer using a model membrane system, containing DMPC (1,2-dimyristoyl-sn-glycero-3-phosphocholine) and cholesterol. We found that the differences in the position and chirality of hydroxyl groups in RvD1 and RvD2 affected their location, orientation, and conformations within the bilayer. Surprisingly, the deprotonation of their carboxyl group caused their orientation and conformation to change from a fully extended one that is oriented in parallel to the membrane plane to a J-shaped bent conformation that is oriented perpendicular to the bilayer plane. Our studies offer valuable insight into the molecular interactions of RvD1/D2 with the lipid bilayer in atomistic details and provide a mechanistic explanation for the observed differences in the encapsulation profiles of RvD1 and RvD2, which may facilitate the rational design of nanovesicle-based therapeutics for treating inflammatory diseases.
Keywords: Resolvins, cell membrane-derived nanovesicles, membrane partitioning, molecular dynamics simulation, drug delivery, nanotechnology
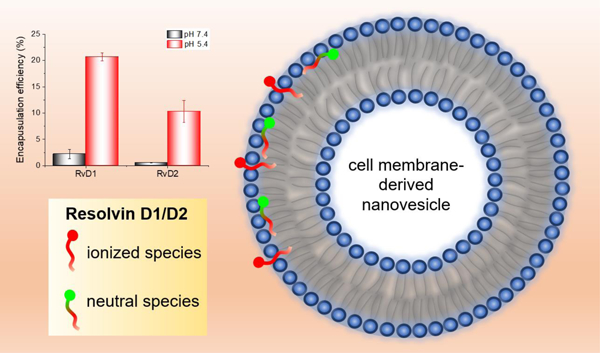
Graphical Abstract
INTRODUCTION
Resolvins belong to the family of specialized pro-resolving lipid mediators (SPMs) that are enzymatically produced from the polyunsaturated fatty acids (PUFAs) such as omega-3 fatty acids in the immune (neutrophils and macrophages) and nonimmune cells (vascular endothelial and smooth muscle cells) in humans1–4. The D- and E-series of resolvins are generated from docosahexaenoic acid and eicosapentaenoic acid, respectively. A growing body of evidence suggests that these SPMs (which also include protectins and maresins) are produced during acute inflammation in response to pathogen invasion or tissue injury and are actively involved in orchestrating key cellular events driving resolution. Resolution of inflammation comprises the ending of neutrophil recruitment, counter-regulation of pro-inflammatory mediators, neutrophil apoptosis, clearance through macrophage phagocytosis (efferocytosis), tissue remodeling, and a return to homeostasis5, 6. Normal homeostasis is maintained by a balance between the pro-inflammatory and pro-resolving pathways. It is increasingly evident that failure to resolving inflammation due to insufficient levels of the SPMs plays an essential role in the pathogenesis and progression of chronic diseases such as arthritis7, asthma, atherosclerosis8–10, diabetes, aging-associated diseases including Alzheimer’s disease11 and many neurological disorders.
Resolvins and other SPMs are potent endogenous regulators of excessive inflammatory responses. They carry out their pro-resolving functions by directly binding to specific GPCRs with high affinity and stereoselectivity. The D-series (1–6) resolvins include the well-studied RvD1 and RvD2, which elicit their resolving effects by signaling through G protein-coupled receptors (GPCRs) FPR2/ALXR, DRV1/GPR32, and DRV2/GPR18, respectively12–14. The E-series (1–4) resolvins include RvE1 and RvE2, which bind to two G protein-coupled receptors, namely ERV1/ChemR23 and BLT115, 16.
RvD1 curbed polymorphonuclear leukocytes recruitment during acute inflammation and produced a significant reduction in the levels of several pro-inflammatory mediators by signaling through FPR2/ALX receptor17. RvD1 has been shown to decrease bone resorption and bone loss in murine arthritis model18. RvD1treatment of mice that were exposed to cigarette smoke for a long-term reduced cigarette smoke-induced emphysema and associated lung inflammation, oxidative stress, and cell death19–21. Recently, several small-molecule RvD1 mimetics have been identified and shown to produce pro-resolving actions through activating DRV1/GPR32 receptor22. Other attempts targeting FPR2/ALXR for the resolution of inflammation, by research groups from both academia and industry have resulted in multiple small-molecules that are currently undergoing clinical trials23, 24.
Resolvin D2 is a potent regulator of leukocytes and controls microbial sepsis25–27. During bacterial infections, RvD2 limited neutrophil infiltration, enhanced phagocyte clearance of bacteria and accelerated resolution14 through stereoselective binding to GPR18. RvD2-stimulated phagocytosis of E. coli and efferocytosis were increased with GPR18 overexpression and significantly reduced by shRNA knockdown of GPR18. A recent study28 revealed that RvD2 increases cellular cAMP to increase intracellular Ca2+ level and stimulates mucin secretions from conjunctival goblet cells. RvD2 may thus aid in the maintenance of the mucous layer of the tear film in preserving ocular surface homeostasis, and potentially be used for the treatment of dry eye disease29.
In adipose tissues, both RvD1 and RvD2 act as potent pro-resolving mediators that counteract local adipokine production and monocyte accumulation in obesity-induced adipose inflammation30. RvD2 also reduced hypothalamic inflammation and rescued mice from diet-induced obesity31. Altogether, these studies suggest that supplementation of resolvins in diseases associated with chronic inflammation resulting from disruption of pro-resolving signaling and metabolic pathways may be therapeutically beneficial to reduce inflammation and progression for diseases of acute and chronic inflammation. However, effective delivery of resolvins in vivo continues to be challenging. As polyunsaturated fatty acids, resolvins are chemically unstable molecules that undergo rapid metabolic inactivation by ubiquitous enzymes such as eicosanoid oxidoreductase in vivo and thus possess short half-lives32–35. In addition, they suffer from poor physicochemical properties such as low aqueous solubility, light sensitivity, and significant degradation at room temperature. Furthermore, some of the resolvins have been shown to be substrates for efflux transporters, which may limit intracellular permeability, causing potential therapeutic failure36,37.
Nanotechnology has been utilized in pharmaceutical industries for several decades as it can significantly improve the bio-availability of administered drugs compared to conventional approaches38. For example, liposome or polymer-based drug delivery systems are successfully used to treat cancers and inflammatory disorders39–43. Due to the challenges associated with synthetic nanoparticle-based drug delivery systems, there are considerable endeavors to harness the bioinspired design of nanoparticles from nature44–46. Cells regularly release cell membrane compartments, called extracellular vesicles (EVs) with the nanoscale dimension, for cell signaling and maintenance of homeostasis47. These vesicles are excellent drug delivery systems due to their exceptional biocompatibility, intrinsic tissue selection, and long circulation times. We have recently developed a nitrogen cavitation technology to efficiently generate cell-membrane nanovesicles similar to EVs from any cell types and demonstrated their potential for translational applications48, 49. We showed that nanovesicles generated from human neutrophils and loaded with an anti-inflammatory drug could specifically target inflamed vasculature and offer increased therapeutic efficacy in acute lung injury and sepsis50.
Also, our recent studies demonstrated that RvD2 loaded into neutrophil membrane-derived nanovesicles can enhance the resolution of inflammation caused by an ischemic stroke in the mouse model, and protect the brain from perfusion surgery during stroke treatment51. In addition, it has been shown that neutrophil-derived nanovesicles containing RvD1 reduced inflammation response significantly in murine peritonitis, and promoted quick wound healing52. These studies collectively demonstrate that cell membrane-derived nanovesicles are efficient vehicles for the in vivo delivery of resolvins. To effectively utilize these nanovesicles for optimal delivery of resolvins, it is important to understand their molecular interactions with constituent lipids of the vesicles, affecting their encapsulation efficiency as well as their release into the target site after administration.
Here we report the encapsulation efficiencies of RvD1 and RvD2 loaded into the neutrophil cell membrane-derived nanovesicles, measured at pH 5.4 and 7.5. Also, we performed microsecond time-scale molecular dynamics (MD) simulations to investigate the molecular interactions of both neutral and ionized species of RvD1 and RvD2 and determined their energetically favorable locations, preferred orientations, and conformations within a model membrane made up of 1,2-dimyristoyl-sn-3-phosphocholine (DMPC) to closely mimic the lipid bilayer of nanovesicles. We used steered MD and Umbrella Sampling simulations53 to determine the free energies of transfer from water to the membrane, partitioning within the membrane, and crossing across the membrane of RvD1 and RvD2. The results from our MD simulations offer useful insight into the experimentally observed encapsulation profiles of RvD1 and D2 in the nanovesicles, assisting in the design and optimization of nanovesicle delivery systems for enhanced delivery, desired pharmacokinetics, and better efficacy of resolvins in treating inflammatory conditions.
EXPERIMENTAL SECTION
MATERIALS AND METHODS
Preparation and Characterization of Membrane-derived Nanovesicles.
Human HL-60 cells (human neutrophil-like cells) were cultured in RPMI1640 medium supplemented with 10% (v/v) FBS and 1% (v/v) pen strep/glutamine. To differentiate HL-60 cells to be neutrophil-like cells, 1.25% (v/v) DMSO was added in the medium, and cells were cultured for 4–6 days. Differentiated HL-60 cells were re-suspended in HBSS, and then nitrogen cavitation at a pressure of 350–400 psi was used to disrupt cells. The resulting suspension was centrifuged at 2,000 g for 30 min to remove nuclei. The supernatant was centrifuged at 100,000 g for 30 min to obtain nanovesicles, and this process was repeated twice. Nanovesicles were lyophilized and stored at −20°C for future use. Nanovesicles were imaged using cryo-electron microscopy (cryo-EM), and the image showed a shell structure with a size of 200 nm in diameter, and the wall thickness was 3–4 nm, which was equivalent to that of the cell membrane48.
The neutrophil membrane-derived nanovesicle suspension was heated to 37°C, and the pH was adjusted to 5.4 or 7.4. RvD1 and RvD2 (purchased from Cayman Chemical, Ann Arbor, MI) were quickly mixed with nanovesicles at a ratio of 2 μg (RvD1 or RvD2)/ml, and 200 μg/mL of nanovesicles followed by sonication for 2 min on ice, and then the suspension was incubated for 30 min at 37 °C. The suspension was centrifuged at 100,000 g to remove free RvD1 or RvD2. Using a Waters ACQUITY UPLC System with Xevo G2-XS QT mass spectroscopy, we determined the amount of RvD1 and RvD2 after they were extracted from the nanovesicles using ethanol. A C18 column (100×2.1 mm) was used at 50°C. There are two solvents used in HPLC measurements: 1) liquid with ammonium formate (5mM) and formic acid (2mM) and 2) acetonitrile. We established the acetonitrile gradient from 35% to 100% before we measured RvD1 or RvD2. UV absorption at 301nm was monitored for RvD1 or RvD2. The mass spectroscopy confirmed RvD1 or RvD2 detection. Finally, the encapsulation efficiencies of RvD1 or RvD2 were calculated as described elsewhere51, 54.
MD simulation protocol.
We employed the following protocol comprising steered molecular dynamics (SMD) and Umbrella Sampling simulations to investigate the membrane partitioning characteristics of both neutral and charged species of RvD1 and RvD2. The 3D structures of the resolvin molecules were built, geometry optimized, and energy minimized in Molecular Operating Environment 55(MOE, Chemical Computing Group). The membrane builder56, 57 module in CHARMM-GUI58 (http://www.charmm-gui.org) was used to generate four model bilayer systems, each containing 64 DMPC lipid molecules per leaflet. The bilayer lipids were solvated on both sides with a 20Å layer of water molecules, and the ionic concentration of the simulated systems was kept to physiologically relevant 0.15 M level by adding enough K+ and Cl− ions. The CHARMM3659 force field parameters and TIP3P water model were used to model lipids and water molecules and their interactions with ions, respectively. The parameters for resolvins were obtained from CGenFF60 force field using Paramchem server61, 62. In each simulation, the studied resolvin molecule was placed in water at 30 Å from the center of the bilayer. The assembled bilayer/resolvin system was subjected to multistage minimization and equilibration protocol (Supplementary Table S2), as described elsewhere63.
Steered MD 64, 65 simulations were used to pull the resolvin molecules from its starting position to the bilayer center along the bilayer normal (z-axis) at a speed of 1 Å per ns with a 1 fs time step. During the pulling simulations, the center of mass (COM) of the resolvin molecule was harmonically restrained with a force constant of 1.5 kcal/mol/Å2. The coordinates of the system with the resolvin molecule located at every 1 Å window over the permeation pathway (a total of 30 windows) were extracted from the simulation trajectories and used as starting structures for subsequent umbrella sampling simulations. These coordinates were equilibrated for an additional 10 ns and umbrella sampling simulations were performed on each window for 50 ns resulting in a total simulation time of ~ 2µs. The z-component of the distance between the center-of-mass of the lipid atoms and the heavy atoms of the resolvin molecules were defined as reaction coordinates. The Colvars module in NAMD was used with a biasing harmonic constraint of 1.5 kcal/mol/Å2. The resulting biased probability distributions were reweighted using the weighted histogram analysis method (WHAM) 66, 67 to obtain the unbiased PMF, providing the free energy of solvation for the studied resolvin molecules.
All MD simulations were performed using the GPU version of NAMD 2.1268 with periodic boundary conditions. The particle mesh Ewald (PME)69 method was used to model long-range electrostatics. The van der Waals interactions were smoothly switched off by force-switching functions70 at 10–12 Å. The covalent bonds involving hydrogen atoms were constrained using the SHAKE algorithm71. The 1fs/step and 2fs/step time steps were used in equilibration and production runs, respectively. The pressure was maintained using a Nose-Hoover Langevin-piston method72 with a piston period of 50 fs and decay of 25 fs. The temperature was controlled at 310.15K using Langevin temperature73 coupling with a friction coefficient of 1 ps−1.
Trajectory analyses
Hydrogen-bond interactions.
Hydrogen bonds between the studied resolvins and the bilayer lipids were monitored only for the section of the trajectory (total 250 ns for each solute) characterizing low energy (well) locations revealed by the PMF curves (Fig. 1). For example, in the case of neutral species of resolvin D1, the low energy well is at |zmin| ≈ 10 Å. Accordingly, the H-bond calculations were performed using trajectories obtained for |z| values 8 Å through 12 Å. The polar carboxyl and three hydroxyl groups of resolvins were monitored for potential H-bonds to phosphate and glyceryl oxygens of the lipid molecules. The H-bond interactions of resolvins with water molecules in the aqueous bulk phase were also calculated. The Hbonds plugin (version 1.2) in VMD software74 was used for the calculation with the following criteria for the formation of an H-bond: the distance between the donor (D) atom and the acceptor (A) atom is less than the cut-off distance of 3.5 Å and the angle D-H-A is less than the cut-off angle 40 degrees.
Fig. 1. Encapsulation efficiency and lipid bilayer partitioning characteristics of resolvins D1 and D2.
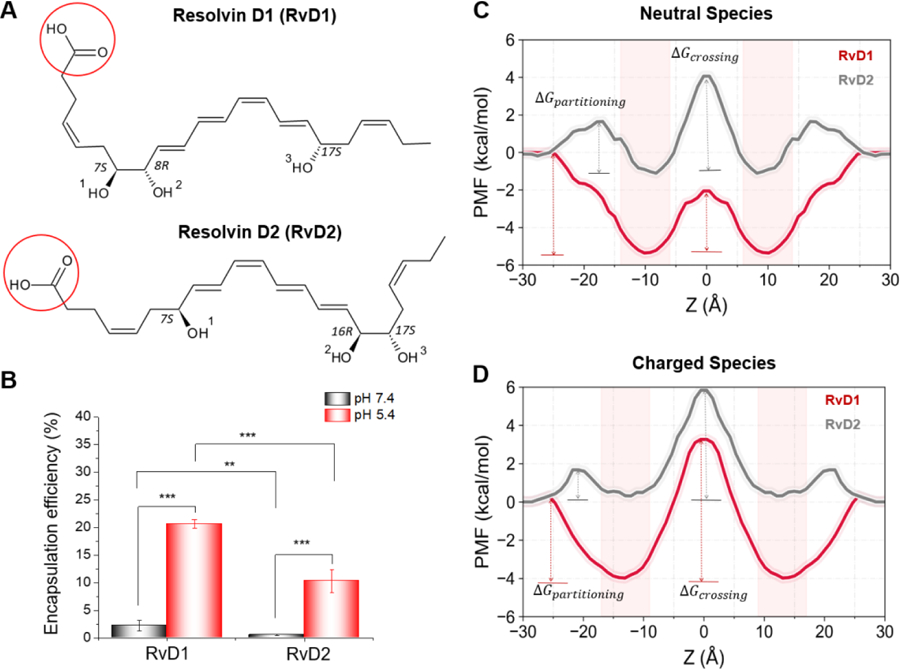
A) 2D structures of neutral species of the studied resolvins D1 and D2 are depicted; ionizable carboxyl (−COOH) groups are circled in red. B) Encapsulation efficiencies of RvD1 and RvD2 determined at pH 7.4, and 5.4 reveal preferable membrane interactions of RvD1 over RvD2; Neutral species have higher encapsulation efficiency than the charged species. C and D) Potential of mean force (PMF) curves describe the differences in the free energies of partitioning and crossing of bilayers for neutral and charged species of RvD1 and RvD2 as well as their energetically favorable time-average positions (highlighted by light-red bands). The free energy profiles were calculated for one bilayer leaflet and were symmetrized to the other side. Statistical analysis was performed using Student’s t-test, and a value of p <0.05 was considered statistically significant (** and *** represent p<0.01 and p<0.001, respectively; data are mean ± SD; n =3).
The number of solute atom contacts with lipid functional groups.
To characterize the major interactions between the studied resolvins and the lipid functional groups as well as to determine molecular orientations of the solutes, the number of contacts between heavy atoms of resolvins, and choline, phosphate, glyceryl, and alkyl groups of the lipids was monitored (Fig. 4A). As described above in H-bond interactions, only trajectories representing low-energy locations in PMF curves were considered for each solute. The heavy atoms of solutes were considered in contact with the lipid groups if the distance between them was less than or equal to 4 Å. Also, the percentage (%) occupancy describing the fraction of simulation time (as a fraction of snapshots) during which the contacts observed (distance cut-off of 4 Å) were calculated between the lipid groups and carboxyl and hydroxyl groups of the solutes. Calculations were performed using an in-house tcl script.
Fig. 4. The molecular interactions of the neutral and charged species of RvD1 and RvD2 with the various functional groups of the DMPC lipids.
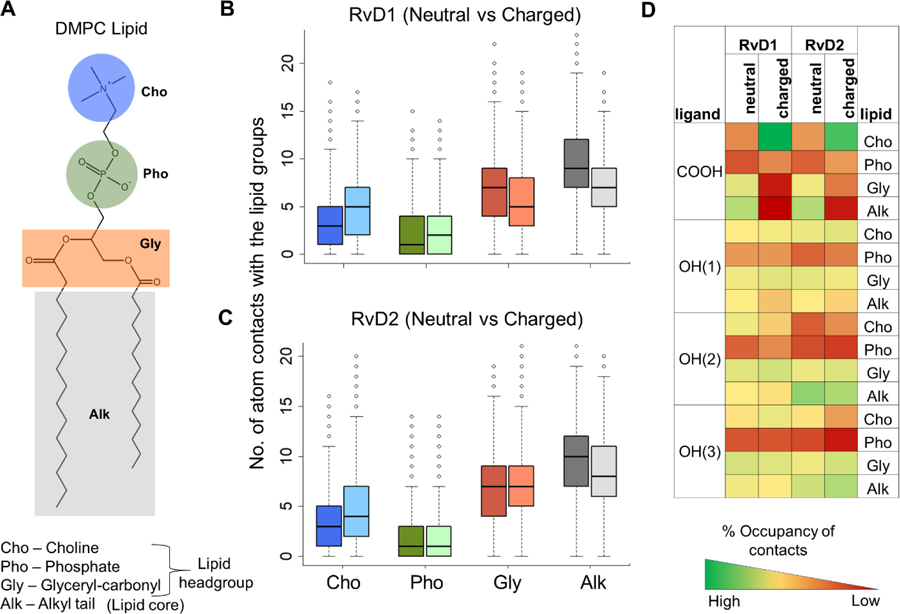
A) A schematic 2D representation of DMPC is shown with its polar head groups choline, phosphate, and glyceryl carbonyls, and nonpolar alkyl tails shaded in blue, green, orange, and gray colors, respectively. B and C) The number of atom contacts of the neutral and charged species of RvD1 and RvD2 with the various functional groups of the DMPC lipids (neutral and charged species are depicted in dark and light colors, respectively). D) The percentage (%) occupancy gives the fraction of simulation time (as a fraction of snapshots) during which the atom contacts were observed between the lipid groups and carboxyl and hydroxyl groups of the solutes. The functional groups were considered in contact if the distance between them was less than or equal to 4 Å.
Resolvins’ orientations with respect to the bilayer normal.
Resolvins are long (spanning up to ~20 Å in length) and highly flexible (14 rotatable bonds) molecules. Most importantly, the ionization state of resolvins’ carboxyl group seems to affect their bilayer orientations and conformations significantly. To characterize resolvins’ orientations within the membrane, we calculated the angle made by the longitudinal vector of the solute with the bilayer normal vector (z-axis) for each solute at their energetically favorable locations (|z|energy-minimum ± 2 Å, as described above for H-bond calculations). For each resolvin molecule, a straight line connecting the center-of-mass of the end carboxyl group at one end (head) and another carbon atom connected to its last -OH group at the other end (origin) represents the longitudinal vector. Calculations were performed using an in-house tcl script.
RESULTS AND DISCUSSION
RvD1 exhibits higher encapsulation efficiency than RvD2.
The nanovesicles were generated from HL-60 cells (human neutrophil-like cells) using nitrogen cavitation, and their sizes were ~200 nm in diameter. RvD1 and RvD2 were loaded in neutrophil-membrane-derived nanovesicles at pH values 5.4 or 7.4 (Fig. 1B). As both RvD1 and RvD2 contain an ionizable carboxylic acid group (pKa of 4.9 ± 0.4, predicted using ACD/Percepta), the pH of the medium dictates the fractions of ionized and neutral species, which in turn significantly affected their interactions with the vesicles and thus their encapsulation efficiencies. The molecules would exist mainly as neutral and ionized species at pH values 5.4 and 7.4, respectively, and they showed significant differences in their encapsulation profiles in these pH values. As expected, both RvD1 and RvD2 showed higher encapsulation efficiencies at pH 5.4, indicating a more favorable membrane partitioning profile for neutral species than the ionized species. Interestingly, at pH 5.4, RvD1 exhibited significantly higher loading efficiency than RvD2. A subtle difference in the positions of hydroxyl groups between RvD1 and RvD2 (Fig. 1) resulted in a significant difference in their encapsulation efficiencies, which are possibly attributed to the differences in molecular interactions affecting their partitioning profiles within the bilayer of nanovesicles. These experimental results are consistent with the free energy profiles and molecular interactions of both neutral and ionized species of RvD1 and RvD2 with the model membrane obtained from MD simulations, as presented below.
RvD1 has a more favorable membrane solvation free-energy profile than RvD2 in the DMPC bilayer.
Molecular dynamics (MD) simulations are efficient techniques to obtain the time-averaged equilibrium position of a solute in a lipid bilayer. The potential of mean force (PMF) curve provides the free energy (ΔG) profile of a solute along the bilayer normal (z-axis) describing its free energy of partitioning from water to membrane, time-average position(s) within membrane regions characterized by energy minima, and resistance to permeation characterized by energy maximum. To the best of our knowledge, the precise positioning (bilayer depth) of RvD1 and RvD2 in lipid bilayers have not yet been determined experimentally. The position-dependent free energy profiles (PMF curves in Fig. 1C and D) obtained from the Umbrella Sampling simulations provide valuable insights to free energies of transfer from water, partitioning within, and crossing across the bilayer of the neutral and charged species of both RvD1 and RvD2 (Fig. 1A and Supplementary Table S1).
Both neutral and charged species of RvD1 show favorable (negative) free energies of transfer from water to the membrane. As the molecules partitioned, the ΔGpartitioning values decreased with the center-of-mass (COM) of the molecules moving towards the bilayer center. The free energy minima positions (of COMs) for neutral and charged RvD1 species are at |zmin| ≈ 10 Å and |zmin| ≈ 13 Å with the ΔGpartitioning values of −5.5 ± 0.01 kcal/mol and −4 ± 0.02 kcal/mol respectively. As the COMs of the molecules moved from their energy minima to the bilayer center (z = 0 Å), the free energies increased to the ΔGcrossing values of 3.28 ± 0.02 kcal/mol and 7.24 ± 0.06 kcal/mol at the bilayer center (z = 0 Å) for the neutral and charged RvD1 species, respectively. With RvD2 (for both the species), there is an energy barrier of ~1.9 kcal/mol for transfer from water to the membrane. The energy minima for the neutral and charged species of RvD2 are at |zmin| ≈ 8 Å and |zmin| ≈ 13 Å with the ΔGpartitioning values of −2.8 ± 0.07 kcal/mol and −1.8 ± 0.02 kcal/mol respectively. At the bilayer center, the ΔGcrossing values are 5.16 ± 0.09 kcal/mol and 5.83 ± 0.12 kcal/mol for neutral and charged species of RvD2 respectively. In all cases, the thermally accessible regions (energy barrier of RT = 0.616 kcal/mol at 310 K) extended for ≈2–3Å both sides around the energy minima (Table 1). The density profiles calculated for the neutral and ionized species of RvD1 and RvD2 (Supplementary Fig. S1) using the entire simulation trajectories support these observations.
Table 1.
Free energy profiles of membrane partitioning of resolvins D1 and D2 in their neutral and charged forms.
| Resolvin | energy-minimum position of COM (bilayer depth) from the center (Å) | bilayer region within reach of thermal motion at 310 K, from the center (Å) | ΔGpartitioning (kcal/mol) | ΔGcrossing (kcal/mol) | |
|---|---|---|---|---|---|
| D1-neutral | 10 | 7 | 13 | −5.5 ± 0.01 | 3.28 ± 0.02 |
| D1-charged | 13 | 10 | 16 | −4 ± 0.02 | 7.24 ± 0.06 |
| D2-neutral | 9 | 7 | 12 | −2.8 ± 0.07 | 5.16 ± 0.09 |
| D2-charged | 13 | 10 | 16 | −1.8 ± 0.02 | 5.83 ± 0.12 |
standard error values were calculated by the Monte Carlo bootstrap error analysis in WHAM66.
The neutral and charged species of RvD1 and RvD2 assume distinct orientations and conformations within the lipid bilayer.
It is not clear how protonation/deprotonation of the carboxyl group on resolvins affects their loading/release from nanovesicles. Therefore, we studied their conformations and orientations in the membrane. The bilayer orientations of the neutral and charged species of RvD1 and RvD2 (Fig. 2) were determined in reference to the bilayer normal (z-axis), the vector perpendicular to the plane of the bilayer with its origin at the center of the bilayer and directed towards the lipid head groups. The angles measured (See Methods section for more details) between the bilayer normal and the longitudinal vectors of resolvins represent the orientational angles. The angles were measured for each species using the trajectories representing their preferred bilayer locations (Table 1) and are presented as probability density graphs (Fig. 2). The neutral species of both RvD1 and RvD2 assume distinct orientations than that of the charged species. The molecules are mostly oriented in parallel to the bilayer plane and are perpendicular to the bilayer normal (The orientation angles are θ = 95° ± 20° for RvD1 and θ = 100° ± 20° for RvD2). In this orientation, both ends of the solute molecules lie at the interfacial region and are in contact with the polar headgroups and the nonpolar alkyl chains of the membrane lipids. In contrast, the charged species assume orientations in which the molecules are seen perpendicular to the bilayer plane and mostly parallel to the bilayer normal (θ = 30° ± 20° for RvD1 and (θ = 60° ± 20° for RvD2). The charged species assume an interesting bent conformations (‘J’ shaped) in which the carboxyl end of the molecule is located near the membrane-water interface, in contact with the choline head groups, whereas the alkyl end is at the interfacial region enabling its hydroxyl groups to interact with the glyceryl polar atoms. Interestingly, the differences in the locations of three -OH groups between RvD1 and RvD2 seem to affect their orientation angles. Compared to RvD1 (θ = 30°), RvD2 molecules (θ = 60°) are less parallel to the bilayer normal due to the presence of two -OH groups at the alkyl end. A careful analysis of atom contacts data (% occupancy, in Fig. 4D and Supplementary Fig. S2) reveal a possible molecular basis for this observed difference. The OH(2), which is located near the carboxyl end in RvD1, resided closer to the choline group and interacted mainly with choline and glyceryl carbonyls. However, in RvD2, the OH(2), due to its location near the alkyl end, was seen relatively less in contact with choline and glyceryl carbonyls and had significantly higher contact near the alkyl tails. Most interestingly, the OH(3) in RvD1 makes contacts in a similar fashion to that of OH(2) despite located at the alkyl end of the molecule. As can be seen in Fig. S2, the % contacts of OH(3) of RvD1 with choline is much higher (31%) than that of RvD2 (18%), which is seen mainly in contact with the alkyl tails (42 % vs. 26%). In addition, this differences in the orientations between neutral and ionized species of RvD1 and RvD2 are clearly captured by the number of water molecules that are within 4Å (Fig. S4). These numbers are significantly higher for ionized species and most interestingly, the ionized species of RvD1 has more water molecules than that of RvD2, which, at least in part, explains the differences in their orientations.
Fig. 2.
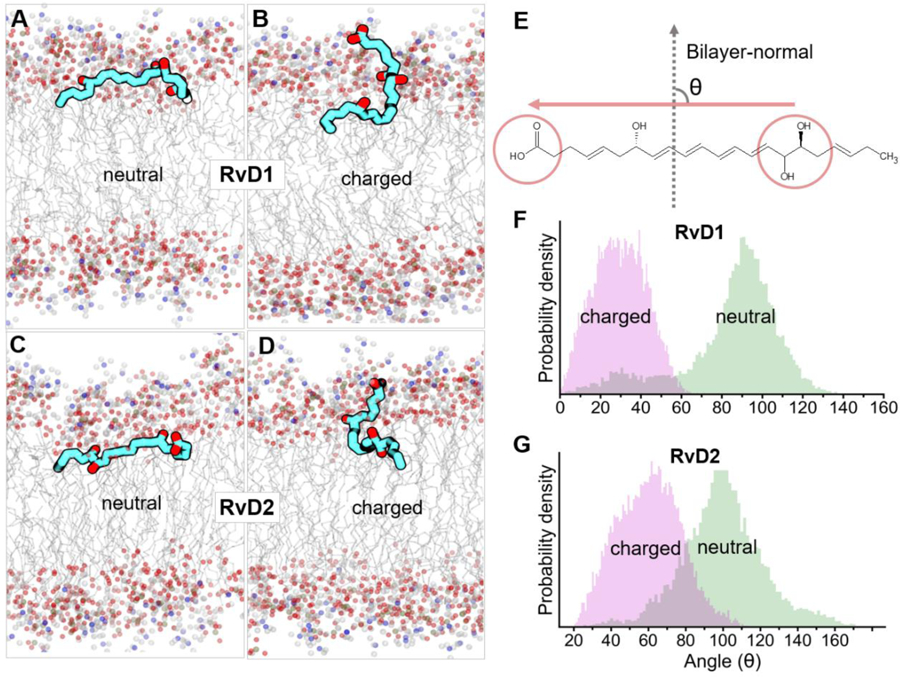
The time-average bilayer orientations of neutral and charged species of RvD1 and RvD2 in the DMPC membrane. A marked difference in the orientations of neutral (A and C) and charged (B and D) species of RvD1 and RvD2 were observed. These represent time-average orientations of the molecules at their energetically favorable bilayer locations (characterized by PMFs in Fig. 1) during 250 ns of simulation for each molecule. The angles between bilayer normal and longitudinal vector of resolvin molecules illustrate that the neutral species of RvD1 and RvD2 orient themselves perpendicular (~100°) to the bilayer normal whereas charged species are approximately parallel (~30° for RvD1 and ~60° for RvD2) to the bilayer normal (F and G). The molecule vector (E) was defined by a straight line connecting two points representing the center-of-mass of the end carboxyl group as head and another carbon atom connected to its last hydroxyl group at the other end as origin, respectively.
To further understand the conformations of the molecules in the membrane, we quantified the distance between the two carbon atoms (carbon atom of the carboxyl group at one end and that of the methyl group at the other end) at both ends. The box and whisker plot (Supplementary Fig. S3A) of these distances indicates that both the charged and neutral species of RvD1 and RvD2 assume extended conformations with slightly varying lengths. In the case of RvD1, the charged species appear to assume a slightly more extended conformation than the neutral species. An opposite trend is observed with RvD2, where the neutral species appear to be in more extended conformations than the charged ones.
Specific molecular interactions of RvD1 and RvD2 with the lipid functional groups.
Hydrogen-bonds.
Both resolvins (RvD1 and RvD2) contain a carboxylic acid group and three hydroxyl groups resulting in a total of four hydrogen bond donors and five hydrogen bond acceptors. The carboxyl group (pKa = 4.9 ± 0.4, predicted using ACD/Percepta) undergoes ionization and gets entirely deprotonated at physiological pH 7.4 (See Supporting info, Table S1) resulting in only three H-bond donors for charged species. Both resolvin molecules, irrespective of their ionization state, seem to form on an average of 2–4 H-bonds (Fig. 3A) while they are located at their preferred bilayer locations as given by the PMF curves (Fig. 1). The maximum number of H-bonds formed by the resolvin molecules were found to be around 12, due to their interactions with multiple lipid species surrounding them. According to the percentage occupancy calculations (Fig. 4D), these H-bonds are formed mainly between the polar carboxyl and hydroxyl groups of resolvins and glyceryl carbonyl oxygens of the lipids. In bulk water (z = 25 to 30 Å), the number of H-bonds formed is much higher for all the species. The ionization state of the molecules appears to affect their H-bonding capacity. Interestingly, the charged species of RvD1 and RvD2 make significantly more H-bond interactions with water than their neutral counterparts. The number of H-bonds formed by the charged species is almost twice (~15 to 18 H-bonds), the number of bonds formed by the neutral species (Fig 3B). This increase agrees with the number of water contacts (number of water molecules within 4 Å of any heavy atoms of the resolvin) quantified surrounding these molecules in the bulk water (Supplementary Fig. S4).
Fig. 3.
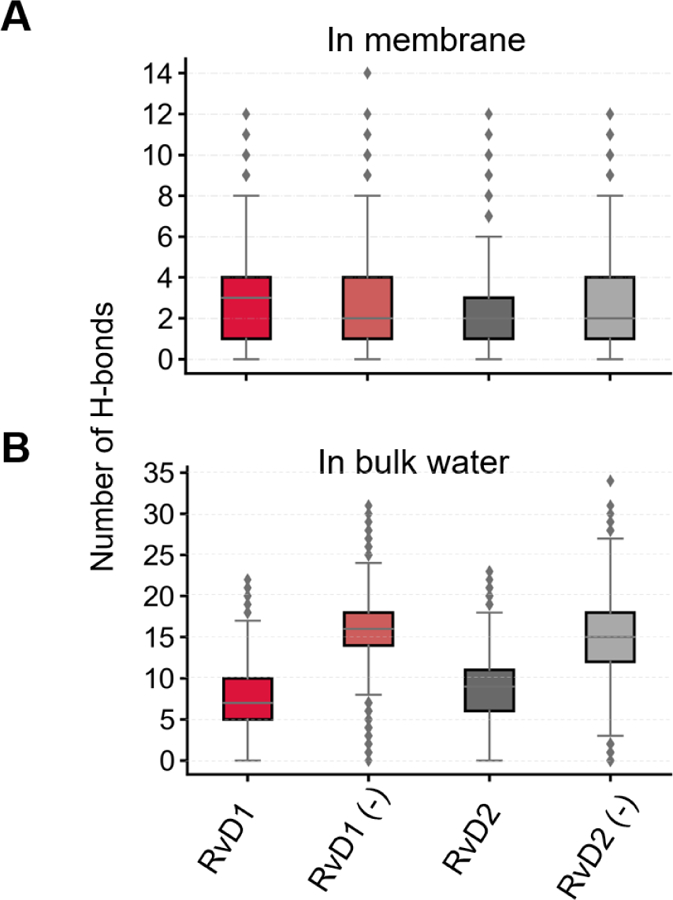
Hydrogen bond interactions of the neutral and charged (denoted by a negative sign) species of RvD1 and RvD2 in the membrane at their preferred locations (A) and in the bulk water (B).
The number of atom contacts as a qualitative measure of intermolecular interactions.
To assess specific molecular interactions accounting for the observed low free energy wells within the membrane (depicted in the PMF curves, in Fig. 1 C and D), we quantified the number of atom contacts between the studied resolvin molecules and individual functional groups of the DMPC lipids (choline, phosphate, glyceryl carbonyls, and alkyl methylene carbons as depicted in Fig. 4A). We also calculated the percentage (%) occupancy of these contacts to assess the stability and longevity of the interactions. The % occupancy presented as heatmap represents the fraction of the simulation time in which these contacts have occurred (Fig. 4D). The simulation trajectories representing |zmin| ± 3Å for the neutral and charged species of RvD1 and RvD2 were used for these calculations (See Methods section for further details). The results indicate that the ionization state of the carboxyl group in RvD1 and RvD2 significantly affect their interactions with the lipids, due to marked differences in their orientations within the membrane. The number of solute atoms near the choline group is significantly (p<0.05) higher for the charged species than the neutral ones (dark and light blue bars in Fig. 4 B and C). The higher numbers are due to the attractive salt-bridge interactions between the positively charged choline groups and the negatively charged carboxyl group of resolvins. The % occupancy of the salt-bridge interactions for RvD1 and RvD2 is 79% and 64%, respectively (green squares on the very first row in Fig. 4D). In the case of neutral species, the protonated carboxyl groups were found mostly at the interface between the glyceryl carbonyls and alkyl tails of the lipids.
The number of atom contacts with phosphate groups is the least in all the resolvin molecules. This observation is also supported by low percentage occupancies observed for all four polar functional groups (red rows in Fig. 4D). A careful observation of the simulation trajectories revealed that phosphate groups are pushed away by preferential interactions of the polar functional groups with the choline and glyceryl carbonyls. Both RvD1 and RvD2 make extensive contacts with both glyceryl and alkyl regions, as they prefer to remain at the headgroup-tail interface. The number of atom contacts with the lipid alkyl region is the highest for all the molecules. Also, these contacts are significantly higher (p <0.001) for the neutral species than the charged ones. The hydroxyl group OH(1) at the carboxyl end of resolvins, was found to be more frequently in contact with choline and glyceryl carbonyls than the lipid alkyl tails. The second hydroxyl group OH(2) of RvD1 and RvD2 differ significantly in % occupancy due to the difference in their locations with respect to OH(1), as can be seen in Fig. 1. In RvD1, being close to OH(1), the OH(2) group has more contact with glyceryl carbonyls than the alkyl tails. On the other hand, the OH(2) of RvD2 is away from OH(1) and is closer to OH(3). It was found more frequently in contact with the lipid alkyl tails than the glyceryl carbonyls. The third hydroxyl OH(3) at the alkyl end of resolvin molecules follows a similar trend as OH(2). Interestingly, the % occupancy of OH(3) with lipid alkyl tails is more for RvD2 compared to RvD1.
CONCLUSIONS
In this study, we determined the encapsulation efficiency of the neutral and ionized species of both resolvin D1 and D2 loaded into the cell membrane-derived nanovesicles at pH values 5.4 and 7.4, respectively. We performed MD simulations to examine the molecular interactions of both neutral and ionized species of resolvin D1 and D2 with the lipid bilayer in atomistic details. The potential of mean force (PMF) curves, characterizing the free energies of solute transfer from water to the bilayer, reveal the energetically favorable locations, orientations, and conformations of the studied molecules within the membrane regions. The ionization state of the carboxyl group significantly affects the molecules’ orientations in the bilayer. The neutral species (protonated carboxyl group) of resolvins assumed fully extended conformations and orientations in which the long axis of the molecules is in parallel to the membrane plane and located mainly at the interface between glyceryl headgroups and lipid alkyl tails. In contrast, the ionized species (deprotonated carboxyl group) of resolvins assume ‘J’ shaped bent conformations and orient themselves in which the long axis of the molecules is perpendicular to the membrane plane with their carboxyl group in close contact with the choline headgroup of the bilayer. The subtle differences in the location of three hydroxyl groups between RvD1 and RvD2 seem to significantly alter the magnitude to which these molecules interact with the different bilayer regions, affecting their free energies of transfer from water to the membrane, which is more favorable for RvD1 than RvD2. Although the MD simulations were performed using a homogenous bilayer made of DMPC lipids, the above findings are in good agreement with the encapsulation efficiencies of RvD1 and RvD2 determined experimentally using the cell membrane-derived nanovesicles. Our results offer valuable insight into the mechanistic details of the molecular interactions of RvD1 and RvD2 with the membrane lipids, characterizing their loading efficiency in nanovesicles and may facilitate the rational design of delivery systems for resolvins in treating inflammatory diseases.
Supplementary Material
ACKNOWLEDGMENTS
Funding: This work was supported, in part, by the National Institutes of Health grants NIGMS/R15 GM131293 to SN and NIBIB/RO1 EB027078 to ZW.
Footnotes
SUPPORTING INFORMATION
Physicochemical properties of RvD1 and RvD2; details of the simulation steps; time-averaged mass density profiles for the neutral and ionized species of RvD1 and RvD2 and the bilayer components; the percentage (%) occupancy of resolvins’ polar groups in contact with the individual lipid functional groups; the end-to-end distance of resolvins, revealing their conformations in the membrane and bulk water; the number of water contacts for RvD1 and RvD2 in the bilayer and bulk water.
References:
- 1.Calder PC Omega-3 polyunsaturated fatty acids and inflammatory processes: nutrition or pharmacology? Br. J. Clin. Pharmacol 2013, 75, 645–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Serhan CN; Clish CB; Brannon J; Colgan SP; Chiang N; Gronert K Novel Functional Sets of Lipid-Derived Mediators with Antiinflammatory Actions Generated from Omega-3 Fatty Acids via Cyclooxygenase 2–Nonsteroidal Antiinflammatory Drugs and Transcellular Processing. J. Exp. Med 2000, 192, 1197–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Serhan CN Pro-resolving lipid mediators are leads for resolution physiology. Nature 2014, 510, 92–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basil MC; Levy BD Specialized pro-resolving mediators: endogenous regulators of infection and inflammation. Nat. Rev. Immunol 2016, 16, 51–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buckley CD; Gilroy DW; Serhan CN; Stockinger B; Tak PP The resolution of inflammation. Nat. Rev. Immunol 2013, 13, 59–66. [DOI] [PubMed] [Google Scholar]
- 6.Serhan CN; Chiang N; Van Dyke TE Resolving inflammation: dual anti-inflammatory and pro-resolution lipid mediators. Nat. Rev. Immunol 2008, 8, 349–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arnardottir HH; Dalli J; Norling LV; Colas RA; Perretti M; Serhan CN Resolvin D3 Is Dysregulated in Arthritis and Reduces Arthritic Inflammation. J. Immunol 2016, 197, 2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conte MS; Desai TA; Wu B; Schaller M; Werlin E Pro-resolving lipid mediators in vascular disease. J. Clin. Invest 2018, 128, 3727–3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spite M; Serhan CN Novel Lipid Mediators Promote Resolution of Acute Inflammation. Circulation Res 2010, 107, 1170–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pirault J; Bäck M Lipoxin and Resolvin Receptors Transducing the Resolution of Inflammation in Cardiovascular Disease. Front. Pharmacol 2018, 9, 1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whittington RA; Planel E; Terrando N Impaired Resolution of Inflammation in Alzheimer’s Disease: A Review. Front. Immunol 2017, 8, 1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krishnamoorthy S; Recchiuti A; Chiang N; Yacoubian S; Lee C-H; Yang R; Petasis NA; Serhan CN Resolvin D1 binds human phagocytes with evidence for proresolving receptors. Proc. Natl. Acad. Sci. U.S.A 2010, 107, 1660–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cooray SN; Gobbetti T; Montero-Melendez T; McArthur S; Thompson D; Clark AJL; Flower RJ; Perretti M Ligand-specific conformational change of the G-protein-coupled receptor ALX/FPR2 determines proresolving functional responses. Proc. Natl. Acad. Sci. U.S.A 2013, 110, 18232–18237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chiang N; Dalli J; Colas RA; Serhan CN Identification of resolvin D2 receptor mediating resolution of infections and organ protection. J. Exp. Med 2015, 212, 1203–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arita M; Bianchini F; Aliberti J; Sher A; Chiang N; Hong S; Yang R; Petasis NA; Serhan CN Stereochemical assignment, antiinflammatory properties, and receptor for the omega-3 lipid mediator resolvin E1. J. Exp. Med 2005, 201, 713–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arita M; Ohira T; Sun Y-P; Elangovan S; Chiang N; Serhan CN Resolvin E1 Selectively Interacts with Leukotriene B4 Receptor BLT1 and ChemR23 to Regulate Inflammation. J. Immunol 2007, 178, 3912. [DOI] [PubMed] [Google Scholar]
- 17.Norling Lucy V; Dalli J; Flower Roderick J; Serhan Charles N; Perretti M Resolvin D1 Limits Polymorphonuclear Leukocyte Recruitment to Inflammatory Loci. Arterioscler. Thromb. Vasc. Biol 2012, 32, 1970–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benabdoun HA; Kulbay M; Rondon E-P; Vallières F; Shi Q; Fernandes J; Fahmi H; Benderdour M In vitro and in vivo assessment of the proresolutive and antiresorptive actions of resolvin D1: relevance to arthritis. Arthritis Res. Ther 2019, 21, 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsiao H-M; Thatcher TH; Colas RA; Serhan CN; Phipps RP; Sime PJ Resolvin D1 Reduces Emphysema and Chronic Inflammation. Am. J. Pathol 2015, 185, 3189–3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsiao H-M; Sapinoro RE; Thatcher TH; Croasdell A; Levy EP; Fulton RA; Olsen KC; Pollock SJ; Serhan CN; Phipps RP; Sime PJ A Novel Anti-Inflammatory and Pro-Resolving Role for Resolvin D1 in Acute Cigarette Smoke-Induced Lung Inflammation. PLoS One 2013, 8, e58258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jannaway M; Torrens C; Warner JA; Sampson AP Resolvin E1, resolvin D1 and resolvin D2 inhibit constriction of rat thoracic aorta and human pulmonary artery induced by the thromboxane mimetic U46619. Br. J. Pharmacol 2018, 175, 1100–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chiang N; Barnaeva E; Hu X; Marugan J; Southall N; Ferrer M; Serhan CN Identification of Chemotype Agonists for Human Resolvin D1 Receptor DRV1 with Pro-Resolving Functions. Cell Chem. Biol 2019, 26, 244–254.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Corminboeuf O; Leroy X FPR2/ALXR Agonists and the Resolution of Inflammation. J. Med. Chem 2015, 58, 537–559. [DOI] [PubMed] [Google Scholar]
- 24.Perretti M; Leroy X; Bland EJ; Montero-Melendez T Resolution Pharmacology: Opportunities for Therapeutic Innovation in Inflammation. Trends Pharmacol. Sci 2015, 36, 737–755. [DOI] [PubMed] [Google Scholar]
- 25.Spite M; Norling LV; Summers L; Yang R; Cooper D; Petasis NA; Flower RJ; Perretti M; Serhan CN Resolvin D2 is a potent regulator of leukocytes and controls microbial sepsis. Nature 2009, 461, 1287–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Campbell EL; Serhan CN; Colgan SP Antimicrobial Aspects of Inflammatory Resolution in the Mucosa: A Role for Proresolving Mediators. J. Immunol 2011, 187, 3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chiang N; de la Rosa X; Libreros S; Serhan CN Novel Resolvin D2 Receptor Axis in Infectious Inflammation. J. Immunol 2017, 198, 842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Botten N; Hodges RR; Li D; Bair JA; Shatos MA; Utheim TP; Serhan CN; Dartt DA Resolvin D2 elevates cAMP to increase intracellular [Ca2+] and stimulate secretion from conjunctival goblet cells. The FASEB Journal 2019, 33, 8468–8478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Connor KM; SanGiovanni JP; Lofqvist C; Aderman CM; Chen J; Higuchi A; Hong S; Pravda EA; Majchrzak S; Carper D; Hellstrom A; Kang JX; Chew EY; Salem N; Serhan CN; Smith LEH Increased dietary intake of ω−3-polyunsaturated fatty acids reduces pathological retinal angiogenesis. Nat. Med 2007, 13, 868–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clària J; Dalli J; Yacoubian S; Gao F; Serhan CN Resolvin D1 and Resolvin D2 Govern Local Inflammatory Tone in Obese Fat. J. Immunol 2012, 189, 2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pascoal LB; Bombassaro B; Ramalho AF; Coope A; Moura RF; Correa-da-Silva F; Ignacio-Souza L; Razolli D; de Oliveira D; Catharino R; Velloso LA Resolvin RvD2 reduces hypothalamic inflammation and rescues mice from diet-induced obesity. J. Neuroinflammation 2017, 14, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pratt DA; Tallman KA; Porter NA Free Radical Oxidation of Polyunsaturated Lipids: New Mechanistic Insights and the Development of Peroxyl Radical Clocks. Acc. Chem. Res 2011, 44, 458–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yin H; Xu L; Porter NA Free Radical Lipid Peroxidation: Mechanisms and Analysis. Chem. Rev 2011, 111, 5944–5972. [DOI] [PubMed] [Google Scholar]
- 34.Sun Y-P; Oh SF; Uddin J; Yang R; Gotlinger K; Campbell E; Colgan SP; Petasis NA; Serhan CN Resolvin D1 and Its Aspirin-triggered 17R Epimer: Stereochemical Assignments, Anti-Inflammatory Properties, and Enzymatic Inactivation. J. Biol. Chem 2007, 282, 9323–9334. [DOI] [PubMed] [Google Scholar]
- 35.Arita M; Oh SF; Chonan T; Hong S; Elangovan S; Sun Y-P; Uddin J; Petasis NA; Serhan CN Metabolic Inactivation of Resolvin E1 and Stabilization of Its Anti-inflammatory Actions. J. Biol. Chem 2006, 281, 22847–22854. [DOI] [PubMed] [Google Scholar]
- 36.Cholkar K; Gilger BC; Mitra AK Topical delivery of aqueous micellar resolvin E1 analog (RX-10045). Int. J. Pharm 2016, 498, 326–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Orr SK; Colas RA; Dalli J; Chiang N; Serhan CN Proresolving actions of a new resolvin D1 analog mimetic qualifies as an immunoresolvent. Am. J. Physiol. Lung Cell. Mol. Physiol 2015, 308, L904–L911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mitragotri S; Burke PA; Langer R Overcoming the challenges in administering biopharmaceuticals: Formulation and delivery strategies. Nat. Re.v Drug Discov 2014, 13, 655–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang CY; Gao J; Wang Z Bioresponsive Nanoparticles Targeted to Infectious Microenvironments for Sepsis Management. Adv. Mater 2018, 30, 1803618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Z; Tiruppathi C; Cho J; Minshall RD; Malik AB Delivery of nanoparticle: Complexed drugs across the vascular endothelial barrier via caveolae. IUBMB Life 2011, 63, 659–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Z; Li J; Cho J; Malik AB Prevention of vascular inflammation by nanoparticle targeting of adherent neutrophils. Nat. Nanotechnol 2014, 9, 204–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barenholz Y Doxil(R)--the first FDA-approved nano-drug: lessons learned. J. Control Release 2012, 160, 117–134. [DOI] [PubMed] [Google Scholar]
- 43.Houdaihed L; Evans JC; Allen C Overcoming the Road Blocks: Advancement of Block Copolymer Micelles for Cancer Therapy in the Clinic. Mol. Pharm 2017, 14, 2503–2517. [DOI] [PubMed] [Google Scholar]
- 44.Hu CM et al. Nanoparticle biointerfacing by platelet membrane cloaking. Nature 2015, 526, 118–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hu CM; Fang RH; Zhang L Erythrocyte-inspired delivery systems. Adv. Healthc. Mater 2012, 1, 537–547. [DOI] [PubMed] [Google Scholar]
- 46.Yurkin ST; Wang Z Cell membrane-derived nanoparticles: emerging clinical opportunities for targeted drug delivery. Nanomedicine (Lond) 2017, 12, 2007–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao J; Dong X; Wang Z Generation, purification and engineering of extracellular vesicles and their biomedical applications. Methods, 2019. S1046–2023 (19) 30219–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gao J, Chu D & Wang Z Cell membrane-formed nanovesicles for disease-targeted delivery. J. Control. Release 2016, 224, 208–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gao J; Wang S; Wang Z High yield, scalable and remotely drug-loaded neutrophil-derived extracellular vesicles (EVs) for anti-inflammation therapy. Biomaterials 2017, 135, 62–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang S; Dong X; Gao J; Wang Z Targeting Inflammatory Vasculature by Extracellular Vesicles. The AAPS Journal 2018, 20, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dong X; Gao J; Zhang CY; Hayworth C; Frank M; Wang Z Neutrophil Membrane-Derived Nanovesicles Alleviate Inflammation To Protect Mouse Brain Injury from Ischemic Stroke. ACS Nano 2019, 13, 1272–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Norling LV; Spite M; Yang R; Flower RJ; Perretti M; Serhan CN Cutting Edge: Humanized Nano-Proresolving Medicines Mimic Inflammation-Resolution and Enhance Wound Healing. J. Immunol 2011, 1003865. [DOI] [PMC free article] [PubMed]
- 53.Tejwani RW; Davis ME; Anderson BD; Stouch TR An Atomic and Molecular View of the Depth Dependence of the Free Energies of Solute Transfer from Water into Lipid Bilayers. Mol. Pharmaceutics 2011, 8, 2204–2215. [DOI] [PubMed] [Google Scholar]
- 54.Zhang CY; Lin W; Gao J; Shi X; Davaritouchaee M; Nielsen AE; Mancini RJ; Wang Z pH-responsive nanoparticles targeted to lungs for improved therapy of acute lung inflammation/injury. ACS Appl. Mater. Interfaces 2019, 11 (18), 16380–16390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Molecular Operating Environment (MOE), 2013.08, Chemical Computing Group ULC: 1010 Sherbrooke St. West, Suite #910, Montreal, QC, Canada, 2019. [Google Scholar]
- 56.Wu EL; Cheng X; Jo S; Rui H; Song KC; Davila-Contreras EM; Qi Y; Lee J; Monje-Galvan V; Venable RM; Klauda JB; Im W CHARMM-GUI Membrane Builder toward realistic biological membrane simulations. J. Comput. Chem 2014, 35, 1997–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jo S; Lim JB; Klauda JB; Im W CHARMM-GUI Membrane Builder for mixed bilayers and its application to yeast membranes. Biophys. J 2009, 97, 50–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee J; Cheng X; Swails JM; Yeom MS; Eastman PK; Lemkul JA; Wei S; Buckner J; Jeong JC; Qi Y; Jo S; Pande VS; Case DA; Brooks CL 3rd; MacKerell AD Jr.; Klauda JB; Im W CHARMM-GUI Input Generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM Simulations Using the CHARMM36 Additive Force Field. J. Chem. Theory Comput 2016, 12, 405–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Klauda JB; Venable RM; Freites JA; O’Connor JW; Tobias DJ; Mondragon-Ramirez C; Vorobyov I; MacKerell AD Jr.; Pastor RW Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B 2010, 114, 7830–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vanommeslaeghe K; Hatcher E; Acharya C; Kundu S; Zhong S; Shim J; Darian E; Guvench O; Lopes P; Vorobyov I; Mackerell AD Jr. CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem 2010, 31, 671–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vanommeslaeghe K; MacKerell AD Jr. Automation of the CHARMM General Force Field (CGenFF) I: bond perception and atom typing. J. Chem. Inf. Model 2012, 52, 3144–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vanommeslaeghe K; Raman EP; MacKerell AD Jr. Automation of the CHARMM General Force Field (CGenFF) II: assignment of bonded parameters and partial atomic charges. J. Chem. Inf. Model 2012, 52, 3155–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pokharel SM; Shil NK; Gc JB; Colburn ZT; Tsai S-Y; Segovia JA; Chang T-H; Bandyopadhyay S; Natesan S; Jones JCR; Bose S Integrin activation by the lipid molecule 25-hydroxycholesterol induces a proinflammatory response. Nat. Commun 2019, 10, 1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Park S; Khalili-Araghi F; Tajkhorshid E; Schulten K Free energy calculation from steered molecular dynamics simulations using Jarzynski’s equality. J. Chem. Phys 2003, 119, 3559–3566. [Google Scholar]
- 65.Park S; Schulten K Calculating potentials of mean force from steered molecular dynamics simulations. J. Chem. Phys 2004, 120, 5946–5961. [DOI] [PubMed] [Google Scholar]
- 66.Kumar S; Rosenberg JM; Bouzida D; Swendsen RH; Kollman PA THE weighted histogram analysis method for free-energy calculations on biomolecules. I. The method. J. Comput. Chem 1992, 13, 1011–1021. [Google Scholar]
- 67.Grossfield A 2.0.9; WHAM: an implementation of the weighted histogram analysis method.
- 68.Phillips JC; Braun R; Wang W; Gumbart J; Tajkhorshid E; Villa E; Chipot C; Skeel RD; Kale L; Schulten K Scalable molecular dynamics with NAMD. J. Comput. Chem 2005, 26, 1781–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ulrich E; Lalith P; Max LB; Tom D; Hsing L; Lee GP A smooth particle mesh Ewald method. J. Chem. Phys 1995, 103, 8577–8593. [Google Scholar]
- 70.Steinbach PJ; Brooks BR New Spherical-Cutoff Methods for Long-Range Forces in Macromolecular Simulation. J. Comput. Chem 1994, 15, 667–683. [Google Scholar]
- 71.Ryckaert J-P; Ciccotti G; Berendsen HJC Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys 1977, 23, 327–341. [Google Scholar]
- 72.Martyna GJ; Tobias DJ; Klein ML Constant-Pressure Molecular-Dynamics Algorithms. J. Chem. Phys 1994, 101, 4177–4189. [Google Scholar]
- 73.Feller SE; Zhang YH; Pastor RW; Brooks BR Constant-Pressure Molecular-Dynamics Simulation - the Langevin Piston Method. J. Chem. Phys 1995, 103, 4613–4621. [Google Scholar]
- 74.Humphrey W; Dalke A; Schulten K VMD: Visual molecular dynamics. J. Mol. Graphics 1996, 14, 33–38. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


