Abstract
During diabetes, renal proximal tubular cells (PTC) are exposed to a combination of high glucose and hypoxic conditions, which plays a relevant role in the development of diabetic kidney disease (DKD). In this work, a time-series proteomic study was performed to analyse the effect of a diabetic-like microenvironment induced changes on HK-2 cells, a human cell line derived from normal proximal tubular epithelial cells. Cells simultaneously exposed to high glucose (25 mM) and hypoxia (1% O2) were compared to cells in control conditions for up to 48 h. Diabetic conditions increased the percentage of death cells after 24 and 48 h, but no differences in the protein/cell ratio were found. The relative protein quantification using dimethyl-labeling and UHPLC-MS/MS analysis allowed the identification of 317, 296 and 259 proteins at 5, 24 and 48 h, respectively. The combination of statistical and time expression profile analyses indicated an increased expression of proteins involved in glycolysis, and a decrease of cytoskeletal-related proteins. The exposure of HK-2 cells to high glucose and hypoxia reproduces some of the effects of diabetes on PTC and, with the limitations inherent to in vitro studies, propose new mechanisms and targets to be considered in the management of DKD.
Introduction
Proteins are key players in many biological processes and cellular signaling events, and the alteration of their abundancy can lead to the development of life threatening diseases [1]. It is also known that many proteins respond to different stimuli changing their expression in a time-dependent manner; therefore, protein expression monitoring over time is important to understand the mechanisms behind these diseases’ development. For protein identification and quantification, high-performance liquid chromatography coupled to mass spectrometry (HPLC-MS) has become a key technology due to its unbiased capacity where there is no need to decide which proteins to target in advance, and because thousands of proteins can be monitored at the same time and remarkable biological information can be accessed [2]. Diabetic kidney disease (DKD) is the most common cause of end-stage renal disease and it develops in approximately 40% of patients with diabetes [3]. Despite this fact, the pathological mechanisms leading to the development and progression of DKD remain poorly understood, and proteomics studies can help shedding some light. It has been demonstrated that increased blood concentrations of glucose and non-esterified fatty acids in diabetes are intracellularly reflected in the kidney, particularly in proximal tubules [4]. It is also evident that the proximal tubule is a prime mover in DKD so that it plays a critical role in the development and progression of DKD [5, 6]. In diabetes, high glucose (HG) content in proximal tubule cells (PTC) results from both increased glucose in plasma and also from excess glucose filtered by the glomeruli, resulting in an excessive glucose load in the tubule [5]. However, although a HG microenvironment is the primary causative factor for the development of DKD, hypoxia also plays a relevant role in the development and progression of this pathology [6–8]. This is particularly true for proximal tubules because they are especially vulnerable to hypoxia, and because PTC are propense to hypoxic injury in diabetes due to three relevant factors: i) diabetes increases two high energy consuming processes, namely gluconeogenesis and sodium reabsorption driven by Na+/K+-ATPase, in PTC (only their Na+/K+-ATPase consumes in basal conditions 80% of the O2 kidney supply) [9]; ii) diabetes leads to impaired O2 utilization because it induces mitochondrial dysfunction and alters substrate delivery in PTC; iii) diabetes reduces O2 delivery to PTC due to microvascular rarefaction [6].
Among others, the hypoxia inducible factor-1α (HIF-1α) protein orchestrates cellular adaptive responses to combat hypoxia, through the expression control of several proteins [10]. However, activation of the HIF system in the hypoxic diabetic kidney has been shown to be suboptimal [7, 11, 12] or even inexistent [8]. This may explain the renoprotective effect in DKD of HIF-1α activation by CoCl2 [8, 13] as well as the fact that a Pro582Ser polymorphism, which confers relative resistance of HIF-1α to the repressive effect of HG, is associated with DKD protection [14]. Activation of the HIF system under hypoxia has also been found to be consistently supressed by HG in PTC [15], and this deficient response seems to be specific for PTC: HIF-1α expression in primary human mesangial cells increased under hypoxia, but a further increase in its expression was observed when these cells were simultaneously exposed to HG and hypoxia [16]. On the other hand, human PTC and HeLa cells did not respond to HG, and in mouse MC, HIF-1α was induced by HG under normoxic conditions, whereas in mouse PTC, that response was absent. In the same way, glucose promotes in cancer cells the expression of HIF-1α [17]. We have previously set up an in vitro model of diabetes-induced proximal tubular dysfunction in which HK-2 cells, a human cell line derived from normal proximal tubular epithelial cells, are exposed to a diabetic-like microenvironment consisting of HG (25 mM) and hypoxia (1% O2). In these conditions, HIF-1α fails to stabilize and accumulate in HK-2 cells (i.e. the same defective response found in the hypoxic diabetic kidney) [18]. Furthermore, impaired HIF-1α regulation in cells exposed to hyperglycemic, hypoxic diabetic-like milieu led to diminished production of vascular endothelial growth factor-A and inhibition of cell migration (responses respectively involved in tubular protection and repair). These effects, as well as impaired HIF-1α regulation, were reproduced in normoglycemia in HK-2 cells incubated with microparticles released by HK-2 cells exposed to hyperglycemic, hypoxic diabetic-like milieu [18]. Finally, neither HG alone nor hypoxia alone affected the migration of HK-2 cells [18] and the release of microparticles under hyperglycemia was greatly diminished in the absence of hypoxia (unpublished results). In summary, HK-2 cells exposed simultaneously to hyperglycemia and hypoxia (i.e. two major conditions of the microenvironment that surrounds proximal tubular cells in diabetes)-but not HK-2 cells exposed separately to either condition- show functional alterations that reproduce some specific effects of diabetes on proximal tubular cells. Given that the monitorization of untargeted protein evolution in HK-2 under hyperglycemic/hypoxic conditions is currently unknown, in the present work we have conducted a shotgun time-series proteomic study based on UHPLC-MS/MS together with stable isotope dimethyl labeling (DML) to relatively quantify the changes in protein expressions over time. The results obtained propose new mechanisms and targets to be investigated in the future for a better understanding of DKD.
Materials and methods
Chemicals and reagents
All reagents herein used were of analytical grade or higher. Acetic acid (AA), ACN, acetone, ethyl-acetate, methanol and urea were obtained from Scharlau (Barcelona, Spain). Formic acid (FA) was from Fisher Scientific (Geel, Belgium). Ammonia solution and NaCl were obtained from Merck (Darmstadt, Germany). β-glycerophosphate, DTT, EDTA, formaldehyde (CH2O) (37%, v/v), iodoacetamide (IAA), n-octyl-β-d-glucopyranoside (BOG), PBS, protease inhibitor cocktail, sodium cyanoborohydride (NaBH3CN), sodium fluoride (NaF), sodium metavanadate (NaVO4), tributylphosphate, triethylammonium bicarbonate (TEAB) and TFA were purchased from Sigma-Aldrich (St. Louis, MO). Trypsin/Lys-C mix (Mass Spec grade V5072) was purchased from Promega (Madison, WI), deuterated formaldehyde (CD2O) (20%, v/v) was obtained from ISOTEC (Miamisburg, OH) and sodium pyrophosphate from Alfa Aesar (Kandel, Germany). Ultrapure water was prepared by Milli-Q water purification system (Millipore, Bedford, MA).
Cell culture conditions
Human proximal tubular epithelial (HK-2) cells were purchased from the American Type Culture Collection (Rockville, MD, USA). HK-2 cells were maintained in DMEM/F12 (ThermoFisher, Grand Island, NY, USA) supplemented with 10% FBS, 1% penicillin/streptomycin/amphoterycin B, 1% glutamine and 1% insulin-transferrine-selenium (ITS) (ThermoFisher, Grand Island, NY, USA). One week before the beginning of the experiments, cell culture medium was changed to DMEM normal glucose (NG, 5.5 mM glucose, ThermoFisher, Grand Island, NY, USA) supplemented as stated above. Cells were routinely maintained in 21% O2 (normoxic conditions) at 37 °C. To perform the experiments, seven p35 cultured plates were seeded with 2.5 x 105 cells (90% confluence) per time and treatment condition. When completely attached, cells were exposed to either medium DMEM NG or DMEM high glucose (HG, 25 mM glucose, ThermoFisher, Grand Island, NY, USA) supplemented with 0.5% FBS, 1% penicillin/streptomycin/amphoterycin B, 1% glutamine and 1% ITS under hypoxic (1% O2) or normoxic conditions (21% O2), for 5, 24 and 48 h. For 48 h experiments, cell culture medium was refreshed at 24 h. Hypoxia experiments were performed in an In vivo200 hypoxia workstation (Ruskinn Technology, West Yorkshire, UK). Cells were manipulated and lysed inside the chamber, and all media and buffers were pre-equilibrated to 1% O2. Three replicates were used to determine the total cell number and the cell viability based on the Trypan Blue dye exclusion assay, and a hemocytometer. Briefly, cells were detached by trypsinization and, together with the detached cells recovered from the culture medium, they were stained with Trypan Blue (1:1). Then, white (viable) and blue (dead) cells were counted using a light microscope and a hemocytometer. For the proteomic study, and after incubation, the growth medium from three culture plates was removed by aspiration, washed with PBS, and proteins were directly extracted from attached cells. An additional plate per time and treatment condition was used to check by Western blot analysis the interaction between HG and hypoxia resulting in specific loss of hypoxia-induced accumulation of HIF-1α in HK-2 cells [18]. Statistical analysis was performed using Statistica software version 7.1 (StatSoft, Inc., USA). One-way ANOVA with LSD Post-hoc test was employed to determine any significant differences between mean values using p < 0.05.
Sample preparation for UHPLC–MS/MS-based proteomics
Cells were lysed with 300 μL of lysis buffer (6 M urea, 1% (m/v) BOG, 0.15 M NaCl, 1.3 mM EDTA, 1 mM NaVO4, 5 mM NaF, 2.5 mM sodium-pyrophospate, and 5 mM β-glycerophosphate in PBS) supplemented with 10 μL of protease inhibitor cocktail to prevent protein degradation. The samples were incubated for 60 min at 4 °C, sonicated for 30 min at 0 °C in a water bath, and then centrifuged at 10000g at 4 °C for 15 min. The supernatants were collected, and protein concentration was quantified using Bradford assay (Bio-Rad Laboratories, Hercules, CA). 20 μg of proteins were mixed with 500 μL of an ice-cold tributylphosphate/acetone/methanol mixture (1:12:1, v/v/v) and incubated at 4 °C for 90 min (mixing every 15 min). The precipitate was centrifuged for 15 min (3000g at 4 °C), washed with 1 mL of cold acetone, and air-dried. Pellets were dissolved in 20 μL of 1% (w/v) BOG with 20% (v/v) ACN in 0.1 M TEAB, and the proteins were reduced with 10 μL of 45 mM DTT at 56 °C for 15 min, cooled down to room temperature and alkylated with 10 μL of 100 mM IAA for 15 min in the dark. Samples were then digested with 0.5 μg of LysC/trypsin solution (5% (w/w), of total protein content) and incubated at 37 °C overnight in darkness. The digests were dried to remove ACN and resuspended in 70 μL of 0.1 M TEAB, and BOG was extracted with water-saturated ethyl-acetate [19]. Dimethyl labelling was performed according to Boersema et al [20]. Briefly, tryptic peptide mixtures were reconstituted in TEAB and NG-Normoxia samples were labelled as light (L) using regular formaldehyde (CH2O, 4% (v/v)), and HG-Hypoxia samples were labelled as medium (M) using deuterated formaldehyde (CD2O, 4% (v/v)). Technical replicates were included for each sample in a swap-labelling experiment form (NG-Normoxia samples labelled as M, and HG-Hypoxia samples labelled as L). To control the labelling efficiency and optimize the UHPLC peptide loading quantity and MS parameters, 120 μg of NG-Normoxia samples were divided into six aliquots of 20 μg, and three aliquots were labelled as L, and the other three as M, following the same procedure as explain above. These samples were considered as “Test” samples. After a brief vortexing, 4 μL of freshly prepared 0.6 M NaBH3CN solution was added to each sample, and the vials were incubated for 60 min at room temperature with mild agitation. The reaction was terminated by adding 1% (v/v) ammonia solution (Merck, Germany), and 5% (v/v) FA (Merck, Germany) to consume the excess labelling reagents. Finally, the real samples labelled as L and M were mixed together and for the “Test” samples, one L sample and one M sample were mixed together, generating 3 “Test” samples. All mixed samples were desalted on Isolute C18 solid-phase extraction columns as specify by the manufacture (1 mL, 50 mg capacity, Biotage, Uppsala, Sweden). After being desalted, peptides were dried in a SpeedVac and redissolved in 2% (v/v) ACN and 0.1% (v/v) TFA prior to UHPLC–MS/MS analysis. Two replicates were analysed for each “Test” condition, and six replicates (considering biological and technical replicates) were analysed for the real samples at each time point, giving a total of 18 LC-MS/MS runs.
UHPLC–MS/MS analysis
Aliquots of 5 μL containing ∼ 1, 2, 4 or 6 μg (for “Test” samples), or ∼ 6 μg (for real samples) tryptic peptides were injected into a LC–MS/MS system consisting of an UltiMate 3000 quaternary UHPLC (Thermo Fisher Scientific, Bremen, Germany) coupled to a Q Exactive Pro Mass Spectrometer (Thermo Fisher Scientific, Bremen, Germany) using a HESI-II probe at 3 kV. Peptide separations were performed on a C18 Ascentis Express column (Sigma, St Louis, USA), having 100 × 2.1 mm i.d. dimensions (fused-core® particles with 0.5 μm thick porous shell and an overall particle size of 2.7 μm), and a guard column (5 × 2.1 mm i.d. × 2.7 μm particle size) of the same composition as the analytical columns. Columns were kept at 55◦C during the analysis of the samples. The separations were performed at a flow rate of 0.3 mL/min with mobile phases A (2% (v/v) ACN, 0.3% (v/v) AA, 0.1% (v/v) FA) and B (80% (v/v) ACN, 0.3% (v/v) AA, 0.1% (v/v) FA). A 60 min gradient from 0.2% B to 46.2% B was used. The mass spectrometer was operated in positive-ion mode and a Top6 data-dependent acquisition (DDA) mode was used. MS1 spectra were acquired within m/z 350–1600 with an automatic gain control (AGC) of 3 x 106 at 35,000 resolution and with a maximum ion time (IT) of 110 ms. Up to 6 of the most abundant precursor ions with ≥2 and ≤8 charges and threshold intensity of 2 x 104 were selected for higher-energy C-trap dissociation (HCD). An isolation window of 2 m/z and normalized collision energy of 28 were used. Ions that were once selected for acquisition were dynamically excluded for 20 s for further fragmentation. MS2 spectra were acquired with an AGC of 1 x 105 at resolution of 35,000 and a maximum IT of 110 ms. A Top12 DDA mode was also tested, keeping the MS1 parameters as for the Top6 but increasing the number of precursor ions to 12, and acquiring the MS2 spectra with an AGC of 1 x 105 at resolution of 17,500 and a maximum IT of 50 ms.
MaxQuant analysis
All MS raw files were collectively processed with MaxQuant (version 1.6.3.3) [21] using the Andromeda search engine [22]. The FDR was set to 1% for both proteins and peptides, and a minimum of two peptides with at least seven amino acid length was specified. MaxQuant scored peptides for identification based on a search with a maximal precursor mass deviation of 20 ppm (first search) and 4.5 ppm (main search), and fragment masses deviations of up to 20 ppm. The Andromeda search engine was used for the MS/MS spectra search against a concatenated forward and reversed version of the Uniprot human database (downloaded on April 24th, 2019 and containing 20,428 entries). Trypsin was selected as digestive enzyme and a maximum of two missed cleavages were allowed. Carbamidomethylation of cysteine residues was set as the fixed modification, and the oxidation of methionine and protein N-acetylation were set as variable modifications. For dimethyl labeling, DimethylLys0 and DimethylNter0 were set as light labels, and DimethylLys4 and DimethylNter4 were set as medium labels. A minimum peptide ratio count of two and at least one “razor peptide” was required for quantification. All mass spectrometry proteomics data and search results have been deposited to the ProteomeXchange Consortium via the PRIDE [23] partner repository with the data set identifier PXD015102.
Database search/data processing
Prior to any statistical analysis, identifications flagged as reverse, potential contaminants, or proteins identified only by site modification were excluded for further analysis, and the normalized relative protein abundance was transformed to the log2 scale. For PCA, only proteins present in all biological and technical replicates across the three time points were considered to avoid imputation of missing values. Thereafter, the values were normalized (subtracting the mean value of each data set and dividing by its standard deviation), and the temporal profiles were clustered using the fuzzy c-means clustering algorithm included in the Mfuzz toolbox [24]. Different combinations of cluster sizes and fuzzification parameters were explored and found optimal partitioning with c = 6 and m = 3. The lists of proteins with Euclidean distance larger than 0.5 in each cluster were searched for enriched gene ontology (GO) terms or pathways using the web-based PANTHER (http://www.pantherdb.org/) [25] or STRING [26]. The corrected p-values were derived from a Fisher's Exact test followed by Benjamini and Hochberg false discovery rate correction, and considered significant when < 0.05. Thereafter, the most significantly altered proteins between hypoxia/hyperglycemia against normoxia/normoglycemia were identified. For this analysis, a protein was included in the analysis when it was quantified in at least four sample replicates at the different time points. In these cases, a p value of < 0.05 (after one sample t-test) and a 1.2-fold change cut-off filter (equivalent to 0.26 and -0.26 in log2 scale) were applied. Finally, the list of significantly altered proteins at 48 h was searched for enriched GO biological processes using PANTHER and STRING softwares using the same parameters as previously specified.
Results
Cell viability and protein content
The main objective of this work was to carry out a time-course proteomic study to investigate the changes induced by a diabetic-like hyperglycemic/hypoxic medium in HK-2 cells, a cell line derived from human proximal tubular cells, as an in vitro model of the effects of the diabetic milieu on proximal tubular cells in the context of DKD. For this study, equal number of cells (5 × 105 per mL) were plated at 90% confluence in p35 cultured dishes and when completely attached, they were treated for 5, 24 and 48 h with HG (25 mM) medium in hypoxic conditions (1% O2), or with NG medium (5.5 mM) in normoxic conditions (21% O2). The interaction between HG and hypoxia resulting in specific loss of hypoxia-induced accumulation of HIF-1α in HK-2 cells, which has important pathological implications in the context of proximal tubular disfunction in DKD [18], was routinely checked (results are not shown).
While the percentage of death cells increased from 7% to 12% after 24 h in HG-Hypoxia compared to NG-Normoxia, and rose up to more than 20% after 48 h (Fig 1A), no differences in the protein/cell ratio between the two experimental groups were found at any time point (Fig 1B).
Fig 1. Percentage of dead cells (A) and protein content/cell (B) after incubation of HK-2 cells in NG (5.5 mM glucose)-normoxia (21% O2) or HG (25 mM glucose)-hypoxia (1% O2).
Error bars are given as the standard deviation of three independent experiments. *Asterisks indicate significant differences between samples after ANOVA with LSD Post-hoc, p-value < 0.05.
Peptide loading quantity and MS acquisition
The proteomic study was performed using preliminary UHPLC and MS conditions as previously reported by Lenčo et al. [27] and some parameters were then optimized. Firstly, the UHPLC peptide loading quantity was optimized based on the total number of identified/quantified proteins after loading 1, 2, 4 and 6 μg of the peptides from the “Test” samples, and the labelling efficiency was checked (Table 1). As shown in this table, the number of identified/quantified proteins increased when more peptides were injected, and the use of the Top6 MS acquisition method allowed the quantification of more than 270 proteins after the injection of 6 μg. Secondly, a Top12 MS acquisition method was evaluated injecting 6 μg of proteins, which did not improve the results obtained using the Top6 method. Moreover, the variability for the protein quantification was similar for all the preparations, indicating that the peptide labelling was adequate. Compared to the work carried out by Lenčo et al. [27] a lower number of quantified proteins was expected as those authors used a faster mass spectrometer and a smaller inner diameter analytical column, which decreases the sample dilution and increases the number of identified proteins. However, they did not quantify and only identified proteins, without specifying the number of peptides used for this identification. In the present study, a restrictive criterion was applied and proteins were identified when at least two peptides belonging to each protein were detected. In addition, a minimum peptide ratio count of two was selected for protein quantification, which means that two peptides for each labelled sample were needed. Despite the fact that the total number of quantified proteins could be increased if more peptides/proteins are injected, the protein quantity was set to 6 μg to analyse the real samples to avoid column overloading and to prevent MS instrument contamination.
Table 1. Loaded peptide quantity, MS detection method, identified/quantified proteins and average quantified proteins in HK-2 cells after UHPLC-Orbitrap-MS/MS analysis.
| Loaded Peptide Quantity (μg) | MS Method | Identified proteins | Quantified proteins | Protein quantification Average ± SD Ratio |
|---|---|---|---|---|
| 1 | Top6 | 51 ± 2 | 32 ± 1 | 0.93 ± 0.13 |
| 2 | Top6 | 131 ± 5 | 81 ± 4 | 0.97 ± 0.18 |
| 4 | Top6 | 265 ± 4 | 188 ± 4 | 0.96 ± 0.14 |
| 6 | Top6 | 321 ± 1 | 278 ± 4 | 0.96 ± 0.15 |
| 6 | Top12 | 207 ± 1 | 137 ± 4 | 0.96 ± 0.16 |
Overview of the temporal expression protein changes
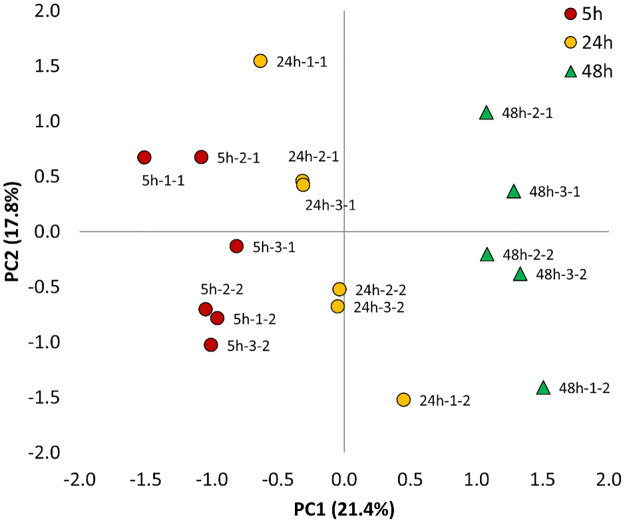
Based on the above information, 6 μg of peptides were injected and the Top6 MS method was selected to study the effect of HG-Hypoxia against NG-Normoxia over time. After applying a FDR of 1%, a total number of 526 proteins were identified, with an average of 317 ± 64, 296 ± 25 and 259 ± 54 proteins at 5, 24 and 48 h, respectively (Table 2 and S1 Table). In addition, a total of 150 proteins were simultaneously quantified in all biological and technical replicates, and a PCA multivariate analysis was carried out to determine, compare and visualize the overall relation between the three conditions studied (Fig 2). This analysis indicates that the combination of the two main components captured almost 40% of the variance of the data, being the principal component 1 the one that clearly separates and groups together the samples representing 5, 24 and 48 h.
Table 2. Number of quantified proteins after the incubation of HK-2 cells in NG-Normoxia and HG-Hypoxia conditions.
| Parameter | 5 h | 24 h | 48 h |
|---|---|---|---|
| Proteins in NG-Normoxia (L)/HG-Hypoxia (H) | 315 | 277 | 250 |
| Proteins in HG-Hypoxia (L)/NG-Normoxia (H) | 319 | 314 | 269 |
| Average proteins (considering 6 replicates) | 317 | 296 | 259 |
| Proteins simultaneously quantified in all replicates | 150 | ||
| Proteins simultaneously quantified in at least four replicates | 286 | 282 | 243 |
| Proteins in at least four replicates with p-val ≤ 0.05 | 35 | 60 | 59 |
| Up-regulated proteinsa | 2 | 3 | 14 |
| Down-regulated proteinsb | 2 | 8 | 13 |
a p-val ≤ 0.05 and log2 fold change ≥ 0.26 between HG-Hypoxia vs NG-Normoxia.
b p-val ≤ 0.05 and log2 fold change ≤ -0.26 between HG-Hypoxia vs NG-Normoxia.
Fig 2. Principal component analysis of the 150 quantified proteins in all biological and technical replicates across the three time points (5, 24 and 48 h) in HK-2 cells treated with HG (25 mM glucose)-hypoxia (1% O2) in comparison to NG (5.5 mM glucose)-normoxia (21% O2).
The first number after the different time points indicates the biological replicate, and the second number indicates the technical replicate.
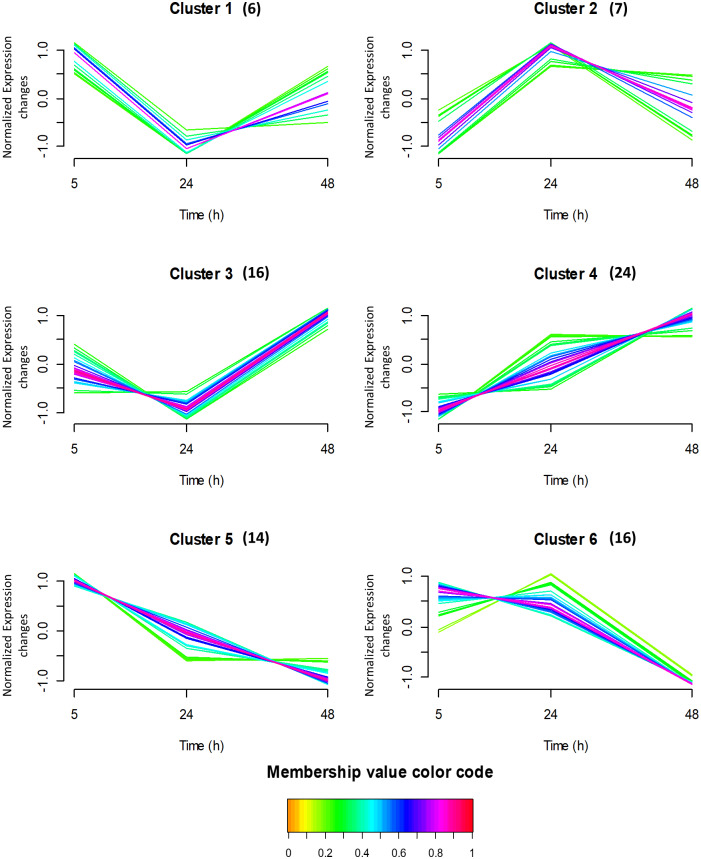
Thereafter, the expression of the same 150 proteins used for PCA were searched for patterns in their time expression profiles using the fuzzy c-means clustering algorithm. This algorithm does not establish a protein expression cut-off filter, but it measures the expression similarity (based on the distance, connectivity, and intensity) between the different proteins across the time, and then groups those proteins accordingly. It has to be noted that each protein can belong to more than one cluster. Using this software, the proteins were clustered in six profiles, containing between 6 and 24 proteins (Fig 3). In the figure, each trace corresponds to a protein, and the colour is coded according to its membership value for the respective cluster (higher values indicate higher similarity to the stablished profile). The proteins in each cluster were then analysed (by functional enrichment analyses) to identify overrepresented GO term and pathways using PANTHER (S2 Table) and STRING (S3 Table) softwares. The proteins considered in cluster 1 (with decreased expression between 5 h and 24 h, but slightly increased between 24 and 48 h) are involved in the translational initiation biological process as some proteins are structural constituent of ribosome. In cluster 2, the protein expression dramatically increased between 5 h and 24 h and then it slightly decreased between 24 and 48 h. These proteins are involved in several terms, but the main molecular function is the RNA binding. The expression of proteins considered in cluster 3 decreased between 5 h and 24 h, but it dramatically increased between 24 and 48 h. These proteins are involved in the broad molecular functions RNA binding and processing. Cluster 4 was the one with the highest number of proteins (24), which abundance continuously increased from 5 to 48 h. These proteins are involved in several biological processes (glycolytic process, ATP generation from ADP, pyruvate biosynthetic process, NADH regeneration, canonical glycolysis), in the Glycolysis/Gluconeogenesis pathways (ENO1, PKM, PGAM1, ALDOA, TPI1, PGK1 and GAPDH) or in the HIF-1 signaling pathway. On the other hand, proteins represented in cluster 5 were highly abundant at 5 h, and their expression constantly decreased with time, being minimal at 48 h. These proteins are involved in the actin-dependent ATPase activity, they are structural constituents of cytoskeleton, or they belong to the ubiquitin-like protein ligase binding, the cadherin binding, or the identical protein binding molecular functions. Some of them also form part of different pathways such as the Cytoskeletal regulation by Rho GTPase. Finally, the expression of proteins in cluster 6 was constant between 5 and 24 h, but it decreased after 48 h. These proteins form part of the structural molecule activity and the RNA binding functions, and in the Eukaryotic Translation Elongation pathway. In summary, this analysis confers an overview of the protein expression profiles and the functions in which the proteins are involved, without losing the information of proteins with small but similar expression patterns.
Fig 3. Time-course protein profiles obtained by fuzzy c-means clustering in HK-2 cells treated with HG (25 mM glucose)-hypoxia (1% O2) in comparison to NG (5.5 mM glucose)-normoxia (21% O2) at 5, 24 and 48 h.
The y axis is log10 transformed and normalized, and the number of proteins considered in each cluster is given in parentheses. Each trace is colour coded according to its membership value for the respective cluster (see colour bar).
Significantly altered proteins at specific time points
In addition to the multivariate and the temporal expression analyses, the most significantly altered proteins between HG-Hypoxia and NG-Normoxia at each specific time point were identified. For this analysis, several filtering criteria were applied (Table 2). Firstly, a protein was selected for further processing when it was identified in at least four sample replicates at the different time points. Taking into account this filtering criteria, 286, 282 and 243 proteins were quantified at 5, 24 and 48 h respectively. Secondly, a p-value cut-off filter <0.05 after one sample t test analysis was applied, and among the proteins quantified at each time point, 35, 60 and 59 passed this criteria at 5, 24 and 48 h, respectively. Finally, and to identify the most significantly altered proteins, a 1.2-fold change cut-off filter (equivalent to 0.26 and -0.26 in log2 scale) was applied and only 4, 11 and 27 proteins (at 5, 24 and 48 h, respectively) passed this criteria. The full list of proteins and their expression values are shown in (Table 3). Additionally, the list of significantly altered proteins at 48 h was searched for enriched GO biological processes using PANTHER and STRING softwares, and the results are shown in Table 4 and Fig 4. The results show that the expression of two ribosomal proteins was decreased (RPL18, RPL34) while the expression of two histone subunits was increased (HIST1H3A, HIST1H4A) after 5 h of treatment. After 24 h, the expression of three additional ribosomal-related proteins was decreased (RPS27, RPL28, UBA52), as well as the expression of different cytoskeletal-related proteins (CNN2, STMN1, TPM1), or other proteins (FN1, STAT1). On the other hand, the levels of two proteins involved in the glycolysis pathway were increased (GPI, LDHA), as well as the rate-limiting enzyme implicated in the reduction of glucose to sorbitol via the polyol pathway (AKR1B1). At 48 h, the levels of two histone subunits already were observed as increased at 5 h (HIST1H3A and HIST1H4A), and the levels of the subunit HIST1H1B, were observed as decreased, while the expression of some proteins observed as decreased at 24 h, decreased even more (FN1, STAT, STMN1). Moreover, the expression of three additional cytoskeletal related proteins were observed as decreased (TMSB4X, TPM4, TUBB), as well as the proteasome subunit PSME1, the ADP/ATP Translocase 2 (SLC25A5) and the ATP Synthase F1 Subunit Beta (ATP5B). On the other hand, most of those proteins with positive ratios are involved in the glycolysis pathway (ALDOA, GAPDH, GPI, LDHA, PGAM1, PGK1, PKM, TPI1), or they are associated with the endoplasmic reticulum (RTN4, HSPA1B, HSPH1). Finally, the expression of AKR1B1 was observed as increased, being its expression higher than at 24 h.
Table 3. Significantly altered proteins after the incubation of HK-2 cells with HG-Hypoxia in comparison to NG-Normoxia for 5, 24 and 48 h.
| 5 h | 24 h | 48 h | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene name | log2FC | N° samples | p-value | log2FC | N° samples | p-value | log2FC | N° samples | p-value |
| AKR1B1 | -0.13 | 6 | 1.9E-03 | 0.27 | 6 | 1.2E-02 | 0.52 | 6 | 2.0E-03 |
| ALDOA | -0.01 | 6 | 5.7E-01 | 0.25 | 6 | 1.2E-05 | 0.45 | 6 | 4.9E-04 |
| ATP5B | -0.10 | 6 | 2.7E-01 | -0.12 | 6 | 9.4E-02 | -0.29 | 6 | 2.2E-02 |
| CNN2 | -0.11 | 6 | 1.8E-01 | -0.27 | 6 | 1.2E-03 | -0.46 | 2 | 3.6E-01 |
| EZR | 0.03 | 6 | 5.7E-01 | 0.08 | 6 | 1.3E-01 | 0.36 | 6 | 6.8E-04 |
| FN1 | -0.03 | 6 | 2.6E-01 | -0.65 | 6 | 5.9E-04 | -0.92 | 6 | 6.7E-07 |
| FUS | -0.02 | 6 | 6.4E-01 | -0.02 | 6 | 4.6E-01 | 0.30 | 5 | 1.3E-03 |
| GAPDH | -0.01 | 6 | 7.4E-01 | 0.07 | 6 | 1.1E-01 | 0.27 | 6 | 1.1E-02 |
| GPI | -0.09 | 6 | 7.5E-02 | 0.28 | 6 | 5.0E-03 | 0.46 | 5 | 2.7E-03 |
| HIST1H1B | 0.03 | 6 | 5.7E-01 | -0.09 | 6 | 2.3E-01 | -0.49 | 5 | 1.1E-02 |
| HIST1H3A | 0.58 | 6 | 4.7E-02 | -0.11 | 6 | 2.5E-01 | -0.66 | 6 | 1.1E-03 |
| HIST1H4A | 0.60 | 6 | 4.0E-02 | -0.11 | 6 | 1.7E-01 | -0.57 | 6 | 9.7E-03 |
| HSPA1B | 0.22 | 6 | 4.1E-04 | 0.23 | 6 | 1.6E-04 | 0.37 | 6 | 4.0E-04 |
| HSPH1 | -0.08 | 2 | 2.7E-01 | 0.53 | 2 | 1.3E-01 | 0.75 | 4 | 2.7E-04 |
| LDHA | -0.01 | 6 | 9.1E-01 | 0.43 | 6 | 1.5E-05 | 0.85 | 6 | 1.5E-05 |
| PGAM1 | -0.01 | 6 | 7.0E-01 | 0.18 | 6 | 4.0E-03 | 0.40 | 6 | 5.0E-04 |
| PGK1 | -0.04 | 6 | 2.0E-01 | 0.23 | 6 | 7.0E-05 | 0.60 | 6 | 4.0E-05 |
| PKM | -0.03 | 6 | 1.3E-01 | 0.12 | 6 | 2.2E-03 | 0.28 | 6 | 2.9E-03 |
| PSME1 | 0.03 | 6 | 7.2E-01 | -0.10 | 6 | 5.3E-02 | -0.29 | 4 | 1.5E-02 |
| RPL18 | -0.30 | 6 | 6.3E-03 | -0.22 | 6 | 3.9E-02 | 0.02 | 5 | 7.4E-01 |
| RPL28 | -0.30 | 2 | 1.4E-01 | -0.40 | 6 | 7.6E-03 | -0.37 | 5 | 1.2E-01 |
| RPL34 | -0.34 | 6 | 2.0E-04 | -0.14 | 6 | 3.3E-02 | -0.01 | 6 | 9.0E-01 |
| RPS27 | 0.10 | 6 | 4.4E-01 | -0.36 | 4 | 3.4E-02 | -0.04 | 3 | 8.8E-01 |
| RTN4 | -0.01 | 6 | 8.6E-01 | 0.14 | 6 | 1.2E-01 | 0.37 | 5 | 1.7E-02 |
| S100A6 | 0.01 | 6 | 9.1E-01 | -0.18 | 6 | 7.6E-02 | -0.30 | 6 | 1.5E-05 |
| SLC25A5 | 0.15 | 6 | 1.4E-02 | -0.03 | 6 | 5.5E-01 | -0.33 | 6 | 8.9E-03 |
| STAT1 | 0.05 | 6 | 1.8E-02 | -0.29 | 6 | 9.0E-04 | -0.79 | 6 | 1.4E-06 |
| STMN1 | -0.14 | 6 | 1.7E-01 | -0.26 | 6 | 4.7E-04 | -0.70 | 6 | 4.3E-03 |
| TMSB4X | 0.09 | 6 | 5.8E-01 | -0.18 | 6 | 2.2E-02 | -0.40 | 6 | 1.7E-03 |
| TPI1 | -0.05 | 6 | 8.7E-02 | 0.21 | 6 | 9.6E-06 | 0.35 | 6 | 4.0E-04 |
| TPM1 | -0.13 | 6 | 1.2E-01 | -0.90 | 4 | 7.8E-06 | - | - | - |
| TPM4 | -0.05 | 6 | 4.8E-01 | -0.10 | 6 | 1.5E-01 | -0.31 | 6 | 3.9E-02 |
| TUBB | -0.15 | 6 | 5.9E-03 | -0.24 | 6 | 5.2E-03 | -0.29 | 6 | 1.0E-03 |
| UBA52 | -0.14 | 6 | 5.2E-02 | -0.33 | 6 | 6.7E-04 | -0.13 | 6 | 3.8E-02 |
* In grey, significantly altered proteins.
Table 4. Overrepresented PANTHER GO biological processes after the incubation of HK-2 cells with HG-Hypoxia in comparison to NG-Normoxia for 48 h.
| PANTHER GO Biological Process | N° proteins (reference) | 48 h | |||
|---|---|---|---|---|---|
| N° proteins | Protein names | Enrichment | FDR | ||
| Glycolytic process | 18 | 5 | PGK1, ALDOA, PKM, GPI, TPI1 | > 100 | 1.4e-07 |
| Glucose metabolic process | 25 | 3 | PGK1, GPI, TPI1 | 96.24 | 7.1e-04 |
| Regulation of proteasomal protein catabolic process | 25 | 2 | HSPA1B, PSME1 | 64.16 | 3.8e-02 |
| Carbohydrate biosynthetic process | 43 | 3 | PGK1, GPI, TPI1 | 55.95 | 2.7e-03 |
| Oxidation-reduction process | 185 | 5 | PGK1, ALDOA, PKM, GPI, TPI1 | 21.67 | 4.9e-04 |
| Regulation of organelle organization | 271 | 4 | HIST1H1B, TMSB4X, HSPA1B, STMN1 | 11.84 | 2.7e-02 |
| Cytoskeleton organization | 587 | 5 | TMSB4X, TPM4, HSPA1B, STMN1, TUBB | 6.83 | 4.7e-02 |
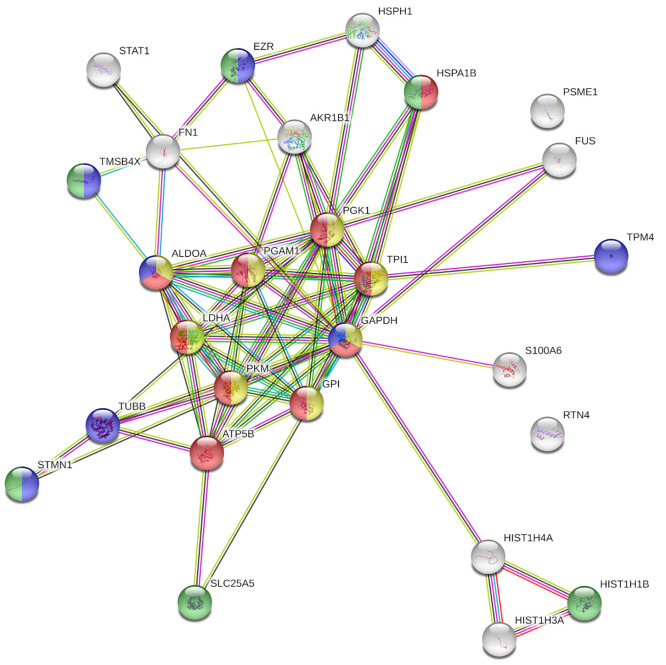
Fig 4. STRING diagram representing protein interaction pathway derived from the 27 proteins significantly altered at 48 h after the incubation of HK-2 cells with HG-Hypoxia in comparison to NG-Normoxia.
The average local clustering coefficient as reported by STRING is 0.703. Proteins in yellow belong to the glycolytic process; proteins in red belong to the ATP metabolic process; proteins in blue belong to the cytoskeleton organization; and proteins in green belong to the regulation of organelle organization.
Discussion
In this study, a UHPLC-MS method was optimized to monitor, for the first time, the protein expression dynamics of HK-2 cells subjected to a diabetic-like microenvironment consisting of HG (25 mM) and hypoxia (1% O2) at different time points (5, 24 and 48 h). One important aspect when studying proteome dynamics is that changes in protein abundance depend on the synthesis of new proteins and/or their degradation, which often requires hours/days. However, the analysis time-frame of these in vitro proteomics studies is usually limited because the control of cell characteristics is commonly lost after 72 hours [28]. The present study demonstrates that short exposure times (5 h) do not significantly affect the overall protein expression, but this expression is significantly affected when the exposure time is sustained for 24 and 48 h, being the changes at 48 h the most remarkable.
As presented in the results section, the most significant protein changes are related to the glycolytic pathway. It is known that oxidation of glucose in the cytosol through glycolysis leads to pyruvate as a final product and renders only two ATP molecules (as compared to 38 ATP molecules generated by complete oxidation of glucose via pyruvate through the tricarboxylic acid cycle and oxidative phosphorylation). Nevertheless, the rate of glucose metabolism through this pathway, in which oxygen is not required, is 10–100 times faster than that through the complete aerobic oxidation of glucose in mitochondria [3]. Therefore, anaerobic glycolysis in the hypoxic kidneys of diabetic patients might contribute to the quick metabolism of glucose from the systemic circulation [29]. In fact, previous studies have shown that the development of DKD is associated with increased anaerobic glycolysis in detriment of complete aerobic oxidation of glucose in mitochondria [30, 31]. In this context, it is not surprising that, in the specific conditions performed in the present study, the glycolysis pathway is the most affected pathway over time in HK-2 cells, being increased from 5 to 48 h. The enhanced expression of 7 glycolytic enzymes (ALDOA, ALDOB, ENO1, GAPDH, PGAM1, PKM2 and TPI1), as well as LDHB and AKR1B1 was identified in a proteomic analysis of post-mortem glomeruli of kidneys from diabetic patients, [31], which is highly coincident with the data obtained in our study. This suggests that our experimental in vitro model of diabetic-like milieu-induced protein expression changes in PTC reproduces important changes in the expression of glucose metabolizing enzymes induced by diabetes at the renal level, and that some of the observed changes in our setting might be beneficial adaptive responses to the diabetic-like microenvironment.
A previous study in db/db type 2 diabetic mice shows little coincidence with our proteomic results. In that study, transcriptomic and metabolomics analyses also indicated that diabetes increases the expression of several glycolytic enzymes in kidney cortex, which is 90% proximal tubules [30], but the enzymes whose mRNA expression was significantly increased were hexokinase, phosphofructokinase and pyruvate kinase (the only enzyme coincident with our study). The little correlation between the proteomic and the transcriptomic profiles has been previously found in cultured skin fibroblasts from type 1 diabetes patients, and suggested to the authors that the expression of glycolytic enzymes is mainly regulated at a post-transcriptional level [32]. A similar discordance between the transcriptome and the metabolome in sciatic nerve from db/db type 2 diabetic mice led the authors to suggest that post-transcriptional or post-translational modifications are important in the development of diabetic nephropathy [30]. Moreover, different transcriptomic studies of kidney biopsy samples from patients with DKD or DN have not identified significant changes in the mRNA levels of glycolytic enzymes [33, 34].
Besides glycolytic enzymes, other proteins whose changes might be considered as part of a beneficial adaptive response to the diabetic-like microenvironment are STAT1 and HSPs. Inhibition of STAT1 (as in our results), a cytosolic transcription factor, has been found to protect against tubulointerstitial injury in animal models of DKD [35,36]. On the other hand, overexpression of several cytoprotective HSPs has been found in animal models of diabetes [37]. In our study, expression of HSP70 isoform 1B and HSP105 in HK-2 cells was increased along the time (being maximum at 48 h) by diabetic-like milieu. HSP105 has been shown as an important element of the HSP70 machinery for refolding of denatured proteins and protection against stress-induced cell death in mammalian cells [38], which suggests that overexpression of HSP105 and HSP70 in HK-2 cells might be a coordinated response against the cellular stress imposed by hypoxia and hyperglycemia.
In our study, the results related with the pathogenesis of DKD can be classified in those involving the mitochondrial reversible proton pump (F1F0-ATP synthase) and those involving changes in the expression of fibrosis-related proteins. Regarding F1F0-ATP synthase, it is well known that diabetes impairs kidney mitochondrial bioenergetics [39] and that decreased levels of ATP5B have been shown to aggravate DKD [40]. Among the different enzymes involved in the oxidative phosphorylation (OxPhos) mechanism, F1F0-ATP synthase is essential to synthesize ATP in the mitochondrion, and synthesized ATP is then exported to the cytosol by the adenine nucleotide translocase [41]. In our study, we have observed a significant decrease of ATP5B levels at 48 h and a decrease in the expression of ATP5A1 over the time, which suggests a shut-down of the OxPhos mechanism leading to a decrease in mitochondrial ATP. These results agree well with previous studies in PTC in which HG exposition reduced the ATP levels and other respiratory parameters [42, 43] or the expression of ATP synthase subunit d (ATP5PD) [44].
With regard to the expression of fibrosis-related proteins, we did not find any increase in the expression of extracellular matrix proteins (fibronectin was even decreased) in HK-2 cells exposed to hypoxia/high glucose conditions. This contrasts with the generally accepted view that deposition of extracellular matrix proteins leading to tubulointerstitial fibrosis is involved in the pathogenesis of DKD [45] and suggests that it is required more than 48 h to increase the expression of extracellular matrix proteins in HK-2 cells under diabetic-like conditions. This view may be supported by the fact that we did not find either any statistically significant difference between cells incubated in control conditions and cells incubated in diabetic-like microenvironment in the total protein/cell ratio (an index of cell hypertrophy) [46] which is expected to be increased because PTC hypertrophy is a consequence of diabetes [47]. Contrarily to extracellular matrix proteins, other changes in proteins related to renal fibrosis, i.e. cytoskeletal-related proteins [48], have been adequately detected in our experimental setting and we speculate that the decrease in the expression of TUBB, CNN2, TPM1 and ACTB in HK-2 cells exposed to the diabetic-like microenvironment might also occur in proximal tubules of diabetic patients, thereby initiating cytoskeletal alterations leading to tubulointerstitial fibrosis. The same hypothesis applies to the decreased expression of STMN1, TMSB4X and MYH9 in HK-2 cells under diabetic-like conditions, because: i) STMN1 is a microtubule-destabilizing phosphoprotein [49] whose deficiency in mice has been connected to the development of renal fibrosis [50]. In addition, STMN1 is a cell cycle regulator and its down-regulation by hypoxia has been linked to renal fibrosis in HK-2 cells through G2/M cell cycle arrest [51]; ii) TMSB4X is a potent regulator of actin polymerization with critical roles in maintaining the cell cytoskeleton [52], and daily TMSB4X treatment for 3 months reduced albuminuria and attenuated renal tubulointerstitial fibrosis in a mouse model of type 2 diabetes mellitus [53]. TMSB4X also alleviates renal tubulointerstitial fibrosis and tubular cell apoptosis in a rat model of tubulointerstitial fibrosis [54], and in tubular epithelial cells from a rat model of renal fibrosis through the TGF-β pathway [55]; iii) MYH9 is an actin regulator via activation of RhoA whose deletion in mice was associated with fibrosis and proteinuria [56]. Another cytoskeleton-related protein that might contribute to tubulointerstitial fibrosis in DKD is ezrin, which links membrane proteins to the actin cytoskeleton, and whose expression increased in HK-2 cells exposed to the diabetic-like milieu. This protein belongs to the ezrin/radixin/moesin protein family that can interact with membrane and F-actin proteins [57]. Ezrin has been suggested to contribute in PTC to the uptake by micropinocytosis of advanced glycation end products (which results from the non-enzymatic glycation of proteins during chronic hyperglycemia and contribute to DKD) [58] and thereby to promote diabetes-induced tubulointerstitial fibrosis [59]. However, further studies are needed to connect all the mentioned cytoskeletal-related changes with fibrosis in a mechanistic fashion.
It should be noted that our work has a number of limitations. We have found that exposure of HK-2 cells to the diabetic-like microenvironment results in a time-dependent increase in cell death, reaching values higher than 20% at 48 h (Fig 1A). Although the presence of some apoptotic tubular cells in both human and experimental DKD is well stablished [60–63], excessive acute cell death is not a feature of DKD. We do not know the reason of the high cell death values found in vitro, but even higher cell death values have been found in a previous work in which HK-2 cells or NRK-52E renal tubular epithelial cells were exposed to 30 mM glucose/1% O2 and apoptosis was quantified [64]. Furthermore, high glucose or hypoxia separately also induce in cultured proximal tubular cells around 15%-25% apoptotic cell death [65–70]. Thus, in DKD studies the in vitro models involving cultured PTC do not adequately reproduce the loss of cell viability found in vivo. In this connection, it should be also noted the limitations of cultured HK-2 cells in particular and of 2-dimension (2D) proximal tubular cell culture in general as models of the proximal tubule. Thus, many of the available model cell lines fail to replicate the differential expression of several uptake and efflux membrane transporters and metabolizing enzymes, which is one of the characteristics of native PTC [71]. Another major shortcoming of PTC culture is the use of metabolic fuels as compared to native proximal tubules: these reabsorb the filtered glucose and are gluconeogenic, but to drive their energy needs, native PTC rely physiologically on oxidative metabolism rather than glycolysis [72–74]. However, when PTC are brought into tissue culture, they revert from oxidative metabolism and gluconeogenesis to high rates of glycolysis. In the case of HK-2 cells, our current results and previous reports [42, 75–77], indicate that they have glycolytic and gluconeogenic activities. Interestingly, in DKD [30] and other pathological conditions, proximal tubule glycolytic flux plays an important pathogenic role [73, 78, 79, 80], which suggests that our results on glycolytic enzymes in HK-2 cells exposed to the diabetic-like milieu might be pathologically relevant. However, specific studies in native proximal tubules should confirm this hypothesis. On the other hand, it is important to note that our model has been specifically conceived to monitor the effects over time of HG and hypoxia on the protein expression in HK-2 cells and, therefore, we refer to these conditions (25 mM glucose/1% O2) as “diabetic-like” microenvironment rather than diabetic microenvironment because the latter is much more complex [5]. Besides, the very large and rapid time changes in glucose and O2 utilized here are far from being physiologic (or even pathologic, excepting in sudden vascular occlusion) and 25 mM glucose is higher than the plasma glucose concentration in most diabetic patients, whereas it is not known whether 1% O2 is the actual degree of hypoxia to which PTC are exposed in the kidneys of the diabetic patients. Furthermore, due to poor penetration of O2 through the medium, the pericellular concentration of O2 in cells grown in culture under atmospheric air may reach the hypoxic range [81]. On the other hand, short term exposure to HG/hypoxia may not reveal all the long-term proteomic alterations in PTC during diabetes, as suggested by our results on the expression of extracellular matrix proteins. Besides these limitations, the novelty of the present study resides on the use of a relatively simple in vitro model that when simultaneously exposed to hyperglycemia and hypoxia, but not exposed separately to either condition, show functional alterations that reproduce important changes induced by diabetes at the renal level in: i) the expression of glucose metabolizing enzymes ii) STAT1 and HSPs and iii) cytoskeletal-related proteins. Because of that, we believe that this model could be used as a previous step before the use of expensive in vivo assays.
In summary, the model we have used allows for a detailed study of the proteomic alterations induced by HG/hypoxia (i.e. two major conditions of the microenvironment that surrounds PTC in diabetes) in HK-2 cells, which may help to identify new mechanisms and targets to be investigated in the future for a better understanding of DKD. But the model has several limitations when compared with the in vivo situation in which native PTC are exposed to the real diabetic environment and therefore further studies in renal samples from diabetic patients or from animal models of diabetes are also required to have a better representation of the proteomic changes induced in PTC by the diabetic milieu.
Conclusion
This is the first time-series proteomic study based on DML and UHPLC-MS/MS technology carried out to analyse the early effects of the diabetic milieu on proximal tubular cells using a model very simplified of the diabetic microambient (i.e. HG and hypoxia) that surrounds proximal tubules in diabetes. The combined exposure to HG and hypoxia conditions increased the percentage of HK-2 dead cells after 24 h without affecting the protein/cell ratio, and it significantly altered the expression of different proteins in a time-dependent manner. Several of these proteins are involved in the glycolysis pathway and their expression control seems to be HIF-1α-independent. Moreover, the abundancy of other proteins involved in the refolding of denatured proteins (HSP105 and HSP70) was also increased with time, which might be a coordinated response against the cellular stress imposed by the conditions used. On the other hand, the expression of different enzymes involved in the oxidative phosphorylation mechanism and the generation of ATP (SLC25A5, ATP5B, ATP5A1) was decreased with time, reflecting a mitochondrial dysfunction that could contribute to the development of DKD. Finally, the time expression profile showed a decrease in the expression of ten cytoskeletal-related proteins (CNN2, TPM1, STMN1 and MYH9 being reported for the first time in connection with DKD), which might be associated to renal fibrosis, but further studies are needed to connect these changes with fibrosis. With the limitations inherent to the very simplified model of the diabetic microenvironment that surrounds proximal tubules in diabetes, the present proteomic study suggests new mechanisms and targets to be investigated in proximal tubular cells in the context of the pathogenesis of DKD.
Supporting information
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
Authors thank the Center for Applied Chemistry and Biotecnology (CQAB) of the University of Alcalá for the availability of UHPLC-Orbitrap equipment.
Data Availability
All raw mass spectrometry proteomics data files, MaxQuant output files and MaxQuant software used for the search are available from the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD015102.
Funding Statement
This work was supported by the Spanish Ministry of Economy and Competitiveness (CTQ2016-76368-P to MLM and MC, SAF2014-53218-R to FJL, FJCI-2016-30741 to AV, and RYC-2013-12688 to MC), by the Comunidad of Madrid (Spain) and the FSE and FEDER European programs (S2018/BAA-4393 - AVANSECAL-II-CM to MLM and B2017/BMD3686 to FJL), and by the University of Alcalá. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Altelaar AF, Munoz J, Heck AJ. Next-generation proteomics: towards an integrative view of proteome dynamics. Nat Rev Genet 2013;14:35–48. 10.1038/nrg3356 [DOI] [PubMed] [Google Scholar]
- 2.Aebersold R, Mann M. Mass-spectrometric exploration of proteome structure and function. Nature. 2016;537:347–355. 10.1038/nature19949 [DOI] [PubMed] [Google Scholar]
- 3.Zhang G, Darshi M, Sharma K. The Warburg effect in diabetic kidney disease. Semin Nephrol 2018;38:111–120. 10.1016/j.semnephrol.2018.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vallon V. The proximal tubule in the pathophysiology of the diabetic kidney. Am J Physiol Regul Integr Comp Physiol 2011;300:R1009–1022. 10.1152/ajpregu.00809.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Slyne J, Slattery C, McMorrow T, Ryan MP. New developments concerning the proximal tubule in diabetic nephropathy: In vitro models and mechanisms. Nephrol Dial Transplant 2015;30:iv60–iv67. 10.1093/ndt/gfv264 [DOI] [PubMed] [Google Scholar]
- 6.Gilbert RE. Proximal tubulopathy: prime mover and key therapeutic target in diabetic kidney disease. Diabetes. 2017;66:791–800. 10.2337/db16-0796 [DOI] [PubMed] [Google Scholar]
- 7.Rosenberger C, Khamaisi M, Abassi Z, Shilo V, Weksler-Zangen S, Goldfarb M, et al. Adaptation to hypoxia in the diabetic rat kidney. Kidney Int 2008;73:34–42. 10.1038/sj.ki.5002567 [DOI] [PubMed] [Google Scholar]
- 8.Nordquist L, Friederich-Persson M, Fasching A, Liss P, Shoji K, Nangaku M, et al. Activation of hypoxia-inducible factors prevents diabetic nephropathy. J Am Soc Nephrol 2015;26:328–338. 10.1681/ASN.2013090990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thaysen JH, Lassen NA, Munck O. Sodium transport and oxygen consumption in the mammalian kidney. Nature 1961;190:919–921. 10.1038/190919a0 [DOI] [PubMed] [Google Scholar]
- 10.Dengler VL, Galbraith M, Espinosa JM. Transcriptional regulation by hypoxia inducible factors. Crit Rev Biochem Mol Biol 2014;49:1–15. 10.3109/10409238.2013.838205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thangarajah H, Yao D, Chang EI, Shi Y, Jazayeri L, Vial IN, et al. The molecular basis for impaired hypoxia-induced VEGF expression in diabetic tissues. Proc Natl Acad Sci USA 2009;106:13505–13510. 10.1073/pnas.0906670106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang ZZ, Zhang AY, Yi FX, Li PL, Zou AP. Redox regulation of HIF-1α levels and HO-1 expression in renal medullary interstitial cells. Am J Physiol Renal Physiol 2003;284:F1207–1215. 10.1152/ajprenal.00017.2002 [DOI] [PubMed] [Google Scholar]
- 13.Ohtomo S, Nangaku M, Izuhara Y, Takizawa S, Strihou CV, Miyata T. Cobalt ameliorates renal injury in an obese, hypertensive type 2 diabetes rat model. Nephrol Dial Transplant 2007;23: 1166–1172. 10.1093/ndt/gfm715 [DOI] [PubMed] [Google Scholar]
- 14.Gu HF, Zheng X, Abu Seman N, Gu T, Botusan IR, Sunkari VG, et al. Impact of the hypoxia-inducible factor-1 α (HIF1A) Pro582Ser polymorphism on diabetes nephropathy. Diabetes Care 2013;36:415–421. 10.2337/dc12-1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Persson P, Palm F. Hypoxia-inducible factor activation in diabetic kidney disease. Curr Opin Nephrol Hypertens 2017;26:345–350. 10.1097/MNH.0000000000000341 [DOI] [PubMed] [Google Scholar]
- 16.Isoe T, Makino Y, Mizumoto K, Sakagami H, Fujita Y, Honjo J, et al. High glucose activates HIF-1-mediated signal transduction in glomerular mesangial cells through a carbohydrate response element binding protein. Kidney Int 2010;78:48–59. 10.1038/ki.2010.99 [DOI] [PubMed] [Google Scholar]
- 17.Lu H, Dalgard CL, Mohyeldin A, McFate T, Tait AS, Verma A. Reversible inactivation of HIF-1 prolyl hydroxylases allows cell metabolism to control basal HIF-1. J Biol Chem 2005;280:41928–41939. 10.1074/jbc.M508718200 [DOI] [PubMed] [Google Scholar]
- 18.García-Pastor C, Benito-Martínez S, Moreno-Manzano V, Fernández-Martínez AB, Lucio-Cazaña FJ. Mechanism and Consequences of The Impaired Hif-1α Response to Hypoxia in Human Proximal Tubular HK-2 Cells Exposed to High Glucose. Sci Rep 2019;9:15868 10.1038/s41598-019-52310-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yeung YG, Stanley ER. Rapid detergent removal from peptide samples with ethyl acetate for mass spectrometry analysis. Curr Protoc Protein Sci 2010;16:16.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boersema PJ, Raijmakers R, Lemeer S, Mohammed S, Heck AJ. Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat Protoc 2009;4:484–494. 10.1038/nprot.2009.21 [DOI] [PubMed] [Google Scholar]
- 21.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 2008;26:1367–1372. 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- 22.Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. Andromeda: A Peptide Search Engine Integrated into the MaxQuant Environment. J Proteome Res 2011;10:1794–1805. 10.1021/pr101065j [DOI] [PubMed] [Google Scholar]
- 23.Vizcaíno JA, Deutsch EW, Wang R, Csordas A, Reisinger F, Ríos D, et al. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat Biotechnol 2014;32:223–226. 10.1038/nbt.2839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Futschik M, Carlisle B. Noise robust clustering of gene expression time-course data. J Bioinform Comput Biol 2005;4:965–988. [DOI] [PubMed] [Google Scholar]
- 25.Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res 2003;13:2129–2141. 10.1101/gr.772403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019;47: D607–D613. 10.1093/nar/gky1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lenčo J, Vajrychová M, Pimková K, Prokšová M, Benková M, Klimentová J, et al. Conventional-Flow Liquid Chromatography-Mass Spectrometry for Exploratory Bottom-Up Proteomic Analyses. Anal Chem 2018;90:5381–5389. 10.1021/acs.analchem.8b00525 [DOI] [PubMed] [Google Scholar]
- 28.Sabatier P, Saei AA, Wang S, Zubarev RA. Dynamic Proteomics Reveals High Plasticity of Cellular Proteome: Growth-Related and Drug-Induced Changes in Cancer Cells are Comparable. Proteomics 2018;18:e1800118 10.1002/pmic.201800118 [DOI] [PubMed] [Google Scholar]
- 29.Takiyama Y, Haneda M. Hypoxia in Diabetic Kidneys. BioMed Res Int 2014;2014:837421 10.1155/2014/837421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sas KM, Kayampilly P, Byun J, Nair V, Hinder LM, Hur J, et al. Tissue-specific metabolic reprogramming drives nutrient flux in diabetic complications. JCI Insight 2016;1:e86976 10.1172/jci.insight.86976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qi W, Keenan HA, Li Q, Ishikado A, Kannt A, Sadowski T, et al. Pyruvate kinase M2 activation may protect against the progression of diabetic glomerular pathology and mitochondrial dysfunction. Nat Med 2017;23:753–762. 10.1038/nm.4328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iori E, Millioni R, Puricelli L, Arrigoni G, Lenzini L, Trevisan R, et al. Glycolytic enzyme expression and pyruvate kinase activity in cultured fibroblasts from type 1 diabetic patients with and without nephropathy. Biochim Biophys Acta 2008;1782:627–633. 10.1016/j.bbadis.2008.08.012 [DOI] [PubMed] [Google Scholar]
- 33.Woroniecka KI, Park AS, Mohtat D, Thomas DB, Pullman JM, Susztak K. Transcriptome analysis of human diabetic kidney disease. Diabetes 2011;60:2354–2369. 10.2337/db10-1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fan Y, Yi Z, D'Agati VD, Sun Z, Zhong F, Zhang W, et al. Comparison of Kidney Transcriptomic Profiles of Early and Advanced Diabetic Nephropathy Reveals Potential New Mechanisms for Disease Progression. Diabetes 2019;68:2301–2314. 10.2337/db19-0204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang SC, Leung JC, Chan LY, Cheng AS, Lan HY, Lai KN. Renoprotection by rosiglitazone in accelerated type 2 diabetic nephropathy: Role of STAT1inhibition and nephrin restoration. Am J Nephrol 2010;32:145–155. 10.1159/000316056 [DOI] [PubMed] [Google Scholar]
- 36.Lopez-Sanz L, Bernal S, Recio C, Lazaro I, Oguiza A, Melgar A, et al. SOCS1-targeted therapy ameliorates renal and vascular oxidative stress in diabetes via STAT1and PI3K inhibition. Lab Invest 2018;98:1276–1290. 10.1038/s41374-018-0043-6 [DOI] [PubMed] [Google Scholar]
- 37.Chevotareva N, Bobkova I, Shilov E. Heat shock proteins and kidney disease: perspectives of HSP therapy. Cell Stress Chaperones 2017;22:319–343. 10.1007/s12192-017-0790-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamagishi N, Yokota M, Yasuda K, Saito Y, Nagata K, Hatayama T. Characterization of stress sensitivity and chaperone activity of Hsp105 in mammalian cells. Biochem Biophys Res Commun 2011;409:90–95. 10.1016/j.bbrc.2011.04.114 [DOI] [PubMed] [Google Scholar]
- 39.Higgins GC, Coughlan MT. Mitochondrial dysfunction and mitophagy: the beginning and end to diabetic nephropathy? Br J Pharmacol 2014;171:1917–1942. 10.1111/bph.12503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guan SS, Sheu ML, Wu CT, Chiang CK, Liu SH. ATP synthase subunit-β down-regulation aggravates diabetic nephropathy. Sci Rep 2015;5:14561 10.1038/srep14561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chevrollier A, Loiseau D, Reynier P, Stepien G. Adenine nucleotide translocase 2 is a key mitochondrial protein in cancer metabolism. Biochim Biophys Acta 2011;1807:562–567. 10.1016/j.bbabio.2010.10.008 [DOI] [PubMed] [Google Scholar]
- 42.Czajka A, Malik AN. Hyperglycemia induced damage to mitochondrial respiration in renal mesangial and tubular cells: Implications for diabetic nephropathy. Redox Biol 2016;10:100–107. 10.1016/j.redox.2016.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Munusamy S, MacMillan-Crow LA. Mitochondrial superoxide plays a crucial role in the development of mitochondrial dysfunction during high glucose exposure in rat renal proximal tubular cells. Free Radic Biol Med 2009;46:1149–1157. 10.1016/j.freeradbiomed.2009.01.022 [DOI] [PubMed] [Google Scholar]
- 44.So EJ, Kim HJ, Kim CW. Proteomic analysis of human proximal tubular cells exposed to high glucose concentrations. Proteomics Clin Appl 2008;2:1118–1126. 10.1002/prca.200780151 [DOI] [PubMed] [Google Scholar]
- 45.Magri CJ, Fava S. The role of tubular injury in diabetic nephropathy. Eur J Intern Med 2009;20:551–555. 10.1016/j.ejim.2008.12.012 [DOI] [PubMed] [Google Scholar]
- 46.Bera A, Das F, Ghosh-Choudhury N, Mariappan MM, Kasinath BS, Ghosh Choudhury G. Reciprocal regulation of miR-214 and PTEN by high glucose regulates renal glomerular mesangial and proximal tubular epithelial cell hypertrophy and matrix expansion. Am J Physiol-Cell Physiol 2017;313:C430–C447. 10.1152/ajpcell.00081.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vallon V, Komers R. Pathophysiology of the diabetic kidney. Compr Physiol 2011;1:1175–1232. 10.1002/cphy.c100049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parrish AR. The cytoskeleton as a novel target for treatment of renal fibrosis. Pharmacol Ther 2016;166:1–8. 10.1016/j.pharmthera.2016.06.006 [DOI] [PubMed] [Google Scholar]
- 49.Belletti B, Baldassarre G. Stathmin: a protein with many tasks. New biomarker and potential target in cancer. Expert Opin Ther Targets 2011;15:1249–1266. 10.1517/14728222.2011.620951 [DOI] [PubMed] [Google Scholar]
- 50.Zahedi K, Revelo MP, Barone S, Wang Z, Tehrani K, Citron DP, et al. Stathmin-deficient mice develop fibrosis and show delayed recovery from ischemic-reperfusion injury. Am J Physiol-Renal Physiol 2006;290:F1559–F1567. 10.1152/ajprenal.00424.2005 [DOI] [PubMed] [Google Scholar]
- 51.Liu T, Liu L, Liu M, Du R, Dang Y, Bai M, et al. MicroRNA-493 targets STMN-1 and promotes hypoxia-induced epithelial cell cycle arrest in G2/M and renal fibrosis. FASEB J 2019;33:1565–1577. 10.1096/fj.201701355RR [DOI] [PubMed] [Google Scholar]
- 52.Sanders MC, Goldstein AL, Wang YL. Thymosin beta 4 (Fx peptide) is a potent regulator of actin polymerization in living cells. Proc Natl Acad Sci USA 1992;89:4678–4682. 10.1073/pnas.89.10.4678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu J, Su LP, Zhou Y, Ye L, Lee KO, Ma JH. Thymosin beta4 attenuates early diabetic nephropathy in a mouse model of type 2 diabetes mellitus. Am J Ther 2015;22:141–146. 10.1097/MJT.0b013e3182785ecc [DOI] [PubMed] [Google Scholar]
- 54.Kumar N, Liao TD, Romero CA, Maheshwari M, Peterson EL, Carretero OA. Thymosin β4 Deficiency Exacerbates Renal and Cardiac Injury in Angiotensin-II-Induced Hypertension. Hypertension. 2018;71:1133–1142. 10.1161/HYPERTENSIONAHA.118.10952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yuan J, Shen Y, Yang X, Xie Y, Lin X, Zeng W, et al. Thymosin β4 alleviates renal fibrosis and tubular cell apoptosis through TGF-β pathway inhibition in UUO rat models. BMC Nephrol 2017;18:314 10.1186/s12882-017-0708-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thelen S, Abouhamed M, Ciarimboli G, Edemir B, Bähler M. Rho GAP myosin IXa is a regulator of kidney tubule function. Am J Physiol-Renal Physiol 2015;309:F501–F513. 10.1152/ajprenal.00220.2014 [DOI] [PubMed] [Google Scholar]
- 57.Fehon RG, McClatchey AI, Bretscher A. Organizing the cell cortex: the role of ERM proteins. Nat Rev Mol Cell Biol 2010;11:276–287. 10.1038/nrm2866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sun YM, Su Y, Li J, Wang LF. Recent advances in understanding the biochemical and molecular mechanism of diabetic nephropathy. Biochem Biophys Res Commun 2013;433:359–361. 10.1016/j.bbrc.2013.02.120 [DOI] [PubMed] [Google Scholar]
- 59.Gallicchio MA, Bach LA. Uptake of advanced glycation end products by proximal tubule. Epithelial cells via macropinocytosis. Biochim Biophys Acta 2013;1833:2922–2933. 10.1016/j.bbamcr.2013.05.024 [DOI] [PubMed] [Google Scholar]
- 60.Kuwagata S, Kume S, Chin-Kanasaki M, Araki H, Araki S, Nakazawa J, et al. MicroRNA148b-3p inhibits mTORC1-dependent apoptosis in diabetes by repressing TNFR2 in proximal tubular cells. Kidney Int 2016;90:1211–1225. 10.1016/j.kint.2016.06.036 [DOI] [PubMed] [Google Scholar]
- 61.Brezniceanu ML, Liu F, Wei CC, Chénier I, Godin N, Zhang SL, et al. Attenuation of Interstitial Fibrosis and Tubular Apoptosis in db/db Transgenic Mice Overexpressing Catalase in Renal Proximal Tubular Cells. Diabetes 2008;57:451–459. 10.2337/db07-0013 [DOI] [PubMed] [Google Scholar]
- 62.Kumar D, Zimpelmann J, Robertson S, Burns KD. Tubular and interstitial cell apoptosis in the streptozotocin-diabetic rat kidney. Nephron Exp Nephrol 2004;96:e77–e88. 10.1159/000076749 [DOI] [PubMed] [Google Scholar]
- 63.Kumar D, Robertson S, Burns KD. Evidence of apoptosis in human diabetic kidney. Mol Cell Biochem 2004;259:67–70. 10.1023/b:mcbi.0000021346.03260.7e [DOI] [PubMed] [Google Scholar]
- 64.Zhang X, Guan T, Yang B, Chi Z, Wan Q, Gu HF. Protective effect of berberine on high glucose and hypoxia-induced apoptosis via the modulation of HIF-1α in renal tubular epithelial cells. Am J Transl Res 2019;11:669–682. [PMC free article] [PubMed] [Google Scholar]
- 65.Wang J, Guo HM. Astragaloside IV ameliorates high glucose-induced HK-2 cell apoptosis and oxidative stress by regulating the Nrf2/ARE signaling pathway. Exp Ther Med 2019;17:4409–4416. 10.3892/etm.2019.7495 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 66.Huang YF, Zhang Y, Liu CX, Huang J, Ding GH. microRNA-125b contributes to high glucose-induced reactive oxygen species generation and apoptosis in HK-2 renal tubular epithelial cells by targeting angiotensin-converting enzyme 2. Eur Rev Med Pharmacol Sci 2016;20:4055–4062. [PubMed] [Google Scholar]
- 67.Guo C, Li Y, Zhang R, Zhang Y, Zhao J, Yao J, et al. Protective Effect of Salidroside Against Diabetic Kidney Disease Through Inhibiting BIM-Mediated Apoptosis of Proximal Renal Tubular Cells in Rats. Front Pharmacol 2018;9:1433 10.3389/fphar.2018.01433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li M, Khan AM, Maderdrut JL, Simon EE, Batuman V. The Effect of PACAP38 on MyD88-Mediated Signal Transduction in Ischemia-/Hypoxia-Induced Acute Kidney Injury. Am J Nephrol 2010;32:522–532. 10.1159/000321491 [DOI] [PubMed] [Google Scholar]
- 69.Mimura I, Hirakawa Y, Kanki Y, Kushida N, Nakaki R, Suzuki Y, et al. Novel lnc RNA regulated by HIF-1 inhibits apoptotic cell death in the renal tubular epithelial cells under hypoxia. Physiol Rep 2017;5:e13203 10.14814/phy2.13203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tanaka T, Hanafusa N, Ingelfinger JR, Ohse T, Fujita T, Nangaku M. Hypoxia induces apoptosis in SV40-immortalized rat proximal tubular cells through the mitochondrial pathways, devoid of HIF1-mediated upregulation of Bax. Biochem Biophys Res Commun 2003;309:222–231. 10.1016/s0006-291x(03)01557-2 [DOI] [PubMed] [Google Scholar]
- 71.Sánchez-Romero N, Schophuizen CM, Giménez I, Masereeuw R. In vitro systems to study nephropharmacology: 2D versus 3D models. Eur J Pharmacol 2016;790:36–45. 10.1016/j.ejphar.2016.07.010 [DOI] [PubMed] [Google Scholar]
- 72.Guder WG, Wagner S, Wirthensohn G. Metabolic fuels along the nephron: pathways and intracellular mechanisms of interaction. Kidney Int 1986;29:41–45. 10.1038/ki.1986.6 [DOI] [PubMed] [Google Scholar]
- 73.Elhamri M, Martin M, Ferrier B, Baverel G. Substrate uptake and utilization by the kidney of fed and starved rats in vivo. Ren Physiol Biochem 1993;16:311–324. 10.1159/000173777 [DOI] [PubMed] [Google Scholar]
- 74.Mandel LJ. Metabolic substrates, cellular energy production, and the regulation of proximal tubular transport. Annu Rev Physiol 1985;47:85–101. 10.1146/annurev.ph.47.030185.000505 [DOI] [PubMed] [Google Scholar]
- 75.Zager RA, Johnson AC, Hanson SY. Proximal tubular cholesterol loading after mitochondrial, but not glycolytic, blockade. Am J Physiol-Renal Physiol 2003;285:F1092–F1099. 10.1152/ajprenal.00187.2003 [DOI] [PubMed] [Google Scholar]
- 76.Li L, Zhao H, Chen B, Fan Z, Li N, Yue J, et al. FXR activation alleviates tacrolimus-induced post-transplant diabetes mellitus by regulating renal gluconeogenesis and glucose uptake. J Transl Med 2019;17:418 10.1186/s12967-019-02170-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sasaki M, Sasako T, Kubota N, Sakurai Y, Takamoto I, Kubota T, et al. Dual Regulation of Gluconeogenesis by Insulin and Glucose in the Proximal Tubules of the Kidney. Diabetes 2017;66:2339–2350. 10.2337/db16-1602 [DOI] [PubMed] [Google Scholar]
- 78.Dickman KG, Mandel LJ. Glycolytic and oxidative metabolism in primary renal proximal tubule cultures. Am J Physiol 1989;257:C333–C340. 10.1152/ajpcell.1989.257.2.C333 [DOI] [PubMed] [Google Scholar]
- 79.Munyon WH, Merchant DJ. The relation between glucose utilisation, lactic acid production and utilization and the growth cycle of L strain fibroblasts. Exp Cell Res 1959;17:490–498. 10.1016/0014-4827(59)90069-2 [DOI] [PubMed] [Google Scholar]
- 80.Sugden MC, Holness MJ, Palmer TN. Fuel selection and carbon flux during the starved-to-fed transition. Biochem J 1989;263:313–323. 10.1042/bj2630313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pavlacky J, Polak J. Technical Feasibility and Physiological Relevance of Hypoxic Cell Culture Models. Front Endocrinol 2020;11:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All raw mass spectrometry proteomics data files, MaxQuant output files and MaxQuant software used for the search are available from the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD015102.