Abstract
The detection of SARS-CoV-2 infection is the premise of quarantine. In many countries or areas, samples need to be shipped or inactivated before SARS-CoV-2 testing. In this study, we checked the influence of sample storage conditions on SARS-CoV-2 nucleic acid testing results, including sample inactivation time, storage temperature, and storage time. All of these conditions caused an increase in the cycle threshold values of the nucleic acid tests and led to the misclassification of at least 10.2% of positive cases as negative or suspected. The results highlight the importance of immediate testing of samples for SARS-CoV-2 nucleic acid detection.
Keywords: influence, nucleic acid detection, SARS-CoV-2, storage, throat swab
COVID-19, which is caused by SARS-CoV-2 infection [1], has become a world pandemic. What is worse is that the pandemic is still accelerating. As of 30 March 2020, the disease was endemic in 200 countries, with a total of 640 000 confirmed cases and a mortality rate of more than 4%, according to WHO data [2].
Testing for SARS-CoV-2 infection is the premise of quarantine. To date, several methods have been developed for COVID-19, including the detection of viral nucleic acids, antigens, or host antibodies [3]. Among these, detection of viral nucleic acids based on reverse transcription polymerase chain reaction (RT-qPCR) is the most popular and specific method. In China, the National Medical Products Administration has approved 10 nucleic acid detection kits for clinical use, mainly based on RT-qPCR techniques. Most of the kits target the ORF1ab and N genes of the virus. A human gene internal standard is always included synchronously. To increase the detection sensitivity, samples should be tested immediately after being collected. However, some hospitals, and even some countries or areas, have no testing capacity or facilities with appropriate biosafety conditions. Many samples need to be stored and/or sent to laboratories with experience in COVID-19 virus testing. Occasionally, in a competent testing laboratory, swab specimens cannot be tested on a daily basis under particular circumstances such as excessive numbers of samples, lack of staff, or facility maintenance. Therefore, it is necessary to investigate the influence of sample storage conditions on nucleic acid testing for SARS-CoV-2. In this study, we checked the effect of sample inactivation, storage temperature, and storage time after collection on SARS-CoV-2 nucleic acid testing.
METHODS
A total of 88 SARS-CoV-2–positive throat swab samples were obtained randomly from Wuhan Huoshenshan Hospital. For collection, sterile synthetic fiber swabs with plastic shafts were used. After collection, the swabs were put into a tube with 3 mL transport medium, which was based on Hank’s solution and contained virus protectants gentamicin, fungal antibiotics, bovine serum albumin, 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), and amino acids (Virus Sampling Kit, Yocon Biotechnology) [4]. The nucleic acid test was carried out within 2 hours after sampling. Throat swabs were treated with a LabServ DNA/RNA Kit (Thermo Fisher Scientific) and total nucleic acids was extracted using a KingFisher Flex Purification System (Thermo Fisher Scientific). RT-qPCR was performed in a 96-well plate using a 2019 Novel Coronavirus (2019-nCoV) Detection Kit (Sansure Biotech). qRT-PCR analysis was conducted using a CFX Real-Time PCR Detection System (Bio-Rad Laboratories). The cycle threshold (Ct) values of the real-time PCR ranged from 23.80 to 39.84 for ORF1ab and 23.23 to 38.73 for N. In 10 samples the Ct was ≤ 30.0, in 31 samples the Ct was > 30.0 to ≤ 35.0, and in 47 samples the Ct was > 35.0 to ≤ 40.0. To demonstrate the effect of different conditions on nucleic acid testing, the selected positive samples were vortex mixed and divided into four 96-well plates. A series of repeated RT-qPCR detections were performed after different storage times (1–2 days), different storage temperatures (2°C–8°C or room temperature), different freeze-thaw times (1–2 times), or different inactivation times (30 and 60 minutes) at 56°C.
According to the virus detection kit manual (Sansure Biotech), only after successful amplification of the internal standard (HEX channel, Ct ≤ 40) can the result be judged as valid. On that premise, if both the ORF1ab (FAM channel) and N genes (ROX channel) were amplified (Ct ≤ 40), the sample was judged SARS-CoV-2 nucleic acid positive. If only the ORF1ab or N gene was amplified (Ct ≤ 40), the sample was judged as suspected. When neither the ORF1ab nor the N gene was amplified significantly (Ct > 40 or no signal), the sample was judged as negative.
RESULTS
After inactivation at 56°C for 30 minutes the detection results of 17.0% (15/88) of positive samples changed (negative 4 and suspected 11). After inactivation at 56°C for 60 minutes, 18.2% (16/88) of the positive results changed (negative 6 and suspected 10). After freezing and thawing once, the results of 10.2% (9/88) of the samples changed (negative 4 and suspected 5), while 19.3% (17/88) changed after a second freeze thaw cycle (negative 5 and suspected 12). After storage at 2°C–8°C for 1 day, 14.8% (13/88) of the sample results changed (negative 7 and suspected 6). After storage at 2°C–8°C for 2 days, 22.7% (20/88) of the sample results changed (negative 6 and suspected 14). After storage at room temperature (approximately 20°C) for 1 day, the positive results of 15.9% (14/88) of the samples changed (negative 5 and suspected 9). After storage at room temperature for 2 days, the positive results of 25% (22/88) of the samples changed (negative 8 and suspected 14).
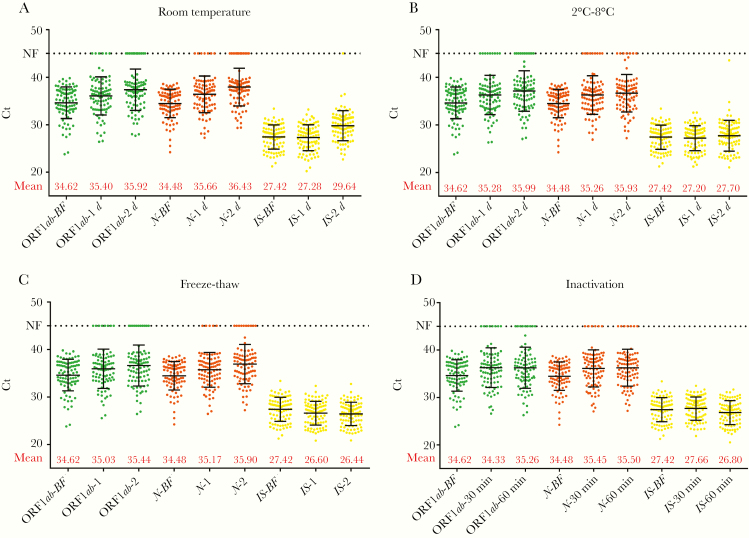
We analyzed the changes in Ct values after storage in different conditions and found that Ct values increased after storage or treatment in all the above conditions. Taking the amplification of ORF1ab as an example, the influence of freeze-thaw cycles was relatively small, with an increase in Ct of 0.41 for 1 freeze-thaw cycle and 0.82 for 2 cycles. When the samples were stored before analysis, the Ct value changed over time, regardless of room temperature (0.78 and 1.30 after 1 and 2 days, respectively) or 4°C storage (0.66 and 1.37 after 1 and 2 days, respectively). For the commonly used inactivation at 56°C for biosafety in SARS-CoV-2 detection, the Ct values also increased by 0.71 and 0.64 for 30 minutes and 60 minutes of inactivation time, respectively (Figure 1).
Figure 1.
Distribution of cycle threshold (Ct) values for SARS-CoV-2 target genes ORF1ab and N before and after different treatments. Distribution of Ct values measured in 88 SARS-CoV-2–positive throat swab samples before and after (A) storage at room temperature for 1 or 2 days; (B) storage at 2°C–8°C for 1 or 2 days; (C) 1 or 2 freeze-thaw cycles; and (D) inactivation at 56°C for 30 or 60 minutes. Bars indicates mean ± SD. Abbreviations: BF, analysis before test conditions; IS, internal standard (human RNase P gene); NF, no fluorescence signal at the maximum number of cycles (45).
Discussion
According to the above results, we suggest that the clinical detection of SARS-CoV-2 should be completed as soon as possible after sample collection. Storage and transportation of samples should be enacted at a low temperature and even in a frozen state as much as possible. If biosecurity is ensured, nucleic acid extraction and RT-qPCR detection are recommended before sample inactivation.
Notes
Financial support. This study was supported by the National Key Research and Development Program of China (grant number 2016YFD0500807).
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Zhu N, Zhang D, Wang W, et al. . A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 2020; 382:727–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. World Health Organization. Coronavirus disease 2019 (COVID-19) situation report-69. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200329-sitrep-69-covid-19.pdf?sfvrsn=8d6620fa_2. Accessed 26 April 2020.
- 3. Corman VM, Landt O, Kaiser M, et al. . Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill 2020; 25:2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Guo ZD, Wang ZY, Zhang SF, et al. . Aerosol and surface distribution of severe acute respiratory syndrome coronavirus 2 in hospital wards, Wuhan, China, 2020 [published online ahead of print 10 April 2020]. Emerg Infect Dis doi: 10.3201/eid2607.200885. [DOI] [PMC free article] [PubMed] [Google Scholar]