Abstract
Objective
To perform antiviral susceptibility monitoring of treated individuals in the community during the 2009 influenza A(H1N1) pandemic in England.
Patients and methods
Between 200 and 400 patients were enrolled daily through the National Pandemic Flu Service (NPFS) and issued with a self-sampling kit. Initially, only persons aged 16 and over were eligible, but from 12 November (week 45), self-sampling was extended to include school-age children (5 years and older). All samples received were screened for influenza A(H1N1)pdm09 as well as seasonal influenza [A(H1N1), A(H3N2) and influenza B] by a combination of RT–PCR and virus isolation methods. Influenza A(H1N1)pdm09 RT–PCR-positive samples were screened for the oseltamivir resistance-inducing H275Y substitution, and a subset of samples also underwent phenotypic antiviral susceptibility testing by enzyme inhibition assay.
Results
We were able to detect virus by RT–PCR in self-taken samples and recovered infectious virus enabling further virological characterization. The majority of influenza A(H1N1)pdm09 RT–PCR-positive NPFS samples (n = 1273) were taken after oseltamivir treatment had begun. No reduction in phenotypic susceptibility to neuraminidase inhibitors was detected, but five cases with minority quasi-species of oseltamivir-resistant virus (an H275Y amino acid substitution in neuraminidase) were detected.
Conclusions
Self-sampling is a useful tool for community surveillance, particularly for the follow-up of drug-treated patients. The virological study of self-taken samples from the NPFS provided a unique opportunity to evaluate the emergence of oseltamivir resistance in treated individuals with mild illness in the community, a target population that may not be captured by traditional sentinel surveillance schemes.
Keywords: influenza virus, surveillance, oseltamivir, zanamivir, pandemic
Introduction
In England, neuraminidase inhibitors (NIs) were prescribed at unprecedented levels throughout the first two waves of the 2009 influenza A(H1N1) pandemic as part of national control and mitigation strategies.1 Oseltamivir was primarily used for the treatment of pandemic influenza cases, with extensive prophylaxis of contacts in the initial containment phase from 27 April to 1 July 2009 that was designed to slow the spread of the virus. Containment was followed by a treatment-only phase, from 2 July 2009 until April 2010, when all suspected cases were offered oseltamivir treatment without a requirement for laboratory confirmation (Figure 1).
Figure 1.
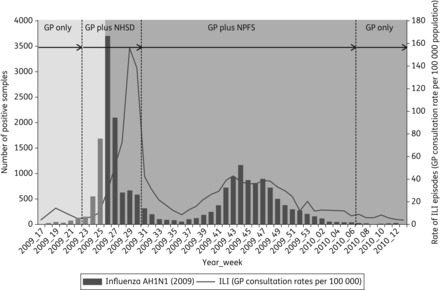
Weekly incidence of ILI per 100 000 population in England and Wales, and influenza A(H1N1)pdm09 laboratory detections from sentinel GP practices (RCGP) and hospitals in England. The containment (pale grey) and treatment-only (dark grey) phases of the UK pandemic strategy are highlighted. Virological sampling sources, through the different community care facilities during the pandemic in England, are also shown—from GP only (pre-pandemic/early first wave) to GP plus NHS Direct (NHSD) (first wave) or GP plus NPFS (second wave), and back to GP only after week 6 of 2010 (after the second wave).
In the containment phase, community access to healthcare assessment was available via a nurse-led multichannel telephone health advice service [National Health Service (NHS) Direct] in addition to traditional primary care facilities [general practitioner (GP) surgeries and emergency departments]. Antiviral drugs were available only by medical prescription.2,3 From 23 July 2009, in the treatment-only phase, the National Pandemic Flu Service (NPFS) was introduced. This was a dedicated telephone and web service managed by NHS Direct for persons with influenza-like illness (ILI) that was designed to manage treatment assessment and antiviral delivery for mild cases of illness, thus alleviating the pressure on the primary care services (Figure 1).4 Patients suspected of having influenza A(H1N1)pdm09 were authorized to collect oseltamivir from one of 2000 NPFS antiviral collection points in pharmacies around England. Until it ceased activity on 12 February 2010, the NPFS made more than 2.4 million patient assessments and authorized 1.6 million courses of oseltamivir, more than 1 million of which were collected for use.5
Monitoring of circulating virus strains for the emergence of resistance was a public health priority during the pandemic.6 Specimens collected by Royal College of General Practitioners (RCGP) sentinel GPs were typically taken at the time of diagnosis, prior to the initiation of any antiviral treatment.7 The NPFS service offered a unique opportunity to monitor drug-treated individuals with mild clinical illness in the community for the emergence of antiviral resistance. A programme of self-sampling was initiated to complement community virological sampling by the existing network of sentinel GPs.7 On a daily basis, a subset of NPFS-assessed individuals were provided a self-sampling kit with instructions to take a nasal swab; samples were returned to the HPA national virological reference facility via the UK postal service, using International Air Transport Association (IATA) 650-compliant packaging (UN3373 biological diagnostic specimens).
We present the results of the virological analysis of self-sampled specimens from NPFS patients during the 2009 pandemic and demonstrate the practicality of a self-sampling approach for monitoring the antiviral susceptibility of influenza.
Patients and methods
NPFS self-sampling virological surveillance scheme
The inclusion criteria for the issue of self-sampling kits to NPFS service users were those assessed to have influenza-like symptoms and generally uncomplicated illness, who were advised to self-care without referral for further medical attention and who were authorized to have antiviral agents. Between 200 and 400 patients, evenly distributed across England, were enrolled daily to the virological sampling study and were issued with a self-sampling kit via the postal service (Table 1). A short epidemiological questionnaire (available as Supplementary data at JAC Online) requesting information on antiviral treatment was included with each kit. Initially, only persons aged 16 and over were eligible, but from 12 November (week 45), self-sampling was extended to include school-age children (5 years and older).
Table 1.
A(H1N1)pdm09-positive patient demographics from the NPFS and sentinel GP schemes
| Patient demographic | NPFS scheme, n (%) (n = 1934 swabs) | Sentinel GP scheme, n (%) (n = 1021 swabs) |
|---|---|---|
| Method of swab | self-taken (or parent/guardian) | taken by healthcare professional |
| Treatment status at time of swab | ||
| no oseltamivir taken | 631 (32.6) | 935 (91.6) |
| mid-oseltamivir treatment (days 1–4) | 1195 (61.8) | 19a (1.9)b |
| oseltamivir treatment completed (day 5+) | 78 (4.0) | |
| no information given | 30 (1.6) | 67 (6.6) |
| Interval between onset date and swab date | ||
| 0 to 1 day | 3 (0.2) | 249 (24.4) |
| 2–3 days | 773 (40.0) | 338 (33.1) |
| 4–5 days | 847 (43.8) | 189 (18.5) |
| 6–7 days | 198 (10.2) | 62 (6.1) |
| 8–9 days | 38 (2.0) | 21 (2.1) |
| 10+ days | 16 (0.8) | 36 (3.5) |
| no onset/swab date given | 59 (3.1) | 126 (12.3) |
| Gender | ||
| female | 1082 (55.9) | 538 (52.7) |
| Age group (years) | ||
| 0–4c | 0 (0) | 124 (12.1) |
| 5–14 | 413 (21.4) | 366 (35.9) |
| 15–24 | 390 (20.2) | 179 (17.5) |
| 25–44 | 779 (40.3) | 248 (24.3) |
| 45–64 | 338 (17.5) | 96 (9.4) |
| 65–74 | 14 (0.7) | 5 (0.5) |
| 75+ | 0 (0) | 3 (0.3) |
| Region | ||
| East Midlands | 223 (11.5) | 96 (9.4) |
| East of England | 233 (12.0) | 103 (10.1) |
| London | 203 (10.5) | 275 (26.9) |
| North East England | 162 (8.4) | 33 (3.2) |
| North West England | 215 (11.1) | 71 (7.0) |
| South East England | 250 (12.9) | 160 (15.7) |
| South West England | 232 (12.0) | 112 (11.0) |
| West Midlands England | 214 (11.1) | 134 (13.1) |
| Yorkshire and Humber England | 202 (10.4) | 37 (3.6) |
aTwo patients had received zanamivir at the time of swabbing, and two patients had not received antivirals but were household contacts of someone receiving oseltamivir.
bFor sentinel GP swabs, the interval from the start of treatment to the swab date was not known, and therefore these 19 patients cannot be separated into the 1–4 day or 5+ day groups.
cThe 0–4 year age group was excluded from the NPFS virological surveillance protocol.
Ethics approval
Self-sampling was undertaken as part of a public health surveillance programme in response to the 2009 influenza pandemic and was carried out under the NHS Act 2006 (section 251), which provides statutory support for disclosure of such data by the NHS, and their processing by the HPA for communicable disease control. As such, no explicit ethical approval was necessary or sought. Only anonymized patient data were used for these analyses.
Virological screening
All samples received were screened for influenza A(H1N1)pdm09 as well as seasonal influenza [A(H1N1), A(H3N2) and influenza B] by a combination of RT–PCR and virus isolation methods. Total nucleic acid was extracted directly from 150 μL clinical specimens followed by reverse transcription and real-time PCR.8,9
Virus isolation and antiviral susceptibility characterization
Influenza A(H1N1)pdm09-positive specimens were screened by pyrosequencing for the H275Y mutation (a CAC to TAC nucleotide substitution) in the viral neuraminidase. Reverse transcription and PCR was performed using the One-Step RT–PCR Kit (Qiagen) and 0.6 μM each of 5′ biotin-labelled forward PCR primer (GGGAAAGATAGTCAAATCAGTCGA) and unlabelled reverse primer (TAGACGATACTGGACCACAACTG) (50°C for 30 min; 95°C for 5 min; 35 cycles of 94°C for 1 min; 62°C for 0.5 min; 72°C for 1 min followed by a final step of 72°C for 10 min). Allele quantification pyrosequencing was performed using a reverse-sense sequencing primer (CAGGAGCATTCCTCA; Qiagen) under standard conditions.
Virus was isolated from a subset of influenza A(H1N1)pdm09-positive samples in cell culture using MDCK and MDCK-SIAT1 cells (stably overexpressing α2,6-sialyltransferase).10 Isolates with sufficient neuraminidase activity were phenotypically analysed for antiviral susceptibility using a fluorescence-based [2′-(4-methylumbelliferyl)-α-D-N-acetylneuraminic acid (MUNANA)] neuraminidase enzyme inhibition assay, as previously described.11
Full-length neuraminidase sequencing was performed by two-step RT–PCR amplification of the full gene, followed by direct sequencing of the product with six primers, according to previously published methodology.12
Statistical analyses
The effect of antiviral use on viral load was assessed within the NPFS scheme by normal error regression on Ct values, adjusting where necessary for the potential confounding factors of the interval from onset to swab, age and gender. To assess the impact of antiviral use on virus recovery, logistic regression was performed with adjustment for the potential confounders of interval from onset to swab, swab to laboratory receipt and Ct value. Finally, antiviral susceptibility was assessed by normal error regression on log 50% inhibitory concentration (IC50) results in both schemes with effects of age, gender, month of swab, antiviral use and swabbing scheme examined. Results were anti-logged to provide fold effects.
Results
We analysed the samples from the NPFS patients to detect influenza virus genome and performed further virological characterization of influenza-positive samples. Between 3 August 2009 (week 31) and 12 February 2010 (week 6), a total of 14 441 swabs were received, of which 1934 were influenza A(H1N1)pdm09 positive (13.4%) (A. Bermingham, unpublished data). There was a variation in the number of doses of oseltamivir taken at the time of self-sampling, as captured by the patient questionnaire.
Considering only the influenza A(H1N1)pdm09-positive swabs (n = 1934), the majority (1195, 61.8%) were taken between 1 and 4 days after starting oseltamivir therapy, which correlates with the typical 2–5 day delay between the date of onset and the swab date (Table 1). A further 631 (32.6%) positive swabs were taken before oseltamivir therapy, and 78 (4.0%) swabs were taken after completion of the full 5 day course of oseltamivir (Table 1). This is in contrast to influenza A(H1N1)pdm09-positive samples taken via the sentinel GP scheme,7 in which only 1.9% of positive samples were taken after or during treatment (Table 1). Insufficient information was given in 30 (1.6%) NPFS cases to determine the stage of therapy when the swab was taken, and these were excluded from further analyses.
Effect of oseltamivir treatment on viral load and virus isolation
Semi-quantitative data based on Ct values from the influenza A(H1N1)pdm09 diagnostic RT–PCR results were taken as an estimation of viral load and used to assign positive samples into high, medium and low viral load groupings (Figure 2). Statistical analyses of individual Ct values showed that the estimated viral load decreased with a longer interval from the date of onset of symptoms to the date of the swab (data not shown; Ct increase of 0.37 per day, 95% CI 0.27–0.50). After adjusting for this confounding factor, there was evidence of a lower viral load in samples taken during the course of antiviral treatment compared with those samples taken without any treatment (difference in Ct 1.28, 95% CI 0.90–1.64). Samples taken after the course of oseltamivir had been completed showed a lower viral load, measured by RT–PCR, than when the sample was taken before antiviral agents, but this was not statistically significant (difference in Ct 0.70, 95% CI –0.26 to 1.65) (Figure 2). It should be noted, however, that the number of samples taken after completion of oseltamivir was low (n = 78).
Figure 2.
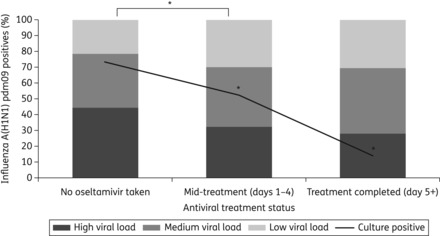
The percentage of samples in each viral load group (shaded columns) is compared with antiviral treatment status, and overlaid with culture positivity rates (black lines). Samples were categorized according to the Ct value from the diagnostic real-time PCR: Ct <30 = high viral load, 30–34 = medium viral load and >34 = low viral load. *Viral loads in samples taken during oseltamivir treatment were significantly lower than those taken when no oseltamivir had been used (difference in Ct 1.28, 95% CI 0.90–1.64) after adjusting for the time from onset of symptoms to the swab date. Virus isolation (culture positive) was significantly reduced from samples taken during or after completion of oseltamivir treatment, compared with samples taken when no oseltamivir had been used.
Based on previous practical experience of the ability to recover infectious virus from PCR-positive material, influenza A(H1N1)pdm09 PCR-positive samples <32 Ct were inoculated into cell culture (n = 735). Virus was successfully isolated from 73.2% of samples that were taken without oseltamivir treatment (186 that were culture positive, from 254 inoculated) compared with 52.5% of samples taken during oseltamivir treatment (228 of which were culture positive, from 434 inoculated) (Figure 2). Virus isolation was successful in 14.3% of samples taken after oseltamivir treatment had been completed (28 samples inoculated, and 4 with virus isolated).
After adjusting for potentially confounding factors affecting the likelihood of successful virus isolation, namely the time between onset and swab date, swab date to date of receipt in the laboratory and viral load (measured by the Ct value), there remained a statistically significant reduction in virus isolation from samples taken after oseltamivir treatment had been initiated or completed compared with those taken before any treatment, with estimates similar to the unadjusted estimates given above.
Genotypic antiviral susceptibility
The most common mechanism of oseltamivir resistance is a single amino acid substitution, histidine to tyrosine at position 275 (H275Y) in the N1 neuraminidase. Screening for this H275Y substitution was performed by pyrosequencing on a total of 1312 influenza A(H1N1)pdm09-positive samples. Of the samples screened, 480 had been taken prior to any dose of oseltamivir, whereas 784 swabs were taken after 1–4 days of oseltamivir treatment and 47 swabs after completion of therapy. Mixed virus populations containing both oseltamivir-susceptible (His-275) and oseltamivir -resistant virus (Tyr-275) were found in five NPFS patient samples, ranging from 13% to 23% Tyr-275; four were taken after 1–4 days of oseltamivir treatment and one from a patient who stated they had not taken any oseltamivir (Table 2). Patients with resistant quasi-species ranged from 5 years old to 50 years old, were from different geographical locations and were detected between week 36 and week 50 (Table 2). To determine the baseline incidence in the community, samples from the sentinel GP scheme were also screened for H275Y (738 samples), one of which had a quasi-species of H275Y at 18% Tyr-275; this had been taken in week 44 from a 15-year-old child who had had no known antiviral treatment or contact (Table 2). The number of patients with a resistant virus quasi-species was not significantly higher in the NPFS scheme than the sentinel GP scheme (P = 0.67, Fisher's exact test).
Table 2.
Patients with oseltamivir-resistant H275Y quasi-species
| Patient | Age (years) | Sample week | Region | Quasi-species percentage with Tyr-275 (±SD) |
Phenotypic susceptibility (IC50, nM ±SD) |
Therapy start to swab (days) | Onset to swab (days) | ||
|---|---|---|---|---|---|---|---|---|---|
| clinical specimen | cultured isolate | OST | ZAN | ||||||
| 1 | 50 | 36 | East of England | 20.4 (±1.34)a | NA | ND | ND | 2 | 5 |
| 2 | 40 | 42 | East of England | 22.9 (±4.52) | 275H only | 1.02 (±0.56) | 0.64 (±0.27) | 3 | 4 |
| 3 | 8 | 46 | East Midlands | 15.9 (±4.82) | 12.2 (±1.48) | 0.62 (±0.13) | 0.43 (±0.12) | 3 | 2 |
| 4 | 5 | 49 | London | 14.8 (±1.80) | 13.4 (±0.85) | 0.98 (±0.24) | 0.51 (±0.11) | 2b | 3 |
| 5 | 5 | 50 | South East | 12.6 (±1.22) | NA | ND | ND | 0 | 4 |
| Sentinel GP | 15 | 44 | West Midlands | 17.8 (±0.85)a | NA | ND | ND | 0 | NK |
NA, not available; ND, not done; NK, not known; OST, oseltamivir; ZAN, zanamivir.
aMean of two tests as there was insufficient material to perform a third.
bThe patient took only one dose and then stopped due to adverse events (vomiting).
Phenotypic antiviral susceptibility testing
All influenza A(H1N1)pdm09 virus isolates with sufficient neuraminidase activity were assessed for NI susceptibility by phenotyping with an enzyme inhibition assay. All isolates tested were susceptible to both oseltamivir and zanamivir, with only five isolates exhibiting IC50 values for oseltamivir higher the normal range for influenza A(H1N1)pdm09 viruses (Figure 3). Of the 219 NPFS isolates tested, 48 were obtained from samples taken prior to any oseltamivir therapy, 165 were from samples taken between 1 and 4 days after starting oseltamivir therapy, and 4 were taken after oseltamivir therapy had been completed. The four statistical outliers from the NPFS scheme were from the ‘during treatment’ group. Full-length neuraminidase sequencing did not identify any amino acid changes between these outlier isolates and their corresponding original clinical material, or any no amino acid substitutions against reference prototypical influenza A(H1N1)pdm09 strain A/California/7/2009 that are known to affect NI susceptibility. Virus was successfully isolated from three of the samples with resistant quasi-species. The minority H275Y quasi-species was maintained in the primary isolates for two of the three samples, but no shift in IC50 was detected (Table 2). This was consistent with previous experience with the fluorescence-based enzyme inhibition assay that quasi-species making up <25% of the total virus population cannot be phenotypically detected.11 Again, to determine any difference from baseline virus NI susceptibility in the untreated community, IC50 values from the NPFS-derived isolates were compared with those from 156 isolates from the sentinel surveillance scheme (untreated) (Figure 3). Overall, the IC50 values of both oseltamivir and zanamivir were similar between the two schemes—NPFS and sentinel primary care practitioners (Figure 3).
Figure 3.
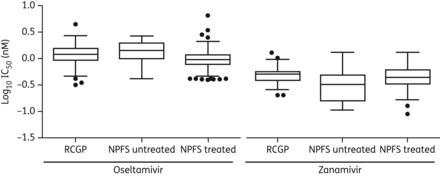
Box and whisker plots comparing IC50 (in vitro susceptibility) values of oseltamivir and zanamivir from sentinel GP (primary care)- and NPFS-derived isolates. Boxes represent the 25th–75th percentile, the central line is the mean, and the whiskers represent 1.5 times the IQR. Circles are individual isolates with IC50 values greater or lower than 1.5 × IQR and are identified as statistical outliers.
Discussion
The NPFS assessed patients using a clinical algorithm, authorized the collection of oseltamivir and provided an opportunity to coordinate self-sampling of suspected influenza A(H1N1)pdm09 cases in the community for virological surveillance. A subset of NPFS patients were issued with self-sampling kits, and the majority of these took their nasal swab after having begun oseltamivir treatment.
We were able to detect virus by RT–PCR in the self-taken samples (NPFS derived) and were also able to recover infectious virus from a high proportion of RT–PCR-positive NPFS samples, enabling further virological characterization, namely genotypic and phenotypic antiviral susceptibility testing, in this study. The issue and return of self-sampling kits via the postal service is therefore a viable system under which to operate a virological surveillance scheme, with the flexibility to respond to changing circumstances. Previously, small-scale clinical studies have used self-sampling as a low-cost means of collecting samples from patients enrolled while admitted to hospital but subsequently discharged.13,14 Self-sampling schemes could therefore be employed in target populations where a sentinel GP network was not available, and they are scalable in response to an outbreak.
The delivery of antivirals through the NPFS enabled the early antiviral treatment of many individuals with symptomatic influenza. NI treatment was found to significantly reduce the likelihood of a severe outcome during the pandemic in those patients who were subsequently admitted to hospital,15 and in some observational studies, early antiviral treatment was shown to reduce the duration of illness and the risk of hospitalization.16,17 Prompt antiviral treatment was also shown to reduce household transmission of the influenza A(H1N1)pdm09 virus during the containment phase of the pandemic in the UK.18 In our study of NPFS-derived samples, significantly lower virus isolation rates were achieved from samples taken after oseltamivir treatment had been initiated or completed compared with those taken before any oseltamivir had been taken, even after adjusting for factors such as the length of time from the onset of symptoms to the swab date to receipt of the sample in the laboratory, and estimated viral load as measured by Ct value. Although this study was not designed to measure the clinical impact of oseltamivir treatment, these results are suggestive of lower live virus shedding in oseltamivir-treated individuals, which would have an effect on the duration of symptoms, illness severity and transmission of the virus.
In New Zealand, oseltamivir has been available without prescription through pharmacies since 2007.19 Despite criticisms of such a system, comparable in some respects to the NPFS system of oseltamivir delivery, they do represent a method of supplying antivirals rapidly to patients when other facets of the country's healthcare system may be under strain due to an increasing epidemic or pandemic.
Emergence of the influenza A(H1N1)pdm09 virus with the H275Y mutation, which confers oseltamivir resistance, did occur under drug pressure in a low percentage of community patients (four patients; 0.3%) and in two patients who were not receiving drug treatment (0.1%), although none contained >30% of resistant virus. The overall rate of detection of resistant virus in the oseltamivir-treated NPFS scheme was not significantly greater than in the untreated population from the sentinel scheme (0.13%, P = 0.67, Fisher's exact test). Two samples containing a minority proportion of the oseltamivir-resistant virus (one from the NPFS scheme and one from the sentinel scheme) were taken from children prior to oseltamivir treatment during the second pandemic wave in the UK. There is no information to suggest these children had been exposed to antivirals, but oseltamivir was being used extensively during this period.
The clinical significance of mixed susceptible/resistant virus populations is not known. Influenza A(H1N1)pdm09 virus with the H275Y substitution does not cause more severe disease than the susceptible virus.20 While the level of transmissibility of the H275Y oseltamivir-resistant A(H1N1)pdm09 strain varies in different in vivo transmission models, none to date has indicated that the resistant strains are any more transmissible than the susceptible strains.21–24 There have, however, been documented episodes of transmission involving the H275Y-containing A(H1HN1)pdm09 viruses in both immunocompromised25,26 and immunocompetent patients.27 These transmission events have all occurred in close-contact settings, specifically hospitals and school camps. More recently, this resistant virus has been identified in otherwise healthy individuals with no epidemiological links,28–30 which highlights the need for robust community surveillance of antiviral susceptibility.
There are <2-fold differences in the range of IC50 values from primary care and the treated/untreated NPFS groups. A WHO laboratory expert working group recently defined a fold-difference of >10 as indicating a reduction in susceptibility in vitro.31 The <2-fold changes we have demonstrated in this study are unlikely to have clinical significance. Phenotypic susceptibility testing remains, however, critical for robust antiviral susceptibility surveillance, as genotypic assays are limited to screening for known and characterized resistance mechanisms.32 Several amino acid substitutions in the influenza viral neuraminidase have been reported to cause reductions in susceptibility to NIs, many of which are subtype dependent. Not all mechanisms of generating resistance are therefore known, particularly in the case of a newly emerging virus, as in the case of a pandemic. One limitation of this study is that we screened for only the H275Y mutation, and other amino acid substitutions that have been shown to cause a reduced susceptibility of the influenza A(H1N1)pdm09 virus to one or more NIs, such as at position 223, were not screened for, due to their comparatively infrequent incidence in global surveillance.33
Community-based virological surveillance schemes typically centre on sentinel GP networks where samples are usually taken on presentation of an ILI to the primary care facility. Usual practice in GP-based sampling is for swabbing to occur prior to any treatment. Surveillance samples from hospital sources can include post-treatment samples and, in some cases, sequential samples over the duration of treatment. However, information such as treatment history and underlying conditions contributing to the severity of the illness is often gathered retrospectively on patients from these sources, which is both difficult and time consuming. The virological study of self-taken samples from the NPFS therefore provided a unique opportunity to evaluate the emergence of oseltamivir resistance in treated individuals in the community with mild illness, a target population that is not typically captured through traditional sentinel GP or hospital-based sampling.
The sentinel RCGP/HPA seasonal sampling scheme has been considered to be the ‘gold standard’ for community-based influenza sampling in England over the last 15 years,9 the advantage of GP sampling being the continuous operation over each winter season, with a capacity for enhancement in special circumstances such as pandemic activity in the summer months. Recent advances in the sentinel sampling scheme now enable the collection of information on comorbidities, antiviral treatments and codified laboratory reports in a systematic fashion, thus facilitating further epidemiological analysis of the data. The development of vaccine effectiveness monitoring programmes, able to collect within-season estimates of vaccine effectiveness, are an example of how, with appropriate development, GP sampling can expand and provide further public health benefit. To maximize the opportunity to gather future data on the effectiveness of the antiviral treatment of seasonal influenza, GPs could be asked to take the samples at a specific point in the course of the therapy, or to take sequential samples over time, but self-sampling offers an alternative opportunity to acquire these specimens without placing a further burden on GP practices.
The combination of self-sampling and the provision of antivirals through this new healthcare delivery system not only enhanced community surveillance to detect influenza A(H1N1)pdm09, but also offered an innovative opportunity to perform antiviral susceptibility monitoring. The key feature of the NPFS scheme was its ability to sample a large number of individuals after treatment had been initiated, giving a cross-sectional community population-based approach to monitoring the emergence of antiviral resistance.
Since the pandemic, an evaluation of the NPFS system has been undertaken from the viewpoint of healthcare professionals and service users, which sets out a number of points for improvement.34 From the laboratory perspective, with particular respect to the surveillance of antiviral susceptibility, the clinical algorithm used to assess patients should be revised to improve clinical safety (e.g. by ruling out other pathogens), and this would need to evolve in any future use, as more is understood about the nature of the pandemic virus and its symptoms. Continual low-level sampling throughout the year would enable the development of baselines for positivity rates and antiviral susceptibility levels to help with interpreting data collected during a pandemic.
For this study, testing of NPFS samples for the detection of virus by RT–PCR was performed as soon as possible after the samples had arrived throughout the pandemic period, whereas the antiviral susceptibility testing was performed retrospectively. As robust laboratory capacity was improved in terms of trained staff and high-throughput equipment, the timeliness of genotypic antiviral susceptibility testing of the NPFS samples increased to within 1–2 weeks after influenza A(H1N1)pdm09 detection had been confirmed, but the more labour-intensive phenotypic testing was beyond the laboratory's capacity until the flow of samples had reduced towards the end of the second pandemic wave. As long as laboratory capacity is such that samples can be processed as they arrive, our study shows that self-sampling schemes like the NPFS are able to function as an early warning system for emerging resistance in treated community patients, despite the 1–2 day delay that results from sample kits being issued to the patient at the point of clinical assessment, rather than the sample being taken directly by the healthcare professional, as would be the case in a sentinel scheme.
It is not envisaged that self-sampling on this scale would be considered for routine seasonal influenza surveillance; however, in the event of a future pandemic where the provision of healthcare services changed, self-sampling could again provide an opportunity to enhance the public health management of the disease.
Funding
This work was supported by the Health Protection Agency (now known as Public Health England), as part of the national influenza surveillance capacity. Virological characterization, including antiviral susceptibility laboratory analyses, was supported by a research grant from the National Institute for Health Research, UK and Health Protection Agency, UK.
Transparency declarations
D. M. F. has received funding to attend influenza related meetings and has received consultancy fees from influenza vaccine manufacturers. A. L. and M. Z. participated in studies with the University of Leicester, funded by Roche. A. L. received funding from Roche for assay development and from Toyama to attend a project meeting. All other authors: none to declare.
Supplementary Material
Acknowledgements
We are grateful for the work and support from many members of staff from the HPA (now Public Health England), Colindale, London, UK, in particular the Respiratory Epidemiology Department, Claudia Rosenow and Shahjahan Miah and other members of the Respiratory Virus Unit, the Virology Specimen Accessions Unit and the High-Throughput Molecular Unit. We thank the participating patients and staff from NHS Direct, the National Pandemic Flu Service, the RCGP Weekly Returns Service scheme and the Specialist Microbiology Network.
References
- 1.Pandemic (H1N1) 2009 briefing note 1. Viruses resistant to oseltamivir (Tamiflu) identified. Wkly Epidemiol Rec. 2009;84:299–399. [PubMed] [Google Scholar]
- 2.Department of Health, UK. 2009. Swine Flu Information Leaflet. http://www.direct.gov.uk/prod_consum_dg/groups/dg_digitalassets/@dg/@en/documents/digitalasset/dg_177903.pdf. (3 June 2013, date last accessed)
- 3.Smith GE, Cooper DL, Loveridge P, et al. A national syndromic surveillance system for England and Wales using calls to a telephone helpline. Euro Surveill. 2006;11 pii=667. [PubMed] [Google Scholar]
- 4.Elliot AJ, Powers C, Thornton A, et al. Monitoring the emergence of community transmission of influenza A/H1N1 2009 in England: a cross sectional opportunistic survey of self sampled telephone callers to NHS Direct. BMJ. 2009;339:b3403. doi: 10.1136/bmj.b3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Health Protection Agency. Epidemiological report of pandemic (H1N1) 2009 in the UK: April 2009 – May 2010. 2010. http://www.hpa.org.uk/webc/HPAwebFile/HPAweb_C/1284475321350. (23 April 2013, date last accessed)
- 6.Lackenby A, Hungnes O, Dudman SG, et al. Emergence of resistance to oseltamivir among influenza A(H1N1) viruses in Europe. Euro Surveill. 2008;13:pii=8026. doi: 10.2807/ese.13.05.08026-en. : [DOI] [PubMed] [Google Scholar]
- 7.Fleming DM, Chakraverty P, Sadler C, et al. Combined clinical and virological surveillance of influenza in winters of 1992 and 1993–4. BMJ. 1995;311:290–1. doi: 10.1136/bmj.311.7000.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ellis J, Iturriza M, Allen R, et al. Evaluation of four real-time PCR assays for detection of influenza A(H1N1)v viruses. Euro Surveill. 2009;14:pii=19230. doi: 10.2807/ese.14.22.19230-en. [DOI] [PubMed] [Google Scholar]
- 9.Stockton J, Ellis JS, Saville M, et al. Multiplex PCR for typing and subtyping influenza and respiratory syncytial viruses. J Clin Microbiol. 1998;36:2990–5. doi: 10.1128/jcm.36.10.2990-2995.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matrosovich M, Matrosovich T, Carr J, et al. Overexpression of the alpha-2,6-sialyltransferase in MDCK cells increases influenza virus sensitivity to neuraminidase inhibitors. J Virol. 2003;77:8418–25. doi: 10.1128/JVI.77.15.8418-8425.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lackenby A, Democratis J, Siqueira MM, et al. Rapid quantitation of neuraminidase inhibitor drug resistance in influenza virus quasispecies. Antivir Ther. 2008;13:809–20. [PubMed] [Google Scholar]
- 12.Baillie GJ, Galiano M, Agapow PM, et al. Evolutionary dynamics of local pandemic H1N1/2009 influenza virus lineages revealed by whole-genome analysis. J Virol. 2012;86:11–8. doi: 10.1128/JVI.05347-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cooper DL, Smith GE, Chinemana F, et al. Linking syndromic surveillance with virological self-sampling. Epidemiol Infect. 2008;136:222–4. doi: 10.1017/S0950268807008412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stephenson I, Democratis J, Lackenby A, et al. Neuraminidase inhibitor resistance after oseltamivir treatment of acute influenza A and B in children. Clin Infect Dis. 2009;48:389–96. doi: 10.1086/596311. [DOI] [PubMed] [Google Scholar]
- 15.Muthuri SG, Myles PR, Venkatesan S, et al. Impact of neuraminidase inhibitor treatment on outcomes of public health importance during the 2009–2010 influenza A(H1N1) pandemic: a systematic review and meta-analysis in hospitalized patients. J Infect Dis. 2013;207:553–63. doi: 10.1093/infdis/jis726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McLean E, Pebody RG, Campbell C, et al. Pandemic (H1N1) 2009 influenza in the UK: clinical and epidemiological findings from the first few hundred (FF100) cases. Epidemiol Infect. 2010;138:1531–41. doi: 10.1017/S0950268810001366. [DOI] [PubMed] [Google Scholar]
- 17.Nguyen-Van-Tam JS, Openshaw PJ, Hashim A, et al. Risk factors for hospitalisation and poor outcome with pandemic A/H1N1 influenza: United Kingdom first wave (May–September 2009) Thorax. 2010;65:645–51. doi: 10.1136/thx.2010.135210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pebody RG, Harris R, Kafatos G, et al. Use of antiviral drugs to reduce household transmission of pandemic (H1N1) 2009, United Kingdom. Emerg Infect Dis. 2011;17:990–9. doi: 10.3201/eid1706.101161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gauld NJ, Jennings LC, Frampton C, et al. Five years of non-prescription oseltamivir: effects on resistance, immunization and stockpiling. J Antimicrob Chemother. 2012;67:2949–56. doi: 10.1093/jac/dks337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Calatayud L, Lackenby A, Reynolds A, et al. Oseltamivir-resistant pandemic (H1N1) 2009 virus infection in England and Scotland, 2009–2010. Emerg Infect Dis. 2011;17:1807–15. doi: 10.3201/eid1710.110117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duan S, Boltz DA, Seiler P, et al. Oseltamivir-resistant pandemic H1N1/2009 influenza virus possesses lower transmissibility and fitness in ferrets. PLoS Pathog. 2010;6:e1001022. doi: 10.1371/journal.ppat.1001022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duan S, Boltz DA, Seiler P, et al. Competitive transmissibility and fitness of oseltamivir sensitive and resistant pandemic influenza H1N1 viruses in ferrets. Influenza Other Resp Viruses. 2010;5(Suppl 1):79–82. [PubMed] [Google Scholar]
- 23.Kiso M, Shinya K, Shimojima M, et al. Characterization of oseltamivir-resistant 2009 H1N1 pandemic influenza A viruses. PLoS Pathog. 2010;6:e1001079. doi: 10.1371/journal.ppat.1001079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seibert CW, Kaminski M, Philipp J, et al. Oseltamivir-resistant variants of the 2009 pandemic H1N1 influenza A virus are not attenuated in the guinea pig and ferret transmission models. J Virol. 2010;84:11219–26. doi: 10.1128/JVI.01424-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen LF, Dailey NJ, Rao AK, et al. Cluster of oseltamivir-resistant 2009 pandemic influenza A (H1N1) virus infections on a hospital ward among immunocompromised patients–North Carolina, 2009. J Infect Dis. 2011;203:838–46. doi: 10.1093/infdis/jiq124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moore C, Galiano M, Lackenby A, et al. Evidence of person-to-person transmission of oseltamivir-resistant pandemic influenza A(H1N1) 2009 virus in a hematology unit. J Infect Dis. 2011;203:18–24. doi: 10.1093/infdis/jiq007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oseltamivir-resistant 2009 pandemic influenza A (H1N1) virus infection in two summer campers receiving prophylaxis–North Carolina: 2009. MMWR Morb Mortal Wkly Rep. 2009;58:969–72. [PubMed] [Google Scholar]
- 28.Hurt AC, Hardie K, Wilson NJ, et al. Community transmission of oseltamivir-resistant A(H1N1)pdm09 influenza. N Engl J Med. 2011;365:2541–2. doi: 10.1056/NEJMc1111078. [DOI] [PubMed] [Google Scholar]
- 29.Lackenby A, Moran Gilad J, Pebody R, et al. Continued emergence and changing epidemiology of oseltamivir-resistant influenza A(H1N1)2009 virus, United Kingdom, winter 2010/11. Euro Surveill. 2011;16:pii=19789. [PubMed] [Google Scholar]
- 30.Meijer A, Jonges M, van Beek P, et al. Oseltamivir-resistant influenza A(H1N1)pdm09 virus in Dutch travellers returning from Spain, August 2012. Euro Surveill. 2012;17:pii=20266. [PubMed] [Google Scholar]
- 31.Meetings of the WHO working group on surveillance of influenza antiviral susceptibility – Geneva, November 2011 and June 2012. Wkly Epidemiol Rec. 2012;87:369–74. [PubMed] [Google Scholar]
- 32.WHO. Laboratory Methodologies for Testing the Antiviral Susceptibility of Influenza Viruses: Neuraminidase Inhibitor (NAI) http://www.who.int/influenza/gisrs_laboratory/antiviral_susceptibility/nai_overview/en/index.html. (23 April 2013, date last accessed) [Google Scholar]
- 33.WHO. Weekly Update on Oseltamivir Resistance to Pandemic Influenza A (H1N1) 2009 Viruses. http://www.who.int/csr/disease/swineflu/oseltamivirresistant20100806.pdf. (23rd April 2013, date last accessed)
- 34.Department of Health. The NPFS: An Evaluation. https://www.gov.uk/government/uploads/system/uploads/attachment_data/file/147395/dh_125338.pdf.pdf. (23 April 2013, date last accessed)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


