Abstract
Pseudomonas aeruginosa colonizes the airways of cystic fibrosis (CF) patients, causing infections that can last for decades. During the course of these infections, P. aeruginosa undergoes a number of genetic adaptations. One such adaptation is the loss of swimming motility functions. Another involves the formation of the rugose small colony variant (RSCV) phenotype, which is characterized by overproduction of the exopolysaccharides Pel and Psl. Here, we provide evidence that the two adaptations are linked. Using random transposon mutagenesis, we discovered that flagellar mutations are linked to the RSCV phenotype. We found that flagellar mutants overexpressed Pel and Psl in a surface-contact dependent manner. Genetic analyses revealed that flagellar mutants were selected for at high frequencies in biofilms, and that Pel and Psl expression provided the primary fitness benefit in this environment. Suppressor mutagenesis of flagellar RSCVs indicated that Psl overexpression required the mot genes, suggesting that the flagellum stator proteins function in a surface-dependent regulatory pathway for exopolysaccharide biosynthesis. Finally, we identified flagellar mutant RSCVs among CF isolates. The CF environment has long been known to select for flagellar mutants, with the classic interpretation being that the fitness benefit gained relates to an impairment of the host immune system to target a bacterium lacking a flagellum. Our new findings lead us to propose that exopolysaccharide production is a key gain-of-function phenotype that offers a new way to interpret the fitness benefits of these mutations.
Author summary
Microbiologists have known for decades that Pseudomonas aeruginosa mutates during chronic respiratory infection of cystic fibrosis (CF) patients. One of the most reported functions lost during these infections is flagellar motility. A long-standing interpretation of this observation is that the flagellum is disadvantageous for the bacterium in the CF environment. We report the surprising finding that mutation of a wide range of flagellar genes results in the overproduction of the biofilm matrix polysaccharides Psl and Pel. We propose, therefore, that flagellar mutations represent a gain-of-function that would help the bacterium to form biofilms and persist in the CF airways.
Introduction
One of the most important CF pathogens is Pseudomonas aeruginosa [1]. During chronic infection, it predictably acquires a number of adaptive mutations that are believed to aid in long-term persistence [2–4]. These adaptations include alginate overproduction [5], amino acid auxotrophy [6], loss of lipopolysaccharide O-antigens [7] and loss of flagellar motility [8]. Although the fitness benefits that these different mutations confer is uncertain, several seem intuitive. For example, amino acid auxotrophy may relieve the metabolic burden of amino acid biosynthesis in the amino acid rich environment of CF airway secretions. Another is the loss of motility functions, which is thought to assist in the evasion of the host immune system by eliminating production of a key antigen and pathogen-associated molecular pattern, flagellin.
Several similar genetic adaptations are observed in laboratory biofilms. This is potentially important because CF airway infections are thought to have biofilm etiology [9, 10]. One P. aeruginosa phenotype that is selected for in laboratory biofilms is the rugose small colony variant (RSCV) [11, 12] (Fig 1A). These variants have also been isolated from CF sputum [12–15]. RSCVs make profuse amounts of the extracellular polysaccharides (EPS) Pel and Psl as well as the extracellular adhesin CdrA [16, 17]. Expression of Pel and Psl is associated with tolerance to some clinically-relevant antibiotics [18, 19], and it safeguards P. aeruginosa against complement-mediated opsonization and neutrophil phagocytosis [20]. Pel, Psl and CdrA are also known to play a role in biofilm aggregate formation [17, 18, 21], which is associated with protection from antimicrobials [22–24] and host defenses [16, 25]. It is not surprising, therefore, that prior reports have associated the appearance of RSCVs in CF sputum to poor patient outcomes [26, 27].
Fig 1. RSCVs are abundant in P. aeruginosa biofilms and among CF isolates.
(A) An aliquot of cells recovered from a single drip-flow reactor biofilm was plated onto LB or VBMM agar. The arrows indicate representative RSCVs from each sample. Each panel represents an area that is approximately 22.9 mm × 16.0 mm. (B) Proportion of RSCV and ancestral smooth colony morphotypes on LB and VBMM agar. Each bar indicates the mean and standard deviation for 6 independent drip-flow reactors. *P<0.05 and ***P ≤ 0.001 colony counts on LB vs. VBMM agar with Student’s t-test. (C) A total of 416 P. aeruginosa isolates acquired from a study of CF respiratory epidemiology were streaked on LB or VBMM-based agar (see S1 Text) and scored for colony morphology. The percentage of isolates with the indicated colony morphologies is illustrated.
Expression of the RSCV morphotype is linked to elevated levels of the second messenger cyclic diguanylate (c-di-GMP) [28–32]. Several loci have been linked to the RSCV phenotype, including the wsp operon [29], the tpb (also denoted aws or yfi) loci [28, 33, 34], amrZ [35, 36], dsbA [37], genes of the rsm signalling pathway [21, 38] and pvrR [30]. Previous work has estimated that RSCVs represent approximately 5% of laboratory grown biofilm populations and that wsp mutations account for a majority of these [16]. RSCVs harbouring mutations in the wsp and yfi loci have also been observed among CF isolates [2, 39].
In this study, we found that the frequency of RSCVs in laboratory biofilms and CF isolates has been grossly underestimated. Using transposon mutagenesis, we sought to identify novel genetic elements linked to the phenotype. We found 22 genes in our screen that we grouped into four classes. Perhaps the most surprising class were mutations in genes encoding flagellum structure and function. Unlike the other classes of RSCVs, flagellar mutants overexpressed Pel and Psl in a surface-contact-dependent fashion. We also found that flagellar mutations are selected for at high frequency in laboratory biofilms, and that increased Pel and Psl expression provides the primary fitness benefit. Suppressor mutagenesis revealed that Psl overexpression by flagellar mutants requires the mot genes as well as components of the Pil-Chp surface-sensing system. Finally, we show that flagellar mutant RSCVs are also found among CF clinical isolates. We propose that gain-of-function phenotypes (Pel and Psl overexpression) offer a new interpretation of the benefits of flagellar mutations in isolates from laboratory biofilms and CF infections.
Results and discussion
P. aeruginosa RSCVs are abundant in laboratory biofilms and CF isolates
P. aeruginosa biofilms grown in vitro rapidly undergo mutation and selection [11]. A manifestation of this is the appearance of diverse colony morphotypes when biofilm bacteria are plated onto solid growth media. Using a growth medium optimized for RSCV detection (Vogel-Bonner minimal medium supplemented with the dyes Congo Red and Coomasie brilliant blue), we found that RSCVs are observed ~9-fold more frequently than on traditional lysogeny broth (LB) agar (Fig 1A and 1B). Many isolates displaying the RSCV phenotype on VBMM had a wild type smooth colony morphotype when re-streaked on LB agar.
A key question for these isolates is whether RSCV-linked traits (elevated Pel and Psl production) are only expressed on VBMM-based agar compared to LB. To test this, Psl and PelC expression were measured for strains that exhibited the RSCV phenotype on VBMM but were smooth when grown on LB. We observed that all these strains still overexpressed PelC and/or Psl on both VBMM and LB (S1 Fig). These data indicate that elevated EPS expression is a feature of these strains regardless of the growth medium or colony morphotype.
We next asked if RSCVs are more common than previously thought among CF sputum isolates (Fig 1C). We examined 416 P. aeruginosa isolates cultured from 46 pediatric CF patients enrolled in a two-year study of respiratory bacterial epidemiology at Seattle Children's Hospital [40]. Only a single isolate had an RSCV phenotype on LB agar. By contrast, 56 (13.5%, Fig 1C) of these isolates (from 40% of the patients) displayed the RSCV phenotype on VBMM-based agar medium. By comparison, perhaps the best-known CF-related P. aeruginosa adaptive colony morphology, mucoidy [41], was observed in 80 (19.2%, Fig 1C) of the isolates (from 49% of the patients).
Identification of genes that are linked to the RSCV phenotype
Work by Starkey and colleagues [16] indicated that wspF mutations account for 70% of the RSCVs that are isolated on LB agar from biofilms. We randomly isolated 25 RSCVs on VBMM agar from biofilms (i.e. 5 RSCVs from each of 5 different drip-flow biofilm reactors in order to increase the likelihood that these RSCVs were non-isogenic) and attempted to complement these mutants with a wild type copy of the wspF gene; however, in only a single instance did this restore the wild type colony morphology of these biofilm isolates. Analogously, transformation of 10 RSCVs (from 10 different patients) recovered on VBMM-based agar from CF sputum with a plasmid bearing wspF restored smooth colony morphology in only 3 of 10 isolates. These data suggest that the majority of RSCVs isolated from biofilms and from CF sputum may be wsp-independent.
To identify wsp-independent genes linked to the RSCV phenotype, we carried out near-saturation transposon mutagenesis of P. aeruginosa PAO1 harboring an engineered deletion of wspR, the diguanylate cyclase of the wsp system [29]. This allowed us to eliminate the isolation of wsp-related mutations that might confer the RSCV phenotype. Here we used the transposon miniTn5Pro, which carries the repressor araC as well as an outward-facing, arabinose-inducible promoter [42]. Approximately 35,000 miniTn5Pro mutants were scored for colony morphology on selective VBMM agar with and without arabinose. This random gene-disruption-gene-activation strategy produced 57 genetically distinct RSCVs in which the transposon was mapped to 22 different genes from 17 different operons (Table 1). Collectively, the identified genetic elements could be placed into four groups based on known and predicted functions: 1) the regulator of secondary metabolism (rsm) signalling pathway, 2) the periplasmic thiol-disulfide interchange protein (dsbA), 3) flagellum biosynthesis, and 4) diguanylate cyclases and phosphodiesterases that synthesize and degrade c-di-GMP, respectively. We also placed oligoribonuclease (orn) in this last category. The orn gene encodes a 3ʹ→5ʹ exoribonuclease that degrades pGpG, an intermediate product produced by EAL-domain phosphodiesterases in the two-step pathway for c-di-GMP degradation [43, 44]. Only a single mutant displayed an arabinose inducible RSCV phenotype, and in this case the transposon insertion point was mapped upstream of the diguanylate cyclase siaD.
Table 1. P. aeruginosa PAO1 ΔwspR transposon mutants with an RSCV phenotype.
| Insertion site1 | # insertions | Function of disrupted gene2 | Operonic structure2 (class3) |
|---|---|---|---|
| Regulator of secondary metabolism (rsm) signalling pathway | |||
| retS | 3 | regulator of exopolysaccharide and type III secretion, sensor histidine kinase | monocistronic |
| Redox homeostasis and protein folding | |||
| dsbA | 2 | thiol-disulfide interchange protein | dsbA-PA5488-PA5487 |
| Flagellum biosynthesis | |||
| flhA | 1 | flagellum export component, membrane target for soluble export complex | monocistronic (II) |
| flhB | 1 | flagellum export component, substrate specificity switch, target for soluble export complex | fliLMNOPQRflhB (II) |
| flhF | 2 | predicted exporter with FHIPEP family motif | flhF-fleN (II) |
| fliF | 5 | flagellum M-ring, outer membrane protein | fliEFGHIJ (II) |
| fliH | 1 | putative flagellum TTSS protein | fliEFGHIJ (II) |
| fliI | 1 | flagellum specific ATP synthase | fliEFGHIJ (II) |
| fliM | 3 | flagellum motor switch protein | fliLMNOPQRflhB (II) |
| fliO | 3 | flagellum export component | fliLMNOPQRflhB (II) |
| fliD | 2 | flagellum capping protein | fliDSSʹfleP (II) |
| flgI | 1 | flagellum basal body P-ring protein | flgFGHIJKL (III) |
| flgJ | 2 | muraminadase | flgFGHIJKL (III) |
| flgK | 3 | flagellum hook-associated protein | flgFGHIJKL (III) |
| flgL | 1 | flagellum hook-associated protein | flgFGHIJKL (III) |
| fliC | 7 | flagellin, type B | fliCfleL (IV) |
| flgN | 2 | chaperone protein, initiation of filament assembly | flgMNZ (IV) |
| Diguanylate cyclases and phosphodiesterases | |||
| orn | 2 | oligoribonuclease | monocistronic |
| siaB1,4 | 1 | hypothetical protein, unknown function, predicted serine phosphatase domain | siaABCD |
| PA18501 | 2 | araC like transcription factor with an amidase domain, predicted DJ-1/ThiJ/PfpI family protein | PA1850-PA1849-PA1848 |
| dipA | 5 | GAF-PAS-ASNEF-EAL protein | dipA-msrA |
| PA5295 | 4 | COG5001-GGDEF-EAL domain protein | monocistronic |
1The outwards facing araC-PBAD of miniTn5Pro was located upstream of siaD and PA1851, which are putative diguanylate cyclases.
2Predicted annotations and operonic structures for flagellar genes were taken from Dasgupta and colleagues [45]. All other annotations and operonic structures were retrieved from the Pseudomonas Genome Database [82] on April 5, 2020.
3Class as defined by the established four-tier flagellar gene transcriptional hierarchy in P. aeruginosa by Dasgupta and colleagues [45].
4This transposon mutant was arabinose responsive.
A limitation of transposon mutagenesis is that it is difficult to predict whether transposon insertions cause polar effects that disrupt the expression of adjacent genes in operons. Therefore, unmarked deletion alleles were constructed for genes chosen to represent each of the four groups of RSCV-linked mutations. As expected, introducing these deletion alleles into the wild type PAO1 (Fig 2) and ΔwspR strains (S2A Fig) duplicated the RSCV phenotype of transposon mutants. Each of these mutants had smooth, and in some instances small, colony morphology on LB agar (S2B Fig). Each mutant strain was then complemented using miniTn7 to insert a single copy of the deleted gene and its native promoter elsewhere on the chromosome (Fig 2). Additionally, precise, in-frame deletion mutations were constructed in ten flagellar genes. Genes were chosen to represent operons from each of the known tiers (Table 1) of the four-tier flagellar gene regulation hierarchy that orders flagellum assembly in P. aeruginosa [45]. Our results show that some but not all disruptions in flagellar operons may result in rugose colony morphology on VBMM (S3 Fig). Moreover, with the exception of ΔfleQ (which is at the first tier of flagellar gene regulation), we noted that RSCV-linked mutations can occur in operons at every tier of the flagellar gene regulation hierarchy. Taken together, these data suggest that the assembly status of the P. aeruginosa flagellum regulates EPS production, which is similar to observations previously reported for other bacterial species [46, 47].
Fig 2. Multiple genes are linked to the RSCV phenotype.
Precisely defined in-frame deletion mutations recreated the RSCV phenotype of transposon mutants. Mutations were complemented by expressing the deleted gene from its native promoter in trans. Here, this was done using a miniTn7 vector to insert a single copy of the deleted gene at the glmS site of the P. aeruginosa chromosome. Insertion of an empty miniTn7 vector into chromosome did not affect colony morphology. In all panels, bacteria were cultured and photographed on VBMM agar containing Congo red and brilliant blue R (see Material and Methods). Each panel represents an area that is approximately 5.0 mm × 3.5 mm. WT, wild type; VC, vector control.
Flagellar mutants overproduce EPS in a surface contact-dependent fashion
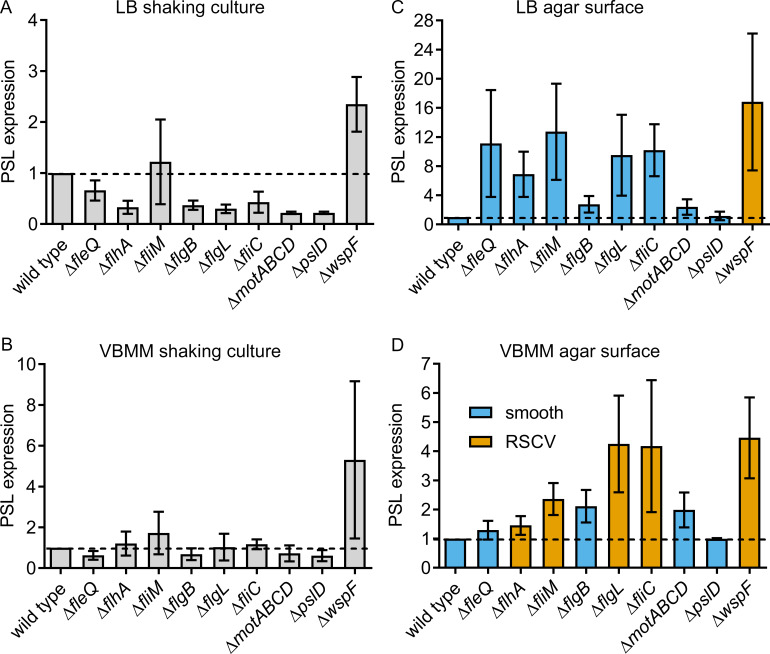
Transcriptional profiling has identified a role for the master flagellar regulator FleQ in repressing the pel and psl operons [45]. However, there is little evidence that other flagellar mutations increase the expression of EPS genes. Here, we assessed molecular markers of EPS production for four flagellar mutants that expressed the RSCV phenotype on VBMM (ΔflhA, ΔfliM, and ΔflgL and ΔfliC) and for three that did not (ΔfleQ, ΔflgB and ΔmotABCD). When grown in shaking liquid culture, none of these mutants displayed increased expression of Psl (Fig 3A and 3B). Moreover, apart from ΔfleQ, none showed overexpression of PelC (S4 Fig). By contrast, nearly all these mutants showed elevated Psl when grown on solid medium (Fig 3C and 3D). Except for the ΔmotABCD strain, nearly all these mutants also overexpressed PelC when grown on VBMM agar (S4 Fig). These data suggest that surface contact might be a stimulus for flagellar mutants to express biofilm matrix polysaccharides. These observations are consistent with reports that the flagellum has a role in mechanosensation that is similar to other bacteria [48], including V. cholerae [49], Vibrio paraheamolyticus [50], Caulobacter crescentus [51], Proteus mirabilis [52] and Bacillus subtilis [53].
Fig 3. Overexpression of Psl by flagellar mutants is surface-contact dependent.
Semi-quantitative dot blots for the Psl polysaccharide from flagellar mutants grown (A and B) in shaken LB or VBMM cultures, respectively, or (C and D) on the surface of LB or VBMM agar, respectively. Each bar indicates the mean and standard deviation for 1 to 3 technical replicates from each of 3 independent biological replicates.
Suppressor mutagenesis reveals a role for the flagellum stator and the Pil-Chp surface-sensing system in regulating EPS production in flagellar mutants
To reveal the regulatory pathway that links flagellar mutations to EPS overexpression, we carried out transposon mutagenesis of the ΔfliC strain. We again utilized minTn5Pro to enable random gene disruption and activation, but this time sought ΔfliC transconjugates that exhibited a smooth colony morphology. Approximately 21,000 ΔfliC miniTn5Pro mutants were evaluated on VBMM agar with or without arabinose, and this analysis yielded 115 genetically distinct mutants in which the transposon was mapped to 70 genes. A subset of 58 candidate second-site suppressor mutations, none of which were arabinose-responsive, mapped to only 26 genes in 17 operons (Table 2).
Table 2. Transposon suppressor mutations of the P. aeruginosa PAO1 ΔfliC RSCV phenotype.
| Insertion site | # insertions | Function of disrupted gene1 | Operonic structure1 (class2) |
|---|---|---|---|
| Regulator of secondary metabolism (rsm) signalling pathway | |||
| gacS | 1 | global activator of cyanide synthesis, sensor histidine kinase, signal transduction | gacSldhA-PA0926-PA0925 |
| Flagellum biosynthesis | |||
| fleQ | 5 | transcriptional regulator | monocistronic (I) |
| fleN | 4 | flagellar synthesis regulator | flhFfleN (II) |
| motA | 4 | flagellum stator protein, exerts torque against motor switch | motAB |
| motB | 5 | flagellum stator protein, converts proton energy into torque | motAB |
| motC | 1 | flagellum stator protein, exerts torque against motor switch | PA1458-PA1459-motCD-PA1462 |
| PA1462 | 1 | unknown function | PA1458-PA1459-motCD-PA1462 |
| C-di-GMP signal transduction | |||
| sadB | 6 | unknown function | monocistronic |
| sadC | 2 | GDDEF domain protein | sadC-PA4331-PA4330 |
| siaA | 3 | hypothetical protein, predicted HAMP domain | siaABCD |
| siaB | 1 | hypothetical protein | siaABCD |
| siaD | 1 | GGDEF domain protein, diguanylate cyclase | siaABCD |
| Type IV pilus | |||
| fimX | 1 | type four pilus biosynthesis, GGDEF-EAL domain protein | fimWfimX |
| pilB | 2 | type four fimbrial biogenesis protein | monocistronic |
| pilC | 1 | type four fimbrial biogenesis protein | pilCDcoaE-PA4530 |
| pill | 1 | component of chemotactic signal transduction system, response regulator | pilGHI |
| pilM | 2 | type four fimbrial biogenesis assembly protein, ATPase | pilMNOPQ |
| pilO | 1 | type four pilus assembly, O-glycosyltransferase | pilMNOPQ |
| pilQ | 1 | type four fimbrial biogenesis outer membrane protein precursor | pilMNOPQ |
| pilR | 1 | two-component response regulator | pilSR |
| pilW | 2 | type four fimbrial biogenesis protein | fimUpilVWXY1Y2E |
| pilY1 | 4 | type four fimbrial biogenesis protein, tip associated adhesin | fimUpilVWXY1Y2E |
| Unknown function | |||
| PA1766 | 2 | hypothetical protein, predicted ATP-grasp domain | PA1768-PA1767-PA1766 |
| PA1767 | 4 | hypothetical protein, predicted cytoplasmic membrane protein | PA1768-PA1767-PA1766 |
| PA1768 | 1 | hypothetical protein, predicted type I export signal | PA1768-PA1767-PA1766 |
| PA1769 | 1 | hypothetical protein | monocistronic |
1Annotations and predicted operonic structures for flagellar genes were taken from Dasgupta and colleagues [45]. All other annotations and predicted operonic structures were retrieved from the Pseudomonas Genome Database [82] on April 5, 2020.
2Class as defined by the established four-tier flagellar gene transcriptional hierarchy in P. aeruginosa by Dasgupta and colleagues [45].
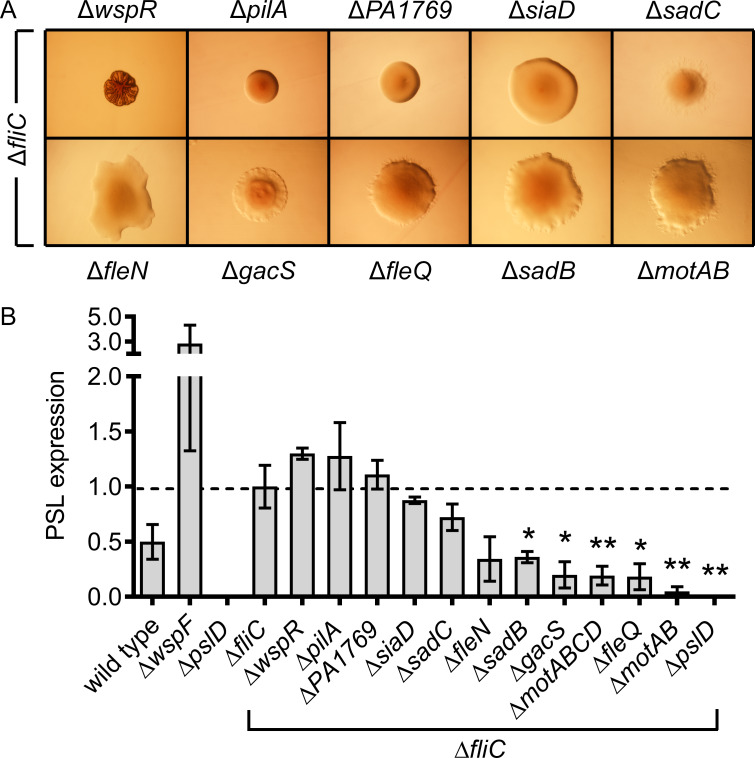
To further investigate these findings, in-frame deletion mutations were constructed in genes representing key suppressor mutations: gacS, fleQ, fleN, motAB, motABCD, sadB, sadC, siaD, pilA, and PA1769. Introducing these deletions into the ΔfliC background in all cases abolished the RSCV-phenotype on VBMM (Fig 4A). To further distinguish suppression of EPS expression from changes in colony morphology, we used an enzyme-linked immunosorbent assay (ELISA) to quantify the amount of Psl produced by each of these double, triple and quintuple mutants. Mutations in gacS, fleQ, sadB, motAB and motABCD signifcantly reduced Psl production by the ΔfliC strain (Fig 4B). Together, motAB or motCD encode a stator complex that provides flagellar torque [54, 55]. Here, we conclude that EPS overexpression by flagellar mutants involves a regulatory pathway that not only requires SadB [56–58], but also involves the flagellum stator proteins. These observations are consistent with a recent report from Baker and colleagues [59] in which the MotCD stator was observed to interact with the diguanylate cyclase SadC to stimulate c-di-GMP production under conditions not permissive to motility. SadC was also identified in our suppressor screen. In addition, we found a number of type IV pilus genes linked to the Pil-Chp surface sensing system, including four separate insertions in pilY1 (Table 2), which encodes a pilus tip protein that has a putative mechanosensory domain [60]. We also found four separate insertions in the pilMONPQ operon (Table 2), which encodes the pilus alignment complex that is thought to transduce stimuli from PilY1 [61]. Because a ΔpilA ΔfliC double mutant did not have decreased levels of Psl production (Fig 4), one interpretation of these data is that type IV pilus-dependent motility might be important for the RSCV phenotype of flagellar mutants; however, another possibility is that the stator-dependent pathway of surface-sensing requires some parts of the Pil-Chp surface-sensing system to stimulate the RSCV phenotype on agar surfaces. While a complete mechanism is not yet evident, a teleological explanation is that altered function of flagellum motor components engages a signal transduction pathway that leads to biofilm formation. These observations might be informative to our understanding of the early stages of biofilm formation.
Fig 4. Suppressor mutagenesis suggests a role for the flagellum stator proteins and the Pil-Chp system in EPS biosynthesis by flagellar mutant RSCVs.
(A) Precisely defined in-frame deletion mutations were engineered into the chromosome of the PAO1 ΔfliC strain. In all panels, bacteria were cultured and photographed on VBMM agar containing Congo red and brilliant blue R (see Material and Methods). Each panel represents an area that is approximately 5.0 × 3.5 mm. (B) Semi-quantitative anti-Psl ELISA assays of the ΔfliC strain bearing second-site suppressor mutations. Each bar represents the mean and standard error of 3 to 6 independent biological replicates. *P<0.05 and **P ≤ 0.005 vs. ΔfliC with Student’s t-test.
These observations are not without precedent: flagellin (flaA) mutants of Vibrio cholerae O139 also display a rugose colony morphology [47]. In V. cholerae, this gain-of-function phenotype depends on the Vibrio polysaccharide (vps) genes, the master regulator of the vps gene cluster (VpsR) and the sodium-driven flagellar motor (MotX) [62]. Work by Wu and colleagues [46] has also identified a stator-dependent signal transduction process that regulates V. cholerae EPS gene expression by modulating intracellular c-di-GMP. This flagellum-dependent biofilm response (FDBR) depends on the activities of at least three different diguanylate cyclases [46]. Under certain conditions and similar to P. aeruginosa [63, 64], V. cholerae O139 flagellar mutants grown on glass coverslips can form thick biofilm monolayers or microcolonies with architectures that are distinct from wild type [47]. Additionally, EPS-producing V. cholerae has an advantage in biofilm competition against isogenic, EPS-deficient strains; however, a cost of EPS-production is an impaired ability to disperse to new locations [65].
Flagellar mutants predictably and reproducibly evolve in laboratory biofilms
One context in which EPS-overproducing flagellar mutants might have an advantage is in biofilms. EPS-mediated adhesion may allow bacteria to better occupy space on the substratum [66] and polymer production may provide better access to oxygen and nutrients as biofilms grow [67]. To test the hypothesis that flagellar mutants might account for RSCVs isolated from in vitro grown biofilms (Fig 1), we began by inoculating five independent reactors with a founding population of wild type PAO1 taken from a single, shaken overnight culture. After 5 d, a random sample of RSCVs was isolated on VBMM agar from each reactor. Genome sequencing revealed that RSCV-linked mutations were found in each isolate that were previously identified by transposon mutagenesis (Table 3). Subsequently, the identified mutant alleles were introduced into the ancestral PAO1 strain, producing the RSCV phenotype (S5 Fig). All these genotypes overproduced PelC (S6A Fig) and Psl (S6B Fig). The most frequent RSCV-linked mutations occurred in genes encoding flagellum biosynthesis and function. Mutants bearing these alleles had a frequency from 23% to 63% in each of the replicate biofilm populations (Table 3). Altogether, these observations indicate that EPS-overproducing flagellar mutants reproducibly evolve during P. aeruginosa biofilm growth.
Table 3. Identity and frequency of RSCV-linked alleles in model biofilms.
| Reactor | Population size (1010 CFU/reactor) |
Est. RSCV frequency | RSCV-linked mutation(s)1 | Description | Est. allele frequency (a/n)2 |
|---|---|---|---|---|---|
| 1 | 10.54 ± 0.05 | 0.51 | fliG128T>G (V43G) | flagellum motor switch protein | 0.22 (15/34) |
| fliG834_844ΔGAAGGTCTTCA | flagellum motor switch protein | 0.12 (8/34) | |||
| 2 | 10.79 ± 0.03 | 0.55 | fliM718C>T (Q240*) | flagellum motor switch protein | 0.23 (40/94) |
| 3 | 10.53 ± 0.03 | 0.46 | wspF777C>A (S259R) | probable methylesterase | 0.01 (1/72) |
| 4 | 10.64 ± 0.08 | 0.60 | tpbB668A>G (D223G) | diguanylate cyclase | 0.01 (2/98) |
| fliH178G>T (E60*) | putative flagellum TTSS protein | 0.33 (54/98) | |||
| 5 | 10.47 ± 0.05 | 0.73 | fliM915delC | flagellum motor switch protein | 0.63 (49/57) |
1Nomenclature extensions to describe complex mutations adapted from Dunnen and Antonarkakis [83].
2n denotes the number of RSCVs isolated in a random sample from the reactor, and a denotes the number of those RSCVs that had the indicated RSCV-linked allele, which was determined by targeted sequencing of the allele in each isolate. Allele frequency in the biofilm population, therefore, was estimated as the ratio of a/n.
EPS overproduction provides flagellar mutants with a biofilm fitness advantage
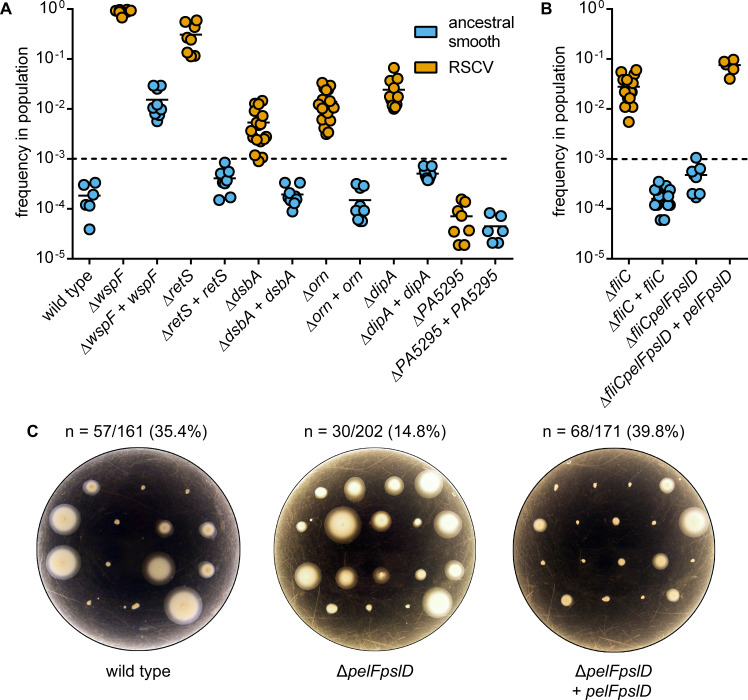
Competitive co-culture was used to assess the fitness of precisely defined genotypes in laboratory-grown biofilms. Here we used a starting ratio of 1 mutant or complemented cell to 1000 ancestral cells. Viable cell counting was enabled by genetically tagging bacteria with an antibiotic resistance gene (aacC1). The aacC1 gene is not selectively neutral, as the wild type PAO1 strain labelled in this manner decreased in frequency approximately 6-fold during competition with its antibiotic-sensitive ancestor (Fig 5A). To account for this change in fitness, all changes in frequency of aacC1-labelled mutant cell lines were calculated relative to the frequency of the aacC1-labelled wild type strain. Correlating with previous reports and with only a single exception (ΔPA5295), we found that acquisition of an RSCV-linked mutation was strongly associated with an increase in biofilm fitness (Fig 5A and 5B).
Fig 5. The biofilm fitness advantage of flagellar mutants depends on Pel and Psl.
(A) Frequency of RSCV cell lines after 3 days of co-culture in biofilm reactors with the ancestral PAO1 strain. Datum points represent technical replicates from each of three independent drip-flow reactors. (B) Frequency of the ΔfliC, ΔfliCΔpelFΔpslD and complemented strains after 3 days of co-culture in biofilm reactors with the ancestral PAO1 strain. Datum points represent technical replicates from each of three independent drip-flow reactors. (C) Frequency at which motility was lost during laboratory evolution of wild type, ΔpelFΔpslD and complemented strains when grown for 5 days in a drip-flow reactor. The hatched line indicates the starting frequency of the mutant cells in the drip-flow reactors. Orange points represent strains that can express the RSCV phenotype, blue points represent strains with ancestral smooth colony morphology.
Subsequently, we focused on flagellar mutants in order to disentangle the roles of EPS production and loss of flagellar function to biofilm fitness. During the course of co-culture, the ΔfliC mutant increased in frequency 150-fold and genetic complementation eliminated the increased biofilm fitness of this genotype (Fig 5B). In contrast to ΔfliC, a ΔfliCpelFpslD triple mutant had greatly diminished biofilm fitness, increasing in frequency only 2.5-fold relative to the aacC1-labelled control (Fig 5B). Genetic complementation of this triple mutant with single copies of pelF and pslD expressed from their native promoters restored the biofilm fitness of this strain, which increased ~410-fold in frequency during co-culture (Fig 5B). These data suggest that Pel and/or Psl are essential for the biofilm fitness of a flagellar mutant. To further test this interpretation, we directly tested for the loss of swimming motility during experimental evolution of wild type and ΔpelFpslD strains in drip-flow reactors (Fig 5C). After 5 days, 35.4% of the biofilm isolates from wild type and 14.9% from the ΔpelFpslD strain were negative for swimming motility. By contrast, repeating experimental evolution with the ΔpelFpslD mutant that had been complemented with pelF and pslD yielded a population where 39.8% of biofilm isolates were negative for swimming motility. Overall, these results suggest that EPS expression is a gain-of-function phenotype that significantly contributes to the biofilm fitness of flagellar mutants.
Flagellar mutants and other classes of RSCVs described here are found among CF isolates
P. aeruginosa that have lost flagellar motility have been reported among CF isolates [8, 68]. Here we examined clonally related isolates from previous studies of P. aeruginosa genetic diversity in CF respiratory infections [2, 40, 69]. Initially, we identified eight closely related pairs of isolates (Fig 6A and S1 Table, each pair recovered from a different, individual CF patient) in which one isolate had smooth colony morphology and the other displayed an RSCV phenotype on VBMM (not on LB; S7 Fig). An additional two pairs of isolates were identified among early and late isolates from prior [2] genome sequencing projects. All the RSCVs displayed upregulation of PelC relative to their related strains with smooth colony morphology (Fig 6A). Data from directed [2] as well as de novo genome sequencing was used to identify putative RSCV-linked mutations. In five cases we successfully identified mutations (retS2078C>A, wspF474_477ΔTTCGinsCAGAC, wspF635_636ΔCG, morA3430C>T, and fleQ364C>T), that when introduced into PAO1, caused the RSCV phenotype (Fig 6B). We were unsure as to why the wspF mutant allele was smooth on LB for two of these clinical isolates. We postulate that this difference could be due to an epistatic interaction in the genetic background of the CF isolates. Regardless, these findings indicate that CF RSCV isolates are associated with similar gain-of-function phenotypes (EPS overexpression), notably including a flagellar mutant.
Fig 6. Multiple genes are linked to the RSCV phenotype of clinical isolates and are associated with PelC overexpression.
(A) Semi-quantitative Western blots for PelC from strains grown on VBMM agar. Each bar indicates the mean and SD for 3 independent biological replicates. (B) Colony morphology of clinical isolates recovered from CF patients. RSCV-linked alleles that were identified in the clinical isolates by genome sequencing were introduced into the P. aeruginosa PAO1 strain. In all panels, bacteria were cultured and photographed on VBMM-based agar containing Congo red and brilliant blue R (see Material and Methods). Each panel represents an area that is approximately 5.0 mm × 3.5 mm.
A new way to interpret the fitness benefits of flagellar mutations
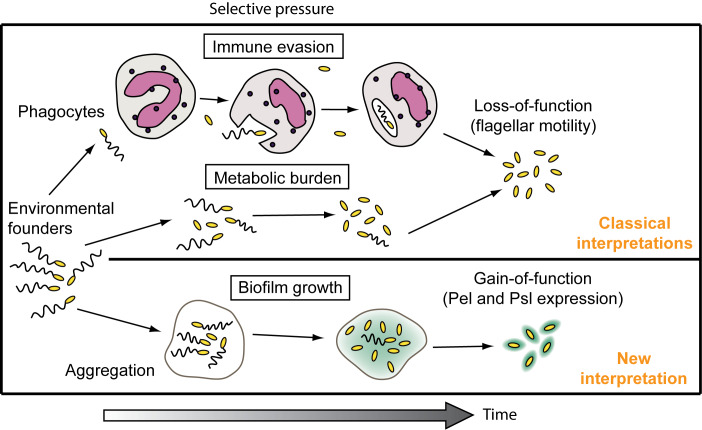
Because an accurate and established small animal model for CF respiratory infection is not available, it is difficult to evaluate what selective forces are at work in vivo. Several factors could lead to bacterial diversification in CF airways, and these pressures might include immune functions, fluctuations in antibiotics, O2 tension or nutrient availability, among others. P. aeruginosa non-mucoid isolates that have lost their flagella have been reported for at least thirty-five [8, 68] years and there are two classic interpretations for the benefits of this adaptation. First, loss of the flagellum confers immune evasion, which could allow P. aeruginosa flagellar mutants to escape negative selection by host immune cells [70]. Second, mutation of flagellar genes could afford a metabolic benefit to mutant cells by eliminating the energetic cost of synthesizing the flagellum, which could provide flagellar mutants with an increased growth rate. Our discovery that flagellar mutants have a gain-of-function phenotype–Pel and Psl expression–offers another perspective on the selective pressures that amplify flagellar gene mutations (Fig 7). One possibility is that positive selection for aggregation and biofilm growth could favor loss of the flagellum; alternatively, immune selection or an increased growth rate could lead to the evolution of non-flagellated bacteria that are also particularly successful at forming biofilms. Because there are multiple parameters that may drive expression of the RSCV phenotype on VBMM agar but not on LB, which might include, for example, nutrition-dependent modulation of c-di-GMP signaling enzymes [71, 72], we have yet to decipher the molecular mechanism for this phenomenon. However, we observed that flagellar mutants display an RSCV phenotype and overexpress PelC and Psl on synthetic cystic fibrosis sputum medium (SCFM) agar (S8 Fig). Staudinger and colleagues have also observed that flagellar mutants aggregate in mucus-based gels and that aggregation decreases the susceptibility of P. aeruginosa flagellar mutants to antibiotics [73]. We remark that our interpretation for how these gain-of-function phenotypes might provide benefits during infection may not be exclusive of other classic interpretations for the benefits of the loss of flagella. Nevertheless, because Pel and Psl production are associated with biofilm fitness, we propose that loss of the flagellum may lead to biofilm growth that contributes to increased bacterial persistence during infection.
Fig 7. Interpreting the benefits of loss of flagellar function in laboratory biofilms and CF.
The classic interpretations for loss of the flagellum view evasion of the host immune system and a decreased doubling time as fitness benefits that could lead to the evolution of non-flagellated bacteria during infection. We propose that selection for gain-of-function phenotypes related to exopolysaccharide production offers a new way for interpreting the causes and consequences of flagellar gene mutation in biofilms and CF infection.
Materials and methods
Bacterial strains and isolates, plasmids and growth conditions
Bacterial strains or isolates and plasmids are listed in S1 Table and S2 Table, respectively. Collection of all clinical isolates was approved by the Seattle Children’s and/or University of Washington Hospital Institutional Review Boards (IRBs). P. aeruginosa was grown at 37°C in lysogeny broth (LB, 10 g L-1 tryptone, 5.0 g L-1 yeast extract, 5.0 g L-1 NaCl) or Vogel-Bonner Minimal Media (VBMM; 0.2 g L-1 MgSO4•7H2O, 2.0 g L-1 citric acid, 3.5 g L-1 NaNH4HPO4•4H2O, 10 g L-1 K2HPO4, pH 7.0) [74] with 10 mM citrate. VBMM with Congo red (CR), brilliant blue R (BB) and 1.0 g L-1 casamino acids (VCBA) was used to grow clinical isolates. Semi-solid plate media were prepared by adding 1.5% w/v Bacto agar to LB or 1.0% noble agar to VBMM. Additional details of growth conditions, antibiotic selection and plasmid construction are described in S1 Text.
Digital photography
An Olympus SZX-ILLK100 stereomicroscope equipped with a C-7070 wide zoom digital camera was used to photograph colonies growing on agar media. Pictures of Petri plates were captured using a Pentax Optio W10 digital camera with tripod. Digital images were adjusted for contrast and brightness using Photoshop CS6 (Adobe).
Transposon mutagenesis
Established protocols were used for mutagenesis of P. aeruginosa with the transposon miniTn5-Pro [42]. Transconjugants were selected on VBMM plates containing 100 μg/ml gentamicin (Gm) + CR/BB. This selection was repeated on plates that additionally contained 0.2% arabinose. Transposon insertion sites were identified by plasmid rescue of the flanking genomic regions followed by DNA sequencing (see S1 Text).
Construction of deletion and site-directed mutants
Deletion alleles were assembled in vitro by removing an in-frame fragment of coding sequence from each gene. This was done by joining PCR products amplified from the adjacent regions of the chromosome by splicing by overlapping extension (SOE) PCR. These DNA fragments were then cloned by restriction into the suicide vector pEX18Gm. Alternatively, cloning of deletion alleles was carried out using Gateway recombination with the suicide vectors pDONRPEX18Gm or pEX18GmGW as previously described [75] (see S1 Text). Alleles containing point mutations that evolved in either biofilm or clinical isolates were directly cloned by PCR using primers specific for the target open reading frames (ORFs) and then introduced into allelic exchange vectors using Gateway technology (see S1 Text). Unmarked deletion and site-directed mutations were then introduced into the P. aeruginosa chromosome by two-step allelic exchange using established procedures [75].
Complementation analysis
PCR primers (S3 Table) targeting wild type alleles and their native promoters were tailed with attB sequences and these PCR products were then cloned by Gateway BP recombination into pDONR223. The plasmid inserts were sequenced using universal forward and reverse M13 primers (S3 Table) and then transferred by LR recombination into pUC18-miniTn7T-Gm-GW [76]. For the purpose of complementation analysis with two ORFs, we used the plasmid pUC18-miniTn7T2.1-Gm-GW [77] and two divergently transcribed ORFs were assembled in multiple steps using SOE-PCR and Multisite Gateway cloning technology (see S1 Text). The miniTn7 constructs were then introduced into the P. aeruginosa chromosome via electroporation with the helper plasmid pTNS1 [78]. Insertion at the neutral attTn7 adjacent to P. aeruginosa glmS was confirmed by PCR using the primers PTn7L, PTn7R, Gm-up and Gm-down (S3 Table) as previously described [76].
Drip-flow biofilm reactors
P. aeruginosa biofilms were grown in drip-flow reactors using 1% tryptic soy broth according to standard protocols [79]. Biofilms were recovered using a cell scraper to dislodge biomass into sterile PBS and these were then disrupted using a tissue homogenizer. Biofilm cells were serially diluted ten-fold in PBS and then plated onto either LB or VBMM + CR/BB agar.
Genome sequencing and bioinformatics
Illumina sequencing technology was used to collect deep-coverage genome sequence data. The procedures for library construction, sequencing, genome assembly and SNP analysis are detailed in S1 Text. Sequencing data have been deposited with links to BioProject accession number PRJNA625996 in the National Center for Biotechnology (NCBI) BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/).
Immunoblots
Chemiluminescent Western and dot blots for PelC and Psl, respectively, were carried out by established methods [80] with slight modification (see S1 Text). Horse-radish peroxidase linked secondary antibodies were used in conjunction with the Pierce SuperSignal West Pico ECL reagent (Thermo Scientific) to visualize proteins bound by primary antibodies and Chemiluminescence was captured and quantified using a FluorChem Q (Alpha Innotech).
ELISA assays
ELISA assays were carried out according to the method of Byrd and colleagues [80] with slight modification using a human monoclonal anti-Psl antibody, Cam-003 (MedImmune) [81] (see S1 Text).
Motility assays
The swimming medium was VBMM that contained 0.3% w/v bacto-agar (Difco). Swim plates were stab inoculated with bacteria from an LB or VBMM agar culture grown overnight at 37°C using a sterile toothpick. Plates were then incubated at 37°C for 16–18 h and photographed.
Supporting information
(PDF)
(PDF)
(PDF)
(PDF)
(A) Semi-quantitative dot blots for the Psl polysaccharide from strains grown on VBMM agar. (B) Semi-quantitative Western blots for PelC from strains grown on VBMM agar. (C) Semi-quantitative dot blots for the Psl polysaccharide from strains grown on LB agar. (D) Semi-quantitative Western blots for PelC from strains grown on LB agar. Each bar indicates the mean and SD for 1 or 2 technical replicates from each of 3 to 4 independent biological replicates.
(TIF)
(A) The diguanylate cyclase WspR is dispensable for the nutrition-dependent RSCV phenotype resulting from RSCV-linked mutations identified in this study. Bacteria in these panels were cultured and photographed on VBMM agar containing Congo red and brilliant blue R (see Material and Methods). (B) These nutrition-dependent RSCV-linked genotypes give rise to bacterial colonies with smooth morphology on LB agar. LB agar was also supplemented with the dyes Congo red and brilliant blue R. Each panel represents an area that is approximately 5.0 × 3.5 mm.
(TIF)
Precisely defined in-frame deletion mutations were introduced into a series of flagellar genes in wild type PAO1 strain. Only some of these mutations caused the RSCV phenotype. In all panels, bacteria were cultured and photographed on VBMM agar containing Congo red and brilliant blue R (see Material and Methods). Each panel represents an area that is approximately 5.0 mm × 3.5 mm.
(TIF)
Semi-quantitative Western blots for PelC from flagellar mutants grown (A and B) in shaken LB or VBMM cultures, or (C and D) on the surface of LB or VBMM agar, respectively. Each bar indicates the mean and standard deviation for 1 or 2 technical replicates from each of 3 to 4 independent biological replicates.
(TIF)
Photographs of agar-grown RSCVs that were isolated from biofilm reactors after experimental evolution. RSCV-linked mutations were identified by whole genome sequencing (top). Colony morphology of mutants in which the RSCV-linked allele from the biofilm isolate was introduced into the ancestral PAO1 strain (bottom). In all panels, bacteria were cultured and photographed on VBMM agar containing Congo red and brilliant blue R (see Material and Methods). Each panel represents an area that is approximately 5.0 mm × 3.5 mm.
(TIF)
(A) Semi-quantitative PelC Western blots for strains grown on VBMM agar. (B) Semi-quantitative Psl dot blots for strains grown on VBMM agar. Each bar indicates the mean and standard deviation for 3 to 6 independent biological replicates.
(TIF)
(A) Colony morphology of isolates on LB and VBMM agar. Each panel represents an area that is approximately 5.0 mm × 3.5 mm. (B) Analysis of pulsed-field gel electrophoresis (PFGE) data indicating that pairs of CF isolates with smooth and RSCV colony phenotypes are close genetic relatives. Phylogenetic relationships were calculated from existing PFGE data for the PASA collection of P. aeruginosa isolates at Seattle Children’s Hospital [40] using Bionumerics Seven (Applied Maths).
(TIF)
(A) Semi-quantitative Western blots for PelC from strains grown on SCFM agar. (B) Semi-quantitative dot blots for the Psl polysaccharide from strains grown on SCFM agar. (C) Colony morphology of flagellum mutants on SCFM agar. Each panel represents an area that is approximately 5.0 mm × 3.5 mm. Each bar indicates the mean and SD for 3 biological replicates. Relative expression levels have been normalized to the ΔwspF strain.
(TIF)
Acknowledgments
The authors thank Michael A. Jacobs, Tam Quach, Natalie A. Gugala, Julie Silverman, Ethan Mann and Fanny Liu for technical assistance, and Dr. George O’Toole for insightful discussions and feedback on the work. The authors would like to acknowledge Benjamin Staudinger and Pradeep K. Singh for the providing clinical isolates from their collections, and Jane L. Burns for providing clinical isolates through the CF Isolate Core at Seattle Children's Research Institute.
Data Availability
All sequencing data have been deposited with links to BioProject accession number PRJNA625996 in the National Center for Biotechnology (NCBI) BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/). All other relevant data are within the manuscript and its Supporting Information files.
Funding Statement
Jane L. Burns provided clinical isolates through the CF Isolate Core at Seattle Children's Research Institute, which has been supported by the National Institute of Health (NIH, Grant P30 DK089507). This work was supported by grants to MRP from the NIH (Grants R01AI077628 and R01AI143916). HA was supported by an Eyes High Postdoctoral Scholar award from the University of Calgary. YI was a University of Washington Cystic Fibrosis Foundation Research and Development Program Fellow. JJH was initially supported by a postdoctoral fellowship from the Natural Sciences and Engineering Research Council (NSERC) of Canada, and later by a Discovery Grant from the NSERC, an infrastructure award from the Canada Foundation for Innovation and a Canada Research Chair from the Canadian Institutes for Health Research (CIHR). This work was supported by the following grant to DJW from the NIH, R01AI134895. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Razvi S, Quittell L, Sewall A, Quinton H, Marshall B, Saiman L. Respiratory microbiology of patients with cystic fibrosis in the United States, 1995–2005. Chest. 2009;136:1554–60. 10.1378/chest.09-0132 [DOI] [PubMed] [Google Scholar]
- 2.Smith EE, Buckley DG, Wu Z, Saenphimmachak C, Hoffman LR, D'Argenio DA, et al. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc Natl Acad Sci U S A. 2006;103:8487–92. 10.1073/pnas.0602138103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Folkesson A, Jelsbak L, Yang L, Johansen HK, Ciofu O, Hoiby N, et al. Adaptation of Pseudomonas aeruginosa to the cystic fibrosis airway: an evolutionary perspective. Nat Rev Microbiol. 2012;10(12):841–51. 10.1038/nrmicro2907 [DOI] [PubMed] [Google Scholar]
- 4.Marvig RL, Sommer LM, Molin S, Johansen HK. Convergent evolution and adaptation of Pseudomonas aeruginosa within patients with cystic fibrosis. Nat Genet. 2015;47(1):57–64. 10.1038/ng.3148 [DOI] [PubMed] [Google Scholar]
- 5.Doggett RG, Harrison GM, Wallis ES. Comparision of some properties of Pseudomonas aeruginosa isolated from infections in persons with and without cysitc fibrosis. J Bacteriol. 1964;87(2):427–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taylor RF, Hodson ME, Pitt TL. Auxotrophy of Pseudomonas aeruginosa in cystic fibrosis. FEMS Microbiol Lett. 1992;71(3):243–6. 10.1016/0378-1097(92)90716-2 [DOI] [PubMed] [Google Scholar]
- 7.Hancock RE, Mutharia LM, Chan L, Darveau RP, Speert DP, Pier GB. Pseudomonas aeruginosa isolates from patients with cystic fibrosis: a class of serum-sensitive, nontypable strains deficient in lipopolysaccharide O side chains. Infect Immun. 1983;42(1):170–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luzar MA, Thomassen MJ, Montie TC. Flagella and motility alterations in Pseudomonas aeruginosa strains from patients with cystic fibrosis: relationship to patient clinical condition. Infect Immun. 1985;50:577–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Worlitzsch D, Tarran R, Ulrich M, Schwab U, Cekici A, Meyer KC, et al. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J Clin Invest. 2002;109(3):317–25. 10.1172/JCI13870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singh PK, Schaefer AL, Parsek MR, Moninger TO, Welsh MJ, Greenberg EP. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature. 2000;407(6805):762–4. 10.1038/35037627 [DOI] [PubMed] [Google Scholar]
- 11.Boles BR, Theondel M, Singh PK. Self-generated diversity produces "insurance effects" in biofilm communities. Proc Natl Acad Sci U S A. 2004;101:16630–5. 10.1073/pnas.0407460101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirisitis MJ, Prost L, Starkey M, Parsek M. Characterization of colony morphology variants isolated from Pseudomonas aeruginosa biofilms. Appl Environ Microbiol. 2005;71:4809–21. 10.1128/AEM.71.8.4809-4821.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Häussler S. Biofilm formation by the small colony variant phenotype of Pseudomonas aeruginosa. Environ Microbiol. 2004;6:546–51. 10.1111/j.1462-2920.2004.00618.x [DOI] [PubMed] [Google Scholar]
- 14.Haussler S, Ziegler I, Lottel A, von Gotz F, Rohde M, Wehmhohner D, et al. Highly adherent small-colony variants of Pseudomonas aeruginosa in cystic fibrosis lung infection. J Med Microbiol. 2003;52(Pt 4):295–301. 10.1099/jmm.0.05069-0 [DOI] [PubMed] [Google Scholar]
- 15.Malone JG. Role of small colony variants in persistence of Pseudomonas aeruginosa infections in cystic fibrosis lungs. Infection and Drug Resistance. 2015;8:237–47. 10.2147/IDR.S68214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Starkey M, Hickman J, Ma L, Zhang N, De Long S, Hinz A, et al. Pseudomonas aeruginosa rugose small colony variants have adaptations that likely promote persistence in the cystic fibrosis lung. J Bacteriol. 2009;191(11):3492–503. 10.1128/JB.00119-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Borlee BR, Goldman AD, Murakami K, Samudrala R, Wozniak DJ, Parsek MR. Pseudomonas aeruginosa uses a cyclic-di-GMP-regulated adhesin to reinforce the biofilm extracellular matrix. Mol Microbiol. 2010;75(4):827–42. 10.1111/j.1365-2958.2009.06991.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Colvin KM, Gordon VD, Murakami K, Borlee BR, Wozniak DJ, Wong GC, et al. The Pel polysaccharide can serve a structural and protective role in the biofilm matrix of Pseudomonas aeruginosa. PLoS Pathog. 2011;7(1):e1001264 10.1371/journal.ppat.1001264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Billings N, Millan M, Caldara M, Rusconi R, Tarasova Y, Stocker R, et al. The extracellular matrix Component Psl provides fast-acting antibiotic defense in Pseudomonas aeruginosa biofilms. PLoS Pathog. 2013;9(8):e1003526 10.1371/journal.ppat.1003526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mishra M, Byrd MS, Sergeant S, Azad AK, Parsek MR, McPhail L, et al. Pseudomonas aeruginosa Psl polysaccharide reduces neutrophil phagocytosis and the oxidative response by limiting complement-mediated opsonization. Cell Microbiol. 2012;14(1):95–106. 10.1111/j.1462-5822.2011.01704.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Irie Y, Starkey M, Edwards AN, Wozniak DJ, Romeo T, Parsek MR. Pseudomonas aeruginosa biofilm matrix polysaccharide Psl is regulated transcriptionally by RpoS and post-transcriptionally by RsmA. Mol Microbiol. 2010;78(1):158–72. 10.1111/j.1365-2958.2010.07320.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tseng BS, Zhang W, Harrison JJ, Quach TP, Song JL, Penterman J, et al. The extracellular matrix protects Pseudomonas aeruginosa biofilms by limiting the penetration of tobramycin. Environ Microbiol. 2013;15(10):2865–78. 10.1111/1462-2920.12155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harrison JJ, Turner RJ, Ceri H. Persister cells, the biofilm matrix and tolerance to metal cations in biofilm and planktonic Pseudomonas aeruginosa. Environ Microbiol. 2005;7(7):981–94. 10.1111/j.1462-2920.2005.00777.x [DOI] [PubMed] [Google Scholar]
- 24.Lebeaux D, Ghigo JM, Beloin C. Biofilm-related infections: bridging the gap between clinical management and fundamental aspects of recalcitrance toward antibiotics. Microbiol Mol Biol Rev. 2014;78(3):510–43. 10.1128/MMBR.00013-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alhede M, Kragh KN, Qvortrup K, Allesen-Holm M, van Gennip M, Christensen LD, et al. Phenotypes of non-attached Pseudomonas aeruginosa aggregates resemble surface attached biofilm. PLoS One. 2011;6(11):e27943 10.1371/journal.pone.0027943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haussler S, Tummler B, Weissbrodt H, Rohde M, Steinmetz I. Small-colony variants of Pseudomonas aeruginosa in cystic fibrosis. Clin Infect Dis. 1999;29(3):621–5. 10.1086/598644 [DOI] [PubMed] [Google Scholar]
- 27.Schneider M, Muhlemann K, Droz S, Couzinet S, Casaulta C, Zimmerli S. Clinical characteristics associated with isolation of small-colony variants of Staphylococcus aureus and Pseudomonas aeruginosa from respiratory secretions of patients with cystic fibrosis. J Clin Microbiol. 2008;46(5):1832–4. 10.1128/JCM.00361-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malone JG, Jaeger T, Spangler C, Ritz D, Spang A, Arrieumerlou C, et al. YfiBNR mediates cyclic-di-GMP dependent small colony variant formation and persistence in Pseudomonas aeruginosa. PLoS Pathog. 2010;6(3):e1000804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hickman JW, Tifrea DF, Harwood CS. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc Natl Acad Sci U S A. 2005;102(40):14422–7. 10.1073/pnas.0507170102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Drenkard E, Ausubel FM. Pseudomonas biofilm formation and antibiotic resistance are linked to phenotypic variation. Nature. 2002;416(6882):740–3. 10.1038/416740a [DOI] [PubMed] [Google Scholar]
- 31.Hengge R. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol. 2009;7:263–73. 10.1038/nrmicro2109 [DOI] [PubMed] [Google Scholar]
- 32.D'Argenio DA, Calfee MW, Rainey PB, Pesci EC. Autolysis and autoaggregation in Pseudomonas aeruginosa colony morphology mutants. J Bacteriol. 2002;184(23):6481–9. 10.1128/jb.184.23.6481-6489.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ueda A, Wood TK. Connecting quorum sensing, c-di-GMP, pel polysaccharide, and biofilm formation in Pseudomonas aeruginosa through tyrosine phosphatase TpbA (PA3885). PLoS Pathog. 2009;5(6):e1000483 10.1371/journal.ppat.1000483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giddens SR, Jackson RW, Moon CD, Jacobs MA, Zhang XX, Gehrig SM, et al. Mutational activation of niche-specific genes provides insight into regulatory networks and bacterial function in a complex environment. Proc Natl Acad Sci U S A. 2007;104(46):18247–52. 10.1073/pnas.0706739104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones CJ, Newsom D, Kelly B, Irie Y, Jennings LK, Xu B, et al. ChIP-Seq and RNA-Seq reveal an AmrZ-mediated mechanism for cyclic di-GMP synthesis and biofilm development by Pseudomonas aeruginosa. PLoS Pathog. 2014;10(3):e1003984 10.1371/journal.ppat.1003984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cabeen MT, Leiman SA, Losick R. Colony-morphology screening uncovers a role for the Pseudomonas aeruginosa nitrogen-related phosphotransferase system in biofilm formation. Mol Microbiol. 2016;99(3):557–70. 10.1111/mmi.13250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Malone JG, Jaeger T, Manfredi P, Dotsch A, Blanka A, Bos R, et al. The YfiBNR signal transduction mechanism reveals novel targets for the evolution of persistent Pseudomonas aeruginosa in cystic fibrosis airways. PLoS Pathog. 2012;8(6):e1002760 10.1371/journal.ppat.1002760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moscoso JA, Mikkelsen H, Heeb S, Williams P, Filloux A. The Pseudomonas aeruginosa sensor RetS switches type III and type VI secretion via c-di-GMP signalling. Environ Microbiol. 2011;13(12):3128–38. 10.1111/j.1462-2920.2011.02595.x [DOI] [PubMed] [Google Scholar]
- 39.Blanka A, Duvel J, Dotsch A, Klinkert B, Abraham WR, Kaever V, et al. Constitutive production of c-di-GMP is associated with mutations in a variant of Pseudomonas aeruginosa with altered membrane composition. Science Signaling. 2015;8(372):ra36 10.1126/scisignal.2005943 [DOI] [PubMed] [Google Scholar]
- 40.Wolter DJ, Emerson JC, McNamara S, Buccat AM, Qin X, Cochrane E, et al. Staphylococcus aureus small-colony variants are independently associated with worse lung disease in children with cystic fibrosis. Clin Infect Dis. 2013;57(3):384–91. 10.1093/cid/cit270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Z, Kosorok MR, Farrell PM, Laxova A, West SE, Green CG, et al. Longitudinal development of mucoid Pseudomonas aeruginosa infection and lung disease progression in children with cystic fibrosis. JAMA. 2005;293(5):581–8. 10.1001/jama.293.5.581 [DOI] [PubMed] [Google Scholar]
- 42.Siehnel R, Traxler B, An DD, Parsek MR, Schaefer AL, Singh PK. A unique regulator controls the activation threshold of quorum-regulated genes in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2010;107(17):7916–21. 10.1073/pnas.0908511107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cohen D, Mechold U, Nevenzal H, Yarmiyhu Y, Randall TE, Bay DC, et al. Oligoribonuclease is a central feature of cyclic diguanylate signaling in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2015;112(36):11359–64. 10.1073/pnas.1421450112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Orr MW, Donaldson GP, Severin GB, Wang J, Sintim HO, Waters CM, et al. Oligoribonuclease is the primary degradative enzyme for pGpG in Pseudomonas aeruginosa that is required for cyclic-di-GMP turnover. Proc Natl Acad Sci U S A. 2015;112(36):E5048–57. 10.1073/pnas.1507245112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dasgupta N, Wolfgang MC, Goodman AL, Arora SK, Jyot J, Lory S, et al. A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol Microbiol. 2003;50(3):809–24. 10.1046/j.1365-2958.2003.03740.x [DOI] [PubMed] [Google Scholar]
- 46.Wu DC, Zamorano-Sánchez D, Pagliai FA, Park JH, Floyd KA, Lee CK, et al. Reciprocal c-di-GMP signaling: Incomplete flagellum biogenesis triggers c-di-GMP signaling pathways that promote biofilm formation. PLoS Genet. 2020;16(3):e1008703 10.1371/journal.pgen.1008703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Watnick PI, Lauriano CM, Klose KE, Croal L, Kolter R. The absence of a flagellum leads to altered colony morphology, biofilm development and virulence in Vibrio cholerae O139. Mol Microbiol. 2001;39(2):223–35. 10.1046/j.1365-2958.2001.02195.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Belas R. Biofilms, flagella, and mechanosensing of surfaces by bacteria. Trends Microbiol. 2014;22(9):517–27. 10.1016/j.tim.2014.05.002 [DOI] [PubMed] [Google Scholar]
- 49.Kawagishi I, Imagawa M, Imae Y, McCarter L, Homma M. The sodium-driven polar flagellar motor of marine Vibrio as the mechanosensor that regulates lateral flagellar expression. Mol Microbiol. 1996;20(4):693–9. 10.1111/j.1365-2958.1996.tb02509.x [DOI] [PubMed] [Google Scholar]
- 50.McCarter L, Hilmen M, Silverman M. Flagellar dynamometer controls swarmer cell differentiation of V. parahaemolyticus. Cell. 1988;54(3):345–51. 10.1016/0092-8674(88)90197-3 [DOI] [PubMed] [Google Scholar]
- 51.Hug I, Deshpande S, Sprecher KS, Pfohl T, Jenal U. Second messenger-mediated tactile response by a bacterial rotary motor. Science. 2017;358(6362):531–4. 10.1126/science.aan5353 [DOI] [PubMed] [Google Scholar]
- 52.Belas R, Suvanasuthi R. The ability of Proteus mirabilis to sense surfaces and regulate virulence gene expression involves FliL, a flagellar basal body protein. J Bacteriol. 2005;187(19):6789–803. 10.1128/JB.187.19.6789-6803.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cairns LS, Marlow VL, Bissett E, Ostrowski A, Stanley-Wall NR. A mechanical signal transmitted by the flagellum controls signalling in Bacillus subtilis. Mol Microbiol. 2013;90(1):6–21. 10.1111/mmi.12342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Doyle TB, Hawkins AC, McCarter LL. The complex flagellar torque generator of Pseudomonas aeruginosa. J Bacteriol. 2004;186(19):6341–50. 10.1128/JB.186.19.6341-6350.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Toutain CM, Zegans ME, O'Toole GA. Evidence for two flagellar stators and their role in the motility of Pseudomonas aeruginosa. J Bacteriol. 2005;187(2):771–7. 10.1128/JB.187.2.771-777.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kuchma SL, Brothers KM, Merritt JH, Liberati NT, Ausubel FM, O'Toole GA. BifA, a cyclic-Di-GMP phosphodiesterase, inversely regulates biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J Bacteriol. 2007;189(22):8165–78. 10.1128/JB.00586-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Caiazza NC, O'Toole GA. SadB is required for the transition from reversible to irreversible attachment during biofilm formation by Pseudomonas aeruginosa PA14. J Bacteriol. 2004;186(14):4476–85. 10.1128/JB.186.14.4476-4485.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Caiazza NC, Merritt JH, Brothers KM, O'Toole GA. Inverse regulation of biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J Bacteriol. 2007;189(9):3603–12. 10.1128/JB.01685-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Baker AE, Webster SS, Diepold A, Kuchma SL, Bordeleau E, Armitage JP, et al. Flagellar stators stimulate c-di-GMP production by Pseudomonas aeruginosa. J Bacteriol. 2019:JB.00741–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Siryaporn A, Kuchma SL, O’Toole GA, Gitai Z. Surface attachment induces Pseudomonas aeruginosa virulence. Proc Natl Acad Sci U S A. 2014;111(47):16860–5. 10.1073/pnas.1415712111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Luo Y, Zhao K, Baker AE, Kuchma SL, Coggan KA, Wolfgang MC, et al. A hierarchical cascade of second messengers regulates Pseudomonas aeruginosa surface behaviors. mBio. 2015;6(1):e02456–14. 10.1128/mBio.02456-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lauriano CM, Ghosh C, Correa NE, Klose KE. The sodium-driven flagellar motor controls exopolysaccharide expression in Vibrio cholerae. J Bacteriol. 2004;186(15):4864–74. 10.1128/JB.186.15.4864-4874.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Klausen M, Heydorn A, Ragas P, Lambertsen L, Aaes-Jorgensen A, Molin S, et al. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol Microbiol. 2003;48(6):1511–24. 10.1046/j.1365-2958.2003.03525.x [DOI] [PubMed] [Google Scholar]
- 64.Barken KB, Pamp SJ, Yang L, Gjermansen M, Bertrand JJ, Klausen M, et al. Roles of type IV pili, flagellum-mediated motility and extracellular DNA in the formation of mature multicellular structures in Pseudomonas aeruginosa biofilms. Environ Microbiol. 2008;10(9):2331–43. 10.1111/j.1462-2920.2008.01658.x [DOI] [PubMed] [Google Scholar]
- 65.Nadell CD, Bassler BL. A fitness trade-off between local competition and dispersal in Vibrio cholerae biofilms. Proc Natl Acad Sci U S A. 2011;108(34):14181–5. 10.1073/pnas.1111147108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schluter J, Nadell CD, Bassler BL, Foster KR. Adhesion as a weapon in microbial competition. The ISME Journal. 2015;9(1):139–49. 10.1038/ismej.2014.174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xavier JB, Foster KR. Cooperation and conflict in microbial biofilms. Proc Natl Acad Sci U S A. 2007;104(3):876–81. 10.1073/pnas.0607651104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mahenthiralingam E, Campbell ME, Speert DP. Nonmotility and phagocytic resistance of Pseudomonas aeruginosa from chronically colonized patients with cystic fibrosis. Infect Immun. 1994;62:596–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jorth P, Staudinger BJ, Wu X, Hisert KB, Hayden H, Garudathri J, et al. Regional isolation drives bacterial diversification within cystic fibrosis lungs. Cell Host Microbe. 2015;18(3):307–19. 10.1016/j.chom.2015.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lovewell RR, Collins RM, Acker JL, O'Toole GA, Wargo MJ, Berwin B. Step-wise loss of bacterial flagellar torsion confers progressive phagocytic evasion. PLoS Pathog. 2011;7(9):e1002253 10.1371/journal.ppat.1002253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Basu Roy A, Sauer K. Diguanylate cyclase NicD-based signalling mechanism of nutrient-induced dispersion by Pseudomonas aeruginosa. Mol Microbiol. 2014;94(4):771–93. 10.1111/mmi.12802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dahlstrom KM, Collins AJ, Doing G, Taroni JN, Gauvin TJ, Greene CS, et al. A Multimodal strategy used by a large c-di-GMP network. J Bacteriol. 2018;200(8):e00703–17. 10.1128/JB.00703-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Staudinger BJ, Muller JF, Halldórsson S, Boles B, Angermeyer A, Nguyen D, et al. Conditions associated with the cystic fibrosis defect promote chronic Pseudomonas aeruginosa infection. Am J Respir Crit Care Med. 2014;189(7):812–24. 10.1164/rccm.201312-2142OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vogel HJ, Bonner DM. Acetylornithinase of Escherichia coli: Partial purification and some properties. J Biol Chem. 1956;218:97–106. [PubMed] [Google Scholar]
- 75.Hmelo LR, Borlee BR, Almblad H, Love ME, Randall TE, Tseng BS, et al. Precision-engineering the Pseudomonas aeruginosa genome with two-step allelic exchange. Nat Protoc. 2015;10(11):1820–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Choi K-H, Schweizer HP. mini-Tn7 insertion in bacteria with single attTn7 sites: example Pseudomonas aeruginosa. Nat Protoc. 2006;1(1):153–61. 10.1038/nprot.2006.24 [DOI] [PubMed] [Google Scholar]
- 77.Zhao K, Tseng BS, Beckerman B, Jin F, Gibiansky ML, Harrison JJ, et al. Psl trails guide exploration and microcolony formation in Pseudomonas aeruginosa biofilms. Nature. 2013;497(7449):388–91. 10.1038/nature12155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Choi K-H, Gaynor JB, White KG, Lopez C, Bosio CM, Karkhoff-Schweizer RR, et al. A Tn7-based broad-range bacterial cloning and expression system. Nat Methods. 2005;2(6):443–8. 10.1038/nmeth765 [DOI] [PubMed] [Google Scholar]
- 79.Goeres DM, Hamilton MA, Beck NA, Buckingham-Meyer K, Hilyard JD, Loetterle LR, et al. A method for growing a biofilm under low shear at the air-liquid interface using the drip flow biofilm reactor. Nat Protoc. 2009;4(5):783–8. 10.1038/nprot.2009.59 [DOI] [PubMed] [Google Scholar]
- 80.Byrd MS, Sadovskaya I, Vinogradov E, Lu H, Sprinkle AB, Richardson SH, et al. Genetic and biochemical analyses of the Pseudomonas aeruginosa Psl exopolysaccharide reveal overlapping roles for polysaccharide synthesis enzymes in Psl and LPS production. Mol Microbiol. 2009;73(4):622–38. 10.1111/j.1365-2958.2009.06795.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.DiGiandomenico A, Warrener P, Hamilton M, Guillard S, Ravn P, Minter R, et al. Identification of broadly protective human antibodies to Pseudomonas aeruginosa exopolysaccharide Psl by phenotypic screening. J Exp Med. 2012;209(7):1273–87. 10.1084/jem.20120033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Winsor GL, Griffiths EJ, Lo R, Dhillon BK, Shay JA, Brinkman Fiona SL. Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res. 2016;44(D1):D646–D53. 10.1093/nar/gkv1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dunnen JTd, Antonarakis SE. Mutation nomenclature extensions and suggestions to describe complex mutations: A discussion. Hum Mutat. 2000;15(1):7–12. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
(PDF)
(PDF)
(A) Semi-quantitative dot blots for the Psl polysaccharide from strains grown on VBMM agar. (B) Semi-quantitative Western blots for PelC from strains grown on VBMM agar. (C) Semi-quantitative dot blots for the Psl polysaccharide from strains grown on LB agar. (D) Semi-quantitative Western blots for PelC from strains grown on LB agar. Each bar indicates the mean and SD for 1 or 2 technical replicates from each of 3 to 4 independent biological replicates.
(TIF)
(A) The diguanylate cyclase WspR is dispensable for the nutrition-dependent RSCV phenotype resulting from RSCV-linked mutations identified in this study. Bacteria in these panels were cultured and photographed on VBMM agar containing Congo red and brilliant blue R (see Material and Methods). (B) These nutrition-dependent RSCV-linked genotypes give rise to bacterial colonies with smooth morphology on LB agar. LB agar was also supplemented with the dyes Congo red and brilliant blue R. Each panel represents an area that is approximately 5.0 × 3.5 mm.
(TIF)
Precisely defined in-frame deletion mutations were introduced into a series of flagellar genes in wild type PAO1 strain. Only some of these mutations caused the RSCV phenotype. In all panels, bacteria were cultured and photographed on VBMM agar containing Congo red and brilliant blue R (see Material and Methods). Each panel represents an area that is approximately 5.0 mm × 3.5 mm.
(TIF)
Semi-quantitative Western blots for PelC from flagellar mutants grown (A and B) in shaken LB or VBMM cultures, or (C and D) on the surface of LB or VBMM agar, respectively. Each bar indicates the mean and standard deviation for 1 or 2 technical replicates from each of 3 to 4 independent biological replicates.
(TIF)
Photographs of agar-grown RSCVs that were isolated from biofilm reactors after experimental evolution. RSCV-linked mutations were identified by whole genome sequencing (top). Colony morphology of mutants in which the RSCV-linked allele from the biofilm isolate was introduced into the ancestral PAO1 strain (bottom). In all panels, bacteria were cultured and photographed on VBMM agar containing Congo red and brilliant blue R (see Material and Methods). Each panel represents an area that is approximately 5.0 mm × 3.5 mm.
(TIF)
(A) Semi-quantitative PelC Western blots for strains grown on VBMM agar. (B) Semi-quantitative Psl dot blots for strains grown on VBMM agar. Each bar indicates the mean and standard deviation for 3 to 6 independent biological replicates.
(TIF)
(A) Colony morphology of isolates on LB and VBMM agar. Each panel represents an area that is approximately 5.0 mm × 3.5 mm. (B) Analysis of pulsed-field gel electrophoresis (PFGE) data indicating that pairs of CF isolates with smooth and RSCV colony phenotypes are close genetic relatives. Phylogenetic relationships were calculated from existing PFGE data for the PASA collection of P. aeruginosa isolates at Seattle Children’s Hospital [40] using Bionumerics Seven (Applied Maths).
(TIF)
(A) Semi-quantitative Western blots for PelC from strains grown on SCFM agar. (B) Semi-quantitative dot blots for the Psl polysaccharide from strains grown on SCFM agar. (C) Colony morphology of flagellum mutants on SCFM agar. Each panel represents an area that is approximately 5.0 mm × 3.5 mm. Each bar indicates the mean and SD for 3 biological replicates. Relative expression levels have been normalized to the ΔwspF strain.
(TIF)
Data Availability Statement
All sequencing data have been deposited with links to BioProject accession number PRJNA625996 in the National Center for Biotechnology (NCBI) BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/). All other relevant data are within the manuscript and its Supporting Information files.