Abstract
Using data for 20 912 patients from 2 large academic health systems, we analyzed the frequency of severe acute respiratory syndrome coronavirus 2 reverse-transcription polymerase chain reaction test discordance among individuals initially testing negative by nasopharyngeal swab who were retested on clinical grounds within 7 days. The frequency of subsequent positivity within this window was 3.5% and was similar across institutions.
Keywords: COVID-19, SARS-CoV-2, RT-PCR, test characteristics, nasopharyngeal
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the etiologic agent of coronavirus disease 2019 (COVID-19). Accurate detection of the virus is essential to strategies endorsed by the Centers for Disease Control and Prevention (CDC) and the World Health Organization. As the availability and speed of SARS-CoV-2 testing platforms improve, results of these tests are increasingly relied upon to inform critical decisions related to therapeutic intervention, use of personal protective equipment, patient isolation, and workforce readiness. While the analytic performance of SARS-CoV-2 reverse-transcription polymerase chain reaction (RT-PCR) tests are well described [1], clinical performance is impacted by several factors that are difficult to measure, such as low levels of shedding during incubation and early infection [2], variability in the site of specimen acquisition [3, 4], and sufficiency of sample collected. In addition, early reports and characterizations in the press have left the medical community and general public with concerns about the reliability of SARS-CoV-2 RT-PCR testing and the interpretation of negative results. Data characterizing the scope of false-negative results observed in the context of current testing practices in the United States (US) are needed to guide clinical protocols and inform the public, but are lacking.
The initial US introduction of COVID-19 through Washington State [5], followed closely by Northern California [6], combined with the early availability of SARS-CoV-2 testing in both regions [7, 8], provides an opportunity to evaluate clinical test performance in a population of repeatedly tested patients. In this study, utilizing data from 2 independent healthcare systems and analyzed by separate research teams, we report the frequency of discordant SARS-CoV-2 RT-PCR results among individuals who initially tested negative and were subsequently retested within 7 days.
MATERIALS AND METHODS
Common Study Methods
The study was conducted at the University of Washington (UW) and Stanford University, involving a total of 23 126 SARS-CoV-2 RT-PCR tests (10 583 UW, 12 543 Stanford) performed on 20 912 eligible patients (8977 UW, 11 935 Stanford), between 2 March and 7 April 2020. Test results through 14 April were extracted from the electronic medical record to allow for a complete 7-day observation period and an additional day for result reporting. Data on cycle threshold values were extracted from the laboratory information system and are interpreted as inversely proportional to the viral load level present in the sample. At both sites, samples were transported in 3 mL of viral or universal transport medium, or phosphate-buffered saline when necessitated due to supply chain shortages, and processed without further dilution.
UW Methods
The UW Virology clinical laboratory serves as the primary testing center for a broad region in the US Pacific Northwest, processing > 60% of all SARS-CoV-2 tests for Washington State during the time period examined. To ensure consistency of clinical data and compliance with patient privacy policies, analysis was limited to adult patients having an established affiliation with UW Medicine. Encounters spanning multiple facilities (eg, outpatient, hospital, and drive-through testing locations) were linked using an unambiguous identifier common to all sites. UW guidelines over the study period for testing included the following: all patients who exhibited 1 or more symptoms of COVID-19 at the time of initial testing per institutional protocol, which involved new symptoms of acute respiratory infection (eg, fever, cough, shortness of breath, myalgias, rhinorrhea, sore throat, anosmia, ageusia), combined with pertinent risk factors (occupation, age, chronic disease status, immunosuppression, contact with confirmed COVID-19 cases, pregnancy, housing stability, exposure to high-risk facilities, or inpatient admission) or based on clinical judgment. A single change to testing criteria occurred during the study period: Beginning 30 March 2020, UW Medicine initiated universal preoperative SARS-CoV-2 screening for all asymptomatic surgical cases, the results of which are included in the primary analysis. Nasopharyngeal (NP) samples were collected according to a standardized institutional protocol that includes bilateral NP sampling. The UW testing platforms included a laboratory-developed 2-target/2-control assay modified from the CDC (target genes N1, N2) operating under a Washington State emergency use authorization [7]; Panther Fusion SARS-CoV-2 assay (Hologic, Marlborough, MA, target genes two conserved regions of ORF1ab); Roche RT-PCR (Basel, Switzerland, target E gene); DiaSorin (Saluggia, Italy, targets ORF1ab and S gene). Inconclusive RT-PCR test results (ie, only 1 of 2 SARS-CoV-2 target genes amplified), which suggest samples with a viral load spanning the lower limit of detection [9], were treated as positive in this analysis in accordance with UW test interpretation guidelines [10] and clinical practice. The UW Institutional Review Board determined this study to be exempt from human subjects review (STUDY00009931).
Stanford Methods
The Stanford Health Care (SHC) Clinical Virology Laboratory is based in Northern California and performed SARS-CoV-2 testing on both adult and pediatric populations. Approximately two-thirds of the samples were from Stanford Medicine facilities and one-third were from medical facilities in Northern California, with the greatest concentration coming from facilities in San Mateo and Santa Clara counties. Stanford guidelines for testing were the same as UW for the initial phase of the study and similarly, beginning 6 April 2020, testing was expanded to include asymptomatic preoperative screening. The NP swabbing protocol at Stanford facilities utilized a unilateral sampling approach but was otherwise comparable. Testing was performed using 1 of 2 assays: (1) SHC Emergency Use Authorization laboratory-developed test (target gene E) [8] or (2) Panther Fusion SARS-CoV-2 assay. This study received approval by the Stanford Institutional Review Board (protocol number 48973), and individual consent was not required.
RESULTS
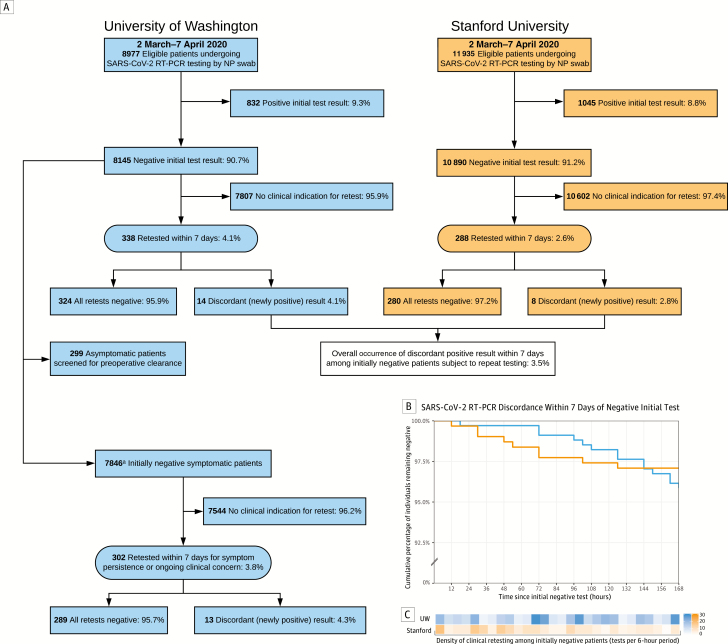
A total of 23 126 SARS-CoV-2 RT-PCR tests (10 583 UW, 12 543 Stanford) were performed in 20 912 eligible patients (8977 UW, 11 935 Stanford) undergoing initial testing by NP swab between 2 March and 7 April 2020. Initial results for 91% (90.7% UW, 91.2% Stanford) of patients were negative (Figure 1A). Characteristics of initially negative patients are shown in Supplementary Table 1. The majority of these patients (95.9% UW, 97.4% Stanford) did not undergo repeat testing within 7 days and did not require subsequent evaluation in the form of outpatient, emergency department, or inpatient encounters (Supplementary Table 1). Several negatively retested patients at both sites were ultimately diagnosed with other viral respiratory illnesses, most commonly influenza A, rhinovirus, RSV, metapneumovirus, and seasonal coronavirus (Supplementary Table 1). However, a small proportion (4.1% UW, 2.6% Stanford) underwent repeat testing within this window despite an initial negative result (Figure 1A). Among those requiring reevaluation, 96.5% (95.9% UW, 97.2% Stanford) remained negative on all repeat tests performed within 7 days.
Figure 1.
Identification of patients initially testing negative for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and outcomes of repeat testing. A, The primary measure was the occurrence of a discordant (newly positive) result within 7 days. aSubgroup analysis excluding asymptomatic patients screened for surgical clearance at the University of Washington (UW) yielded similar results. B, Among patients initially testing negative for SARS-CoV-2 by reverse-transcription polymerase chain reaction (RT-PCR) of a nasopharyngeal (NP) swab, > 95% of patients at both UW and Stanford subjected to retesting remained negative on subsequent tests performed within 7 days. C, Retesting of initially negative individuals occurred at varied intervals across the 7-day period of observation.
It was observed that 3.5% (4.1% UW, 2.8% Stanford) of patients subjected to retesting on clinical grounds within 7 days were subsequently found to be positive during this period, suggesting a false-negative initial result. The timing of clinical retesting and occurrence of newly discordant positive results among these patients is shown by site in Figure 1B and Figure 1C, respectively. The clinical contexts and testing parameters of the 22 patients with discordant results are summarized in Supplementary Table 2. In this group, the mean interval between initial negative test and first positive retest was 4.0 days (standard deviation [SD], 2.0 days). RT-PCR cycle threshold values of newly positive results averaged 28.5 (SD, 8.0).
At UW, the use of standardized testing algorithms enabled subgroup analysis based on testing indication (Figure 1A). A total of 299 asymptomatic individuals who were tested as part of universal screening for preoperative clearance were excluded, leaving 7846 symptomatic individuals who tested negative at the time of initial presentation for analysis. Of the 302 individuals in this group with persistent or worsening symptoms warranting additional testing within 7 days, 4.3% converted from negative to positive and 95.7% remained negative on all subsequent SARS-CoV-2 tests performed within this window.
DISCUSSION
In this report, 2 independent research teams describe that, among patients initially testing negative by SARS-CoV-2 RT-PCR of NP swabs, repeat testing within 7 days yielded a positive result in 3.5% of cases; the majority (96.5%) of those warranting additional testing for any reason remained negative on all subsequent tests within this window. Among the subgroup of UW patients confirmed to have symptoms prior to an initial negative result who were retested for persistent or worsening symptoms, a similar proportion (4.3%) was subsequently found to be positive within 7 days. These observations suggest that false-negative NP SARS-CoV-2 RT-PCR results do occur, but potentially at a lower frequency than is currently believed.
Results from each research group have limitations. Neither team was able to calculate a true clinical sensitivity or false-negative proportion due to the absence of retesting in all initially negative patients and the lack of a gold-standard confirmatory mechanism. The cause of false-negative initial results also cannot be determined with confidence. However, the range of cycle threshold values observed in subsequent positive assays suggests that both sampling inefficiencies and low viral load (in cases of adequate sampling) may be contributing factors in this population. Additionally, it cannot be ruled out that some discordant test results in this cohort may be due to newly acquired infection. By limiting the scope of retesting considered to a 7-day period, the likelihood of this scenario is minimized, but not eliminated. Finally, we were unable to ascertain the disease status of the individuals who initially tested negative for COVID-19 but did not undergo repeat testing; in most cases this likely reflects the absence of an indication for retesting (eg, alternative diagnosis or resolution of symptoms), but could also be the result of limited access to care.
The intention of this report is not to definitively quantify the clinical performance of NP SARS-CoV-2 RT-PCR testing, which will likely require orthogonal approaches such as serology. Rather, by characterizing the experience of 2 large US health systems on the short-term occurrence of newly positive SARS-CoV-2 results among initially test-negative patients, we provide data on a topic of practical significance that should be used in combination with other reports to guide the use and interpretation of this common testing modality.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. The authors thank Christine Fong and the Center for Perioperative and Pain Initiatives in Quality Safety Outcome at the University of Washington, Seattle, for assistance with the data extract analyzed in the present work.
Financial support. This work was supported by the National Institute of General Medical Sciences (grant number T32 GM086270-11 to D. R. L.) and the National Institute on Drug Abuse (K23 DA046686 to J. E. S.) at the National Institutes of Health as well as the National Science Foundation (1914873 and 1812559 to J. E. S.).
Potential conflicts of interest. A. L. G. reports personal fees from Abbott Molecular, outside the submitted work. All other authors report no potential conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. US Food and Drug Administration. Coronavirus disease 2019 (COVID-19) EUA information: in vitro diagnostic products Available at: https://www.fda.gov/emergency-preparedness-and-response/mcm-legal-regulatory-and-policy-framework/emergency-use-authorization#2019-ncov. Accessed 3 April 2020.
- 2. Wölfel R, Corman VM, Guggemos W, et al. Virological assessment of hospitalized patients with COVID-2019. Nature 2020; 581:465–9. [DOI] [PubMed] [Google Scholar]
- 3. Wang W, Xu Y, Gao R, et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA 2020; 323:1843–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fang Y, Zhang H, Xie J, et al. Sensitivity of chest CT for COVID-19: comparison to RT-PCR [published online ahead of print 19 February 2020]. Radiology 2020. doi:10.1148/radiol.2020200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Holshue ML, DeBolt C, Lindquist S, et al. First case of 2019 novel coronavirus in the United States. N Engl J Med 2020; 382:929–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hogan CA, Sahoo MK, Pinsky BA. Sample pooling as a strategy to detect community transmission of SARS-CoV-2. JAMA 2020; 323:1967–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Casto AM, Huang M-L, Nalla A, et al. Comparative performance of SARS-CoV-2 detection assays using seven different primer/probe sets and one assay kit. J Clin Microbiol 2020; 58:e00557–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. US Food and Drug Administration. Stanford Health Care clinical virology laboratory SARS-CoV-2 test: EUA summary 2020. Available at: https://www.fda.gov/media/136818/download. Accessed 10 April 2020.
- 9. Lu X, Wang L, Sakthivel SK, et al. US CDC real-time reverse transcription PCR panel for detection of severe acute respiratory syndrome coronavirus 2 [manuscript published online ahead of print 12 May 2020]. Emerg Infect Dis 2020. doi:10.3201/eid2608.201246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. University of Washington Laboratory Medicine. Frequently asked questions about COVID-19 testing. Available at: https://testguide.labmed.uw.edu/public/guideline/covid_faq. Accessed 25 May 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.