Abstract
Objectives
To evaluate the clinical performance of 3 molecular assays for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).
Methods
We used 184 nasopharyngeal swab specimens to compare Abbott ID NOW COVID-19 (Abbott ID NOW), DiaSorin Molecular Simplexa COVID-19 Direct (DiaSorin Simplexa), and Roche cobas 6800 SARS-CoV-2 (Roche cobas) assays. In a separate analysis, 3 specimens (nasopharyngeal, oropharyngeal, and nasal) were collected from 182 unique patients presenting to the emergency department with suspicion of coronavirus disease 2019 and were tested utilizing Abbott ID NOW. To further characterize each assay, relative limits of detection were evaluated utilizing positive nasopharyngeal patient samples.
Results
The positive percent agreement was 91% (95% confidence interval [CI], 0.76-0.97) for Abbott ID NOW and 100% (95% CI, 0.90-1.00) for DiaSorin Simplexa and Roche cobas. The negative percent agreement was 100% (95% CI, 0.98-1.00) for all 3 assays. All swab types tested with the Abbott assay produced concordant results. Polymerase chain reaction assays had approximately 10 to 100 times lower limits of detection than Abbott ID NOW.
Conclusions
Based on these evaluations, a multiplatform testing approach is proposed, depending on patient population and assay sensitivity, to address testing needs during a public health emergency.
Keywords: SARS-CoV-2, Abbott ID NOW, Molecular diagnostics, COVID-19, Coronavirus
Key Points.
Polymerase chain reaction assays had approximately 10 to 100 times lower limits of detection than Abbott ID NOW.
Matched dry oropharyngeal and dry nasal swabs performed equally to the nasopharyngeal swab collected in viral transport media when tested using the Abbott ID NOW SARS-CoV-2 assay.
A multiplatform testing approach is proposed to meet the patient testing needs during this public health emergency.
In December 2019, an outbreak of viral pneumonia with severe acute respiratory syndrome coronavirus-like (SARS-CoV) symptoms emerged in the Hubei province of China. The unknown virus spread rapidly throughout mainland China and ultimately to nearly every country in the world.1,2 The etiological agent of this outbreak was later identified as a novel human virus, named SARS-CoV-2, causing coronavirus disease 2019 (COVID-19).2,3 SARS-CoV-2 (COVID-19) was declared to be a pandemic by the World Health Organization and has caused more than 3 million infections and 210,000 deaths globally4,5 at the time of this study. During the early stages of the pandemic, China, Italy, and Iran suffered from the greatest number of SARS-CoV-2 infections and subsequently had high numbers of casualties.
The virus traveled to the United States (US) early on during the outbreak, with the first reported cases coming from the state of Washington on January 19, 2020.6 Due to the rapid spread of the virus in the US, the Secretary of Health and Human Services declared a public health emergency on February 4, 2020, which facilitated the development and emergency use authorization (EUA) of in vitro diagnostic tests for SARS-CoV-2. SARS-CoV-2 has not spread uniformly across the US, although all 50 states have reported cases of COVID-19 to the Centers for Disease Control and Prevention (CDC).7 The coastal regions, including New York, California, and Louisiana, became epicenters of infection, likely due to community spread exacerbated by the high population densities in those areas. The rapid spread of COVID-19 coupled with the need for widespread testing has resulted in shortages of essential components of COVID-19 test kits, including reagents, nasopharyngeal swabs, and universal viral transport medium (UVT).
The clinical presentation of COVID-19 encompasses a broad spectrum of illness, ranging from the asymptomatic to those with mild to severe respiratory illness. Symptoms at the onset can include fever, cough, shortness of breath, chills, and loss of taste or smell.8 While it is thought that the majority of cases are asymptomatic or mild, the infection can progress rapidly and cause severe disease, especially in older adults, patients with serious underlying medical conditions, and those requiring mechanical ventilation.9 Although other human coronaviruses have been capable of causing devastating outbreaks, including SARS-CoV and the Middle East respiratory syndrome coronavirus (MERS-CoV), SARS-CoV-2 seems to be more efficiently and rapidly transmitted between humans, resulting in a massive global pandemic.10-13
Timely and accurate laboratory diagnosis of SARS-CoV-2 can significantly impact patient management, which is critically important to infection control measures aimed to curb the pandemic within communities and hospitals. Due to the rapid timeframe for assay development under the Food and Drug Administration (FDA) EUA and the lack of reference materials, it remains unclear how each assay performs relative to the others. Many clinical laboratories have implemented multiple assays for molecular testing of SARS-CoV-2 in an effort to meet testing demand. It is left to each laboratory to understand the differences between any testing platforms that they choose to implement, and to guide patient care based on the respective results.
In the present study, we evaluated the clinical performance of 3 SARS-CoV-2 assays: Abbott ID NOW COVID-19 (Abbott ID NOW), DiaSorin Molecular Simplexa COVID-19 Direct (DiaSorin Simplexa), and Roche cobas 6800 SARS-CoV-2 (Roche cobas) using nasopharyngeal swabs from symptomatic patients. The equivalency of samples collected in UVT vs dry swabs tested on Abbott ID NOW was also assessed. In addition, we analyzed the analytical sensitivity of those 3 assays. This information may aid health care providers and public health officials as they decide how and where to implement the testing platforms available within their systems. The importance of obtaining rapid, reliable results is more critical than ever and will have a large impact on states across the country that are in danger of becoming COVID-19 epicenters.
Materials and Methods
Specimen Collection and Storage
Flocked swabs were used to collect nasopharyngeal specimens from symptomatic patients suspected of COVID-19 that met criteria for testing, either presenting to the emergency department or as inpatients at OhioHealth Riverside Methodist Hospital. After collection, the swabs were placed into 3 mL of sterile UVT (Becton Dickinson). Specimens were tested as soon as possible after collection, or if testing was delayed, were stored for up to 72 hours at 2°C to 8°C. Following routine testing, samples were stored frozen (≤–80°C) until comparator testing with the Roche cobas assay could be completed.
Study Design
Clinical performance of the 3 assays was evaluated using a total of 184 prospective nasopharyngeal specimens collected from patients of all ages and both genders presenting with signs and/or symptoms of COVID-19 infection. The samples were originally submitted for routine COVID-19 testing at OhioHealth Laboratories on the DiaSorin Simplexa assay. Subsequent testing on the additional platforms was based on availability of samples and meeting storage requirement criteria for testing on each platform according to the package insert requirements at the time of the study.
To compare the equivalence of nasopharyngeal, oropharyngeal, and nasal swabs using the Abbott ID NOW assay, a quality improvement study approved by the OhioHealth Research Institute was conducted where 3 specimens were collected from patients that presented to the emergency department. A total of 182 prospective specimen sets were collected. The nasopharyngeal swab was collected as part of standard of care testing, while oropharyngeal and nasal swabs were collected upon consent from the patient. Nasal swabs were collected according to the CDC instructions using a single swab to sample both nares. All 3 samples were tested using the Abbott ID NOW assay, while the nasopharyngeal samples collected in UVT were also tested using the DiaSorin Simplexa assay.
DiaSorin Molecular Simplexa COVID-19 Direct EUA
The DiaSorin Molecular Simplexa COVID-19 Direct EUA assay (DiaSorin Molecular) was performed according to the manufacturer’s instructions for use. Briefly, 50 μL of Simplexa COVID-19 Direct Kit reaction mix (MOL4150) was added to the “R” well of the 8-well Direct Amplification Disc followed by adding 50 μL of nonextracted nasopharyngeal swab sample (collected in approximately 3 mL of UVT) to the “SAMPLE” well. Tests were run on the LIAISON MDX system and data collection and analysis was performed with LIAISON MDX Studio software. The assay targets 2 different regions of the SARS-CoV-2 genome, the S gene and ORF1ab, differentiated with FAM and JOE fluorescent probes. An RNA internal control (Q670 probe) is used to detect reverse transcription polymerase chain reaction (RT-PCR) failure and/or inhibition. The result interpretation algorithm for reporting a positive specimen requires only 1 of the 2 targets to be detected (S or ORF1ab gene).
Abbott ID NOW COVID-19 EUA
The Abbott ID NOW COVID-19 assay (Abbott Diagnostics) was performed according to the manufacturer’s instructions for use. Briefly, the orange test base was inserted into the appropriate orange color-coded receptacle, followed by placing the sample receiver into the corresponding blue color-coded receptacle. The nasopharyngeal swab was placed into UVT, while the oropharyngeal and nasal dry swabs were placed directly into the sample receiver. The swab was vigorously mixed in the sample receiver buffer for 10 seconds for direct testing. For specimens in UVT, 200 µL was pipetted into the sample receiver and mixed. The assay uses isothermal nucleic acid amplification and targets the RdRp region of the SARS-CoV-2 genome. Abbott ID NOW contains an internal control that has been designed to control for sample inhibition, amplification, and assay reagent function.
Roche cobas 6800 SARS-CoV-2
The Roche cobas 6800 SARS-CoV-2 assay (Roche Diagnostics) was performed according to the manufacturer’s instructions for use. The test uses a minimum required sample volume of 600 µL. The sample preparation is fully automated (nucleic acid extraction and purification) followed by PCR amplification and detection. The assay targets the ORF1 a/b nonstructural region that is unique to SARS-CoV-2. Additionally, the assay targets a conserved region in the structural protein envelope E-gene with pan-sarbecovirus detection that will also detect SARS-CoV-2 virus. The result was interpreted as positive if both targets were detected and presumptive positive if 1 of 2 targets was detected.
Analytical Sensitivity
Relative analytic sensitivity was determined by evaluating serial dilutions from 10 clinical specimens containing SARS-CoV-2. Ten-fold dilutions of each clinical sample were tested on all 3 platforms. The relative limit of detection (LOD) was evaluated qualitatively using all 10 clinical samples.
Statistical Methods
Positive percent agreement (PPA), and negative percent agreement (NPA) between all assays were calculated with two-sided (upper/lower) 95% confidence intervals (CIs) using the Evidence-Based Medicine Toolbox, Knowledge Translation Program. The PPA was calculated as the fraction and percentage of cases that agree with a consensus standard of 2 out of 3 results when the specified technique produced a positive test. The NPA was similarly calculated among cases for which the specified technique produced a negative test.
Results
Clinical Performance of Three EUA SARS-CoV-2 (COVID-19) Molecular Assays
For evaluation of clinical performance 184 nasopharyngeal specimens in UVT were tested on all 3 assays and compared. DiaSorin Simplexa and Roche cobas molecular assays demonstrated a PPA of 100% (33/33), while Abbott ID NOW showed a PPA of 91% (30/33) compared to the consensus standard. NPA was 100% (151/151) among all three assays Table 1.
Table 1.
Clinical Performance Comparison of Abbott ID NOW, DiaSorin Simplexa, and Roche cobas in Nasopharyngeal Swab Specimens Collected in Universal Viral Transport Medium (n = 184)
| Molecular Assay | Consensus Standarda | |||
|---|---|---|---|---|
| Positive | Negative | PPA (± 95% CI) | NPA (± 95% CI) | |
| Abbott ID NOW | ||||
| Positive | 30 | 0 | 91% | 100% |
| Negative | 3b | 151 | (0.76-0.97) | (0.98-1.00) |
| DiaSorin Simplexa | ||||
| Positive | 33 | 0 | 100% | 100% |
| Negative | 0 | 151 | (0.90-1.00) | (0.98-1.00) |
| Roche cobas | ||||
| Positive | 33 | 0 | 100% | 100% |
| Negative | 0 | 151 | (0.90-1.00) | (0.98-1.00) |
CI, confidence interval; NPA, negative percent agreement; PPA, positive percent agreement.
aThe consensus standard was defined as the result obtained from at least 2 of the 3 assays.
bCycle threshold not available.
Comparison of Matched Nasopharyngeal Specimens in UVT With Dry Oropharyngeal and Nasal Swabs
Specimen type comparisons (nasopharyngeal in UVT, dry oropharyngeal, and dry nasal) from 182 patients tested by Abbott ID NOW produced 100% concordance for positive (12/12/12) and negative patients (170/170/170). As a comparator, each nasopharyngeal specimen was also tested on DiaSorin Simplexa. All 12 positive specimens were correctly identified by DiaSorin Simplexa; however, 1 additional positive specimen was identified. The discordant sample was misidentified as negative on all 3 specimen types when tested on the Abbott ID NOW instrument Table 2.
Table 2.
Clinical Comparison of Matched Nasopharyngeal, Oropharyngeal, and Nasal Swab Specimens Collected From Patients Presenting to the Emergency Department Using Abbott ID NOW (n = 182)
| Molecular Assay | DiaSorin Simplexaa | |||
|---|---|---|---|---|
| Positive | Negative | PPA (± 95% CI) | NPA (± 95% CI) | |
| Abbott ID NOW (NPS UVT) | ||||
| Positive | 12 | 0 | 92% | 100% |
| Negative | 1b | 169 | (0.67-0.99) | (0.98-1.00) |
| Abbott ID NOW (Dry OPS) | ||||
| Positive | 12 | 0 | 92% | 100% |
| Negative | 1b | 169 | (0.67-0.99) | (0.98-1.00) |
| Abbott ID NOW (Dry NS) | ||||
| Positive | 12 | 0 | 92% | 100% |
| Negative | 1b | 169 | (0.67-0.99) | (0.98-1.00) |
CI, confidence interval; NPA, negative percent agreement; NPS, nasopharyngeal swabs; NS, nasal swabs; OPS, oropharyngeal swabs; PPA, positive percent agreement; UVT, universal viral transport medium.
aDiaSorin Simplexa was considered the comparator and was only performed on NPS collected in UVT.
bCycle threshold not available
Relative Analytical Sensitivity of SARS-CoV-2 (COVID-19) in 10 Patient Samples
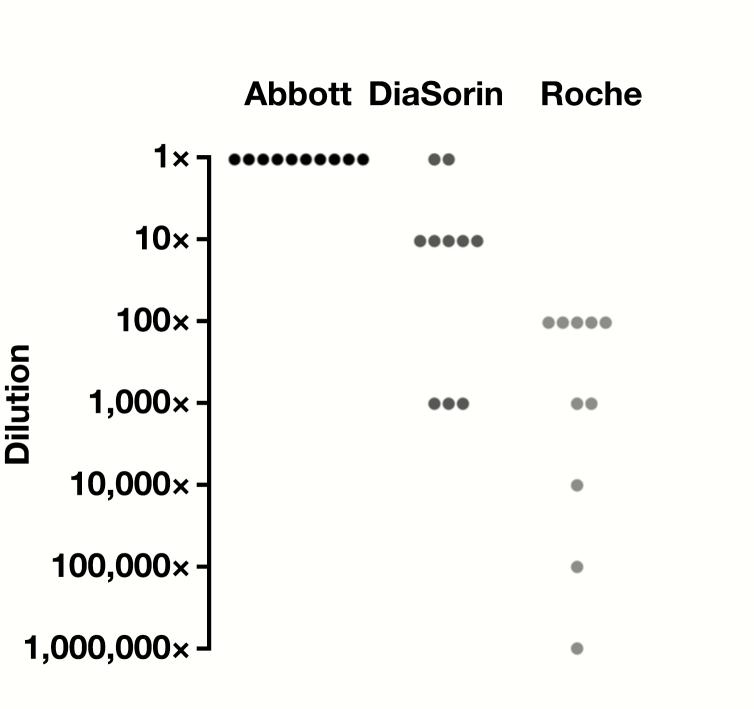
Ten positive nasopharyngeal specimens were serially diluted in UVT and all dilutions were tested by Abbott ID NOW, DiaSorin Simplexa, and Roche cobas assays Figure 1. For each series, results were normalized to the last positive dilution resulted on the Abbott ID NOW to account for different starting concentrations. Relative to Abbott ID NOW, half of the specimens detected SARS-CoV-2 at 10× lower concentrations by DiaSorin Simplexa, indicating approximately 10-fold lower LOD for this assay. The Roche cobas assay produced 5 of 10 positive reactions at 100× lower relative dilution, indicating at least 100-fold lower LOD than Abbott ID NOW.
Figure 1.
Relative limits of detection of Abbott ID NOW, DiaSorin Simplexa, and Roche cobas using 10 nasopharyngeal swab–positive patient samples collected in universal viral transport medium. A 10-fold dilution series was made for each patient sample and tested on all 3 instruments until a negative result was obtained. For each patient series, results were normalized to the lowest detected dilution of Abbott ID NOW (1×) and the last detectable reaction below that dilution is indicated for each instrument. Each dot represents the lowest detected dilution for each patient relative to Abbott ID NOW. All data used for this analysis can be found in the supplemental dataset (all supplemental material is available at American Journal of Clinical Pathology online).
Discussion
Accurate results for SARS-CoV-2 testing are extremely important since they affect not only decision-making in the clinical setting but also have public health implications for the community. In addition, turnaround time for results can be critical when treating an isolated patient suspected of having COVID-19. Use of personal protective equipment required by health care professionals might be spared along with the use of limited space in isolation rooms and other vital resources such as personnel to care for those patients. The consequences of a false-negative result can be serious since the virus is so transmissible and can have devastating effects in vulnerable patient populations. A false-positive result is also undesirable, as it may have a negative impact on the health of patients (eg, not seeking care for another disease) or that of health care workers, as they will have to abstain from their duties when they are most needed.
A number of different studies have been published regarding the clinical sensitivity of various RT-PCR COVID-19 assays.14-16 Published reports have demonstrated 94% PPA of the Abbott ID NOW and 96% PPA of the DiaSorin Simplexa assays compared to a modified CDC assay,17 while another study comparing nasal and nasopharyngeal specimens tested on Abbott ID NOW and mAbbott 2000 showed 75% PPA between the two assays.18 In our study we compared the PPA and NPA of the Abbott ID NOW to the DiaSorin Simplexa and the Roche cobas assays. In our area, the positivity rate based on reporting of positive tests was 7% at the time of testing.19 The variability in PPA observed in different reports likely depends on the amount of virus present in samples at the time of testing. This suggests the importance of choosing the right assay for each patient population.
In our institution only the DiaSorin Simplexa platform was available initially, followed soon after by the Abbott ID NOW and then the Roche cobas. Because of questions surrounding the use of UVT on the Abbott ID NOW assay we performed an additional study to evaluate the equivalency of the nasopharyngeal swab in UVT to that of dry swabs (either oropharyngeal or nasal). This was critical, as Abbott modified the assay instructions for use to include only dry swabs. In our study the use of UVT was equivalent to that of dry swabs when using the Abbott ID NOW. During the same time period, a large educational effort was underway by our nursing educators for proper nasopharyngeal specimen collections. All the specimen collectors in our system were reeducated and their competency reverified by direct observation. Given the emphasis put on properly collected specimens, we believe the difference observed in the PPA between assays was not due to specimen collection but rather due to the analytical limits of the assays.
Due to the lack of verified reference material to evaluate the LOD across the assays available in our institution, we examined the relative LOD using real patient specimens. By performing serial dilutions on patient samples with varying degrees of positivity (moderately to low positive samples as indicated by the cycle threshold values obtained by the DiaSorin Simplexa assay), we directly tested the LOD relative to one another. Roche cobas was the most sensitive assay as expected, since it uses a larger volume of sample for extraction (600 µL), and an RNA extraction step takes place followed by the RT-PCR assay. DiaSorin Simplexa appears to have a higher LOD than the Roche cobas assay; this test utilizes a smaller sample volume (50 µL) than Roche cobas and does not have a separate extraction step. Abbott ID NOW has an LOD approximately 10- to 100-fold higher than the PCR assays in our study. The assay uses 200 µL of UVT sample or uses a dry swab placed into the sample receiver buffer. Also, the Abbott ID NOW assay only amplifies one target for detection of SARS-CoV-2 compared to the other assays where 2 targets are used. However, its overall clinical sensitivity was about 10% lower when compared to DiaSorin Simplexa and Roche cobas. This suggests that the performance of the assay will suffer when it is used in patient populations that have low viral loads.
High testing demand coupled with limited availability of testing kits makes a multiplatform approach an attractive method for laboratories to meet the high demands for testing. Laboratories are in need of every testing option available but they must gain an understanding of the limitations of each. It is crucial to optimize utilization within the patient populations that can benefit most from each platform’s unique capabilities and limitations. Our institution is a large, multihospital network, and based on our experience with a variety of patient populations, turnaround time demands, and instrument/reagent availability, we propose a model for the use of each type of assay Table 3. Currently, none of the assays are FDA EUA approved for testing of asymptomatic individuals. However, a highly sensitive test is needed to assess patients as hospitals open up for elective and other procedures, particularly those that may involve aerosolization. In this scenario, a highly sensitive assay with a longer turnaround time may be most appropriate. For asymptomatic patients being admitted to the hospital, or for inpatients that had previously tested negative but have clinical symptoms consistent with COVID-19, a fast and sensitive test is needed to appropriately identify and stratify them in hospital units. Finally, for symptomatic patients with high clinical suspicion and high pretest probability presenting to the emergency department, a rapid point-of-care option may be appropriate.
Table 3.
SARS-CoV-2 Testing Platform Prioritization Matrix for Patient Testinga
| Patient Population | ||||
|---|---|---|---|---|
| Symptomatic: Rule out COVID-19 | Symptomatic: Prior COVID-19 Negative | Asymptomatic: Inpatient Admission | Asymptomatic: Preadmission | |
| TAT Requirement (h) | 1 | 1-12 | 1-12 | 24-72 |
| Consequences of incorrect test | Lower, patient kept in isolation due to high clinical index of suspicion | High, patient may be removed from isolation due to negative test result | High, exposure risk to others | High, exposure risk to others |
| Assay-based method | POCT | Rapid RT-PCR | Rapid RT-PCR | Batched RT-PCR |
COVID-19, coronavirus disease 2019; POCT, point-of-care test; RT-PCR, reverse transcription polymerase chain reaction; TAT, turnaround time.
aDepending on the presentation of patients and the TAT requirement, the matrix uses each available platform based on the patient population.
This is a single center study with a relatively small sample size and low positivity rate in our population. However, these studies were performed after a large educational effort was put in place in our system regarding specimen collection, therefore minimizing the impact of sample quality in our study. Additionally, all the matched samples were tested within 2 hours of collection so as not to compromise sample integrity for the dry collected swabs as defined by the manufacturer. A dry paired nasopharyngeal swab was not collected since the patients were already contributing 3 different specimens for this study. Finally, all samples were collected from patients meeting testing criteria for COVID-19 as described by the CDC recommendations. Due to limited availability of inactivated viral material, a formal evaluation of the limits of detection was not possible so the assessment of these assays was restricted to the relative limit of detection experiment.
In summary, we evaluated the Abbott ID NOW point-of-care testing system using nasopharyngeal samples collected in UVT and compared its analytical sensitivity and clinical performance to 2 other molecular assays. Also, by analyzing matched specimens we found no difference in the performance of the Abbott ID NOW between swabs collected in UVT and dry swab collection. Finally, the data suggest that the molecular point-of-care testing using isothermal amplification is not as sensitive as other molecular assays included in the study that utilize PCR amplification. DiaSorin Simplexa and Roche cobas assays appear to have LODs at least 10× and 100× lower than Abbott ID NOW, respectively. Laboratories are faced with the challenge of limited testing supplies yet immense testing needs. Taking into consideration all parameters for each testing platform and patient care needs, laboratories should assess what the best test is for the diagnosis of each patient.
Supplementary Material
References
- 1. World Health Organization. Coronavirus disease (COVID-19) outbreak 2020. https://www.who.int/emergencies/diseases/novel-coronavirus-2019. Accessed April 20, 2020.
- 2. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020. pii: S0140-6736(20)30183–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chan JF, Yuan S, Kok KH, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Centers for Disease Control and Prevention. Locations with confirmed COVID-19 cases, global map 2020. https://www.cdc.gov/coronavirus/2019-nco. Accessed April 20, 2020.
- 5. Coronavirus COVID-19 global cases by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (JHU) https://gisanddata.maps.arcgis.com/apps/opsdashboard/index.html#/bda7594740fd40299423467b48e9ecf6. Accessed April 20, 2020.
- 6. Holshue ML, DeBolt C, Lindquist S, et al. ; Washington State 2019-nCoV Case Investigation Team First case of 2019 novel coronavirus in the United States. N Engl J Med. 2020;382:929-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Centers for Disease Control and Prevention. Coronavirus disease 2019 (COVID-19): situation summary https://www.cdc.gov/coronavirus/2019-ncov/cases-updates/summary.html. Accessed April 28, 2020.
- 8. Centers for Disease Control and Prevention. Coronavirus disease 2019 (COVID-19): symptoms of coronavirus https://www.cdc.gov/coronavirus/2019-ncov/symptoms-testing/symptoms.html. Accessed April 28, 2020.
- 9. Bhatraju PK, Ghassemieh BJ, Nichols M, et al. Covid-19 in critically ill patients in the Seattle region - case series. N Engl J Med. 2020;382:2012-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cheng VC, Lau SK, Woo PC, et al. Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin Microbiol Rev. 2007;20:660-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chan JF, Lau SK, To KK, et al. Middle East respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS-like disease. Clin Microbiol Rev. 2015;28: 465-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fauci AS, Lane HC, Redfield RR. Covid-19 - navigating the uncharted. N Engl J Med. 2020;382:1268-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zou L, Ruan F, Huang M, et al. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382:1177-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Craney AR, Velu P, Satlin MJ, et al. Comparison of two high-throughput reverse transcription-polymerase chain reaction systems for the detection of severe acute respiratory syndrome coronavirus 2. J Clin Microbiol. 2020. doi: 10.1128/JCM.00890-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lieberman JA, Pepper G, Naccache SN, et al. Comparison of commercially available and laboratory developed assays for in vitro detection of SARS-CoV-2 in clinical laboratories. J Clin Microbiol. 2020. doi: 10.1128/JCM.00821-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhen W, Manji R, Smith E, et al. Comparison of four molecular in vitro diagnostic assays for the detection of SARS-CoV-2 in nasopharyngeal specimens. J Clin Microbiol. 2020. doi: 10.1128/JCM.00743-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rhoads DD, Cherian SS, Roman K, et al. Comparison of Abbott ID Now, DiaSorin Simplexa, and CDC FDA EUA methods for the detection of SARS-CoV-2 from nasopharyngeal and nasal swabs from individuals diagnosed with COVID-19. J Clin Microbiol. 2020. doi: 10.1128/JCM.00760-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Harrington A, Cox B, Snowden J, et al. Comparison of Abbott ID Now and Abbott m2000 methods for the detection of SARS-CoV-2 from nasopharyngeal and nasal swabs from symptomatic patients. J Clin Microbiol. 2020.doi: 10.1128/JCM.00798-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. The COVID tracking project https://covidtracking.com/data#state-oh. Accessed April 29, 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.