Abstract
Objectives
Several studies suggest the sensitivity of chest computed tomography (CT) is far greater than that of reverse transcription polymerase chain reaction (RT-PCR) in diagnosing COVID-19 patients, and therefore, CT should be included as a primary diagnostic tool. This systematic review aims to stratify studies as high or low risk of bias to determine the true sensitivity of CT for severe acute respiratory syndrome coronavirus-2 infection according to the unbiased (low risk) studies, a topic of particular importance given the insufficient quantity of RT-PCR kits in many countries. We focus on sensitivity as that is the chief advantage perceived of CT.
Materials and Methods
This systematic review involved searching the PubMed and Google Scholar databases for articles conducted and published between January 1 and April 15, 2020. The quality assessment tool QUADAS-2 was used to stratify studies according to their risk of bias, and exclusion criteria included not providing the information deemed relevant for such a stratification, such as not indicating if the patients were symptomatic or asymptomatic, or identifying the source of the specimen for the reference standard, RT-PCR (eg, nasal, oropharyngeal, etc). Sensitivity values were then extracted, and random effects meta-analyses were performed.
Results
Of 641 search results, 37 studies (n = 9610 patients) were included in the analysis. The mean sensitivity of RT-PCR for COVID-19 reported by the biased studies was 70% (n = 5409/7 studies; 95% confidence interval [CI], 43–97; I2 = 99.1%), compared with 78% by unbiased studies (n = 534/4 studies; 95% CI, 69–87, I2 = 89.9%). For chest CT, the mean sensitivity reported by biased studies was 94% (n = 3371 patients/24 studies; 95% CI, 92–96; I2 = 93.1%), compared with 75% by unbiased studies (n = 957/10 studies; 95% CI, 67–83; I2 = 89.5%).
Conclusions
The difference between the sensitivities of CT and RT-PCR for severe acute respiratory syndrome coronavirus-2 infection is lower than previously thought, as after stratifying the studies, the true sensitivity for CT based on the unbiased studies is limited.
Key Words: COVID-19, SARS-CoV-2, RT-PCR, chest CT, sensitivity, specificity
Coronavirus disease 2019 (COVID-19) now affects 210 countries and all 6 World Health Organization regions to date.1 COVID-19 represents a multisystem disease caused by the novel coronavirus strain severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), which was first detected in Wuhan, China, in December 2019.2 The US Centers for Disease Control and Prevention reported 690,714 coronavirus cases and 35,443 coronavirus deaths as of April 19, 2020.3 Globally, COVID-19 has infected more than 2.3 million people and led to 163,000 deaths.2 Like 2002 SARS-CoV and 2012 MERS-CoV, SARS-CoV-2 is a β-coronavirus suspected to be phylogenetically derived from the bat gene pool.4
Current Centers for Disease Control and Prevention criteria for detecting SARS-CoV-2 and diagnosing COVID-19 champion real-time reverse transcription polymerase chain reaction (RT-PCR) identification of SARS-CoV-2 RNA. However, new evidence indicates that negative RT-PCR results do not rule out SARS-CoV-2 infection.5–7 In fact, a number of independent studies communicate consistently higher sensitivities using computed tomography (CT) (97.7%) compared with RT-PCR (75.9%) for detecting SARS-CoV-2 infection (n = 87).5,8 In addition, Fang et al9 reported an initial negative and subsequent positive RT-PCR result in 29.4% and CT result in 2% of patients later confirmed for COVID-19, suggesting that CT abnormalities may predate RT-PCR positivity (n = 51). Likewise, Ai et al10 concluded a 1014-patient study by nominating chest CT for primary COVID-19 detection. Others are more critical of CT-guided diagnosis and instead argue for its usage strictly as an adjunct.11,12
With more than 60,000 deaths in the United States alone, COVID-19 is likely to continue putting strain on health care systems and economies throughout the world for the foreseeable future.13 Thus, optimizing the effectiveness and efficiency of diagnostic measures is of paramount importance. Although both CT and RT-PCR have clear diagnostic value for COVID-19, the question of which method is preferable remains unanswered. In this systematic review, we investigate the risk of bias in existing studies on CT and RT-PCR for COVID-19 and help shed light on the true sensitivity of CT. Although there is more to clinical utility than diagnostic parameters like sensitivity and specificity, such as cost and access, we focus on the question of sensitivity as that is the chief advantage perceived of chest CT. In addition, we do not aim to provide a head-to-head comparison as RT-PCR is functioning as the reference standard. However, as initial RT-PCR tests can provide false-negatives (FN) in cohorts later confirmed to be positive on RT-PCR, sensitivity values of RT-PCR are also extracted to provide context on the sensitivity of this reference standard. Instead, our aim is to determine the true sensitivity of chest CT and, in doing so, demonstrate how biased methodologies have affected the literature.
MATERIALS AND METHODS
Data Sources and Searches
We used Medical Subject Headings search terms (sensitivity and specificity, AND RT-PCR, AND coronavirus, AND SARS-CoV-2) OR the presence of keywords (CT AND COVID-19 OR severe acute respiratory syndrome coronavirus 2) in the title, abstract, or full-text to find publications in the PubMed database published between January 1, 2020, and April 15, 2020 (n = 201). The Google Scholar database was searched using the keywords “COVID-19” AND “SARS-CoV-2” AND “sensitivity” AND “coronavirus” AND “RT-PCR” AND “chest CT” AND “imaging” anywhere in the text articles published between January 1, 2020, and April 15, 2020 (n = 478). Of the total 641 search results, 37 studies were ultimately included in the data synthesis. Studies were evaluated by authors JW and KL, and any disagreements were resolved by the senior author MH. Additional articles (n = 13), either from government or scientific organizations or our original 641 search results, were referenced to provide background and contextual information.1–7,11,13–17
Study Selection and Quality Assessment
Of the total search results identified (n = 679), 38 were duplicates. Of these nonduplicate searches (n = 641), 590 were screened out and 14 were subsequently excluded after being assessed for eligibility, leaving 37 articles to be included in the systematic review for discussion. Collectively, the 37 included articles provided the most relevant data on the clinical utility of CT and RT-PCR in diagnosing COVID-19. Articles that documented the same cohort of patients as another study were excluded.
We stratified studies as high or low risk of bias using QUADAS-2, a quality assessment tool for studies of diagnostic accuracy.14 QUADAS-2 is used in systematic reviews to assess the risk of bias in studies based on 4 key factors:
Patient selection. To be considered low risk of bias, patient cohorts must have included both symptomatic and asymptomatic patients, as just one or the other would not be reflective of the broader SARS-CoV-2-infected population. The inappropriate exclusion of symptomatic or asymptomatic patients may have introduced bias. In addition, several studies included only pediatric patients and were thus classified as high risk of bias due to the exclusion of adult patients.
Index test. Studies involving chest CT scans that were interpreted as positive for COVID-19 in the presence of any abnormal finding without confirming with RT-PCR were considered high risk for bias. Patient cohorts must consist of RT-PCR confirmed cases to be considered low risk of bias. This is in accordance with the American College of Radiology's recommendation that RT-PCR serve as the primary diagnostic tool for COVID-19. Nevertheless, studies involving subjects that tested positive on CT, but did not test positive on RT-PCR, were categorized as high risk of bias, as opposed to being excluded from the review, as this group includes the clinically relevant group of patients in which it was not clear if they had COVID-19.
Reference standard. QUADAS-2 states the conduct of the reference standard may have introduced bias. For the reference standard of RT-PCR, a proper swab must have been taken (from the upper or lower respiratory tract, in accordance with American College of Radiology guidelines). Because RT-PCR cannot meaningfully serve as both the reference standard for CT and a comparator test, this review focuses on assessing the sensitivity of CT alone as a potential independent diagnostic tool instead of comparing it to RT-PCR. In addition, the accuracies of RT-PCR and CT are both dependent on the clinical stage and time point during the course of disease and thus have different clinical applications.
Flow and timing. Studies that did not include all patients in their analysis were considered high risk for bias, as well as studies in which not all patients received the same reference standard, RT-PCR, conducted in the same way (swabs taken from the same place).
To stratify the studies based on these factors, the studies had to provide the relevant information. Thus, exclusion criteria included not indicating if the patients were symptomatic or asymptomatic, not indicating if the patients were adults or children, not indicating whether the patient cohort involved RT-PCR confirmed SARS-CoV-2 infection, and not identifying the source of the specimen for RT-PCR (eg, nasal, oropharyngeal, etc). In addition, preprints were not sought out as they have not passed the scrutiny of peer review. The number of articles excluded for each exclusion criteria are as follows: 92 for not providing information relevant to QUADAS-2 patient selection, 39 for index test, 21 for reference standard, and 9 for flow and timing. The criteria for exclusion criteria are summarized in Figure 1. The reason for exclusion during the full-text screen involved reported redundant information, such as review studies that summarized information already extracted from other included studies.
FIGURE 1.
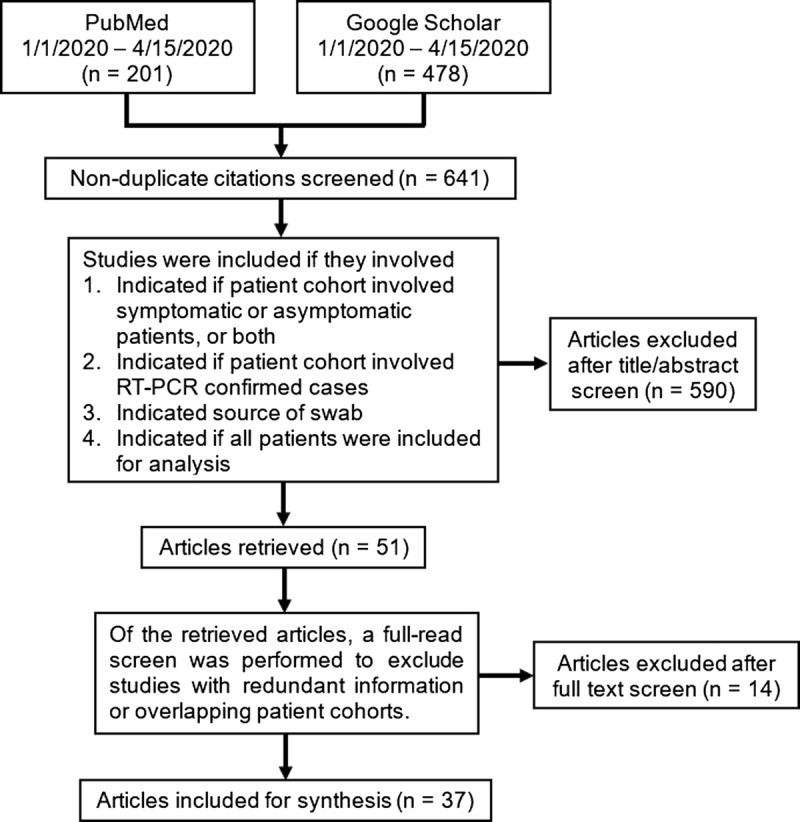
Literature search and exclusion criteria. RT-PCR indicates reverse transcription polymerase chain reaction.
Data Synthesis and Analysis
Computed tomography has demonstrated a poor specificity of 37% for COVID-19, in addition to a relatively high cost and limited access of scans across the world, as compared with the cheap and high-throughput RT-PCR.15 The rationale for using CT as a primary diagnostic tool, therefore, has been proposed to be its superior sensitivity. Thus, this review is focused primarily on sensitivity, as opposed to other diagnostic factors like specificity, likelihood ratios, true positive, FN, etc. Again, this focus on sensitivity of CT is because of its perception as its chief advantage. Thus, the qualitative synthesis involved aggregating and summarizing the data included in the selected articles, with a focus on sensitivity data.
Data on the number of positive results (CT or RT-PCR) out of the number of total tests conducted were extracted from each eligible study. Random effects meta-analyses were performed on these proportions. Random effects models were used to control for within and between study variability. Studies were divided by biased (or high risk of bias) and unbiased (or low risk of bias) as classified by the authors; the analysis included these subgroups as well as overall results. I2 was calculated to examine heterogeneity between studies. Finally, forest plots were created to show the sensitivity findings of CT and RT-PCR in each study based on these random effect models and controlling for heterogeneity.
Role of the Funding Source
The authors received no financial support for the research, authorship, or publication of this article.
RESULTS
In total, 641 studies were assessed for eligibility in this meta-analysis. Of these, 604 were excluded for the criteria shown in Figure 1. Our analyses therefore included a total of 37 studies. The results for each modality are discussed below.
RT-PCR as a Reference Standard to Detect SARS-CoV-2 Infection
Methodological Biases
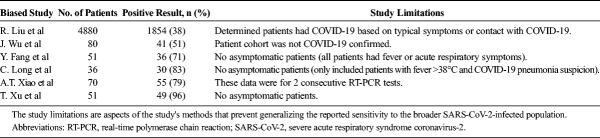
As stated previously, of 641 nonduplicate search results, 37 studies were ultimately included in the review after being assessed for eligibility, and data reported on sensitivity were extracted. Before delving into the results of CT studies, we must first provide context by assessing the reference standard, RT-PCR, as studies using this diagnostic tool are not all free from risk of bias. In fact, although RT-PCR is the putative diagnostic tool for COVID-19, a considerable portion of the data regarding its sensitivity and specificity is marred by methodological biases. A recent study by Li and colleagues18 tested a cohort of patients assumed to have SARS-CoV-2 infection owing to CT findings consistent with a viral pneumonia and reported an RT-PCR sensitivity of 27.5% (n = 610). Importantly, the assumption that all patients with viral pneumonia findings on chest CT have COVID-19 likely contributed to an inaccurate RT-PCR sensitivity. Similarly, Wu et al,19 who also included suspected but not confirmed COVID-19 patients in their cohort, reported 51% sensitivity of RT-PCR for SARS-CoV-2 infection (n = 80) A separate study of 4880 suspected COVID-19 patients by Liu et al20 determined the sensitivity of RT-PCR to be 38.25% when using nasal and oropharyngeal swabs. Like the cohorts in the Li and Wu studies, these patients were not all confirmed to have COVID-19. Instead, the cohort consisted of patients with nonspecific respiratory symptoms or potential contact with COVID-19 in addition to those with a positive RT-PCR result.
In addition, Xiao et al21 found at least 1 positive result in 78.6% (n = 70) for 2 consecutive SARS-CoV-2 RT-PCR results; however, RT-PCR sensitivity after the initial test was not provided. Because the sensitivity of the initial RT-PCR test is highly likely to be lower than that of 2 consecutive tests, this finding cannot be reliably generalized to all SARS-CoV-2-infected patients. Furthermore, a recent study by Fang et al9 observed an RT-PCR sensitivity of 71% in a cohort that lacked asymptomatic patients and instead consisted entirely of patients with fever or nonspecific acute respiratory symptoms (n = 51, P < 0.001). Several other studies also included patient cohorts consisting solely of symptomatic patients8,22 (Table 1). Overall, analyses of patient cohorts consisting of suspected rather than confirmed COVID-19 patients likely resulted in an underestimation of the sensitivity of RT-PCR for COVID-19.
TABLE 1.
Sensitivities of Initial RT-PCR for Diagnosing COVID-19 Infection in Biased Studies
Unbiased Studies
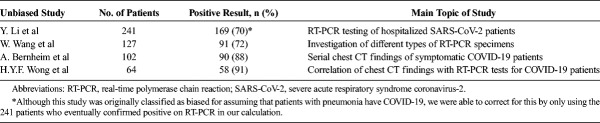
Because RT-PCR assay testing is used to confirm COVID-19, its sensitivity may be appropriately determined by retrospectively evaluating the initial RT-PCR results of patients who were later confirmed positive. Even in unbiased studies, the sensitivity of RT-PCR is not 100% as initial tests can report as FN. In these studies, the entire cohort got a repeat RT-PCR test several days later (the exact time delay when reported varied but was often not presented and thus there is not enough available information to assess this time delay) if initial results were negative regardless of symptoms. Bernheim et al,23 for example, found the RT-PCR sensitivity for SARS-CoV-2 infection to be 88% (n = 102). Another study by Wang et al,24 using a similar methodology, reported a 71.7% sensitivity of RT-PCR (n = 127). Interestingly, this study found that RT-PCR sensitivity varied by specimen type, noting a 93% sensitivity for bronchoalveolar lavage fluid specimens (n = 15) compared with 72% for sputum (n = 104), 63% for nasal swabs (n = 8), and 32% for pharyngeal swab samples (n = 398, 1–3 days after admission; 95% confidence interval [CI], 31.2%–33.1%).24 Although limited in sample size, these findings suggest that bronchoalveolar lavage fluid, sputum, and nasal swab specimens may be more effective than pharyngeal swabs for RT-PCR testing. Other studies, which were also categorized as low risk of bias, enabled the results to be generalized more appropriately to the broader SARS-CoV-2-infected population and found sensitivities of RT-PCR to range from 70% to 91% (Table 2, Fig. 2).24,25 The heterogeneity in results in Figure 2 (and Fig. 3) may be attributed to differences in patient selection (variation in degree of symptomatology) or the time the test was taken relative to the onset of symptoms (not enough studies provided this information to adequately take this into account in the analysis).
TABLE 2.
Sensitivities of Initial RT-PCR for Diagnosing COVID-19 Infection in Unbiased Studies
FIGURE 2.
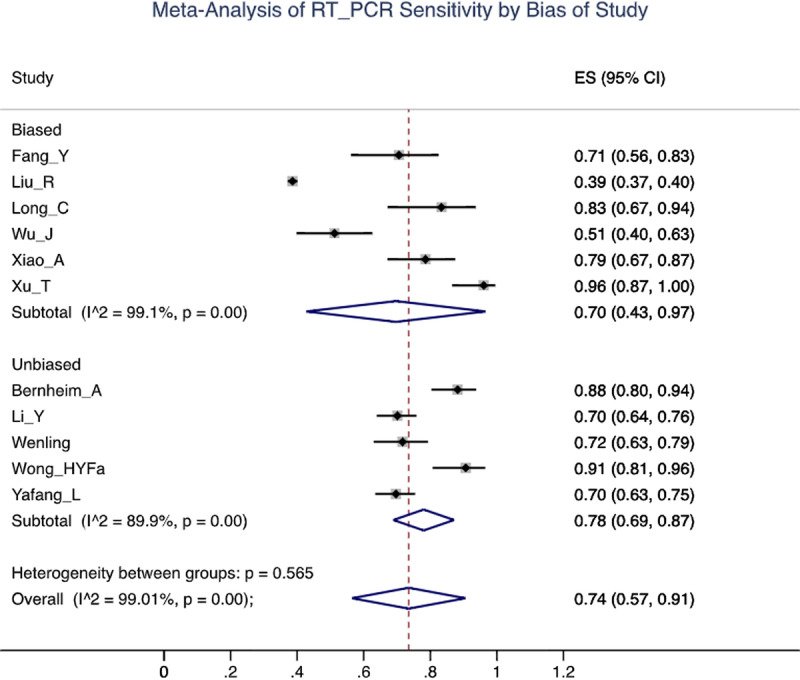
Forest plot of reverse transcription polymerase chain reaction (RT-PCR) studies showing the sensitivity of each study using a random effects model to control for heterogeneity and showing subgroups by bias in the studies.
FIGURE 3.
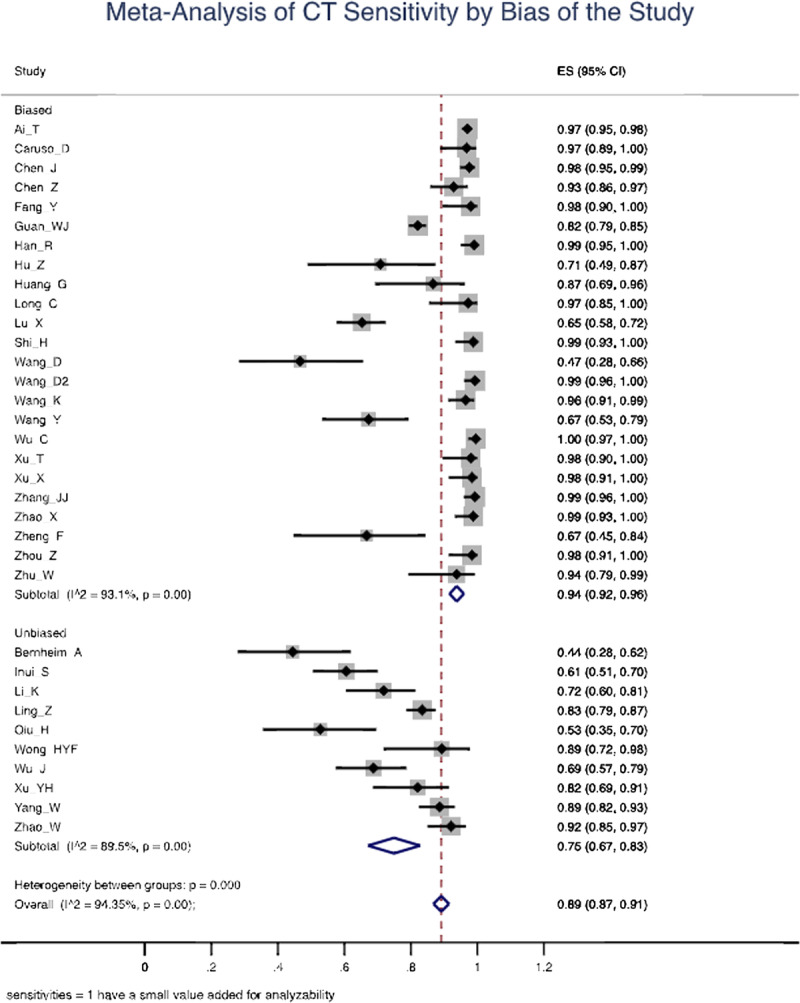
Forest plot of computed tomography (CT) studies showing the sensitivity of each study using a random effects model to control for heterogeneity and showing subgroups by bias in the studies.
The Sensitivity of Chest CT for SARS-CoV-2 Infection
Methodological Biases
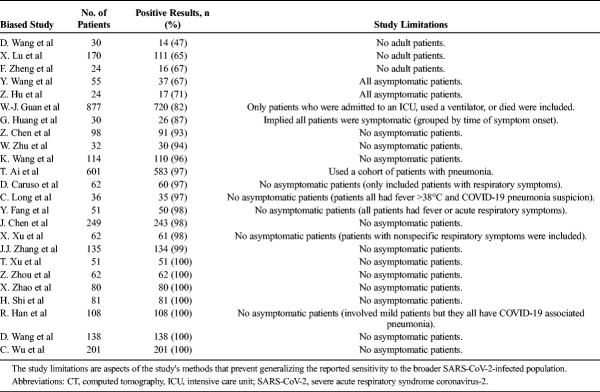
Several studies involving CT have results with a limited ability to be generalized owing to biased patient cohorts. For example, the 97% (n = 601; 95% CI, 95%–98%) sensitivity of CT for detecting SARS-CoV-2 infection when RT-PCR is used as a reference reported by Ai et al may be overestimated owing to the cohort consisting of patients with pneumonia.10 In contrast, Guan et al26 used only confirmed COVID-19 patients in their patient cohort and reported a CT sensitivity of 82.1% (n = 877). However, this finding should also be interpreted with caution because the Guan et al cohort comprised patients with adverse outcomes (eg, intensive care unit admission, mechanical ventilation, or death). Therefore, the reported sensitivity in both studies may be higher than the true sensitivity.
A lack of asymptomatic patients was the most common methodological feature found in studies, which limits the generalizability of their reported CT sensitivities for SARS-CoV-2 infection.8,9,27–40 For example, 1 study included only patients with respiratory symptoms and found a high sensitivity of 97% (n = 62).31 Another study also observed a high CT sensitivity of 97% (n = 36) but included only patients with a fever higher than 38°C and COVID-19-suspected pneumonia.8 Additional studies reporting near-perfect sensitivities (98%–100%) targeted patients with fever, respiratory symptoms, and/or pneumonia.9,33,38 Interestingly, literature citing much lower sensitivities (47%–67% and 67%–71%) used patient cohorts that were entirely pediatric39,41,42 and asymptomatic patients, respectively (Table 3).43,44 A study by Inui et al,12 which assessed the CT results of 104 cases from the Diamond Princess cruise ship, found a significantly higher degree of CT sensitivity for SARS-CoV-2 infection in symptomatic (79%, n = 28) compared with asymptomatic patients (54%, n = 76, P = 0.023). Because many individuals infected with SARS-CoV-2 are asymptomatic, excluding them may yield a sensitivity that does not accurately reflect the ability of CT to detect the disease among the broader SARS-CoV-2-infected population.
TABLE 3.
Sensitivities of Initial Chest CT for Diagnosing COVID-19 Infection in Biased Studies
Unbiased Studies
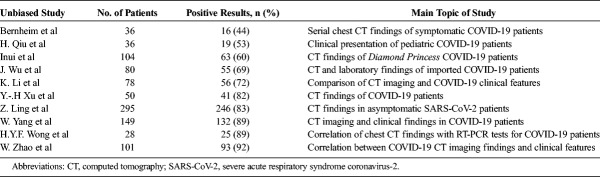
Studies that were deemed unbiased showed lower overall sensitivities of chest CT for SARS-CoV-2 infection. One study found that CT scans were normal in 21% (n = 28) of symptomatic patients and 46% (n = 76) of asymptomatic patients (P = 0.023).12 Another retrospective study found that 56% (n = 36; 95% CI, 47%–65%) of chest CT scans were normal among COVID-19-confirmed patients when taken within 2 days of the onset of symptoms.23 Only 1 of these patients had a negative initial RT-PCR test. A panel of radiologists referenced this study in suggesting that as many as 50% of CT scans may be normal when taken within the first 2 days of COVID-19 symptom onset.11
Other studies that included COVID-19-confirmed symptomatic and asymptomatic patients showed CT sensitivities (53%–72%) comparable with Inui et al and Bernheim et al (Table 4).19,45,46 Although many other generalizable studies yielded higher CT sensitivities (82%–92%), even these findings are considerably lower than those previously described from studies with weaker patient selection criteria.25,47–50 Overall, the mean sensitivity of chest CT reported was 94% (n = 3371 patients/24 studies; 95% CI, 92–96) in biased studies and 75% (n = 957/10 studies; 95% CI, 67–83) in unbiased studies. These findings are summarized in Figure 3.
TABLE 4.
Sensitivities of Initial Chest CT for Diagnosing COVID-19 Infection in Unbiased Studies
DISCUSSION
Several methodological approaches made in the biased studies contributed to the overestimation of the true sensitivity of CT. For example, although studies using purely symptomatic cohorts provide useful clinical information regarding the sensitivity of CT for symptomatic patients, many SARS-CoV-2-infected individuals are asymptomatic. As a result, the overall sensitivity of CT for all infected individuals cannot be reliably determined from such studies. In addition, overestimation of chest CT sensitivity is evident when patients with pneumonia are assumed to have COVID-19. Such assumptions limit the broad generalizability of positive results found.10 Similarly, the underestimation of sensitivity of RT-PCR is also apparent when studies assume that patients have COVID-19 if they have typical symptoms or have been in contact with a confirmed COVID-19-positive individual, as these patients are not necessarily all infected with SARS-CoV-2.20
Reverse transcription-PCR has played a pivotal role in the diagnosis and detection of COVID-19 on an individual basis; however, challenges remain with its widespread use. In addition to the apparent shortage of testing kits present in some parts of the world, RT-PCR may also require an extended waiting period of several hours before results are processed and available for analysis.16 On the other hand, the high specificity of RT-PCR speaks to its advantage in accuracy and precision as a diagnostic tool as compared with chest CT. In addition, the transmissibility of SARS-CoV-2 should also be kept in mind when making clinical decisions, as CT presents an intrinsic risk of exposure to radiology personnel. Although there are yet other factors relevant in the clinical utility of a diagnostic tool, such as likelihood ratios and cost, this review focuses on sensitivity as it is the primary advantage proponents of CT refer to.
Overall, sensitivity ratings for both chest CT scans and RT-PCR differ among the literature, yet a trend is present in which CT sensitivity is overall greater than that of RT-PCR. A previous meta-analysis found the pooled sensitivity for chest CT to be 94% (95% CI, 91%–96%) and 89% (95% CI, 81%–94%) for RT-PCR.15 This study provided meaningful data but did not attempt to categorize studies based on risk of bias. We categorized studies as such and, after excluding biased studies, such as studies that only involved symptomatic patients, their sensitivities appear to be similar, with a 75% (95% CI, 67%–83%) sensitivity for CT and a 78% (95% CI, 69%–87%) sensitivity for RT-PCR. Although the meaning of this comparison is limited as RT-PCR is serving as the reference standard, it does demonstrate not only that the difference in sensitivities are likely less than previously thought, but that the sensitivity of CT for SARS-CoV-2 infection is limited as according to the unbiased studies.
Limitations
The diagnostic utility of a test is not just dependent on its sensitivity. Although our review focuses on sensitivity due to it being the chief advantage proponents of chest CT refer to, our conclusions are limited in that without discussing specificity, precision, or accuracy, an overall assessment of the performance of CT as a diagnostic test cannot be made. Even factors aside from diagnostic parameters are clinically relevant and warrant discussion in future research as well, such as costs, availability, and need for personal protective equipment. The current literature does not yet allow for such a comprehensive analysis that takes into account these diagnostic parameters and other factors, as few papers assess the specificity of CT for COVID-19, for example. In addition, the sensitivity of CT (and RT-PCR) is also related to the time of disease onset and severity of symptoms, which was discussed when the studies provided that information, but many studies did not provide that information and did not allow for this to be thoroughly accounted for in the analysis. Thus, without the sufficient data, we could not test how changes in these parameters would affect our overall result through a sensitivity analysis, which would have involved changing 1 parameter and keeping others constant to quantify the impact of these parameters in the overall sensitivity. Finally, only using patients with a positive RT-PCR in unbiased studies may have biased our results to some extent against CT, as CT cannot identify subjects in whom RT-PCR was falsely negative. That is why we chose to treat RT-PCR primarily as the reference standard, and although we present the sensitivity of this reference standard to provide context, we stray from presenting the data as a head-to-head comparison with CT, and thus, we did not run any statistical tests to directly compare their sensitivities. Instead, the focus of this review is an assessment of the chief benefit associated with chest CT, its sensitivity, to help paint a clearer picture of its diagnostic utility.
CONCLUSION
Multiple studies demonstrate a higher sensitivity report for chest CT when compared with RT-PCR, but the methods of these reports involve biases and confounding variables that detract from their generalizability. Researchers and providers should be aware of these various methodologies so that their results can be interpreted appropriately. While we cannot state with certainty that the differences in sensitivity of RT-PCR and CT for SARS-CoV-2 infection are not significantly different, as RT-PCR is serving as the reference standard as opposed to comparator test, our results indicate that the difference between their sensitivities is likely not as large as previously thought and warrant large-scale comparator investigations using unbiased methodologies. This review stratified studies based on their risk of bias and demonstrated how biases affected the literature and that the sensitivity of CT for SARS-CoV-2, its chief perceived advantage, is in fact actually limited. We conclude that RT-PCR should continue to serve as the primary diagnostic tool for COVID-19. This suggestion is in line with the recommendations provided by the American College of Radiologists.17 We go further by recommending chest CT be considered as a supplemental tool for diagnosing COVID-19 in symptomatic patients.
Footnotes
Conflicts of interest and sources of funding: none declared.
REFERENCES
- 1.World Map. Centers for Disease Control and Prevention. 2020. https://www.cdc.gov/coronavirus/2019-ncov/cases-updates/world-map.html. Accessed April 19, 2020.
- 2.Dong E, Du H, Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis. 2020;20:533–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. Cases in U.S. 2020. https://www.cdc.gov/coronavirus/2019-ncov/cases-updates/cases-in-us.html. Accessed April 19, 2020.
- 4.Forster P, Forster L, Renfrew C, et al. Phylogenetic network analysis of SARS-CoV-2 genomes. Proc Natl Acad Sci. 2020;202004999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lippi G, Simundic AM, Plebani M. Potential preanalytical and analytical vulnerabilities in the laboratory diagnosis of coronavirus disease 2019 (COVID-19) [published online ahead of print]. Clin Chem Lab Med. [DOI] [PubMed] [Google Scholar]
- 6.Hao W, Li M. Clinical diagnostic value of CT imaging in COVID-19 with multiple negative RT-PCR testing. Travel Med Infect Dis. 2020;34:101627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng H, Liu Y, Lv M, et al. A case report of COVID-19 with false negative RT-PCR test: necessity of chest CT. Jpn J Radiol. 2020;38:409–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Long C, Xu H, Shen Q, et al. Diagnosis of the coronavirus disease (COVID-19): rRT-PCR or CT? Eur J Radiol. 2020;126:108961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fang Y, Zhang H, Xie J, et al. Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology. 2020;200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ai T, Yang Z, Hou H, et al. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020;200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kanne JP, Little BP, Chung JH, et al. Essentials for radiologists on COVID-19: An update-radiology scientific expert panel. Radiology. 2020;200527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Inui S, Fujikawa A, Jitsu M, et al. Chest CT findings in cases from the cruise ship “diamond princess” with coronavirus disease 2019 (COVID-19). Radiol Cardiothorac Imaging. 2020;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Institute for Health Metrics and Evaluation. IHME: COVID-19 projections. https://covid19.healthdata.org/united-states-of-america. Accessed April 30, 2020.
- 14.Quadas-2. University of Bristol, Population Health Sciences. 2019. Bristol.ac.uk.
- 15.Kim H, Hong H, Yoon SH. Diagnostic performance of CT and reverse transcriptase-polymerase chain reaction for coronavirus disease 2019: a meta-analysis. Radiology. 2020:201343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.COVID-19 CSC. Chinese Clinical Guidance for COVID-19 Pneumonia Diagnosis and Treatment (7th edition) Fighting New Coronary Pneumonia. http://kjfy.meetingchina.org/msite/news/show/cn/3337.html. Accessed April 17, 2020.
- 17.American College of Radiology. ACR recommendations for the use of chest radiography and computed tomography (CT) for suspected COVID-19 infection. https://www.acr.org/Advocacy-and-Economics/ACR-Position-Statements/Recommendations-for-Chest-Radiography-and-CT-for-Suspected-COVID19-Infection. Accessed April 18, 2020.
- 18.Li Y, Yao L, Li J, et al. Stability issues of RT-PCR testing of SARS-CoV-2 for hospitalized patients clinically diagnosed with COVID-19. J Med Virol. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu J, Liu J, Zhao X, et al. Clinical characteristics of imported cases of coronavirus disease 2019 (COVID-19) in Jiangsu Province: a multicenter descriptive study. Clin Infect Dis. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu R, Han H, Liu F, et al. Positive rate of RT-PCR detection of SARS-CoV-2 infection in 4880 cases from one hospital in Wuhan, China, from Jan to Feb 2020. Clin Chim Acta. 2020;505:172–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiao AT, Tong YX, Zhang S. False-negative of RT-PCR and prolonged nucleic acid conversion in COVID-19: rather than recurrence. J Med Virol. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu T, Chen C, Zhu Z, et al. Clinical features and dynamics of viral load in imported and non-imported patients with COVID-19. Int J Infect Dis. 2020;94:68–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bernheim A, Mei X, Huang M, et al. Chest CT findings in coronavirus disease-19 (COVID-19): relationship to duration of infection. Radiology. 2020;200463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang W, Xu Y, Gao R, et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wong HYF, Lam HYS, Fong AH, et al. Frequency and distribution of chest radiographic findings in COVID-19 positive patients. Radiology. 2019;201160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guan WJ, Ni ZY, Hu Y, et al. , China Medical Treatment Expert Group for COVID-19. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382:1708–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang G, Gong T, Wang G, et al. Timely diagnosis and treatment shortens the time to resolution of coronavirus disease (COVID-19) pneumonia and lowers the highest and last CT scores from sequential chest CT. Am J Roentgenol. 2020;1–7. [DOI] [PubMed] [Google Scholar]
- 28.Chen Z, Fan H, Cai J, et al. High-resolution computed tomography manifestations of COVID-19 infections in patients of different ages. Eur J Radiol. 2020;126:108972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu W, Xie K, Lu H, et al. Initial clinical features of suspected coronavirus disease 2019 in two emergency departments outside of Hubei, China. J Med Virol. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang K, Kang S, Tian R, et al. Imaging manifestations and diagnostic value of chest CT of coronavirus disease 2019 (COVID-19) in the Xiaogan area. Clin Radiol. 2020;75:341–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caruso D, Zerunian M, Polici M, et al. Chest CT features of COVID-19 in Rome, Italy. Radiology. 2020;201237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen J, Qi T, Liu L, et al. Clinical progression of patients with COVID-19 in Shanghai, China. J Infect. 2020;80:e1–e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu XW, Wu XX, Jiang XG, et al. Clinical findings in a group of patients infected with the 2019 novel coronavirus (SARS-Cov-2) outside of Wuhan, China: retrospective case series. BMJ. 2020;368:m606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang JJ, Dong X, Cao YY, et al. Clinical characteristics of 140 patients infected with SARS-CoV-2 in Wuhan, China. Allergy. 2020. [DOI] [PubMed] [Google Scholar]
- 35.Zhou Z, Guo D, Li C, et al. Coronavirus disease 2019: initial chest CT findings. Eur Radiol. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao X, Liu B, Yu Y, et al. The characteristics and clinical value of chest CT images of novel coronavirus pneumonia. Clin Radiol. 2020;75:335–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi H, Han X, Jiang N, et al. Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: a descriptive study. Lancet Infect Dis. 2020;20:425–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Han R, Huang L, Jiang H, et al. Early clinical and CT manifestations of coronavirus disease 2019 (COVID-19) pneumonia. AJR Am J Roentgenol. 2020:1–6. [DOI] [PubMed] [Google Scholar]
- 39.Wang D, Hu B, Hu C, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323:1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu C, Chen X, Cai Y, et al. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern Med. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu X, Zhang L, Du H, et al. SARS-CoV-2 infection in children. N Engl J Med. 2020;382:1663–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng F, Liao C, Fan QH, et al. Clinical characteristics of children with coronavirus disease 2019 in Hubei, China. Curr Med Sci. 2020;40:275–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Y, Liu Y, Liu L, et al. Clinical outcomes in 55 patients with severe acute respiratory syndrome coronavirus 2 who were asymptomatic at hospital admission in Shenzhen, China. J Infect Dis. 2020;221:1770–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu Z, Song C, Xu C, et al. Clinical characteristics of 24 asymptomatic infections with COVID-19 screened among close contacts in Nanjing, China. Sci China Life Sci. 2020;63:706–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qiu H, Wu J, Hong L, et al. Clinical and epidemiological features of 36 children with coronavirus disease 2019 (COVID-19) in Zhejiang, China: an observational cohort study. Lancet Infect Dis. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li K, Fang Y, Li W, et al. CT image visual quantitative evaluation and clinical classification of coronavirus disease (COVID-19). Eur Radiol. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu YH, Dong JH, An WM, et al. Clinical and computed tomographic imaging features of novel coronavirus pneumonia caused by SARS-CoV-2. J Infect. 2020;80:394–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ling Z, Xu X, Gan Q, et al. Asymptomatic SARS-CoV-2 infected patients with persistent negative CT findings. Eur J Radiol. 2020;126:108956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang W, Cao Q, Qin L, et al. Clinical characteristics and imaging manifestations of the 2019 novel coronavirus disease (COVID-19):a multi-center study in Wenzhou city, Zhejiang, China. J Infect. 2020;80:388–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao W, Zhong Z, Xie X, et al. Relation between chest CT findings and clinical conditions of coronavirus disease (COVID-19) pneumonia: a multicenter study. Am J Roentgenol. 2020;214:1072–1077. [DOI] [PubMed] [Google Scholar]