Abstract
Two different members of the fatty acid‐binding protein (FABP) family are found in enterocyte cells of the gastrointestinal system, namely liver‐type and intestinal fatty acid‐binding proteins (LFABP and IFABP, also called FABP1 and FABP2, respectively). Striking phenotypic differences have been observed in knockout mice for either protein, for example, high fat‐fed IFABP‐null mice remained lean, whereas LFABP‐null mice were obese, correlating with differences in food intake. This finding prompted us to investigate the role each protein plays in directing the specificity of binding to ligands involved in appetite regulation, such as fatty acid ethanolamides and related endocannabinoids. We determined the binding affinities for nine structurally related ligands using a fluorescence competition assay, revealing tighter binding to IFABP than LFABP for all ligands tested. We found that the head group of the ligand had more impact on binding affinity than the alkyl chain, with the strongest binding observed for the carboxyl group, followed by the amide, and then the glycerol ester. These trends were confirmed using two‐dimensional 1H–15N nuclear magnetic resonance (NMR) to monitor chemical shift perturbation of the protein backbone resonances upon titration with ligand. Interestingly, the NMR data revealed that different residues of IFABP were involved in the coordination of endocannabinoids than those implicated for fatty acids, whereas the same residues of LFABP were involved for both classes of ligand. In addition, we identified residues that are uniquely affected by binding of all types of ligand to IFABP, suggesting a rationale for its tighter binding affinity compared with LFABP.
Keywords: chemical shift perturbation, endocannabinoids, endocannabinoid ligand binding affinity, fatty acid ethanolamide, fatty acid‐binding protein, fatty acids, fluorescence, intestinal FABP, liver‐type FABP, NMR
Abbreviations
- 2‐AG
2‐arachidonoylglycerol, 2‐OG, 2‐oleoylglycerol
- 2‐PG
2‐palmitoylglycerol
- ADIFABP
acrylodan labeled intestinal fatty acid binding protein
- AEA
arachidonylethanolamide or anandamide
- ANS
8‐anilinonaphthalene‐1‐sulfonic acid
- ARA
arachidonate
- CSP
chemical shift perturbation
- DAUDA
11‐(dansylamino)undecanoic acid
- EC
endocannabinoids
- HSQC
2D 1H–15N heteronuclear single quantum coherence spectroscopy
- IFABP
intestinal fatty acid‐binding protein
- ITC
isothermal titration calorimetry
- LFABP
liver‐type fatty acid‐binding protein
- NBD
nitrobenzoxadiazole
- NMR
nuclear magnetic resonance
- OEA
oleoylethanolamide
- OLA
oleate
- PAL
palmitate
- PEA
palmitoylethanolamide
1. INTRODUCTION
Fatty acids are the precursors of lipids that make up cellular membranes and play significant roles in mammalian signal transduction pathways. 1 Their transport within the cytoplasm is thought to be facilitated by fatty acid‐binding proteins (FABPs). 2 , 3 , 4 The FABP family consists of more than ten different structurally similar proteins that have tissue‐specific distributions. 5 The common structural characteristics of this superfamily of proteins are a barrel‐like, central cavity formed by networks of β‐strands and a cap‐like helical portal structure. 3 , 5 The interior of the barrel contains structured water molecules but is also lined with hydrophobic amino acid sidechains that can accommodate hydrophobic ligands such as fatty acids. The features that distinguish different members within this family are the breadth and volume of the binding cavity, as well as the positioning and amino acid sequence of the cap. Indeed, differences in the helical cap domain have been shown to impart striking differences in the mechanism of ligand transfer to and from membranes. 6 , 7 , 8 , 9 , 10 Variations in the FABP structures are thought to modulate the stoichiometry of ligand binding as well as the ligand specificity.
Two different FABPs are expressed in enterocytes, the absorptive epithelial cells in the small intestine: intestinal FABP (IFABP; FABP2) and liver‐type FABP (LFABP; LFABP1). Comparisons of the structural features of IFABP and LFABP have been described in detail previously. 11 , 12 , 13 , 14 , 15 Briefly, whereas both proteins have the characteristic FABP family β‐barrel architecture that is capped by a helix‐turn‐helix hinged region and display conformational dynamics that may facilitate ligand entry, 16 , 17 one distinguishing feature of LFABP is a broader cavity due to the formation of an additional beta strand that results from nontraditional hydrogen bonding patterns. 14 The wider cavity accommodates two ligand molecules, with one molecule situated at the base of the cavity, forming a U‐shaped structure referred to as the primary binding site. 14 , 15 Once this molecule is bound, it creates a more hydrophobic binding pocket and the second ligand binds in a roughly perpendicular orientation to it, with the alkyl chain inserted into the curve of the U‐formation and the headgroup near the portal region. 14 , 15 IFABP binds a single molecule of fatty acid oriented similarly to the first LFABP ligand. Some of the amino acid side chains that are potentially involved in binding different ligands have been identified using NMR and x‐ray crystallography, for instance as seen for LFABP with oleate 14 , 15 , 18 and IFABP with palmitate. 19
In order to determine if IFABP and LFABP have different functions within the small intestine, we previously examined mice with either one or the other gene ablated and found a striking phenotypic divergence: after feeding with a high fat diet, the IFABP−/− mice exhibited a lean phenotype whereas the LFABP−/− mice exhibited an obese phenotype. 20 Of note, there was no difference in fecal fat content between the knockout mice and wild type, suggesting no specific alterations in intestinal lipid absorption 21 ; their appetites, as measured by food intake, corresponded to their respective phenotypes. 20
Appetite has been shown to be regulated by stimulation of cannabinoid receptors, which bind to both cannabinoids (exogenous) and endocannabinoids (EC, endogenous). The latter class is exemplified by the arachidonic acid‐derived compounds 2‐arachidonoylglycerol (2‐AG) and anandamide (AEA). 22 , 23 Endocannabinoid ligands are hydrophobic and require a cytosolic protein to bind and carry them to intracellular locations; this property is illustrated by our previous report demonstrating high affinity interactions between LFABP and 2‐monoacylgylcerols. 24 The role of LFABP in cytosolic transport of ECs was initially indicated by elevated mucosal levels of 2‐AG in LFABP null mice. 20 AEA and 2‐AG levels were also increased in the brain of LFABP knockout mice. 25 These findings provided further support for the critical function of LFABP in EC homeostasis. 2‐AG has been shown to be related to stimulation of appetite, 26 and thus the higher levels of 2‐AG in LFABP null mice are consistent with their increased food intake and relative weight gain. 20 In contrast, IFABP null mice had lower levels of mucosal 2‐AG, consistent with their reduced appetite and leaner phenotype.
In order to better understand the molecular interactions of endocannabinoid neurotransmitters with their transport partners, we adopted a biochemical and biophysical approach. To this end, we characterized the interactions between IFABP and LFABP and a panel of EC (AEA and 2‐AG) and EC‐like ligands (OEA, PEA, 2‐OG, 2‐PG) that varied in head group or alkyl chain (Figure 1). We determined the binding affinities of each ligand using a fluorescence displacement assay, and their respective molecular regions of contact with the two transport proteins using solution‐state NMR. The results demonstrate distinctive binding affinities and sites of protein‐ligand interaction, which likely underlie their differential functions at the cellular and systemic levels.
FIGURE 1.
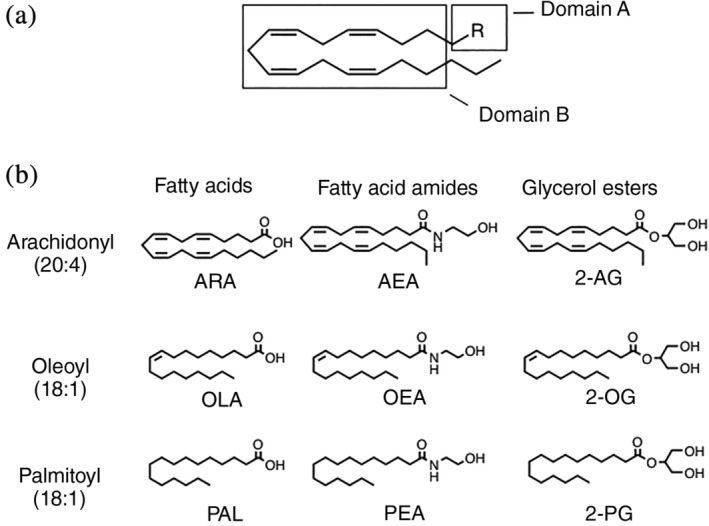
Structural features of ligands used in this study. (a) Using the 20:4 aliphatic chain as a model, shown are the two structural domains that were varied systematically. Domain A is the head group that can be carboxyl, amide, or glycerol ester. Domain B is the alkyl group that was varied as 20:4, 18:1, or 16:0. (b) From left to right are comparisons of ligands with different Domain A (functional head groups). From top to bottom are comparisons of ligands with different Domain B (saturation and length of the alkyl chain)
2. RESULTS
2.1. Ligand binding affinities differ for IFABP and LFABP
In order to relate the phenotypic differences seen in IFABP−/− versus LFABP−/− mice to interactions between the proteins and ligand, we determined the binding affinities of purified recombinant IFABP and LFABP proteins for the panel of ligands shown in Figure 1. Structurally, each ligand can be divided into two domains. Domain A represents the head group, which was either a carboxyl, amide, or glycerol ester. Domain B is the alkyl chain, which varies in saturation and length: 20:4 (arachidonyl), 18:1 (oleoyl), or 16:0 (palmitoyl).
To determine binding affinities, we chose to use a modification of the 11‐(dansylamino)undecanoic acid (DAUDA) displacement assay, 27 , 28 , 29 , 30 , 31 in which we determined the effect of ligand concentration on K d(DAUDA) in order to calculate K d(LIGAND), similar to calculating binding affinity in other systems. 32 The value for K d(LIGAND) that we calculated is a binding constant and is theoretically the same regardless of the fluorescent fatty acid used for displacement (e.g., anilinonaphthalene‐1‐sulfonic acid, ANS). A complete description of the derivation of K d(LIGAND) by this method and representative data can be found in Supporting Information (Figure S1). We note that because the binding affinities we determine by this method inherently incorporate the saturation of binding by ligand, the resulting value reflects a global binding event and does not distinguish between the two sites within LFABP.
Notably, the K d(LIGAND) values were lower for IFABP than LFABP for all ligands tested (Table 1). For fatty acid and monoacylglycerol ligands, this preference is reversed from what was previously reported using other assays. In retrospect, this finding is not surprising given the inherent limitations of these assays (Section 3). 33 , 34 , 35 Also, as noted above, the current method describes total, overall binding affinity of LFABP, and does not distinguish binding to its high affinity site, as defined using other methods.
TABLE 1.
Equilibrium dissociation constants (K d(LIGAND)) for ligands in this study a
| Fatty acids | Fatty acid amides | Glycerol esters | |
|---|---|---|---|
| ARA | AEA | 2‐AG | |
| IFABP | 4.3 ± 3.1 | 23.0 ± 2.7 | 183.8 ± 14.5 |
| LFABP | 18.9 ± 10.8 | 239.7 ± 46.6 | 475.4 ± 52.7 |
| OLA | OEA | 2‐OG | |
| IFABP | 0.9 ± 0.4 | 48.4 ± 11.4 | 160.4 ± 25.7 |
| LFABP | 7.7 ± 5.5 | 168.9 ± 27.1 | 333.9 ± 51.7 |
| PAL | PEA | 2‐PG | |
| IFABP | 3.3 ± 2.0 | 150.8 ± 16.6 | 106.0 ± 25.9 |
| LFABP | 21.6 ± 10.6 | 585.1 ± 9.7 | 1,536.5 ± 131.5 |
Note: In order to calculate K d(LIGAND), entire binding curves were performed in duplicate and then were repeated on at least two different days, for a minimum of four measurements. Calculation of K d(LIGAND) is dependent on mathematical curve fitting which can result in high values for SEM. The defined trends in affinity are within the limits of the reported error.
Values are given as μM ± SEM.
Interestingly, Domain A, the head group, had more impact on K d(LIGAND) than the alkyl chain. For example, ligands with carboxylate head groups have the lowest values of K d(LIGAND) (~0.9–21.6 μM), followed by ligands with ethanolamide head groups (K d(LIGAND) ~23–585 μM), and finally ligands with glycerol ester head groups had the highest values of K d(LIGAND) (~106–1,536 μM). These results paralleled our determinations using isothermal titration calorimetry (ITC, see Figure S2). Previous reports have indicated that positively charged residues inside the ligand binding pocket (e.g., the Arg122 of rat LFABP) are in position to interact with the carboxyl group of oleate, 14 , 36 and thus the lower K d(LIGAND) (i.e., tighter binding) we determined for ligands with carboxyl head groups are consistent with charged amino acids directing the binding of fatty acids. The values we report here for K d(LIGAND) are within the range of values reported previously for fatty acids such as oleate, arachidonate, and palmitate. 37 , 38 , 39 , 40 , 41 , 42 , 43
We also used 2D 1H−15N heteronuclear single quantum coherence (HSQC) NMR to assess the interactions of the proteins with ligand. NMR spectra were obtained in the presence of increasing concentrations of ligand in order to calculate binding affinities by probing the amide NH groups of amino acid residues that are perturbed by ligand‐induced changes in their respective magnetic environments. The determination of binding affinity by NMR requires detection of changes in chemical shift (Δδ) for a range of ligand concentrations that approximate the value of K d. Ligands that bind very tightly (K d < μM) will likely have bound and unbound forms of the protein in slow exchange (k ex < Δδ) and require lower concentrations of each partner than are feasible to detect by NMR. 44 , 45 , 46 Conversely, ligands that have a high K d (>100 μM), will typically exhibit fast exchange between the bound and unbound forms (k ex > Δδ), yielding protein chemical shifts that are weighted averages of the two. However, these latter measurements can often require ligand and/or protein concentrations that are problematic in terms of solubility or aggregation. Thus, it was not possible to monitor the chemical shift changes needed to calculate K d(LIGAND) for ligands with carboxyl head groups that are in slow exchange on the NMR timescale, nor for ligands with glycerol ester head groups that had limited solubility and binding affinities that were too weak. However, we were successful in measuring NMR spectra as a function of concentration for those endocannabinoid ligands with ethanolamide head groups. Of the ethanolamides, we focused our detailed analysis on AEA owing to the considerable physiological effects of this acylethanolamide, and because the spectra with OEA and PEA were complicated by significant line broadening (k ex ~ Δδ) (data not shown).
As an example, Figure 2a shows changes in the contour plots of 2D 1H–15N HSQC NMR spectra of IFABP and LFABP upon binding AEA under a single saturating condition (ratio of AEA:protein of 3:1). Binding affinities were calculated after titrating in ligand at the indicated molar ratios (Figure 2b). The effect of ligand concentration on the average chemical shift perturbations (CSPs) for those residues exhibiting fast exchange behavior between bound and unbound states was graphed in order to determine K d(LIGAND) (Figure 2c). The data were fit to an allosteric model of binding, indicating a binding stoichiometry greater than 1:1, for both IFABP and LFABP. However, we note that this calculation is based on an average of specific residues in each protein: residues I58, D59, F68, I76, E77, F93, V96, L102, I103, and R106 were used to determine the K d(LIGAND) for IFABP, whereas residues D34, I35, E40, I41, H43, E44, N111, and R122 were used for LFABP; thus, an individual residue with a particularly high affinity would artificially introduce an appearance of nonstoichiometric binding to the entire curve.
FIGURE 2.
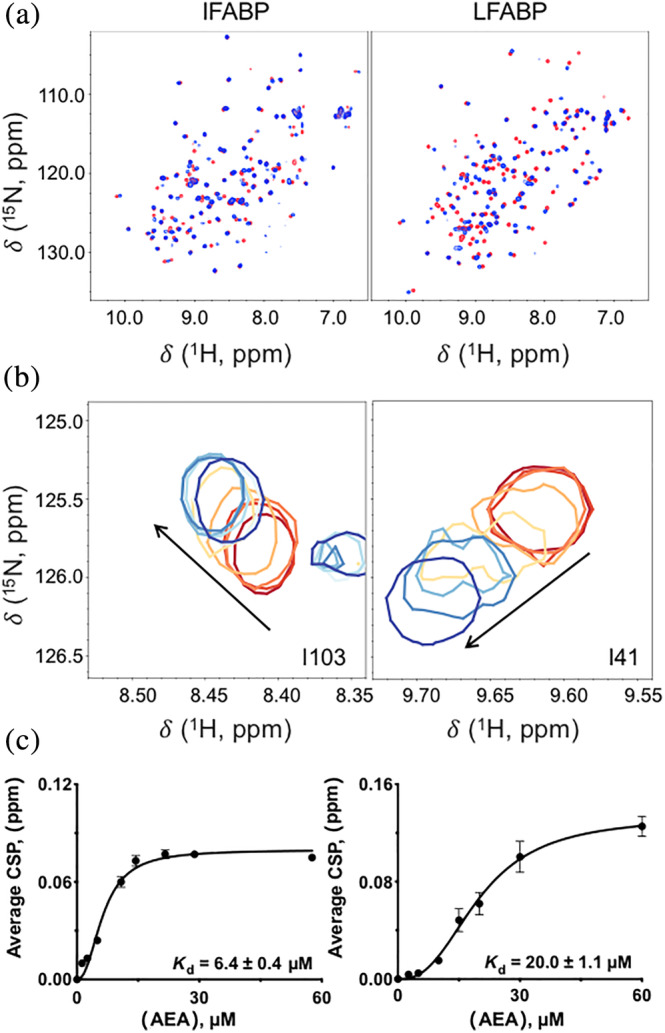
NMR‐based determination of K d(ligand). (a) 1H–15N HSQC NMR contour plots for IFABP and LFABP in the absence of ligand (in red) superimposed on AEA‐bound FABP (blue). Ligand was freshly made and added immediately prior to data acquisition. The final molar ratio of FABP to AEA in the ligand‐bound spectra is 1:3. (b) Resonances of IFABP and LFABP exhibited fast exchange on the NMR timescale during titration of AEA. A subset of residues with titratable chemical shifts is shown at protein:ligand ratios of 1:n, where n has values of 0 (in red), 0.13, 0.25, 0.55, 0.73, 1.1, 1.5, or 3.0 (blue, highest tested). (c) Dependence of chemical shift perturbation (CSP) on concentration of ligand. The CSPs from several residues were averaged and plotted against ligand concentration in order to calculate K d(LIGAND). K d(LIGAND) values are given as ±SEM
The trend in binding affinities determined by this method (6.4 ± 0.4 μM for IFABP‐AEA; 20 ± 1.1 μM for LFABP‐AEA) was consistent with the results of our fluorescence displacement assays (21.2 ± 1.2 μM for IFABP‐AEA and 202.4 ± 9.4 μM for LFABP‐AEA); however, the actual affinities were 3–10 times higher, most likely due to differences in the method used. Specifically, for the NMR analyses, the value of K d(LIGAND) is derived from the individual sites chosen for their variations in Δδ with ligand concentration; thus, the value is based on binding to a defined set of residues which are, by definition, involved in interacting with ligand. In contrast, the values determined from the fluorescence displacement assay are based on the measurement of whole‐molecule binding, and thus reflect the binding affinity of the whole protein rather than just selected residues.
The interaction of anandamide with FABP5 (skin FABP) and FABP7 (brain FABP) has been reported relatively recently, but detailed binding affinities were not included in those studies. 47 , 48 In those studies, nitrobenzoxadiazole (NBD)‐fluorescently conjugated AEA was used to determine the binding affinity for AEA binding to LFABP, but only one molecule of the fluorescent ligand was bound 49 and it is unclear if the mode of binding was affected by the bulky, polar fluorescent label.
2.2. The head group directs interactions between protein and ligand
Analysis of the IFABP and LFABP CSPs for each backbone residue was conducted at a saturating concentration of each ligand of our nine‐member panel. Calculation of CSPs from 2D 1H–15N HSQC NMR spectra was based on previously described methods. 45 CSPs were considered significant if they exceeded the sum of the average CSP and its SD. This significance threshold was determined after eliminating CSPs that were greater than 3 SDs above the mean value, in order to avoid artificial skewing of the mean by large outliers that would result in the omission of some perturbed residues. Complete CSP profiles are shown in Figures S3 and S4. For clarity, each residue that displayed a significant perturbation is represented with a dash in Figure 3; broadened residues that indicated intermediate exchange with bound states were also scored as significant and plotted accordingly. Note that the binding of hydrophobic ligands within the core of FABP is expected to involve nonspecific as well as specific interactions, so that instead of analyzing the CSPs quantitatively, we evaluated the significant CSPs qualitatively.
FIGURE 3.
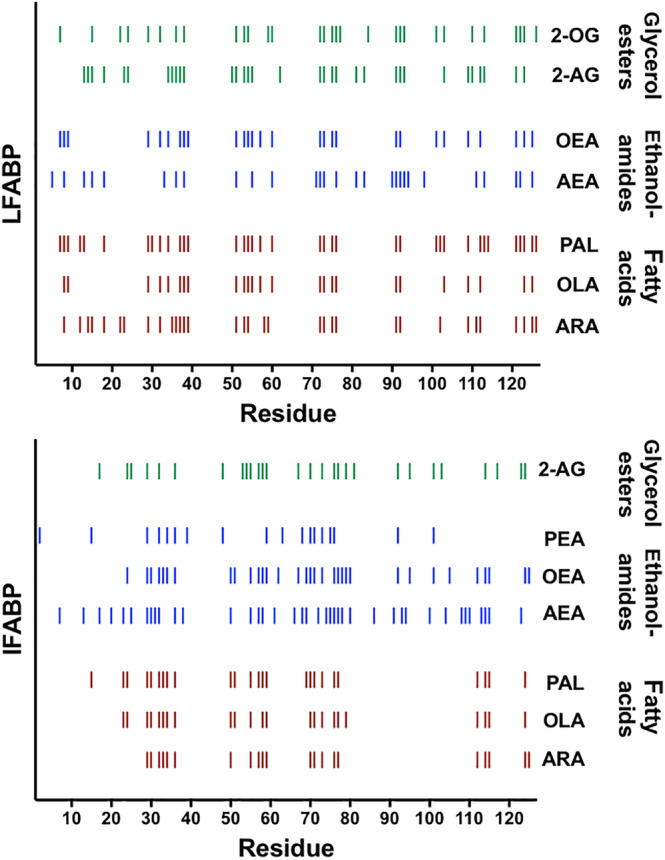
Residues with significant chemical shift perturbations (CSPs). Fatty acids are shown in red‐brown, ethanolamides in blue, and glycerol esters in green. Patterns in binding appear when ligands are grouped according to head group; specifically, the periodicity in significant CSPs, a gap in CSPs between residues 80–110 for fatty acids with IFABP, and additional significant CSPs between residues 66–70 for all the ligands with IFABP
In Figure 3, we represent those residues with significant CSPs with dashes that are color‐coded and grouped according to head group, with fatty acids as red‐brown, ethanolamides as blue, and glycerol esters as green. When we group the residues this way, we see strikingly similar patterns of perturbation and a noticeable periodicity that is common to both proteins and for ligands that have a given head group. However, when we group the CSPs according to alkyl chain (Figure S5), the profiles are more disparate. These comparisons indicate that the head group (Domain A) has a more substantial impact on interactions with the protein than the alkyl chain (Domain B), and that the chain does not direct binding of ligand within the pocket. This trend is consistent with our determinations of equilibrium dissociation constants described above.
Note that addition of PEA and 2‐PG resulted in no observable CSPs with LFABP, most likely because the ligand concentrations needed to saturate the protein were unachievable due to limited ligand solubility. Similarly, the high K d(LIGAND) of 2‐OG with IFABP made it problematic to retain solubility of this ligand at the concentrations needed to obtain saturation of the protein. Thus, we did not pursue investigation of these ligands further.
2.3. Fatty acids are coordinated differently from endocannabinoids
Most strikingly, IFABP exhibits a noticeable gap in significant CSPs spanning residues 80–110 upon binding of the fatty acids, whereas this is not the case when it binds ECs (Figure 3). Thus, the 80–110 region of IFABP is more engaged in binding ECs than fatty acids. By contrast, the analogous region in LFABP displayed significant CSPs for all types of ligands studied, indicative of differences in the mode of binding for the two proteins (Figure 5). The known co‐crystal structure of IFABP with palmitate (PDB 2IFB12) shows the potential for electrostatic interactions between Arg106 and the carboxylate head group of the ligand. Although we detected a slight change in the chemical shift of Arg106 (IFABP) that was marginally above the average CSP in the presence of each fatty acid, it did not meet our cutoff for significance as described above. Notably, a different arginine (Arg122), located outside of this region, was identified previously as being important for binding fatty acid to LFABP. Interestingly, mutation of Arg122 does not completely inhibit binding of fatty acid; instead the double mutation T102Q/R122Q is required. 41 In the alignment of residues 80–110 shown in Figure 4, Thr102 of LFABP corresponds to Arg106 of IFABP, indicating that these residues may serve parallel functions in coordinating fatty acids.
FIGURE 4.

Alignment of the region defined by residues 80–110 of IFABP and LFABP. Alignment was generated by Clustal Omega. 50 Lines indicate a fully conserved residue, colon indicates residues with strongly similar properties, period indicates residues with weakly similar properties. Arrows indicate the alignment of Arg106 of IFABP, which is thought to be responsible for binding the carboxylate of the ligand, to Thr102 of LFABP which was shown to have a significant role in binding fatty acid. Strikingly, LFABP has five threonine residues within this region, whereas IFABP has only two. Threonines are indicated in red
Of note, LFABP has 5 threonine residues in this region (residues 80–110), whereas IFABP has only 2 (Figure 4). The importance of a specific threonine residue (Thr54) in binding fatty acid by IFABP has been noted previously. 51 It is likely that the threonines in this region of LFABP create a local polar environment that is responsible for binding fatty acids to LFABP, and that the lack of analogous threonine residues in IFABP is the major factor accounting for the difference in CSPs in this region. Unexpectedly, in our global determination, LFABP displays a higher dissociation constant (i.e., binds fatty acids less tightly) than IFABP despite LFABP having these additional contacts; thus, it is possible that the conformational changes required to accommodate ligand in LFABP may actually impose an energy barrier to binding.
2.4. IFABP has additional interactions with ECs compared to LFABP
Significant CSPs for all categories of ligands were identified in a region of IFABP defined by five amino acids, specifically residues 66–70 (Figure 5a), whereas the analogous residues in LFABP were not involved in binding any of the tested ligands.
FIGURE 5.
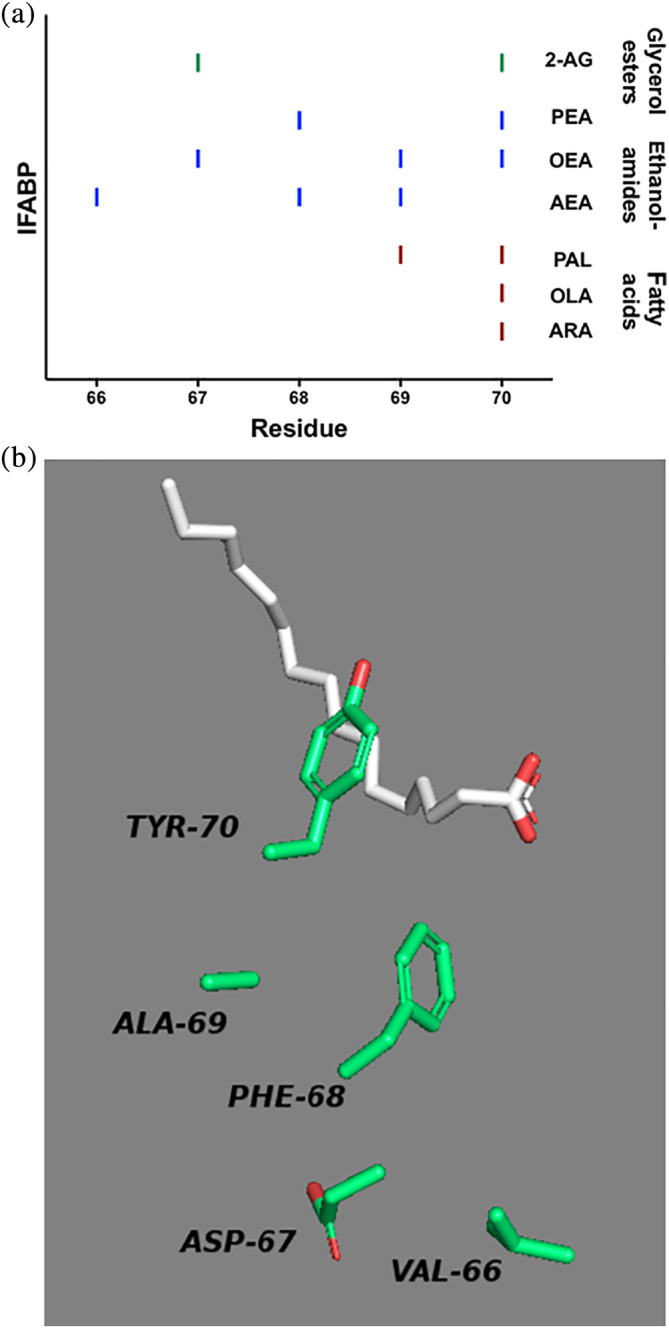
Ligand binding to IFABP involves unique residues that are not used for binding to LFABP. (a) Significant chemical shift perturbations (CSPs) are indicated with a dash for IFABP bound to the indicated ligand. Residues in this region of LFABP were not significantly perturbed. (b) Amino acids 66–70 of the co‐crystal of IFABP with palmitate (PDB 2IFB) are shown. Residues 69 and 70 are involved in binding fatty acid, and all five residues are involved in binding ECs
For IFABP, binding of ligand involved the entire five‐residue region: residue 70 was perturbed in the presence of all three fatty acids, whereas the presence of PAL perturbed both residues 69 and 70. Presence of ECs resulted in perturbation of all five residues (66–70) in IFABP, specifically AEA perturbed residues 66, 68, and 69, while 2‐AG perturbed residues 67 and 70. The EC‐like ligands OEA and PEA perturbed residues in this region as well: for OEA, residues 67, 69, 70; and for PEA, residues 68 and 70. These additional contacts with IFABP that are not found in ligand‐bound LFABP may contribute to the tighter binding affinity of all ligands for IFABP.
Focusing on just these residues in the previously solved co‐crystal of IFABP with palmitate (PDB 2IFB12) (Figure 5b), we see that residue 70 in IFABP is a tyrosine that could be involved in hydrogen bonding with the fatty acid head group to enhance binding. Residues that are involved in EC binding but not fatty acid binding (residues 66–68) are located deeper within the binding cavity and are primarily hydrophobic.
3. DISCUSSION
We found several differences in the interactions of IFABP and LFABP with physiologically important hydrophobic ligands. First, IFABP displayed a smaller binding dissociation constant (tighter binding) than LFABP for all of the tested ligands. This trend indicates that within the enterocyte, where both proteins are found, IFABP is more likely to be fully saturated with ligand than LFABP. Notably, LFABP has two binding sites, with occupancy of the first site required to form the second binding site. 52 This two‐step binding scheme may then contribute to the overall binding affinity we measure in our assays for LFABP, and is likely responsible for the different relative affinities that we find herein, relative to prior reports that indicated higher affinity binding of LFABP compared to IFABP for some ligands, such as long‐chain fatty acids. 24 , 37 , 53 , 54
Further, the discrepancy between these results and the previously reported findings can also be attributed to the variety of methods used to determine their value. In choosing the most suitable assay for our purpose, we considered the following: (a) The acrylodan‐labeled intestinal fatty acid binding protein (ADIFAB) has been used as a fluorescent reporter of unbound fatty acid concentration. This method, however, is inherently dependent on binding of ligand to the modified IFABP, and so accuracy is diminished for ligands that do not bind to ADIFABP with higher affinity than to the protein being tested. 55 (b) Another method uses ligands that are anthroyloxy‐ or nitrobenzoxadiazole (NBD)‐labeled in order to measure changes in fluorescence upon binding. However, NBD‐labeled ligand bound to LFABP in an atypical manner, with only one molecule in the binding cavity instead of two, suggesting that the large fluorophore may limit the association of ligand within the binding cavity. 34 (c) The lipophilic Lipidex 1000 resin has been used to separate bound from unbound radioactively labeled ligand; however, affinity for Lipidex has been shown to be dependent on the type of lipid. 56 In addition, it was demonstrated that this method is complicated by technical issues, such as lipid and FABP binding to the tubes. 57 It is also possible that the protein associates with resin, resulting in an overestimation of unbound ligand. (d) An additional method measures relative binding affinities by the displacement of fluorescent fatty acid (either ANS or DAUDA) 28 , 29 , 30 , 31 at a single saturating concentration of the reporter. However, these values are dependent on the binding affinity of the fluorescent reporter and therefore do not reflect an intrinsic binding constant. Overall, after extensive review of the literature, we identified the current work as the first report in which this full panel of ligands was studied consistently using the same assay and experimental conditions to determine binding affinities.
For both proteins, the head group had more effect on binding affinity than the alkyl chain, with the preference for head group as follows: carboxylate > amide > glycerol ester. The patterns of chemical shift changes measured from 2D 1H–15N HSQC NMR profiles for both proteins upon ligand binding revealed a similar dependence on head group, confirming that identity of the head group had more impact on protein interactions than the alkyl chain.
It is important to note that a lower binding constant may not directly correlate with the number of residues perturbed upon binding; rather, the residues involved in binding need to be analyzed to determine the nature of the binding interaction. Residues that form tight bonds may contribute to lower K d(LIGAND), whereas more global conformational changes may increase binding energy and therefore result in higher K d(LIGAND). The NMR analysis revealed several key molecular features that could direct how these ligands interact with their protein chaperones.
3.1. Binding of ECs involves all regions of the proteins, in contrast to the coordination of fatty acids
Perturbations were extensively distributed throughout the whole protein for both IFABP and LFABP when they were bound to ECs, whereas the points of contact were more discrete and localized when bound to fatty acids. The CSP patterns associated with EC binding indicate global protein conformational changes rather than the more defined binding interface observed for FA ligands. It is likely that the specific contacts with FAs contribute stronger interactions, in turn producing tighter binding than for ECs that must overcome more energetically unfavorable conformational changes to accommodate a ligand.
3.2. Fatty acids are coordinated differently in IFABP with respect to LFABP
The difference in how fatty acids are coordinated was particularly noticeable with IFABP, which showed a large region (residues 80–110) that was not significantly perturbed in the presence of fatty acid (Figure 6a). In contrast, the analogous region of LFABP exhibited discrete interactions with fatty acids (Figure 6b).
FIGURE 6.
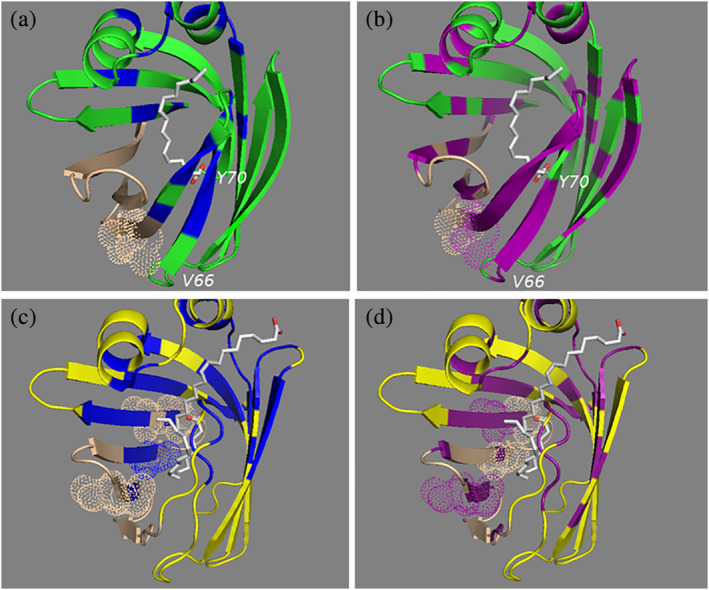
Structural representation of unique regions that interact with ligand. IFABP (pdb 2IFB) is shown with a green ribbon structure, and LFABP (pdb 1lfo) is shown with a yellow backbone. Residues that are significantly perturbed in the presence of ligand are shown in blue for FAs (OLA, ARA, PAL) and purple for ECs (AEA, 2‐AG). The residues of region 80–110 that are not significantly perturbed in the presence of ligand are shown in tan. (a) Shows that this region has no interactions with FAs for IFABP. IFABP has fewer threonine residues in this region than LFABP (shown with dot representations). (a, b) Also, the presence of ligand resulted in significant perturbations of IFABP in the region defined by residues 66–70, but the analogous region in LFABP was not involved in binding ligand (c, d)
3.3. IFABP has additional contacts with ECs that are not seen in LFABP
We identified a region defined by five amino acid residues (66–70) that is involved in binding ligand only for IFABP (Figure 6b). These additional interactions with ligand that are not found with LFABP may be responsible for the tighter binding to IFABP.
The binding results described here are in line with reports that LFABP knockout mice exhibit higher cytosolic levels of ECs. The most striking phenotypic difference in these mice was their body mass after eating a high‐fat diet. IFABP null mice were lean, whereas LFABP null mice were obese, and this trend correlated to their food intake and apparent appetite. Appetite has been attributed to fatty acid ethanolamides (FAEs) and related endocannabinoids. 22 , 23 , 58 , 59 LFABP has been implicated in the regulation of levels of these signaling molecules by transporting them to their downstream metabolic fates including enzymatic degradation by fatty acid amide hydrolase (FAAH) or monoacylglycerol lipase (MAGL), 60 , 61 , 62 , 63 or through activation of nuclear receptors such as PPARα. 64 , 65 In this way, LFABP may mark the ligand for processing or degradation, consistent with the observation that the absence of LFABP results in elevated levels of cytosolic 2‐AG. 20 , 25
The results presented here indicate that IFABP has a higher affinity relative to LFABP, suggesting that ligand will be bound preferentially to IFABP, which in turn indicates that the presence of IFABP will lower the access of ligand to LFABP and may limit downstream processing, depending upon the absolute level of unbound ligand available. This proposal is supported by the increased levels of AEA and 2‐AG in LFABP knockout mice. 20 , 25 Conversely, the absence of IFABP will make more ligand accessible for LFABP‐mediated degradation. This latter scheme is supported by the decreased levels of 2‐AG in IFABP knockout mice. 20 , 25 These studies have enhanced our understanding of the different roles of IFABP and LFABP in intestinal enterocytes, supporting their functions as nutrient sensors which contribute to the regulation of systemic energy homeostasis.
4. MATERIALS AND METHODS
4.1. Expression and purification of IFABP and LFABP
Expression of cleavable his‐tagged rat IFABP and LFABP from a Gateway® pDEST17expression vector in BL21(AI) cells was achieved after induction with arabinose (0.2%) in M9 medium; for uniformly labeled protein, ammonium chloride salt (1 g/L) was added as the sole source of 15N. The cell paste was resuspended in 50 ml of chilled lysis buffer (20 mM Tris–HCl, 300 mM NaCl, 1 mM PMSF, 0.5 mg of DNase, 50 μM of MgSO4, pH 8) and sonicated for 30 min in an ice bath. Upon sonication, the lysate was clarified by ultracentrifugation at ~30,000g (16,000 rpm in a Beckman JA25.5 rotor) at 4°C for 30 min. The lysate was then loaded onto a 5 ml HiTrap HP Ni column at 1 ml/min and washed extensively with buffer (20 mM Tris–HCl, 300 mM NaCl, 25 mM imidazole, pH 8). Bound protein was then treated with TEV protease to remove the His‐tag: 1.5 mg of TEV protease was added to the bound protein on the column and circulated at a low flow rate (~0.1 ml/min) at room temperature for 24–48 hr. Cleaved protein eluted in the unbound fractions. Both IFABP and LFABP proteins were purified using the same procedure.
4.2. Delipidation of IFABP and LFABP
For subsequent ligand binding assays, it is essential to ensure complete removal of lipids that may have co‐purified with the protein. Delipidation of IFABP was achieved by incubating the protein twice with pre‐swelled HAP‐Dextran at a 5:1 ratio as described previously. 66 Each incubation was carried out at 37°C for 3 hr, with shaking at 225 rpm. After each incubation, the protein–resin mixture was poured into an empty Gravity Kontes FlexColumn and lipid‐free protein was eluted with buffer. The protein was concentrated to ~300 μM using an Amicon Ultra‐15 Centrifugal Filter Unit with a 10 kDa molecular weight cutoff (cat. No. UFC901024).
We used organic extraction for the delipidation of LFABP. In this protocol, 1/3 volume of n‐butanol was added to the protein, mixed thoroughly, and allowed to agitate for 15 min. This mixture was then centrifuged to achieve phase separation and the aqueous protein‐containing phase was removed. This procedure was repeated at least three times to ensure complete lipid extraction. The sample was washed extensively using an Amicon Ultra‐0.5 ml Centrifugal Filter Unit with 3 kDa molecular weight cutoff (cat. No. UFC500396) to remove any residual n‐butanol. HSQC NMR spectra were acquired for the delipidated proteins to confirm the apo state by reference to published spectra. 15
4.3. Ligand competition binding assay
A modification of a standard displacement assay 41 was used to determine binding affinities of FABP for a panel of ligands. Binding affinity of ligand was calculated based on the effect of the ligand on the binding affinity of the fluorescent fatty acid, DAUDA. DAUDA fluorescence at 500 nm is greatly enhanced when bound to protein, so that increasing concentrations of added ligand produce decreases in fluorescence intensity as the ligand displaces the DAUDA. The effect of increasing concentrations of ligand (0–50 μM) on a standard binding curve for DAUDA was measured, and the binding affinity of ligand was derived as described in Supporting Information. In order to achieve similar maximum fluorescence values for DAUDA, we used 1 μM LFABP and 2 μM IFABP. IFABP required twofold higher concentrations of protein to reach a similar maximum fluorescence as LFABP, most likely because the binding cavity of LFABP can accommodate two molecules of ligand.
Controls containing only ligand and DAUDA in the absence of protein were run in parallel and did not significantly contribute to the observed fluorescence; these readings were subtracted as background. It is important to note that all ligands were present at concentrations below those that would contribute to protein‐independent background fluorescence, a phenomenon that has been shown previously to interfere with interpreting results from these types of assays. 38
4.4. Preparation of ligand solutions
Sodium palmitate and sodium oleate (cat Nos. P9767 and O7501) were purchased from Sigma‐Aldrich. Sodium arachidonic acid (cat No. 10006607), AEA (cat No. 90050), OEA (cat No. 90265), PEA (cat No. 90350), 2‐AG (cat No. 62160), 2‐OG (cat No. 16537), 2‐PG (cat No. 17882), and DAUDA (cat No. 10005188) were purchased from Cayman Chemical. The ligands were dissolved in HPLC‐grade ethanol or methanol to prepare stock solutions ranging from 10 to 15 mM.
4.5. Solution‐state NMR spectroscopy
The two‐dimensional NMR experiments were performed on Bruker Avance I or III spectrometers operating at 1H frequencies of 600 or 800 MHz, each equipped with a 5‐mm TCI cryoprobe. The spectra for IFABP were acquired at 30°C, whereas the spectra for LFABP were obtained at 25°C. The resulting data were processed using NMRPipe software 67 and analyzed by NMRViewJ software. 68
Combined CSPs were calculated using the following equation 45 :
NMR experiments with [U‐15N]‐labeled apo‐IFABP were performed in NMR buffer A (20 mM phosphate buffer, 50 mM KCl, 5% D2O, pH 7.2), whereas NMR experiments with [U‐15N]‐apo‐LFABP were performed in NMR buffer B (20 mM phosphate buffer, 50 mM KCl, 5% D2O, pH 7.0). Residue assignments were based on previously published data for both LFABP 15 and IFABP. 19
For NMR titration experiments, we prepared 20 μM [U‐15N]‐apo‐IFABP in NMR buffer A and 20 μM [U‐15N]‐apo‐LFABP in NMR buffer B, respectively. We then mixed 5 μl of serially diluted AEA solutions with 495 μl of the protein solutions and transferred each of the resulting samples to clean NMR tubes. The serially diluted AEA solutions were derived from a 6 mM AEA stock solution and prepared as twofold serial dilutions using ethanol. The final molar equivalents of AEA were 0, 0.13, 0.25, 0.55, 0.73, 1.1, 1.5, and 3.0 times the concentration of FABP. All samples contained the same percentage of ethanol, which was no more than 1%.
For 1H–15N NMR experiments to determine chemical shifts at saturating ligand levels, we mixed 5 μl of stock ligand solution (20–26 mM) with 495 μl of protein solution (100 μM). The final concentration of organic solvent, ethanol or acetonitrile was no more than 1% in these samples.
AUTHOR CONTRIBUTIONS
May Lai: Data curation; formal analysis; investigation; validation; writing‐original draft; writing‐review and editing. Francine S. Katz: Data curation (supporting); formal analysis (supporting); methodology (supporting); supervision (lead); writing‐original draft (supporting); writing‐review & editing (lead). Cedric Bernard: Supervision; writing‐review and editing. Judith Storch: Conceptualization; funding acquisition; writing‐review and editing. Ruth Stark: Conceptualization; funding acquisition; methodology; supervision; writing‐review and editing.
Supporting information
Appendix S1. Supporting information
ACKNOWLEDGMENTS
This research was supported by the National Institutes of Health (5R01‐DK038389‐31), with infrastructural assistance provided by the National Institutes of Health through the National Institute on Minority Health and Health Disparities (5G12‐MD007603‐30), the U.S. National Science Foundation (MCB‐1411984), and the New Jersey Agricultural Experiment Station (NJ14115). Institutional support came from the CUNY Institute for Macromolecular Assemblies and the CUNY Advanced Science Research Center. R. E. S. is a member of the New York Structural Biology Center (NYSBC); data collection at that facility was made possible by a grant from the New York State Office of Science, Technology and Academic Research and an NIH Office of Research Infrastructure Program Facility Improvement grant (CO6RR015495). We gratefully acknowledge Drs Hsin Wang, James Aramini, and Shibani Bhattacharya for assistance with the NMR experiments and Prof David Jeruzalmi for access to the fluorescence‐equipped plate reader.
Lai MP, Katz FS, Bernard C, Storch J, Stark RE. Two fatty acid‐binding proteins expressed in the intestine interact differently with endocannabinoids. Protein Science. 2020;29:1606–1617. 10.1002/pro.3875
Funding information CUNY Advanced Science Research Center; CUNY Institute for Macromolecular Assemblies; National Institute on Minority Health and Health Disparities, Grant/Award Number: 5G12‐MD007603‐30; National Institutes of Health, Grant/Award Number: 5R01‐DK038389‐31; New Jersey Agricultural Experiment Station, Grant/Award Number: NJ14115; NIH Office of Research Infrastructure Program Facility Improvement, Grant/Award Number: CO6RR015495; U.S. National Science Foundation, Grant/Award Number: MCB‐1411984
Contributor Information
Francine S. Katz, Email: rstark@ccny.cuny.edu.
Ruth E. Stark, Email: fkatz@ccny.cuny.edu.
REFERENCES
- 1. Hulbert AJ, Turner N, Storlien LH, Else PL. Dietary fats and membrane function: Implications for metabolism and disease. Biol Rev Camb Philos Soc. 2005;80:155–169. [DOI] [PubMed] [Google Scholar]
- 2. Storch J, Corsico B. The emerging functions and mechanisms of mammalian fatty acid‐binding proteins. Annu Rev Nutr. 2008;28:73–95. [DOI] [PubMed] [Google Scholar]
- 3. Banaszak L, Winter N, Xu Z, Bernlohr DA, Cowan S, Jones TA. Lipid‐binding proteins: A family of fatty acid and retinoid transport proteins. Adv Protein Chem. 1994;45:89–151. [DOI] [PubMed] [Google Scholar]
- 4. Bass NM. The cellular fatty acid binding proteins: Aspects of structure, regulation, and function. Int Rev Cytol. 1988;111:143–184. [DOI] [PubMed] [Google Scholar]
- 5. Storch J, McDermott L. Structural and functional analysis of fatty acid‐binding proteins. J Lipid Res. 2009;50(Suppl):S126–S131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liou HL, Kahn PC, Storch J. Role of the helical domain in fatty acid transfer from adipocyte and heart fatty acid‐binding proteins to membranes: Analysis of chimeric proteins. J Biol Chem. 2002;277:1806–1815. [DOI] [PubMed] [Google Scholar]
- 7. Hsu KT, Storch J. Fatty acid transfer from liver and intestinal fatty acid‐binding proteins to membranes occurs by different mechanisms. J Biol Chem. 1996;271:13317–13323. [DOI] [PubMed] [Google Scholar]
- 8. Corsico B, Cistola DP, Frieden C, Storch J. The helical domain of intestinal fatty acid binding protein is critical for collisional transfer of fatty acids to phospholipid membranes. Proc Natl Acad Sci USA. 1998;95:12174–12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wu F, Corsico B, Flach CR, Cistola DP, Storch J, Mendelsohn R. Deletion of the helical motif in the intestinal fatty acid‐binding protein reduces its interactions with membrane monolayers: Brewster angle microscopy, IR reflection‐absorption spectroscopy, and surface pressure studies. Biochemistry. 2001;40:1976–1983. [DOI] [PubMed] [Google Scholar]
- 10. Corsico B, Liou HL, Storch J. The alpha‐helical domain of liver fatty acid binding protein is responsible for the diffusion‐mediated transfer of fatty acids to phospholipid membranes. Biochemistry. 2004;43:3600–3607. [DOI] [PubMed] [Google Scholar]
- 11. Sacchettini JC, Gordon JI, Banaszak LJ. The structure of crystalline Escherichia coli‐derived rat intestinal fatty acid‐binding protein at 2.5‐A resolution. J Biol Chem. 1988;263:5815–5819. [PubMed] [Google Scholar]
- 12. Sacchettini JC, Gordon JI, Banaszak LJ. Crystal structure of rat intestinal fatty‐acid‐binding protein. Refinement and analysis of the Escherichia coli‐derived protein with bound palmitate. J Mol Biol. 1989;208:327–339. [DOI] [PubMed] [Google Scholar]
- 13. Sacchettini JC, Gordon JI, Banaszak LJ. Refined apoprotein structure of rat intestinal fatty acid binding protein produced in Escherichia coli . Proc Natl Acad Sci USA. 1989;86:7736–7740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Thompson J, Winter N, Terwey D, Bratt J, Banaszak L. The crystal structure of the liver fatty acid‐binding protein. A complex with two bound oleates. J Biol Chem. 1997;272:7140–7150. [DOI] [PubMed] [Google Scholar]
- 15. He Y, Yang X, Wang H, et al. Solution‐state molecular structure of apo and oleate‐liganded liver fatty acid‐binding protein. Biochemistry. 2007;46:12543–12556. [DOI] [PubMed] [Google Scholar]
- 16. Cheng P, Liu D, Chee PX, Yang D, Long D. Atomistic insights into the functional instability of the second helix of fatty acid binding protein. Biophys J. 2019;117:239–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hodsdon ME, Cistola DP. Discrete backbone disorder in the nuclear magnetic resonance structure of apo intestinal fatty acid‐binding protein: Implications for the mechanism of ligand entry. Biochemistry. 1997;36:1450–1460. [DOI] [PubMed] [Google Scholar]
- 18. D'Onofrio M, Barracchia CG, Bortot A, Munari F, Zanzoni S, Assfalg M. Molecular differences between human liver fatty acid binding protein and its T94A variant in their unbound and lipid‐bound states. Biochim Biophys Acta Proteins Proteom. 2017;1865:1152–1159. [DOI] [PubMed] [Google Scholar]
- 19. Hodsdon ME, Ponder JW, Cistola DP. The NMR solution structure of intestinal fatty acid‐binding protein complexed with palmitate: Application of a novel distance geometry algorithm. J Mol Biol. 1996;264:585–602. [DOI] [PubMed] [Google Scholar]
- 20. Gajda AM, Zhou YX, Agellon LB, et al. Direct comparison of mice null for liver or intestinal fatty acid‐binding proteins reveals highly divergent phenotypic responses to high fat feeding. J Biol Chem. 2013;288:30330–30344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lackey AI, Chen T, Zhou YX, et al. Mechanisms underlying reduced weight gain in intestinal fatty acid‐binding protein (IFABP) null mice. Am J Physiol. 2019;318:G518–G530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gamage TF, Lichtman AH. The endocannabinoid system: Role in energy regulation. Pediatr Blood Cancer. 2012;58:144–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Watkins BA, Kim J. The endocannabinoid system: Directing eating behavior and macronutrient metabolism. Front Psychol. 2014;5:1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lagakos WS, Guan X, Ho SY, et al. Liver fatty acid‐binding protein binds monoacylglycerol in vitro and in mouse liver cytosol. J Biol Chem. 2013;288:19805–19815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Martin GG, Chung S, Landrock D, et al. FABP‐1 gene ablation impacts brain endocannabinoid system in male mice. J Neurochem. 2016;138:407–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kirkham TC, Williams CM, Fezza F, Di Marzo V. Endocannabinoid levels in rat limbic forebrain and hypothalamus in relation to fasting, feeding and satiation: Stimulation of eating by 2‐arachidonoyl glycerol. Br J Pharmacol. 2002;136:550–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Maatman RG, van Moerkerk HT, Nooren IM, van Zoelen EJ, Veerkamp JH. Expression of human liver fatty acid‐binding protein in Escherichia coli and comparative analysis of its binding characteristics with muscle fatty acid‐binding protein. Biochim Biophys Acta. 1994;1214:1–10. [DOI] [PubMed] [Google Scholar]
- 28. Veerkamp JH, van Moerkerk HT, Prinsen CF, van Kuppevelt TH. Structural and functional studies on different human FABP types. Mol Cell Biochem. 1999;192:137–142. [PubMed] [Google Scholar]
- 29. Wilkinson TC, Wilton DC. Studies on fatty acid‐binding proteins. The detection and quantification of the protein from rat liver by using a fluorescent fatty acid analogue. Biochem J. 1986;238:419–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Peeters RA, Groen MA, de Moel MP, van Moerkerk HT, Veerkamp JH. The binding affinity of fatty acid‐binding proteins from human, pig and rat liver for different fluorescent fatty acids and other ligands. Int J Biochem. 1989;21:407–418. [DOI] [PubMed] [Google Scholar]
- 31. Kane CD, Bernlohr DA. A simple assay for intracellular lipid‐binding proteins using displacement of 1‐anilinonaphthalene 8‐sulfonic acid. Anal Biochem. 1996;233:197–204. [DOI] [PubMed] [Google Scholar]
- 32. Ascenzi P, Menegatti E, Guarneri M, Amiconi G. Active‐site titration of serine proteinases acting selectively on cationic substrates by N‐alpha‐carbobenzoxy‐L‐arginine p‐nitrophenyl ester and N‐alpha‐carbobenzoxy‐L‐lysine p‐nitrophenyl ester; determination of active enzyme concentration. Biochim Biophys Acta. 1987;915:421–425. [DOI] [PubMed] [Google Scholar]
- 33. Storch J. Diversity of fatty acid‐binding protein structure and function: Studies with fluorescent ligands. Mol Cell Biochem. 1993;123:45–53. [DOI] [PubMed] [Google Scholar]
- 34. Schroeder F, McIntosh AL, Martin GG, et al. Fatty acid binding protein‐1 (FABP1) and the human FABP1 T94A variant: Roles in the endocannabinoid system and dyslipidemias. Lipids. 2016;51:655–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Richieri GV, Ogata RT, Kleinfeld AM. The measurement of free fatty acid concentration with the fluorescent probe ADIFAB: A practical guide for the use of the ADIFAB probe. Mol Cell Biochem. 1999;192:87–94. [PubMed] [Google Scholar]
- 36. Cheng YY, Huang YF, Lin HH, Chang WW, Lyu PC. The ligand‐mediated affinity of brain‐type fatty acid‐binding protein for membranes determines the directionality of lipophilic cargo transport. Biochim Biophys Acta Mol Cell Biol Lipids. 2019;1864:158506. [DOI] [PubMed] [Google Scholar]
- 37. Richieri GV, Ogata RT, Kleinfeld AM. Equilibrium constants for the binding of fatty acids with fatty acid‐binding proteins from adipocyte, intestine, heart, and liver measured with the fluorescent probe ADIFAB. J Biol Chem. 1994;269:23918–23930. [PubMed] [Google Scholar]
- 38. Norris A, Spector A. Very long chain n‐3 and n‐6 polysaturated fatty acids bind strongly to liver fatty acid‐binding protein. J Lipid Res. 2002;43:646–653. [PubMed] [Google Scholar]
- 39. Richieri GV, Ogata RT, Kleinfeld AM. Fatty acid interactions with native and mutant fatty acid binding proteins. Mol Cell Biochem. 1999;192:77–85. [PubMed] [Google Scholar]
- 40. Kurian E, Kirk WR, Prendergast FG. Affinity of fatty acid for rRat intestinal fatty acid binding protein: Further examination. Biochemistry. 1998;37:6614. [DOI] [PubMed] [Google Scholar]
- 41. Thumser AE, Evans C, Worrall AF, Wilton DC. Effect on ligand binding of arginine mutations in recombinant rat liver fatty acid‐binding protein. Biochem J. 1994;297:103–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Velkov T, Horne J, Laguerre A, Jones E, Scanlon MJ, Porter CJ. Examination of the role of intestinal fatty acid‐binding protein in drug absorption using a parallel artificial membrane permeability assay. Chem Biol. 2007;14:453–465. [DOI] [PubMed] [Google Scholar]
- 43. Santambrogio C, Favretto F, D'Onofrio M, Assfalg M, Grandori R, Molinari H. Mass spectrometry and NMR analysis of ligand binding by human liver fatty acid binding protein. J Mass Spectrom. 2013;48:895–903. [DOI] [PubMed] [Google Scholar]
- 44. Schumann FH, Riepl H, Maurer T, Gronwald W, Neidig KP, Kalbitzer HR. Combined chemical shift changes and amino acid specific chemical shift mapping of protein‐protein interactions. J Biomol NMR. 2007;39:275–289. [DOI] [PubMed] [Google Scholar]
- 45. Williamson MP. Using chemical shift perturbation to characterise ligand binding. Prog Nucl Magn Reson Spectrosc. 2013;73:1–16. [DOI] [PubMed] [Google Scholar]
- 46. Kleckner IR, Foster MP. An introduction to NMR‐based approaches for measuring protein dynamics. Biochim Biophys Acta. 2011;1814:942–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kaczocha M, Glaser ST, Deutsch DG. Identification of intracellular carriers for the endocannabinoid anandamide. Proc Natl Acad Sci USA. 2009;106:6375–6380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kaczocha M, Vivieca S, Sun J, Glaser ST, Deutsch DG. Fatty acid‐binding proteins transport N‐acylethanolamines to nuclear receptors and are targets of endocannabinoid transport inhibitors. J Biol Chem. 2012;287:3415–3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Huang H, McIntosh AL, Martin GG, et al. FABP1: A novel hepatic endocannabinoid and cannabinoid binding protein. Biochemistry. 2016;55:5243–5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sievers F, Wilm A, Dineen D, et al. Fast, scalable generation of high‐quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Storch J, Thumser AE. Tissue‐specific functions in the fatty acid‐binding protein family. J Biol Chem. 2010;285:32679–32683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sharma A, Sharma A. Fatty acid induced remodeling within the human liver fatty acid‐binding protein. J Biol Chem. 2011;286:31924–31928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Richieri GV, Ogata RT, Zimmerman AW, Veerkamp JH, Kleinfeld AM. Fatty acid binding proteins from different tissues show distinct patterns of fatty acid interactions. Biochemistry. 2000;39:7197–7204. [DOI] [PubMed] [Google Scholar]
- 54. Gajda AM, Storch J. Enterocyte fatty acid‐binding proteins (FABPs): Different functions of liver and intestinal FABPs in the intestine. Prostaglandins Leukot Essent Fatty Acids. 2015;93:9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Huber AH, Kampf JP, Kwan T, Zhu B, Kleinfeld AM. Fatty acid‐specific fluorescent probes and their use in resolving mixtures of unbound free fatty acids in equilibrium with albumin. Biochemistry. 2006;45:14263–14274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nemecz G, Schroeder F. Selective binding of cholesterol by recombinant fatty acid binding proteins. J Biol Chem. 1991;266:17180–17186. [PubMed] [Google Scholar]
- 57. Vork MM, Glatz JF, Surtel DA, van der Vusse GJ. Assay of the binding of fatty acids by proteins: Evaluation of the Lipidex 1000 procedure. Mol Cell Biochem. 1990;98:111–117. [DOI] [PubMed] [Google Scholar]
- 58. Rodriguez de Fonseca F, Navarro M, Gomez R, et al. An anorexic lipid mediator regulated by feeding. Nature. 2001;414:209–212. [DOI] [PubMed] [Google Scholar]
- 59. Jamshidi N, Taylor DA. Anandamide administration into the ventromedial hypothalamus stimulates appetite in rats. Brit J Pharmacol. 2001;134:1151–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Deutsch DG. A personal retrospective: Elevating anandamide (AEA) by targeting fatty acid amide hydrolase (FAAH) and the fatty acid binding proteins (FABPs). Front Pharmacol. 2016;7:370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Berger WT, Ralph BP, Kaczocha M, et al. Targeting fatty acid binding protein (FABP) anandamide transporters—A novel strategy for development of anti‐inflammatory and anti‐nociceptive drugs. PLoS One. 2012;7:e50968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kaczocha M, Rebecchi MJ, Ralph BP, et al. Inhibition of fatty acid binding proteins elevates brain anandamide levels and produces analgesia. PLoS One. 2014;9:e94200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. McIntosh AL, Huang H, Landrock D, et al. Impact of Fabp1 gene ablation on uptake and degradation of endocannabinoids in mouse hepatocytes. Lipids. 2018;53:561–580. [DOI] [PubMed] [Google Scholar]
- 64. Hostetler HA, McIntosh AL, Atshaves BP, et al. L‐FABP directly interacts with PPARalpha in cultured primary hepatocytes. J Lipid Res. 2009;50:1663–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Velkov T. Interactions between human liver fatty acid binding protein and peroxisome proliferator activated receptor selective drugs. PPAR Res. 2013;2013:938401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wang Q, Rizk S, Bernard C, et al. Protocols and pitfalls in obtaining fatty acid‐binding proteins for biophysical studies of ligand‐protein and protein‐protein interactions. Biochem Biophys Rep. 2017;10:318–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. [DOI] [PubMed] [Google Scholar]
- 68. Johnson BA, Blevins RA. NMR View: A computer program for the visualization and analysis of NMR data. J Biomol NMR. 1994;4:603–614. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1. Supporting information


