Abstract
The interaction between the transcriptional coactivators CREBBP/p300 and NCOA is governed by two intrinsically disordered domains called NCBD and CID, respectively. The CID domain emerged within the NCOA protein in deuterostome animals (including vertebrates) after their split from the protostomes (molluscs, worms, and arthropods). However, it has not been clear at which point a high affinity interaction evolved within the deuterostome clade and whether all present‐day deuterostome animals have a high affinity NCBD:CID interaction. We have here expressed and measured affinity for NCBD and CID domains from animal species representing different evolutionary branches of the deuterostome tree. While all vertebrate species have high‐affinity NCBD:CID interactions we found that the interaction in the echinoderm purple sea urchin is of similar affinity as that of the proposed ancestral domains. Our findings demonstrate that the high‐affinity NCBD:CID interaction likely evolved in the vertebrate branch and question whether the interaction between CREBBP/p300 and NCOA is essential in nonvertebrate deuterostomes. The data provide an example of evolution of transcriptional regulation through protein‐domain based inventions.
Keywords: ACTR, CBP, CID, CREBBP, echinoderm, intrinsically disordered protein, NCBD, p300, protein evolution, protein–protein interaction
1. INTRODUCTION
Evolution of multicellular life involved gene duplications and reuse of existing proteins for new function. One key event in the evolution of present day chordates was the two whole genome duplications, 1 which occurred around 450 million years ago (Myr). The resulting four copies of each gene enabled sub‐ and neofunctionalization of several genes involved in cell signaling and development. While a large fraction of the duplicated genes was subsequently lost several were subject to positive selection and can be found as paralogs in most present‐day chordates. We have previously studied in detail the evolution of the interaction between two of these proteins,2, 3 which are transcriptional coactivators: (a) CREB‐binding protein (CREBBP or CBP) and its paralog p300, 4 and (b) Nuclear coactivator (NCOA) (or p160 steroid receptor coactivator) 1, 2, and 3 (also called Src1, TIF2, and ACTR, respectively). 5 More specifically, the interaction is between two small intrinsically disordered domains present in the respective protein, Nuclear coactivator binding domain (NCBD) in CREBBP/p300 and CREBBP‐binding domain (CID) in NCOA1, 2, and 3 (Figure 1). CID is an intrinsically disordered protein domain whereas NCBD can be described as molten‐globule like, that is, it has a hydrophobic core but very dynamic properties. 6 Most animals (metazoa) have bilaterian (two‐sided) symmetry and are divided into two major groups: deuterostomes (including chordates, hemichordates, and echinoderms) and protostomes (including molluscs, worms, and arthropods). The NCBD domain of CREBBP/p300 is present in all animal phyla, including the nonbilaterian cnidarians (e.g., jellyfish). However, the CID domain of the NCOA paralogs could only be identified in genes from deuterostome animals. Thus, we previously concluded that the interaction between CREBBP/p300 and NCOA originated in the deuterostome lineage by evolution of a CID domain within the ancestral NCOA protein between 450 and 550 Myr.2, 3 Furthermore, ancestral variants of NCBD and CID from different evolutionary time points were reconstructed and the binding affinities for different ancestral complexes showed that this interaction most likely emerged as a low‐affinity interaction (3–10 μM), which evolved into the high‐affinity interaction (0.1 μM) that is observed in extant humans.7, 8 Finally, the respective CID from human NCOA1, 2, and 3 all bind with high affinity to human NCBD, but also to NCBD from other chordate species (zebra fish and the more distantly related sea lamprey, a jawless fish), 2 suggesting that all chordate NCOAs display high affinity for CREBBP/p300.
FIGURE 1.
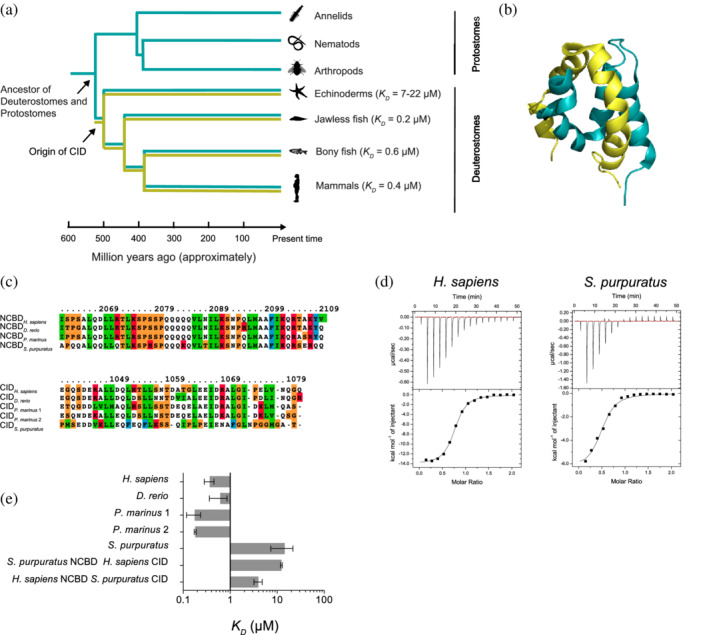
NCBD and CID: evolution, structure, and affinities. (a) A schematic phylogenetic tree showing the evolutionary relationship between extant species along with binding affinities of NCBD:CID complexes determined by ITC. Blue‐green color denotes the evolutionary tree for NCBD and olive denotes the evolution of CID. The schematic animals were downloaded from http://phylopic.org. (b) Solution structure of the human complex consisting of NCBD from CREBBP (blue‐green) and CID from NCOA3/ACTR (olive; PDB entry 6ES7). (c) NCBD (top) and CID (bottom) sequences from four species. Two CID paralogues were identified in Petromyzon marinus. The sequence alignments were performed using PRALINE and the colour coding denotes residue type. (d) Example ITC thermograms for the human (Homo sapiens) complex (left) and purple sea urchin (S. purpuratus) complex (right). (e) The binding affinities of the NCBD:CID complexes in different animals. Each experiment was performed at least twice and the error bars denote the SEM for replicate or triplicate experiments
We have now expressed, purified, and measured affinities for NCBD and CID from several present‐day animals to refine the evolutionary history of the interaction. We find that the interaction indeed has high affinity in all vertebrate animals but not in the echinoderm Strongylocentrotus purpuratus (purple sea urchin), suggesting that evolution of a stronger interaction between CREBBP/p300 and NCOA played a role in the development of vertebrate animals.
2. RESULTS
In order to gain further insight into the evolution of the NCBD:CID protein–protein interaction, we used isothermal titration calorimetry (ITC) to investigate binding affinities of NCBD:CID complexes from four extant animals representing distinct lineages. These were Homo sapiens (human) from mammals, Danio rerio (zebra fish) from bony fish, Petromyzon marinus (sea lamprey) from jawless fish, and S. purpuratus (purple sea urchin) from echinoderms. Circular dichroism spectra confirmed that NCBD variants were folded and that all CIDs were highly disordered (Figure S1), similarly to previously characterized NCBD and CID domains. S. purpuratus CID appeared slightly more ordered than the other ones and this is possibly related to the apparent dimerization observed in the ITC experiments (see below). The H. sapiens NCBD:CID complex, consisting of NCBD from CREBBP and CID from NCOA3 (ACTR), displayed a K D‐value of 0.4 μM (Figures 1, S1 and Table S1). Note that several of our previous studies on the human NCBD:CID interaction reports a lower K D‐value (around 0.1 μM). This is because a longer construct was used in accordance with the original studies on this interaction.7, 9 However, in our evolutionary studies2, 3 we had to trim down in particular the CID domain to a length corresponding to the interaction surface between the two proteins, since any sequence similarity is lost outside of the conserved interaction regions. This shorter CID gives a slightly lower affinity. In the present study, all NCBD and CID proteins correspond to the shorter constructs (Figure 1a) and their affinities can be directly compared. The binding affinities of NCBD:CID complexes from D. rerio and P. marinus were in the same order as the human one, with K D‐values ranging from 0.2–0.6 μM. P. marinus has two NCOA proteins, and we found that both contain CID domains with high affinity for NCBD (KD = 0.2 μM). It is not fully clear if the jawless fishes originated before or after the two whole genome duplications in vertebrates. Thus, while CID from NCOA1, 2, and 3 in fish and mammals are paralogs from the whole genome duplications, the two NCOAs in jawless fish could have originated from a local gene duplication. Nevertheless, all interactions between human CREBBP/p300 NCBD and NCOA1, 2, and 3 variants, 2 and the native interactions in other vertebrate species presented here are of high affinity. For D. rerio we only tested CID from NCOA3. However, the sequence identity between human and D. rerio CIDs from NCOA1 and NCOA2 are high 2 and their interactions with NCBD most likely of similar high affinity as the human ones. On the other hand, the affinity of the NCBD:CID complex from S. purpuratus was >20‐fold lower, displaying a K D‐value in the range of 7–22 μM. Interestingly, we noted that the stoichiometry of the S. purpuratus NCBD:CID complex was 1:2 according to the obtained n‐value of 1.7 when CID was titrated to NCBD, suggesting a different binding mechanism as compared with the other NCBD:CID complexes. Titration of high concentrations (mM) of S. purpuratus CID into buffer resulted in an ITC thermogram characteristic of dimerization and the background signal from dimerization was subtracted from the overall signal of the binding experiment before fitting to the model. In order to keep the concentrations of CID low, and thus minimize the signal from dimerization of CID, we performed an ITC experiment where S. purpuratus NCBD was titrated into S. purpuratus CID. This experiment gave an n‐value of 0.48 in accordance with a 1:2 binding model of the S. purpuratus NCBD:CID complex. In either case, the K D‐value for the S. purpuratus complex was high (7–22 μM depending on whether NCBD or CID was the titrant). In order to deduce whether it is mainly variations in NCBD or CID that is responsible for the low affinity in S. purpuratus, we cross‐tested S. purpuratus NCBD with H. sapiens CID (KD = 13 μM) and H. sapiens NCBD with S. purpuratus CID (KD = 4.8 μM). The similar affinities to the native S. purpuratus complex suggest that variations in both S. purpuratus NCBD and CID contributes to the lower affinity observed in this animal, although NCBD contributes more to this effect. Furthermore, S. purpuratus CID displayed the same 2:1 stochiometry in complex with H. sapiens NCBD as with S. purpuratus NCBD.
This low affinity in the echinoderm lineage corroborates our previous hypothesis that the interaction was of low affinity in an early deuterostome ancestor and suggests that a high‐affinity interaction was established in the vertebrate lineage after the divergence of echinoderms but before the split between jawed and jawless fish (Figure 1).
3. DISCUSSION
The evolution and divergence of species is tightly linked to protein interaction networks regulating transcription and cell signaling and regulation. The interaction between CREBBP/p300 and the NCOA family of proteins is one example of how two transcriptional coregulators form a new interaction, which was initially of low affinity but beneficially enough to evolve into a higher‐affinity interaction in a vertebrate ancestor and conserved in present day vertebrates. 2 Furthermore, two and three variants, respectively, of CREBBP/p300 and NCOA have been conserved in present day mammals, reptiles, and birds. In bony fish, a third whole genome duplication occurred 10 and CREBBP is present in two copies with similar NCBD domains. This redundancy resulting from gene duplication events is found for several other protein interactions, likely contributing stable networks of protein–protein interactions during the radiation of vertebrate animals. In nonvertebrate animals, only one copy of CREBBP/p300 and NCOA, respectively, are usually present since they diverged before the whole‐genome duplications. We previously investigated the cross reactivity of human CID domains and NCBD from different species. Consistent with the present data we found that NCBD from Drosophila melanogaster (fruit fly) bound to human CID with low affinity (5–40 μM depending on which of the three human CIDs was used in the experiment). 2 Thus, the similar and low affinities of ancestral NCBD and extant D. melanogaster and S. purpuratus NCBD for CID is consistent with a scenario in which DNA encoding an ancestral CID domain emerged within the NCOA gene after the divergence of protostomes and deuterostomes (~600 Myr). Subsequently, there was positive selection for higher affinity in early vertebrates (~525 Myr) while the NCBD:CID interaction remained low affinity in echinoderms, a nonvertebrate deuterostome phylum, here represented by S. purpuratus. Our findings show quantitatively how evolution could work by invention of a new interaction motif, the intrinsically disordered CID domain, and how this could lead to improved function (in this case in transcriptional regulation). On a molecular level evolution operates through point mutation, but also through recombination of DNA encoding whole or parts of protein domains. It is not clear exactly how the CID domain originated. It could for example have been via extensive point mutation but also through local rearrangements or horizontal gene transfer, or a combination. What is the structural basis of the higher affinity that evolved in chordate NCBD:CID interactions? We know from previous studies that Tyr2108 in chordate NCBDs is one key position contributing to higher affinity (Figure 1). 2 This position is Gln in low affinity ancestral NCBD as well as in protostome NCBDs (e.g., D. melanogaster) and in nonchordate deuterostome NCBDs, including that from S. purpuratus. Furthermore, CIDs from nonchordate deuterostomes lacks a positively charged Arg1069 (or Lys) in the highly conserved DRALGI (or DKALGI) motif. This residue appears to interact with a Glu1066 to stabilize helix 2 in CID and therefore likely contributes to the higher affinity in chordate NCBD:CID complexes. However, the NCBD:CID interaction appears plastic both with regard to bound ground states2, 3 and the binding pathway. 11 Thus, the higher affinity cannot be linked to one particular residue, but rather to a combination of substitutions and the resulting structure and dynamics. As mentioned in Section 2, another factor contributing to affinity is flanking regions of the actual binding interface, as we have observed for the human NCBD:CID interaction. Hence, we cannot rule out that flanking regions in the low affinity NCBD:CID complexes contribute to affinity. However, we find it unlikely that they would contribute more to the affinity than observed for the human complex (fourfold) and so account for the large difference in affinity between low‐ and high‐affinity NCBD:CID interactions.
4. MATERIALS AND METHODS
4.1. Protein expression and purification
cDNA sequences encoding the protein variants were purchased from GenScript and included an N‐terminal tag (6xHis‐Lipo domain). Plasmids encoding the protein constructs were transformed into Escherichia coli BL‐21 DE3 pLysS (Invitrogen) and distributed on LB agar plates containing 35 μg chloramphenicol/ml and 100 μg ampicillin/ml. Colonies were used to inoculate LB media containing 50 μg ampicillin/ml. The cultures were grown at 37°C to OD600 0.6–0.7 at which point expression was induced by addition of 1 mM isopropyl β‐d‐1‐thiogalactopyranoside followed by overnight expression at 18°C. Cells were harvested by centrifugation, lysed by sonication and centrifuged again at approximately 50,000 g to remove cell debris. The resulting lysate was filtered (0.45 μm) and applied onto a Ni Sepharose 6 Fast Flow (GE Healthcare) column equilibrated with 30 mM Tris–HCl pH 8.0, 500 mM NaCl. After washing, bound proteins were eluted with 30 mM Tris–HCl pH 8.0, 500 mM NaCl, 250 mM imidazol. The 6xHis‐Lipo tag was cleaved off using Thrombin (GE Healthcare) according to the instructions of the manufacturer and the sample was subsequently loaded onto the same column as described above to remove the cleaved tag. Finally, the proteins were separated on a RESOURCE™ reversed phase chromatography column (GE Healthcare) using a 0–70% acetonitrile gradient in 0.1% trifluoroacetic acid/H2O. The purity of the respective protein was verified by the single‐peak appearance on the reversed phase chromatogram or by SDS‐PAGE and the identity was verified by MALDI‐TOF mass spectrometry. Fractions containing pure protein were lyophilized. Before experiments, the proteins were dissolved in buffer and the concentration measured by absorption spectrometry at 280 nm for variants that contained a Tyr or Trp residue. The concentration of proteins (CID) lacking a Tyr or Trp residue, were determined by absorption at 205 nm using extinction coefficients calculated at http://nickanthis.com/tools/a205.html. 12
4.2. CD spectroscopy
Far‐UV CD spectra were recorded using a J‐1500 spectrophotometer (JASCO) in 20 mM sodium phosphate buffer pH 7.4, 150 mM NaCl at 25°C to check that the NCBD variants were folded. The bandwidth was 1 nm, scanning speed 50 nm/min and data pitch 1 nm. The concentration of protein was between 20 and 40 μM and each spectrum was an average of 2–3 individual spectra.
4.3. Isothermal titration calorimetry
ITC experiments were performed at 25°C in a MicroCal iTC200 System (GE Healthcare). Proteins were dialyzed against the same experimental buffer (20 mM sodium phosphate pH 7.4, 150 mM NaCl) to reduce buffer mismatch. The concentration of NCBD or CID in the cell was 15–150 μM (depending on variant) and the concentration of the titrant in the syringe was between 150 and 1,500 μM, such that a 1:2 stoichiometry was achieved at the end of each experiment. When S. purpuratus CID was titrated into S. purpuratus NCBD, the syringe was refilled such that a final stoichiometry of approximately 1:4 was achieved. The data were fitted using the built‐in software to a two‐state binding model with manual correction of the endpoint baseline.
AUTHOR CONTRIBUTIONS
Elin Karlsson: Conceptualization; formal analysis; investigation; methodology; visualization; writing‐original draft; writing‐review and editing. Amanda Lindberg: Formal analysis; investigation; methodology. Eva Andersson: Formal analysis; investigation; methodology. Per Jemth: Conceptualization; formal analysis; funding acquisition; project administration; resources; supervision; writing‐original draft; writing‐review and editing.
Supporting information
Figure S1 Circular dichroism and isothermal titration calorimetry experiments. (a) Normalized circular dichroism (CD) spectra of nuclear coactivator binding domain (NCBD) variants (left panel) and CREBBP‐binding domain (CID) variants (right panel). (b) ITC experiments of NCBD and CID from four different deuterostome animals. Titration of Strongylocentrotus purpuratus CID to S. purpuratus NCBD (lower left panel) resulted in a strong background signal, presumably arising from dimerization of S. purpuratus CID in the syringe. An experiment where S. purpuratus CID was titrated into buffer was therefore subtracted from the binding data prior to fitting. See Table S1 for all fitted parameters.
Table S1 Thermodynamic parameters obtained in the ITC measurements. The measurements were conducted in 20 mM sodium phosphate, pH 7.4, 150 mM NaCl at 25°C and the affinity of each pairwise interaction was repeated two or three times. Homo sapiens and Danio rerio CID were from NCOA3.
ACKNOWLEDGMENT
We thank Chris Johnson for comments on the ITC data. This work was funded by the Swedish Research Council, Grant Number 2016‐04965 (to P. J.).
Karlsson E, Lindberg A, Andersson E, Jemth P. High affinity between CREBBP/p300 and NCOA evolved in vertebrates. Protein Science. 2020;29:1687–1691. 10.1002/pro.3868
Funding information Swedish Research Council, Grant/Award Number: 2016‐04965
REFERENCES
- 1. McLysaght A, Hokamp K, Wolfe KH. Extensive genomic duplication during early chordate evolution. Nat Genet. 2002;31:200–204. [DOI] [PubMed] [Google Scholar]
- 2. Hultqvist G, Åberg E, Camilloni C, et al. Emergence and evolution of an interaction between intrinsically disordered proteins. Elife. 2017;6:e16059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jemth P, Karlsson E, Vögeli B, et al. Structure and dynamics conspire in the evolution of affinity between intrinsically disordered proteins. Sci Adv. 2018;4:eaau4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Goodman RH, Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000;14:1553–1577. [PubMed] [Google Scholar]
- 5. Xu J, Li Q. Review of the in vivo functions of the p160 steroid receptor coactivator family. Mol Endocrinol. 2003;17:1681–1692. [DOI] [PubMed] [Google Scholar]
- 6. Kjaergaard M, Teilum K, Poulsen FM. Conformational selection in the molten globule state of the nuclear coactivator binding domain of CBP. Proc Natl Acad Sci U S A. 2010;107:12535–12540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Demarest SJ, Deechongkit S, Dyson HJ, Evans RM, Wright PE. Packing, specificity, and mutability at the binding interface between the p160 coactivator and CREB‐binding protein. Protein Sci. 2004;13:203–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dogan J, Schmidt T, Mu X, Engström Å, Jemth P. Fast association and slow transitions in the interaction between two intrinsically disordered protein domains. J Biol Chem. 2012;287:34316–34324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Demarest SJ, Martinez‐Yamout M, Chung J, et al. Mutual synergistic folding in recruitment of CBP/p300 by p160 nuclear receptor coactivators. Nature. 2002;415:549–553. [DOI] [PubMed] [Google Scholar]
- 10. Christoffels A, Koh EGL, Chia J‐M, Brenner S, Aparicio S, Venkatesh B. Fugu genome analysis provides evidence for a whole‐genome duplication early during the evolution of ray‐finned fishes. Mol Biol Evol. 2004;21:1146–1151. [DOI] [PubMed] [Google Scholar]
- 11. Karlsson E, Andersson E, Dogan J, Gianni S, Jemth P, Camilloni C. A structurally heterogeneous transition state underlies coupled binding and folding of disordered proteins. J Biol Chem. 2019;294:1230–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Anthis NJ, Clore GM. Sequence‐specific determination of protein and peptide concentrations by absorbance at 205 nm. Protein Sci. 2013;22:851–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Circular dichroism and isothermal titration calorimetry experiments. (a) Normalized circular dichroism (CD) spectra of nuclear coactivator binding domain (NCBD) variants (left panel) and CREBBP‐binding domain (CID) variants (right panel). (b) ITC experiments of NCBD and CID from four different deuterostome animals. Titration of Strongylocentrotus purpuratus CID to S. purpuratus NCBD (lower left panel) resulted in a strong background signal, presumably arising from dimerization of S. purpuratus CID in the syringe. An experiment where S. purpuratus CID was titrated into buffer was therefore subtracted from the binding data prior to fitting. See Table S1 for all fitted parameters.
Table S1 Thermodynamic parameters obtained in the ITC measurements. The measurements were conducted in 20 mM sodium phosphate, pH 7.4, 150 mM NaCl at 25°C and the affinity of each pairwise interaction was repeated two or three times. Homo sapiens and Danio rerio CID were from NCOA3.


