Single yeast cell nanomotions monitored by optical microscopy reveal cellular activity and antifungal drug response.
Abstract
Living single yeast cells show a specific cellular motion at the nanometer scale with a magnitude that is proportional to the cellular activity of the cell. We characterized this cellular nanomotion pattern of nonattached single yeast cells using classical optical microscopy. The distribution of the cellular displacements over a short time period is distinct from random motion. The range and shape of such nanomotion displacement distributions change substantially according to the metabolic state of the cell. The analysis of the nanomotion frequency pattern demonstrated that single living yeast cells oscillate at relatively low frequencies of around 2 hertz. The simplicity of the technique should open the way to numerous applications among which antifungal susceptibility tests seem the most straightforward.
INTRODUCTION
Our team developed previously an atomic force microscopy (AFM)–based assay to assess the effects of chemicals on the viability of bacteria (1). The detection is based on the observation that living organisms oscillate at a nanometric scale and transfer these oscillations to the AFM cantilever onto which they are attached. These oscillations stop as soon as the viability of the cells is compromised. We demonstrated that these oscillations are present in living bacteria, yeasts, plant, and mammalian cells (2). The nanomotions of living bacterial cells that are attached to a surface have been confirmed by using other detection methods such as plasmonic imaging of the z motion of bacteria (3), tracking the submicron scale x-y motion of attached bacteria (4), sensing of attached bacterial vibrations with phase noise of a resonant crystal (5), and subcellular fluctuation imaging (6).
We recently noticed that an optical microscope equipped with a video camera can detect the motions of single yeast cells that are sedimented on a glass surface. In this work, we explored the nanomotion of nonattached yeast cells using this optical nanomotion detection (ONMD) method. The cellular x-y motions were monitored by recording 12-s-long movies (1000 frames) taken at a magnification of ×400 (Fig. 1). By periodically recording these movies, temporal behavior of the cells was characterized as a function of different chemical and physical stimuli (Fig. 1, A and B). To track the cellular motions of single cells, we used a cross-correlation image registration algorithm (7). The algorithm is based on the initial estimation of the cross-correlation peak between the first and every subsequent frame. It provides a numerical value for the image translation with a subpixel (submicrometer) resolution. The cell displacement for each frame, the trajectories of tracked cells (Fig. 1C), and the root mean square of the total displacement (Fig. 1F) were calculated. Single-cell nanomotions were characterized by plotting the distribution of the displacements per frame as a violin plot (Fig. 1D). The motions of the set of 20 cells were characterized by plotting grouped cellular displacements per frame as violin plots and the total displacements of 20 cells over 1000 frames as box-and-whisker plots (Fig. 1E).
Fig. 1. Overview of the optical nanomotion method to detect cellular nanomotions.
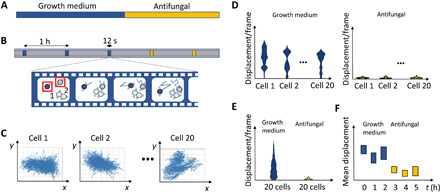
(A) Time period of cells in growth medium followed by, e.g., an antifungal treatment. (B) At different time points, movies are recorded of 1000 frames. Cell movements within the box are detected and analyzed. (C) The x-y displacements of individual cells (typically 20 cells) are calculated using the cross-correlation algorithm. (D) For each cell, the displacement per frame is calculated, and this distribution is represented by a combined violin and box plot. (E) The displacement per frame for all cells for a condition/sampling point is represented as a combined violin plot and box plot. (F) The mean of the total displacements of 20 cells is calculated for each condition/sampling point and represented in a box plot.
RESULTS
First, we compared single-cell nanomotions of Saccharomyces cerevisiae cells that were grown in the presence of nutrients [by growing them in yeast extract, peptone, and dextrose (YPD) growth medium] to cells that were in a nutrient-free physiological phosphate-buffered saline (PBS) buffer. Single-cell displacements were recorded every hour during 4 hours (Fig. 2, A and B, and fig. S1, A and B). Actively growing single cells showed a large distribution of displacements. The distribution of the displacements is not symmetric, and this reflects the nonrandom behavior of the cells (as could also be observed from the x-y displacements graphs in Fig. 3), i.e., cells can make jumps from time to time. This motion behavior is also reflected in the shape of the violin plots that represents the displacements distribution. In this set, a few cells (one to three) display a very small displacement distribution and can be classified as inactive. In contrast, significantly more inactive cells were present in the absence of nutrients, especially after 3 to 4 hours of incubation (Fig. 2B and fig. S1B). This behavior is also reflected in the grouped displacements violin plots (Fig. 2, A and B, bottom) and the total displacements boxplots (fig. S1, C and D). In these last plots, the adaptation of the cells to the new growth condition can clearly be observed, i.e., a significant increase of the total displacement after 1 hour, in contrast with the measurements obtained in PBS.
Fig. 2. Effect of the nutritional environment and the temperature on the cellular nanomotions of yeast cells.
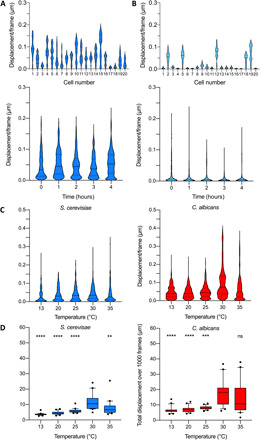
(A) The distribution of the displacements per frame of 20 S. cerevisiae BY4742 cells growing in YPD growth medium after 2 hours (top). Time evolution of the merged distributions of the displacements for 20 cells (bottom). (B) The distribution of the displacements per frame of 20 S. cerevisiae BY4742 cells present in PBS after 2 hours (top). Time evolution of the merged distributions of the displacements for 20 cells (bottom). (C) Time evolution of grouped displacements in growth medium. Effect of the temperature on the displacement distribution (20 cells) for S. cerevisiae BY4742 and C. albicans DSY294. (D) Effect of the temperature on the total displacement of S. cerevisiae BY4742 and C. albicans DSY294. Wilcoxon test: ****P < 0.0001; ***P < 0.001; **P < 0.01; ns, not significant.
Fig. 3. Monitoring of life-death transition by observing cellular nanomotions of dying yeast cells in the presence of ethanol or antifungal.
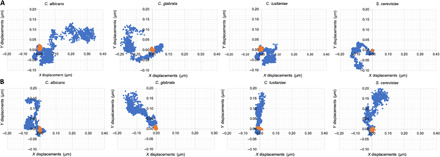
Effect of (A) ethanol (70%, v/v) and (B) caspofungin (100 μg/ml) on the x-y displacements of C. albicans DSY294, C. glabrata DSY562, C. lusitaniae DSY4606, and S. cerevisiae BY4742 cells during 12 s (1000 frames, 83 fps). Blue dots represent the position of the cell without ethanol or caspofungin treatment; the orange dots represent the positions after the treatment.
The cellular nanomotions were also compared to the motions of silica beads recorded in the same conditions (fig. S2, A to D). The distributions of the displacements were symmetric. The magnitudes of the motions were much reduced compared to living cells and were of the same order of dead cells (fig. S2E).
To assess the effect of the temperature on the nanomotion pattern of yeast, we monitored the cellular oscillations at different temperatures in the range of 13° to 35°C (Fig. 2, C and D, and fig. S3). Each strain is characterized by a different distribution of grouped displacement distributions. For both yeast strains, a maximal activity was detected at around 30°C. This value is in concordance with the documented optimal growth temperature of 30° to 35°C for S. cerevisiae BY4742 (8) and 33° to 38°C for Candida albicans (9). These experiments demonstrate that the magnitude of the cellular activity is proportional to the magnitude of the distribution of the displacements and the total displacement over 1000 frames.
Next, we characterized the cellular motions of Candida and S. cerevisiae cells when they are exposed to a killing agent. The Candida species can be involved in candidiasis, which is a human fungal infection that can be hard to treat due to the acquired resistance (10). Some of the evaluated yeast strains were hypersusceptible or resistant to the applied antifungal drugs to challenge their viability. First, we explored the effect of a high ethanol concentration (70%) on C. albicans, Candida glabrata, Candida lusitaniae, and S. cerevisiae cells. The x-y displacements are quickly and markedly reduced after adding ethanol (Fig. 3 and movies S1 and S2). The displacement distributions of the single cells (Fig. 4), the grouped displacements of 20 cells [Fig. 4, E and F (top)], and the total displacements [Fig. 4, E and F (bottom)] were reduced. Ten minutes after the addition of ethanol, a reduction of the displacements in the set of 20 cells for all strains is observed. C. glabrata and—in a lesser extent—S. cerevisiae are somewhat more ethanol tolerant than C. glabrata and C. lusitaniae, since a significant decrease of the total displacement was only observed after 60 min. Ethanol tolerance is strain dependent (11, 12), affects the growth rate, and will impair the cell membrane integrity (13), which results in ionic species permeability and leakage of metabolites (14); and freely diffuses inside the cell, where it directly perturb and denature intracellular proteins (15).
Fig. 4. Life-death transition by observing cellular nanomotions of yeast cells in the presence of ethanol.
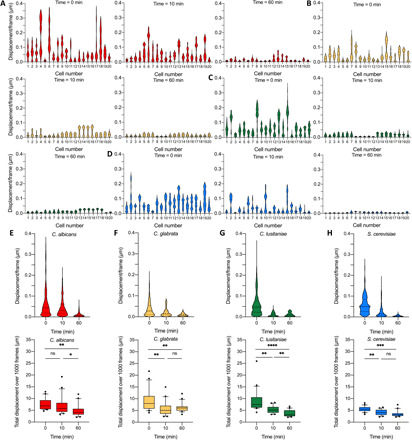
The distribution of the displacements of 20 cells at time 0 min (left) and 60 min (right) for (A) C. albicans DSY294, (B) C. glabrata DSY562, (C) C. lusitaniae DSY4606, and (D) S. cerevisiae BY4742. Time evolution of displacements/frame per frame of 20 cells (top) for (E) C. albicans DSY294, (F) C. glabrata DSY562, (G) C. lusitaniae DSY4606, and (H) S. cerevisiae BY4742 and the corresponding graphs of the total displacement during 12 s measurement as a function of time (bottom). Wilcoxon test: ****P < 0.0001; ***P < 0.001; **P < 0.01; *P < 0.1.
Second, we assessed the effect of killing of cells on the change in cellular nanomotions by exposing them to different concentrations of various antifungals. The x-y displacements were significantly reduced after treatment of the cells with antifungals (Fig. 3B). The effect of amphotericin B, caspofungin, and fluconazole on the cellular motions of C. albicans DSY294 wild-type strain is shown in Fig. 5 (A to C) and fig. S4 (A to C). The polyene amphotericin B selectively binds to ergosterol in the cell membrane and causes the formation of pores (which results in a quicker dead), whereas the azole fluconazole selectively inhibits cytochrome P450–dependent lanosterol 14-α-demethylase, and the echinocandin caspofungin inhibits fungal β-1,3-glucan synthase (16). For all three antifungals, a significant decrease in the cellular displacements and total displacement was detected after 1-hour treatment. Amphotericin B at a high concentration is very effective, since after already 1 hour, the cellular nanomotion decreased to values close to those recorded on dead cells in the whole set of 20 cells (fig. S4A). Caspofungin did not affect the nanomotion of the candin-resistant C. albicans DSY4614 clinical strain (Fig. 5E) but did kill the hypersusceptible (mutant for efflux systems) C. albicans DSY1024 strain (Fig. 5D and table S1). The reported caspofungin minimal inhibitory concentration (MIC) values for C. albicans are in the range of 0.03 to 8 μg/ml (17–19). Fluconazole decreased the cellular nanomotions of C. albicans DSY294 significantly after 1 hour (Fig. 5C).
Fig. 5. Effect of antifungals on the cellular nanomotion of C. albicans.
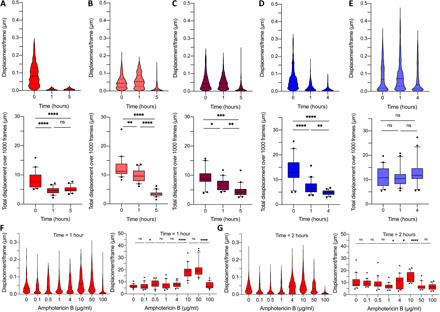
(A) Effect of amphotericin B (500 μg/ml), (B) caspofungin (100 μg/ml), and (C) fluconazole (400 μg/ml) on C. albicans DSY294. Time evolution of displacement distributions of 20 cells of (top) and corresponding graphs of the total displacement during 12 s (bottom). Effect of caspofungin (10 μg/ml) on (D) the hypersusceptible C. albicans DSY1024 and (E) the candin-resistant C. albicans DSY4614. Time evolution of displacement distributions of 20 cells of (top) and corresponding graphs of the total displacement (bottom). (F) Effect of increasing amphotericin B concentrations on C. albicans DSY294: cellular displacements (left) and the total displacement for 20 cells (right) after 1-hour treatment and (G) after 2-hour treatment. Wilcoxon test: ****P < 0.0001; ***P < 0.001; **P < 0.01; *P < 0.1.
To observe the life-death transition, we exposed C. albicans DSY294 clinical wild-type strain to lower amphotericin concentrations, including the MIC (which has been reported for C. albicans DSY strains as 0.5 μg/ml) (20, 21). The total displacements curves show that there is an increase after 1 hour for a concentration of 10 μg/ml (Fig. 5F); the increase is reduced after 2-hour treatment (Fig. 5G). A similar amphotericin B response was recorded for the C. albicans CAF2-1 wild-type strain (fig. S6). On the single-cell level, a larger number of cells show a significantly reduced motion for amphotericin B concentration in the range of 0.1 to 0.5 μg/ml (figs. S5 and S6), which corresponds to the reported MIC value of 0.3 μg/ml for this strain (17).
To evaluate whether the cells interact significantly with the glass surface, which could influence the measured displacements, we compared the cellular nanomotions on a cell-repellent surface [poly-l-lysine (PLL)–polyethylene glycol (PEG)–coated glass surface] to the ones on a nontreated glass surface. Comparable results were obtained for C. albicans DSY294 and DSY1024 treated with caspofungin (fig. S7). The motion of silica beads on a PLL-PEG–coated surface showed that there was a small effect on the displacement distribution and the total displacement (fig. S2, A to D). When the Candida cells were adhered to the glass surface by concanavalin A, the displacements were reduced strongly and no significant difference between caspofungin treated and nontreated cells could be detected (fig. S2, F to K).
To highlight differences in the oscillation pattern occurring during the life-death transition, we performed numerical analysis in the frequency domain (fig. S8). Every frequency range is characterized by its low- and high-frequency limit. With the present method of recording, the low-frequency limit is determined as the reciprocal value of the signal duration, which equals to 0.083 Hz for 12 s. Calculating the high limit is more complex and varies between different cells. We named the high-frequency limit as critical frequency (fcrit) and devised a procedure for its calculation. These analyses revealed that untreated resistant C. albicans cells show a maximum activity in a frequency range (0.083-fcrit) where fcrit varied, varying from 0.7 to 1.5 Hz. C. albicans DSY294 had also an fcrit at a higher frequency (2.5 to 3.0 Hz) (fig. S8C), whereas caspofungin-treated cells presented a broadening of their fcrit, which was most extended for the hypersusceptible DSY1024 cells (fig. S8D). In that case, we also observed an additional fcrit around 2 Hz. These results show that dying cells exposed to low antifungal concentrations are characterized by an increased cellular oscillation frequency, and the largest difference of the oscillation pattern in the frequency domain between living and death yeast cells is located in the very low-frequency range.
The data processing was further accelerated by using a deep learning algorithm that detects individual yeast cells (fig. S9A and movies S3 and S4). Deep learning models have been widely successful in automated objects (22, 23) and cells detection (24, 25) tasks. Compared to a manual selection of cells, this approach could permit to analyze automatically a significantly larger number of cells (100 to 1000). The developed algorithm is based on a medium-sized shallow You Only Look Once (YOLO) (24) architecture. As a proof of principle, we reanalyzed the video data demonstrating the influence of the temperature (fig. S9, B and C). The increase in the samples size for the analysis resulted in a smaller distribution of the total displacements.
DISCUSSION
The newly developed ONMD method is based on basic laboratory material, i.e., an optical microscope, a camera, and a computer. The technique is label free and does not require the attachment of the living sample onto a substrate. The method can address the nanomotion pattern of single cells and cellular populations. ONMD has the potential to detect a single resistant cell in a large population. A limitation of the current ONMD method is that only the x-y motions are recorded. The measurement envelope could be extended by considering also the z axis motion (3), which will further increase the sensitivity of the method.
We could link the cellular nanomotions of single yeast cells to its metabolic activity by comparing the nanomotions of the cells in the presence and absence of nutrients, as well as by detecting a maximum cellular motion at the optimal growth temperature. Living single-cell nanomotions show a nonrandom behavior as was clear from the x-y displacements graphs and the distribution of the displacements during 1000 frames.
The nanomotion analysis of increasing amphotericin B concentration on C. albicans DSY294 and C. albicans CAF2-1 showed that the analysis based on the distribution of the displacements per frame of single cells seems to be more sensitive than those based on the total displacement of the whole cellular population. The effect of concentrations as low as the MIC of this antifungal became noticeable in a population of 20 cells. In addition, the results showed that at amphotericin B concentrations of around 10 times the MIC, the cellular nanomotion is increased. This indicates that the mechanism of action of amphotericin B (which binds selectively to ergosterol in the cell membrane and causes the formation of pores) (26) increases the motion of the cells, i.e., the antifungal action increases the metabolic activity of the cells, probably due to an increased activity of the efflux pumps. An increase of the nanomotion (measured by the AFM cantilever method) of the bacterium Bordetella pertussis for the antibiotic was also previously observed (27).
The analysis of the nanomotion frequency pattern demonstrated that single living yeast cells oscillate at relatively low frequencies of around 2 Hz. These results complement those published by Gimzewski and coworkers (28) who highlighted a periodical motion of the S. cerevisiae cell wall in the range of 0.8 to 1.6 kHz. These measurements were accomplished by AFM on yeast cells that were mechanically trapped into a filter pore. An ultrasonic excitation and interferometric motion detection permitted to detect resonance frequencies of single S. cerevisiae cells in the range of 330 kHz, which correspond to rigid body oscillations of the cell (29). These high-frequency ranges are too high to be measured with our optical microscopy setup. Therefore, the low-frequency oscillations that we observed probably correspond to the whole-body displacements of single yeast cells. Future experiments involving high-speed optical microscopy and AFM-based measurements should highlight the full spectrum of cellular oscillations and a possible contribution of low-frequency cell wall oscillations.
The molecular processes that could cause the observed oscillations have not been investigated yet in detail. Additional experiments consisting in blocking or activating molecular actors (processes) would permit to better understand the observed phenomena. The nanomotion signal is made of vibrations arising from many metabolically related sources that combine energy consumption with local movement or molecule redistributions (30). Cellular nanomotion could arise from processes such as DNA replication, DNA transcription, protein assembly, cytoskeleton rearrangement, ionic pumps activity, organelle transport, etc. The involvement of the cytoskeleton has already been demonstrated by depolymerizing the actin cytoskeleton of osteoblasts by cytochalasin, which resulted in a reduced cellular motion (as measured by the AFM cantilever method) (2). In addition, conformational changes of proteins [as was demonstrated for human topoisomerase II (31)] could contribute to nanomotion.
By automatizing the cell recognition using a deep learning algorithm, we could avoid manual cell detection, extend the number of analyzed cells, and reduce the processing time. In addition, this opens the way to analyze a larger population of cells. Future developments will include dedicated microfluidic chip development and software optimization to run the acquisition and/or the data processing steps onto a low-end computer. These developments could eventually lead to an easy operational mobile device that can be directly implemented in hospitals or even in remote doctor’s practices in developing world countries where it will allow to perform antifungal susceptibility testing in the earliest possible treatment stage and make the appropriate decision for a personalized effective antifungal therapy.
MATERIALS AND METHODS
Strains and cell growth
All yeast strains (table S1) were cultured by inoculating 10 ml of YPD [yeast extract (10 g/liter), peptone (20 g/liter), and dextrose (20 g/liter)] medium with a colony from a YPD agar (20 g/liter) plate. The cultures were grown overnight in Erlenmeyer flasks [30°C and 200 revolutions per minute (rpm)]. The overnight cultures were 10- to 20-fold diluted in 5-ml YPD medium to obtain an optical density at 600 nm (OD600nm) of 0.5 and were then allowed to grow in Erlenmeyer flasks for 1 hour at 30°C and 200 rpm. The cultures were further diluted afterward, depending on the cell concentration (OD600nm value) to obtain an optimal number of cells for visualization.
Optical nanomotion experiment
Ten microliters of each yeast cell culture was dispensed in one of the microwells, using a 4 Well FulTrac micro-Insert (Ibidi, Germany) in an imaging micro-dish (Ibidi, Germany). The yeast cells were allowed to sediment for a period of 10 min before starting the measurement. The motion of cells was observed by taking movies of 1000 frames with a framerate of 84 frames per second (fps) using an electron multiplying charge-coupled device camera (Andor iXon, Oxford Instruments) using a Nikon TE-2000 microscope with a 40× objective. The petri dish was kept at 30°C using a microscope stage top incubator (Ibidi, Germany).
For the experiments where the cell activity in PBS [NaCl (8 mg/ml), KCl (0.20 mg/ml), Na2HPO4 (1.44 mg/ml), and KH2PO4 (0.24 mg/ml)] was compared to YPD growth medium, the overnight cultured cells were 1000-fold diluted in either YPD medium or PBS and immediately dispensed in the microwells. The nanomotions of yeast cells were measured every hour during 4 hours. For the experiments where the effect of the temperature on the metabolism was evaluated, the temperature inside the microwells was controlled by adapting the temperature of water (from a recirculating water bath) circulating around the microwells. The temperature was successively adapted from 13° to 20°, 25°, 30°, and lastly, 35°C. The yeast cells were allowed to adapt during 20 min to each temperature before measuring the nanomotion of the cells. For the experiments with glass surface treatment, the glass surface was coated with concanavalin A (2 mg/ml; Sigma) or PLL-g-PEG (0.1 mg/ml; SuSoS AG, Switzerland) by incubating the glass surface for 30 min with the coating solutions.
The effect of ethanol on the viability of the cells was compared with cells grown in YPD medium. Therefore, first, cell nanomotion videos were recorded, and next (after approximately 1 min), 200 μl of ethanol (70%, v/v) was added to the top chamber of the four micro-Insert wells. Videos were recorded every hour during a period of 5 hours. Caspofungin, amphotericin B, and fluconazole were used as antifungals to assess their effect on cell viability. Before starting the treatment with the antifungal, reference (no treatment) videos were recorded of yeast cells in YPD medium (time = 0 hours). Then (at time = 1 min), 200 μl of a certain concentration of antifungal was added to the chamber (200 μl) above the four microwells.
As abiotic reference particles, silica microbeads (monodisperse silica standard, Whitehouse Scientific) with a diameter of 3 μm were used. The beads were dissolved in YPD medium, and the measurements were performed at 30°C.
Nanomotion detection software
The ONMD algorithm calculates the cell displacement for each frame and saves the trajectories of tracked cells as well as the root mean square of the displacement to a MS Excel file. The main part of the program is based on the algorithm of Guizar-Sicairos et al. (7), ported from MATLAB to Python in the open-source image processing library sci-kit image (32). The Python package nd2 reader (Verweij R, online: www.lighthacking.nl/nd2reader/) was used to import the videos in the ND2 Nikon format.
Deep learning cell detection
The described nanomotion analysis previously described starts from the position of each individual cell, which is currently provided through a manual selection of the bounding box of a cell in the first video frame. However, the number of videos and cells present in the videos can be considerably large, especially when determining the MIC or the impact of different temperature conditions. Providing cell detection at every few frames instead of only in the first video frame allows rectifying the position of those cells that drifts away from their initial positions. In these cases, manual detection efforts could span over several hours for multiframe annotations and thus must be replaced by an automatic detection process of the cells. Therefore, we decided to use a deep learning algorithm, i.e., a medium-sized YOLO (24), to automatize cell detection. The training process is performed using a set of 50,000 synthetic cell images randomly generated. These synthetic images, obtained using a phase-contrast imaging model we previously proposed (33), look very similar compared to the cell images obtained with the microscope in terms of cells distribution, illumination, and imaging artefacts. Once the YOLO model has been trained, it is then used to automatically detect cells in real microscopic images, and each detection is then used as initial position to calculate the optical nanomotion with the cross-correlation algorithm previously described. The overall processing pipeline starts from a bulk of video sequences and performs a per-frame automated single-cell detection. Next, the results are refined by averaging the cells position and detection confidence across frames to correct abrupt changes between consecutive frames. Last, the position of cells detected with high confidence (i.e., >0.6) is provided as input to the nanomotion analysis algorithm (fig. S11A and movies S1 to S5).
Automation of the process
The overall process consists of reading a sequence of frames as images containing the cells and applying the above described cell detection process every 10 frames to obtain a set of bounding boxes indicating the location of each individual cell automatically detected. On the basis of their positions, the cells are tracked across time as the video analysis process unfolds. At the end, in a subsequence refinement stage, we find the contours of cells (34) indicated by the bounding boxes to confirm that their center roughly corresponds to that of the bounding boxes, and any mismatch is then used to penalize the cells detection confidence proportionally. In addition, the bounding boxes position, and detection confidence is averaged across frames to avoid sharp changes the cells detection positions.
Calculation of the frequency region of optically recorded cell movements and the critical frequency
The approach based on calculating fast Fourier transform (FFT) amplitude spectrum, computed for two signals of cell movements—one in the horizontal (“x”) and the other in the vertical (“y”) direction—was developed to calculate the frequency region and critical frequency of optically recorded yeast cell movements. We named this method Double Region Interpolation Method. The idea is to perform two linear interpolations on these averaged amplitudes, in two different frequency regions. One should be performed on the wideband noise part of the spectrum, sufficiently distant from the cell activity region (e.g., 5 to 10 Hz). The other is performed on the low-frequency region, where the cell is active. Crossing point between these two straight lines determines the upper critical frequency of the cell activity region.
Our approach to determine the frequency range of the cell movements is based on the direct application of the FFT algorithm on two detrended signals: one obtained for movements in the horizontal (x) and the other in the vertical (y) direction. Both x and y spectra have similar profiles; however, some amplitudes were slightly different. We therefore derived their frequency-by-frequency average to equally incorporate frequencies of cell movements in both directions. Since signals were recorded with a sampling frequency of 83 fps, we visually inspected frequencies up to 41 Hz and confirmed that cell movements are confined to a few Hz only (fig. S8A, inset). As each signal contained 1000 samples (duration of 12.0482 s), each FFT spectrum was taken from the first 12 s, resulting in a frequency resolution of 0.083 Hz. To maintain this resolution, FFT was applied to each 12-s signal as a whole, thus avoiding any shorter moving windows. The task of determining the frequency range of cell movements implies assessing both its upper and lower critical frequencies.
Statistical analysis
Violin and box-and-whisker (10th to 90th percentile) plots were created with Prism8 (GraphPad). A Wilcoxon matched-pairs signed-rank test was performed to determine the significant differences between conditions (boxplots of mean total displacements over 1000 frames).
Supplementary Material
Acknowledgments
The Research Council of the Vrije Universiteit Brussel (Belgium) and the University of Ghent (Belgium) are acknowledged to support the Alliance Research Group VUB-UGhent NanoMicrobiology (NAMI). We also would like to thank R. Behrend from Geneva Observatory for help in developing the cells tracking software. Funding: This work was supported by the Belgian Federal Science Policy Office (Belspo) and the European Space Agency (ESA) PRODEX program (Yeast Bioreactor project). The Research Council of the Vrije Universiteit Brussel (Belgium) support the International Joint Research Group (IJRG) VUB-EPFL BioNanotechnology & NanoMedicine (NANO) and the Strategic Research Program (project SRP11-M3D2). Swiss members of the team were funded by the Swiss National Grants 200021-144321, 407240-167137, and CRSII5_173863; the Gebert Rüf Stiftung GRS-024/14; and NASA NNH16ZDA001N-CLDTCH. Serbian members of the team were funded by the grant 173017 from the Ministry of Education, Science and Technological Development of the Republic of Serbia. Author contributions: R.G.W. and S.K. conceived the idea for the research, designed the experiments, and supervised the writing of the manuscript. P.V.B. performed the optical nanomotion experiments. R.G.W., S.K., P.V.B., A.M., M.A.-P., D.S., and G.D. interpreted the nanomotion results and/or contributed to the optimization of the method. A.M. and S.K. introduced the cross-correlation method to analyze the data. A.M. developed the cross-correlation analysis program. R.G.W. and S.K. introduced the displacement/frame distribution analysis (violin plot analysis). R.G.W., S.K., and M.I.V. contributed to the violin plot analyses. A.M., P.V.B., K.R., A.K., and M.A.-P. contributed to the different steps of the data analyses. H.S. and M.A.-P. introduced the AI cell recognition/detection method. M.A.-P. wrote the cell detection program. K.R., D.B., and A.K. contributed via the analysis of the motion signal and related frequency analyses. All authors discussed the results and contributed to and commented on the manuscript. R.G.W. and S.K. conceived the overall project and supervised the research. Competing interests: R.G.W., S.K., A.M., P.V.B., M.A.-P., and H.S. are authors on a patent application filed by Vrije Universiteit Brussel and Ecole Polytechnique Fédérale de Lausanne (no. PCT/EP 2020/062143. Submitted April 2019). The authors declare that they have no other competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/26/eaba3139/DC1
REFERENCES AND NOTES
- 1.Longo G., Alonso-Sarduy L., Marques Rio L., Bizzini A., Trampuz A., Notz J., Dietler G., Kasas S., Rapid detection of bacterial resistance to antibiotics using AFM cantilevers as nanomechanical sensors. Nat. Nanotechnol. 8, 522–526 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Kasas S., Ruggeri F. S., Benadiba C., Maillard C., Stupar P., Tournu H., Dietler G., Longo G., Detecting nanoscale vibrations as signature of life. Proc. Natl. Acad. Sci. U.S.A. 112, 378–381 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Syal K., Iriya R., Yang Y., Yu H., Wang S., Haydel S. E., Chen H.-Y., Tao N., Antimicrobial susceptibility test with plasmonic imaging and tracking of single bacterial motions on nanometer scale. ACS Nano 10, 845–852 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Syal K., Shen S., Yang Y., Wang S., Haydel S. E., Tao N., Rapid antibiotic susceptibility testing of uropathogenic E. coli by tracking submicron scale motion of single bacterial cells. ACS Sens. 2, 1231–1239 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Johnson W. L., France D. C., Rentz N. S., Cordell W. T., Walls F. L., Sensing bacterial vibrations and early response to antibiotics with phase noise of a resonant crystal. Sci. Rep. 7, 12138 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bermingham C. R., Murillo I., Payot A. D. J., Balram K. C., Kloucek M. B., Hanna S., Redmond N. M., Baxter H., Oulton R., Avison M. B., Antognozzi M., Imaging of sub-cellular fluctuations provides a rapid way to observe bacterial viability and response to antibiotics. bioRxiv , 460139 (2018). [Google Scholar]
- 7.Guizar-Sicairos M., Thurman S. T., Fienup J. R., Efficient subpixel image registration algorithms. Opt. Lett. 33, 156–158 (2008). [DOI] [PubMed] [Google Scholar]
- 8.Lee J.-S., Park E.-H., Kim J.-W., Yeo S.-H., Kim M.-D., Growth and fermentation characteristics of Saccharomyces cerevisiae NK28 isolated from kiwi fruit. J. Microbiol. Biotechnol. 23, 1253–1259 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Lemos-Carolino M., Madeira-Lopes A., Van Uden N., The temperature profile of the pathogenic yeast Candida albicans. Z. Allg. Mikrobiol. 22, 705–709 (1982). [DOI] [PubMed] [Google Scholar]
- 10.Sanglard D., Emerging threats in antifungal-resistant fungal pathogens. Front. Med. 3, 11 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zeuthen M. L., Dabrowa N., Aniebo C. M., Howard D. H., Ethanol tolerance and the induction of stress proteins by ethanol in Candida albicans. J. Gen. Microbiol. 134, 1375–1384 (1988). [DOI] [PubMed] [Google Scholar]
- 12.Pandey A. K., Kumar M., Kumari S., Kumari P., Yusuf F., Jakeer S., Naz S., Chandna P., Bhatnagar I., Gaur N. A., Evaluation of divergent yeast genera for fermentation-associated stresses and identification of a robust sugarcane distillery waste isolate Saccharomyces cerevisiae NGY10 for lignocellulosic ethanol production in SHF and SSF. Biotechnol. Biofuels 12, 40 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schiavone M., Formosa-Dague C., Elsztein C., Teste M.-A., Martin-Yken H., De Morais M. A. Jr., Dague E., François J. M., Evidence for a role for the plasma membrane in the nanomechanical properties of the cell wall as revealed by an atomic force microscopy study of the response of Saccharomyces cerevisiae to ethanol stress. Appl. Environ. Microbiol. 82, 4789–4801 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stanley D., Bandara A., Fraser S., Chambers P. J., Stanley G. A., The ethanol stress response and ethanol tolerance of Saccharomyces cerevisiae. J. Appl. Microbiol. 109, 13–24 (2010). [DOI] [PubMed] [Google Scholar]
- 15.Henderson C. M., Block D. E., Examining the role of membrane lipid composition in determining the ethanol tolerance of Saccharomyces cerevisiae. Appl. Environ. Microbiol. 80, 2966–2972 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Willaert R. G., Micro- and nanoscale approaches in antifungal drug discovery. Fermentation 4, 43 (2018). [Google Scholar]
- 17.Sanglard D., Ischer F., Marchetti O., Entenza J., Bille J., Calcineurin A of Candida albicans: Involvement in antifungal tolerance, cell morphogenesis and virulence. Mol. Microbiol. 48, 959–976 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Wiederhold N. P., Grabinski J. L., Garcia-Effron G., Perlin D. S., Lee S. A., Pyrosequencing to detect mutations in FKS1 that confer reduced echinocandin susceptibility in Candida albicans. Antimicrob. Agents Chemother. 52, 4145–4148 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cordeiro R. A., Teixeira C. E. C., Brilhante R. S. N., Castelo-Branco D. S. C. M., Paiva M. A. N., Giffoni Leite J. J., Lima D. T., Monteiro A. J., Sidrim J. J. C., Rocha M. F. G., Minimum inhibitory concentrations of amphotericin B, azoles and caspofungin against Candida species are reduced by farnesol. Med. Mycol. 51, 53–59 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Sanglard D., Ischer F., Calabrese D., Majcherczyk P. A., Bille J., The ATP binding cassette transporter gene CgCDR1 from Candida glabrata is involved in the resistance of clinical isolates to azole antifungal agents. Antimicrob. Agents Chemother. 43, 2753–2765 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vale-Silva L., Beaudoing E., Tran V. D. T., Sanglard D., Comparative genomics of two sequential Candida glabrata clinical isolates. G3 7, 2413–2426 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.L. Castrejón, K. Kundu, R. Urtasun, S. Fidler, Annotating object instances with a Polygon-RNN, IEEE Conference on Computer Vision and Pattern Recognition (CVPR, 2017), pp. 4485–4493. [Google Scholar]
- 23.Russakovsky O., Deng J., Su H., Krause J., Satheesh S., Ma S., Huang Z., Karpathy A., Khosla A., Bernstein M., Berg A. C., Fei-Fei L., ImageNet large scale visual recognition challenge. Int. J. Comput. Vis. 115, 211–252 (2015). [Google Scholar]
- 24.J. Redmon, S. Divvala, R. Girshick, A. Farhadi, You only look once: Unified, real-time object detection, IEEE Conference on Computer Vision and Pattern Recognition (CVPR, 2016), pp. 779–788. [Google Scholar]
- 25.O. Ronneberger, P. Fischer, T. Brox, in Lecture Notes in Computer Science (Springer Verlag, 2015), vol. 9351, pp. 234–241. [Google Scholar]
- 26.Brajtburg J., Powderly W. G., Kobayashi G. S., Medoff G., Amphotericin B: Current understanding of mechanisms of action. Antimicrob. Agents Chemother. 34, 183–188 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Villalba M. I., Stupar P., Chomicki W., Bertacchi M., Dietler G., Arnal L., Vela M. E., Yantorno O., Kasas S., Nanomotion detection method for testing antibiotic resistance and susceptibility of slow-growing bacteria. Small 14, 1702671 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Pelling A. E., Sehati S., Gralla E. B., Valentine J. S., Gimzewski J. K., Local nanomechanical motion of the cell wall of Saccharomyces cerevisiae. Science 305, 1147–1150 (2004). [DOI] [PubMed] [Google Scholar]
- 29.Farzi B., Cetinkaya C., Micromechanical and surface adhesive properties of single Saccharomyces cerevisiae cells. J. Phys. D Appl. Phys. 50, 375401 (2017). [Google Scholar]
- 30.Kohler A. C., Venturelli L., Longo G., Dietler G., Kasas S., Nanomotion detection based on atomic force microscopy cantilevers. Cell Surf. 5, 100021 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alonso-Sarduy L., De Los Rios P., Benedetti F., Vobornik D., Dietler G., Kasas S., Longo G., Real-time monitoring of protein conformational changes using a nano-mechanical sensor. PLOS One 9, e103674 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van der Walt S., Schönberger J. L., Nunez-Iglesias J., Boulogne F., Warner J. D., Yager N., Gouillart E., Yu T.; scikit-image contributors , Scikit-image: Image processing in python. PeerJ 2, e453 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.M. Alioscha-Perez, R. Willaert, H. Tournu, P. Van Dijck, H. Sahli, in Lecture Notes in Computer Science (Springer, Berlin, Heidelberg, 2013), vol. 8259, pp. 25–32. [Google Scholar]
- 34.Trujillo-Pino A., Krissian K., Alemán-Flores M., Santana-Cedrés D., Accurate subpixel edge location based on partial area effect. Image Vis. Comput. 31, 72–90 (2013). [Google Scholar]
- 35.Fonzi W. A., Irwin M. Y., Isogenic strain construction and gene mapping in Candida albicans. Genetics 134, 717–728 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marchetti O., Majcherczyk P. A., Glauser M. P., Bille J., Moreillon P., Sanglard D., Sensitive bioassay for determination of fluconazole concentrations in plasma using a Candida albicans mutant hypersusceptible to azoles. Antimicrob. Agents Chemother. 45, 696–700 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coste A. T., Karababa M., Ischer F., Bille J., Sanglard D., TAC1, transcriptional activator of CDR genes, is a new transcription factor involved in the regulation of Candida albicans ABC transporters CDR1 and CDR2. Eukaryot. Cell 3, 1639–1652 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brachmann C. B., Davies A., Cost G. J., Caputo E., Li J., Hieter P., Boeke J. D., Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14, 115–132 (1998). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/26/eaba3139/DC1


